Phototropism, the ability of plants to grow in the direction of light, is dependent on the microtubule cytoskeleton, which is also central to cell division and differentiation. In no other kingdom are microtubules more intimately connected with organismal shape and growth than in plants, which pattern cellulose deposition using the underlying microtubule array. The article by Lindeboom et al. on page * of this issue (1), together with several recently published studies (2, 3) provides fundamental insights into the mechanism used by plants to switch the orientation of their cortical microtubule array in response to light.
During cell elongation, cortical microtubules in the Arabidopsis hypocotyl are arranged in parallel arrays with a predominant orientation almost perpendicular to the axis of expansion. In response to blue light, microtubules undergo a rapid 90° reorientation, thereby changing the cellulose deposition pattern. In an imaging tour-de-force, Lindeboom et al. now show that this reorientation of the microtubule array proceeds through two phases: the first involves γ-tubulin dependent nucleation of new microtubules at high angles from the existing transverse “seed” microtubule, while the second involves microtubule number amplification through severing at microtubule crossovers. Microtubule crossovers have been previously shown to be hotspots for severing in cortical arrays (4). The newly generated microtubule end grows at a shallow angle, maintaining the overall orientation of the “seed” microtubule, and thereby serves to not only amplify microtubule number, but also maintain the overall orientation of the seed. This reorientation of the microtubule track by severing and regrowth at an angle is reminiscent of a railroad turntable where the subunits of the incoming train are separated from each other and then allowed to progress at different angles (see figure). The architecture of the molecular players that promote microtubule branching likely determines the angle of the new growing microtubule, but the broad distribution of angles observed also suggests mechanical feedback from the preexisting microtubule array.
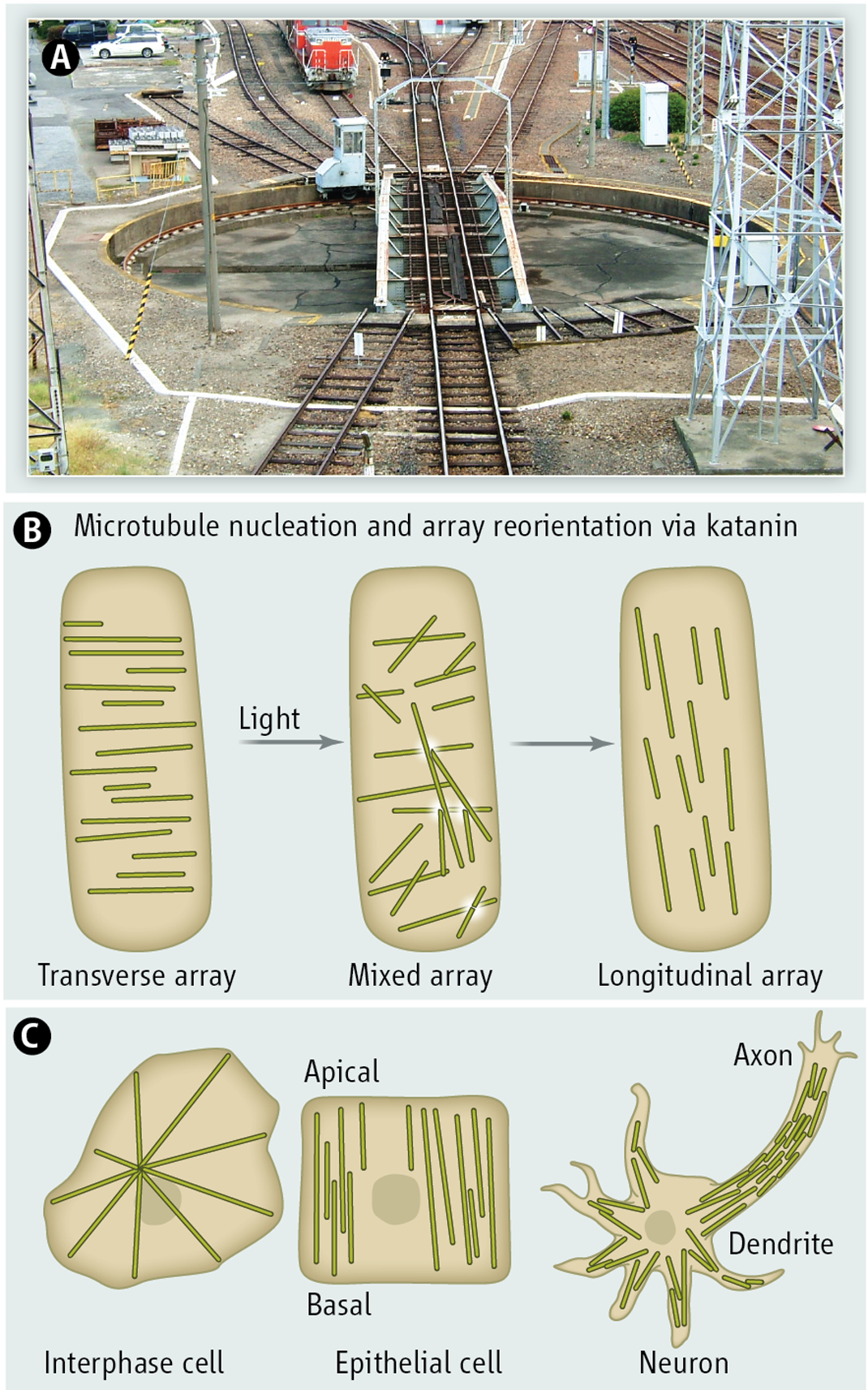
(Top panel)
Severing dependent microtubule growth is analogous to a railroad turntable that splits two tracks
(Middle panel)
Microtubule amplification and array reorientation driven by katanin severing in the plant cortical array
(Lowe panel)
Microtubules are arranged in diverse patterns best adapted to cell type and organism; interphase cell, radial; epithelial cell, parallel, neuron, tiled.
The key player in this process is the microtubule severing enzyme katanin, a AAA ATPase that is thought, by analogy with its family member spastin, to sever the microtubule through extraction of tubulin subunits from the lattice (5, 6). Lindeboom et al. show that GFP-labeled katanin is recruited to microtubule crossovers and precedes all plus-end generation events. A katanin null mutant shows no creation of new microtubule plus-ends, and impaired reorientation of the array upon blue light exposure. Since the new ends created by severing do not depolymerize, but appear to be stable, severing has a constructive role and contributes to the net growth of the microtubule array. One implication of this crossover-activated severing mechanism is that as the array gradually orients and the population of parallel microtubules increases, the frequency of severing decreases (since there are fewer crossovers), thus dampening new microtubule generation and stabilizing the array in its new orientation. Interestingly, a complementary study by Zhang et al., which makes similar observations, proposes that the dominant outcome of severing in the transverse hypocotyl array is depolymerization, with severing ultimately serving to eliminate unaligned, discordant cortical microtubules. Future studies will likely address the balance between these two outcomes.
How does katanin distinguish crossover sites from bundled or single microtubules? How nanometer-scale proteins sense and respond to cellular architecture at the micrometer scale is a central question in cell biology. In vitro, katanin severs microtubules along their length (7); however, experiments with dynamic microtubules or complex microtubule geometries more closely resembling those in cells have not been performed and the fate of the new microtubule end generated by a microtubule severing enzyme in vitro is still not known. Katanin consists of a catalytic subunit (p60) that uses ATP hydrolysis to disassemble the microtubule, and a regulatory subunit (p80) that enhances severing activity and targets the enzyme to specific subcellular locations (6). It is possible that the regulatory subunit, of which Arabidopsis encodes four, senses microtubule crossovers. Future studies will address whether p60 targeting to crossovers is impaired in p80 mutants. Additional factors could also sense the crossing-over microtubule and regulate katanin function. A recent study in Arabidopsis pavement and petiole cells revealed that severing frequency at microtubule crossovers inversely correlates with the presence of the microtubule-associated protein SPIRAL2 which itself induces microtubule crossovers (3). The phenotypes of katanin and spriral2 mutants are diametrically opposed: the katanin mutant displays complex crossovers formed by multiple microtubules, the spiral2 mutant has very few. How SPIRAL2 itself senses crossovers is not clear, but it might be actively transported along the microtubule to crossover sites (3). Future in vitro studies with microtubule arrays of diverse geometries and in the presence of various microtubule-associated proteins will be essential in understanding the feedback between array architecture and katanin function.
A severing-dependent mechanism for microtubule amplification and array reorientation was postulated to be important not only in the morphogenesis of noncentrosomal plant cortical arrays (8), but also in spindles, epithelial cells and neurons (9–12); however, it was not directly observed in vivo until now, partly because the high microtubule density in these systems had made direct observation of severing challenging. Centrosomes can be limiting in a polarized cell because they pattern an isotropic microtubule array. Most cells in our body do not have radial microtubule arrays and many cells lack centrosomes altogether (11). For example, the majority of microtubules in neurons or epithelial cells are disconnected from the centrosome (see figure). Microtubule number amplification by severing is an attractive mechanism for generating the large microtubule mass needed to fill long neuronal processes and offers the possibility of fast reorientation of the microtubule array in response to local stimuli. Thus the mechanistic insights from the studies by Lindeboom et al and Zhang et al are likely relevant in other systems and firmly establish katanin as a driving force in the making and remodeling of noncentrosomal ordered arrays.
Acknowledgments
This work was supported by the intramural program of the National Institute of Neurological Disorders and Stroke
References
- 1.Lindeboom JJ et al. , A Mechanism for Reorientation of Cortical Microtubule Arrays Driven by Microtubule Severing. Science, (November 7, 2013). [DOI] [PubMed] [Google Scholar]
- 2.Zhang Q, Fishel E, Bertroche T, Dixit R, Microtubule severing at crossover sites by katanin generates ordered cortical microtubule arrays in Arabidopsis. Current biology : CB 23, 2191 (November 4, 2013). [DOI] [PubMed] [Google Scholar]
- 3.Wightman R, Chomicki G, Kumar M, Carr P, Turner SR, SPIRAL2 Determines Plant Microtubule Organization by Modulating Microtubule Severing. Current biology : CB 23, 1902 (October 7, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wightman R, Turner SR, Severing at sites of microtubule crossover contributes to microtubule alignment in cortical arrays. The Plant journal : for cell and molecular biology 52, 742 (November, 2007). [DOI] [PubMed] [Google Scholar]
- 5.Roll-Mecak A, Vale RD, Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature 451, 363 (January 17, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roll-Mecak A, McNally FJ, Microtubule-severing enzymes. Current opinion in cell biology 22, 96 (February, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNally FJ, Vale RD, Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75, 419 (November 5, 1993). [DOI] [PubMed] [Google Scholar]
- 8.Wasteneys GO, Microtubule organization in the green kingdom: chaos or self-order? Journal of cell science 115, 1345 (April 1, 2002). [DOI] [PubMed] [Google Scholar]
- 9.Srayko M, O’Toole E T, Hyman AA, Muller-Reichert T, Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Current biology : CB 16, 1944 (October 10, 2006). [DOI] [PubMed] [Google Scholar]
- 10.Roll-Mecak A, Vale RD, Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? The Journal of cell biology 175, 849 (December 18, 2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartolini F, Gundersen GG, Generation of noncentrosomal microtubule arrays. Journal of cell science 119, 4155 (October 15, 2006). [DOI] [PubMed] [Google Scholar]
- 12.Ahmad FJ, Yu W, McNally FJ, Baas PW, An essential role for katanin in severing microtubules in the neuron. The Journal of cell biology 145, 305 (April 19, 1999). [DOI] [PMC free article] [PubMed] [Google Scholar]


