Abstract
Purpose
Deficiency of adenosine deaminase 2 (DADA2) is an autosomal recessive disorder that manifests with fever, early-onset vasculitis, strokes, and hematologic dysfunction. This study aimed to identify disease-causing variants by conventional Sanger and whole exome sequencing in two families suspected to have DADA2 and non-confirmatory genotypes. ADA2 enzymatic assay confirmed the clinical diagnosis of DADA2. Molecular diagnosis was important to accurately identify other family members at risk.
Methods
We used a variety of sequencing technologies, ADA2 enzymatic testing, and molecular methods including qRT-PCR, MLPA.
Results
Exome sequencing identified heterozygosity for the known pathogenic variant ADA2: c.1358A>G, p.Y453C in a 14-year-old female with a history of ischemic strokes, livedo, and vasculitis. No second pathogenic variant could be identified. ADA2 enzymatic testing in combination with quantitative RT-PCR suggested a loss-of-function allele. Subsequent genome sequencing identified a canonical splice site variant, c.−47+2T>C, within the 5’UTR of ADA2. Two of her unaffected siblings were found to carry the same two pathogenic variants.
A homozygous 800bp duplication comprising exon 7 of ADA2 was identified in a 5-year-old female with features consistent with Diamond-Blackfan anemia (DBA). The duplication was missed by Sanger sequencing of ADA2, chromosomal microarray, and exome sequencing but was detected by MLPA in combination with long-read PCR sequencing. The exon 7 duplication was also identified in her non-symptomatic father and younger sister.
Conclusions
ADA2 pathogenic variants may not be detected by conventional sequencing and genetic testing and may require the incorporation of additional diagnostic methods. A definitive molecular diagnosis is crucial for all family members to make informed treatment decisions.
Keywords: Exome sequencing, Genome sequencing, Deficiency of Adenosine Deaminase 2, Loss-of-function variants
INTRODUCTION
Deficiency of adenosine deaminase type 2 is an autosomal recessive disease that is caused by loss-of-function (LOF) variants in ADA2 [1, 2]. DADA2 was initially reported in patients presenting with periodic fever, livedoid rash, hepatosplenomegaly, early-onset vasculopathy/vasculitis and susceptibility to ischemic and hemorrhagic stroke involving the deep-brain nuclei and the brain stem. Over the last 5 years, additional reports of patients have broadened the phenotypic spectrum of DADA2 to now also comprise immune dysregulation and hematologic dysfunction [3–7]. Some patients with DADA2 present with pure red cell aplasia, lineage-specific cytopenias, or trilineage marrow failure. Patients have also been described with lymphoproliferative diseases [8, 9].
The DADA2 phenotype demonstrates a significant degree of variable expressivity and reduced penetrance has been described in several patients [10, 11]. Even patients homozygous for the same founder pathogenic variant or with the same biallelic compound heterozygous pathogenic variant may have variable age of presentation, frequency, and severity of symptoms [11–13].
Disease-causing variants in ADA2 are located all over the entire coding region affecting all domains of ADA2 and to date no mutational hotspots have been identified. Most pathogenic variants are missense changes, however, copy number variations (CNVs) comprising single or multiple exons and frameshift, nonsense, or splicing variants have also been identified [14]. The description of mutation-negative patients with a clinical and enzymatic phenotype consistent with DADA2 suggests the presence of variants within intronic regions and non-coding regulatory elements that have not yet been described [15]. Most pathogenic variants in ADA2 are either novel or found at a low allele frequency (1:1000) in population databases. Some variants exhibit notably higher allele frequencies in specific populations, such as p.Gly47Arg in the Georgian-Jewish population or p.Arg169Gln in Northern European populations [16].
Standard clinical testing for DADA2 consists of enzymatic testing in combination with sequence analysis of the coding region of ADA2. Genetic molecular testing for DADA2 is done either by conventional Sanger sequencing, NGS-based targeted panels, or exome sequencing, and may include a single nucleotide polymorphism (SNP) microarray to detect larger CNVs (>10–25kb). Multiplex ligation-dependent probe amplification (MLPA) analysis used for the detection of small genomic deletions/duplications or single exon CNVs (>50bp) is not part of routine testing for DADA2. ADA2 enzymatic testing in blood samples is confirmatory for DADA2 and it has been found that patients have low (less than 5% of normal) or undetectable ADA2 catalytic activity depending on the genotype [17, 18].
Treatment for DADA2 includes corticosteroids, cytokine inhibitors, and hematopoietic stem cell transplantation in patients presenting with severe hematological manifestations [4, 19, 20]. Several recent studies have demonstrated the efficacy of TNF?-inhibitors in preventing strokes, normalizing inflammatory markers, and improving vascular integrity [21, 22].
The present study describes the identification of disease-causing variants in two unrelated families with probands diagnosed with DADA2. The causal variants were initially missed by standard testing methods, including Sanger sequencing, chromosomal microarray, and exome sequencing. Genetic testing revealed biallelic pathogenic variants in seemingly unaffected family members, prompting in-depth evaluation to look for subclinical effects of the disease.
PATIENTS AND METHODS
Patients
Patients were evaluated at the NIH Clinical Center and/or Johns Hopkins University. Written informed consent was obtained from all patients and healthy relatives. If the patient was a minor, written informed consent was obtained from the parents. Consents were approved by the NIDDK/NIAMS and Johns Hopkins Medicine Institutional Review Board.
ADA2 Sanger sequencing
Genomic DNA samples were extracted from peripheral blood using the Maxwell® 16 Blood DNA Purification Kit (Promega). Coding regions as well as flanking intronic regions of the ADA2 gene (RefSeq: NM_001282225.2) were amplified using AmpliTaq Gold Fast PCR Master Mix (Thermo Fisher Scientific) and sequenced on a 3130xl Genetic Analyzer (Applied Biosystems).
Long-range PCR
Long range PCR for the identification of the exon 7 duplication in ADA2 was performed using LA Taq DNA Polymerase (Takara, Kyoto, Japan) according to the manufacturer’s manual. Primers are available upon request. Multiplex Ligation-dependent Probe Amplification (MLPA) CNVs with in the ADA2 gene were assessed using a MLPA according to the manufacturer’s manual (MRC Holland, Amsterdam, Netherlands).
ADA2 enzymatic testing
Serum ADA2 activity relative to a control group was determined at the NIH by a spectrophotometric assay using a commercially available kit (Diazyme Laboratories, Poway, CA, USA) by adding ADA1-inhibitor EHNA (erythro-9-Amino-b-hexyl-a-methyl9H-purine-9-ethanol hydrochloride) (Sigma-Aldrich, Zwijndrecht, Netherlands) according to the manufacturer’s protocol.
ADA2 enzymatic activity was measured by a CLIA -laboratory at Duke University, Durham, NC, on plasma for a subset of individuals of this study (reference ranges established by the testing laboratory are provided in supplementary table 2). HPLC was used to monitor the deamination of adenosine (10 mM) in the presence of EHNA (100 μM) as previously described in [3], except that the incubation was performed in 100 mM Tris-acetate, pH 7.0 instead of phosphate buffer. Results are presented as milliunits (nanomol/min) of adenosine deaminated per mL plasma (mU/mL).
Real-time quantitative reverse transcription PCR (qRT-PCR)
RNA was isolated using PAXgene Blood RNA system (Qiagen). 100ng of total RNA was reverse transcribed using SuperScript™III Reverse Transcriptase (Thermo Fisher Scientific), and qRT-PCR using PowerUP SYBR Green was performed (Thermo Fisher Scientific). Three distinct primer sets were used. Samples were run in duplicate on a ViiATM 7 Real-Time PCR System, and data were analyzed using the same system (Applied Biosystems).
Haplotype Analysis
Haplotype reconstruction for each family member from case 1 were performed by MERLIN [23]. For the haplotype analysis 19 SNPs within and around the ADA2 gene locus were used.
Chromosomal Microarray
Illumina HumanCoreExome-24v1–0 Microarray, containing over 550,000 markers (mean spacing 5.28 kb) (Illumina, Inc. USA) using Human Genome Build 37/hg19 was utilized according to the manufacturer’s protocol. The array detects copy number gains and losses consistent with deletions and duplications of the genomic regions represented in the array and long continuous stretches of homozygosity.
Genome and exome sequencing
Genomic DNA samples were extracted from peripheral blood using the Maxwell® 16 Blood DNA Purification Kit (Promega). Genome sequencing libraries were prepared using the TruSeq DNA PCR-free Library Preparation Kit (Illumina) with at least 500ng DNA input following Illumina’s recommended protocol with modifications for automated robotic liquid handling. Sequencing libraries were assessed for size distribution and absence of adapter dimers on the Fragment Analyzer. The libraries were quantified by real-time PCR and sequencing was performed on a HiSeq X platform with single library, single lane topography, at 2 × 150 bp read length.
Exome sequence libraries were prepared using Roche SeqCap EZ Human Exome Probes v3.0 (case 1) and Agilent SureSelect HumanAllExonV4_51MbKit (case 2).
Paired-end sequencing was performed on an Illumina HiSeq 2500 platform using TruSeq Rapid PE Cluster Kit-HS and TruSeq Rapid SBS-HS kits. Genome and exome sequencing data was analyzed according to GATK Best Practices recommendations. Exome sequencing data for case 2 was analyzed using BWA mem 0.7.8, GATK 3.1–1 joint calling with HaplotypeCaller tools. The read-depth-based CNV detection algorithms XHMM, CNVnator, and Atlas-CNV were included in the exome sequencing data analysis pipeline. Gemini and exomiser software packages were applied to further prioritize variants. The effect of splice site variants was assessed using Human Splicing Finder, GeneSplicer, NNSplice, and SpliceSiteFinder.
Deep RNA-Sequencing
RNA sequencing was performed in order to functionally assess the impact of the potential splice variants. Messenger RNA was isolated using the Qiagen (Hilden, Germany) RNeasy kit according to standard manufacturer protocols. Following isolation, RNA was fragmented and sequenced on an Illumina NextSeq instrument to a mean depth of 128 million reads per individual using paired-end PE150 base pair chemistry. Post-sequencing, the raw reads were aligned to the hg 19 reference genome build using a two-pass enabled STAR 2.6 short read RNA-seq alignment algorithm using Snakemake pipeline scripts and the NIH Biowulf Supercomputer. Each individual was assessed for altered splicing impact using the Integrative Genomics Viewer (IGV 2.3) for analysis and visualization.
Long-range sequencing
The Pacific Biosciences protocol “Preparing Amplicon Libraries using PacBio Barcoded Adapters for Multiplex SMRT Sequencing” was used for the generation of PacBio sequencing libraries. For each library 200 ng of amplicon DNA was used and a unique DNA barcode was attached. The libraries were pooled in an equimolar ratio. Sequencing was performed using one SMRT cell on a Sequel sequencer (Pacific Biosciences) using version 3 SMRT cells and sequencing reagents with 20-hour movies. Barcoded library reads were separated using PacBio SMRTLink v6.0 Demultiplex Barcodes pipeline. Polished consensus sequences of phased haplotypes from each demultiplexed library were generated using PacBio SMRTLink v6.0 Long Amplicon Analysis (LAA) pipeline. Selected representative consensus sequences were aligned to hg19 using NGMLR [24].
Statistical analysis
Differences between the groups were tested for statistical significance using the Mann-Whitney U test or the chi-square test as appropriate. A p-value of less than 0.05 was considered statistically significant. Spectrophotometric ADA2 enzyme assay data are presented as mean fold change relative to the control group±± SEM.
RESULTS
Case 1
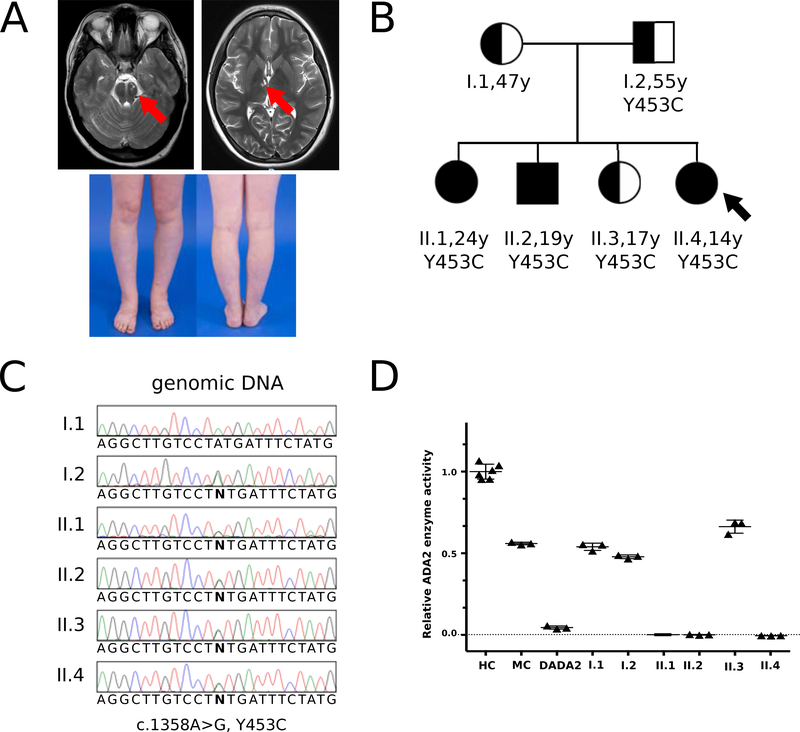
A 14-year-old female evaluated at the NIH Clinical Center presented with a history of unexplained fevers occurring irregularly starting at age 2. She also had multiple ischemic strokes (occurring at 8 and 11 years), livedo racemosa, and a nodular rash that when biopsied revealed granulomatous vasculitis (Fig. 1A). She had been treated with varying doses of glucocorticoids, baby aspirin, clopidogrel, and rituximab without complete suppression of her inflammation. Notably, she had a stroke while taking baby aspirin. Laboratory studies were remarkable for an elevated CRP of 44 mg/L despite taking prednisone (15 mg daily), hypogammaglobulinemia and mild enlargement of the spleen.
Figure 1.
Clinical presentation, family pedigree, ADA2 Sanger sequencing, and ADA2 enzymatic testing in the family of patient 1.
A: Magnetic resonance imaging (MRI) shows location of two strokes and lower extremities showing livedo rashes in patient II.4.
B: Pedigree of the two-generation family showing carrier individuals (I.1, I.2, II.3) and affected individuals (II.1, II.2, II.4 [index patient]); Arrow points at index patient.
C: Sanger sequencing electropherograms depict the pathogenic variant ADA2: c.1358A>G; p.Y453C present in the father (I.2) and in the four children (II.1, II.2, II.3, II.4), and not in the mother (I.1).
D: ADA2 enzymatic activity is strongly reduced/absent in II.1, II.2, and the index patient II.4, while ADA2 enzyme levels are in the range of monoallelic carriers in I.1, I.2, and II.3. Relative ADA2 enzyme activity was determined using a commercial kit (see methods). HC, healthy controls; MC, monoallelic carriers; DADA2, patients with biallelic pathogenic variants.
Exome sequencing was performed on a sample from the proband which identified a known pathogenic variant in ADA2: c.1358A>G; p.Y453C. Segregation analysis using targeted Sanger sequencing demonstrated that the proband (II.4) and each of her three presumed unaffected siblings (II.1, II.2, II.3) inherited the variant from their healthy father (I.2) (Fig. 1B, C). ADA2 enzymatic activity testing revealed that the proband and two of unaffected siblings (at the time) had low to absent ADA2 enzyme activity, comparable to levels seen in patients with DADA2 (Fig. 1D). Her parents and third sibling (II.3) exhibited ADA2 enzyme levels in the range of monoallelic carriers.
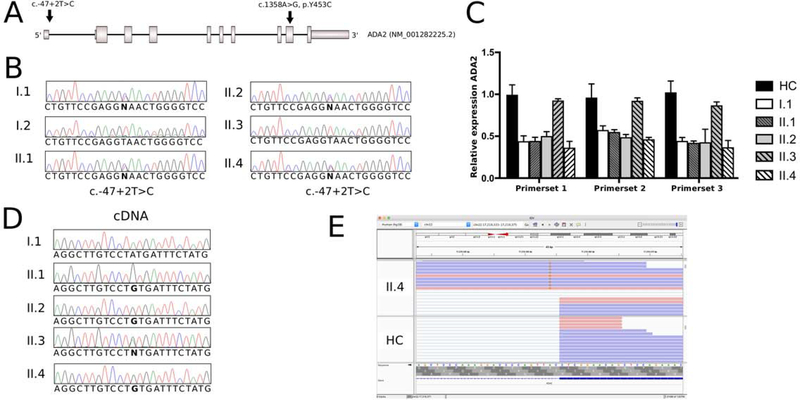
Haplotype analyses revealed that the haplotype carrying the p.Y453C pathogenic variant among all four children is inherited from the father (Fig. S1A). All three children with low ADA2 enzyme activity (the index patient II.4, II.1, and II.2) inherited the same haplotype from the mother, carrying a second at the time unknown pathogenic variant. Sibling II.3 with carrier range of ADA2 enzyme activity inherited the maternal wild type allele from the haplotype denoted in yellow. ADA2 CNV analysis using MLPA was negative (Fig. S1B). Genome sequencing of the mother and the three affected children identified the novel canonical donor splice site variant ADA2: c.−47+2T>C (Chr22(GRCh37):g.17700244A>G) in the 5’-UTR of ADA2 (RefSeq: NM_001282225.2) present in all four individuals (Fig. 2A). Sanger sequencing analysis confirmed the segregation of this variant within the family (Fig. 2B). The c.−47+2T>C variant is absent from population databases (gnomAD v2.1.1) and is predicted to effect splicing according to several in silico algorithms. Quantitative RT-RNA expression analysis showed that the variant has a LOF effect on gene expression leading to an approximately 50% reduction in ADA2 mRNA levels in all individuals carrying the variant (Fig. 2C). Sanger sequencing of complementary ADA2 (cDNA) showed that only the paternal allele carrying the pathogenic missense variant p.Y453C is e;xpressed in the affected individuals (Fig. 2D). Deep sequencing using patient’s peripheral blood RNA confirmed the almost complete lack of ADA2 transcript carrying the splice site variant and further showed splicing defects in the residual pre-mRNAs (Fig. 2E).
Figure 2.
Identification and confirmation of the second pathogenic variant in ADA2 in patient 1.
A: Schematic representation of the ADA2 gene locus (RefSeq: NM_001282225.2) showing the location of two pathogenic variants (c.−47+2T>C and c.1358A>G; p.Y453C) identified in patients 11.1, II.2, and II.4 (index patient).
B: Sanger sequencing identified the c.−47+2T>C canonical splice site variant in the mother (1.1), 11.1, II.2, and II.4.
C: Quantitative reverse transcription (qRT) PCR of ADA2 using three distinct sets of primers demonstrates an approximately 50% reduction in gene expression of ADA2 in 1.1, 11.1, II.2, and index patient II.4 compared to healthy controls. Individual II.3, who is not a carrier, shows no reduction in expression. HC; healthy control.
D: Sanger sequencing of cDNA demonstrates that affected individuals II.1, II.2, and II.4 solely express solely the paternal allele carrying the pathogenic variant ADA2: c. 1358A>G (p.Y453C).
E: Representation of deep RNA sequencing data using Integrative Genomics Viewer (IGV, version 2.5.2) shows aberrant RNA-splicing of residual mutant transcript in individual II.4 as compared to healthy control (HC).
The index patient II.4 initiated treatment with a TNF inhibitor, etanercept, in February 2016, and since then has not had any further ischemic strokes. Additionally, there has been no recurrence of her nodular rash and her inflammatory markers have normalized, allowing her prednisone to be discontinued. The two asymptomatic siblings who had identical genotypes to the patient II.4 underwent comprehensive evaluation as well. Her adult sister (II.1) was found to have low iron with mild microcytic anemia and upon ultrasound of the liver was found to have mild coarse hepatic echotexture, suggestive of mild liver disease. Her adult brother (II.2) was found to have a mildly elevated CRP of 8.9 mg/L, a positive anti-SSB antibody, a low IgM, and mild splenomegaly upon abdominal ultrasound. Although offered, her sister and brother declined treatment with anti-TNF agents. Ten months after initial evaluation, her brother presented to a local emergency room with a 24-hour history of hiccups and mild right facial weakness. MRI revealed a stroke of the posterior limb of the left internal capsule. Etanercept was initiated and he has not had any recurrence of ischemic strokes. After this event, the other affected sibling (II.1) decided to initiate treatment with etanercept. She, too, remains in good health.
Case 2
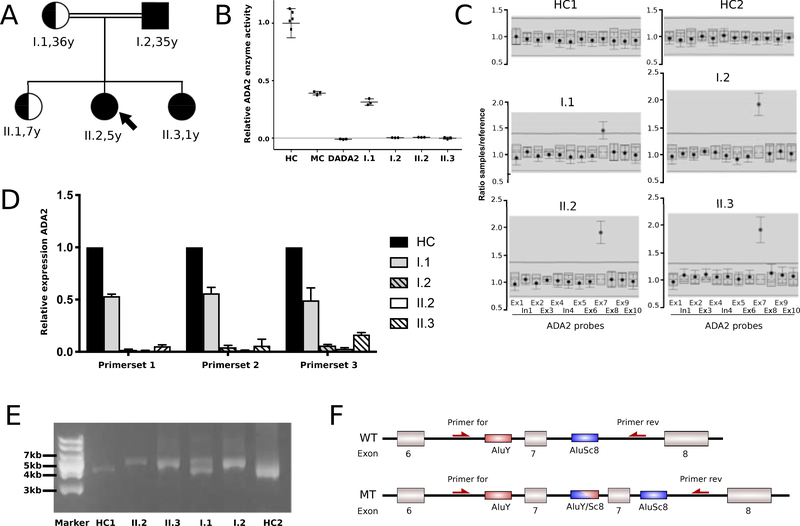
A 5-year-old female, born to a consanguineous Pakistani family, was followed at the Johns Hopkins University Hospital and the NIH Clinical Center. The proband presents with congenital anemia with low hemoglobin (3 g/dl), low reticulocyte count (0.4%), and has received red blood cell transfusion once per month since age 3 months. There was no history of inflammatory episodes nor was there a significant infection history. Laboratory evaluation revealed her to have hypogammaglobulinemia and a decreased absolute neutrophil count of 550 K/uL. Her hepatic transaminases and ferritin were elevated which was thought to be secondary to iron overload.
Chromosomal breakage studies to evaluate for Fanconi anemia were negative. Sequence analysis of genes associated with Diamond-Blackfan Anemia (RPL35A, RPL11, RPL5, RPS10, RPS17, RPS19, RPS24, RPS26, RPS7), including CNV analysis of RPL35A, failed to identify any pathogenic variants. Exome sequencing for the patient and her parents was performed but did not reveal potentially disease-associated single-nucleotide variants (SNVs) or CNVs. Chromosomal microarray analysis showed no significant copy number changes and, in agreement with the consanguinity in her family, several long continuous stretches of homozygosity (each > 3 Mb) were detected ( Supplementary table 1). The total size of these homozygous regions in the proband represents approximately 7.6% of the genome. One of these regions of homozygosity comprises the ADA2 gene locus and DADA2 was considered in her differential diagnosis based on the observation that some patients can present with hematologic findings including pure red cell aplasia [3, 5]. ADA2 Sanger sequencing was negative for pathogenic variants, however, ADA2 enzymatic testing showed almost complete absence of ADA2 enzyme activity in the proband and her unaffected father (I.2) and at the time unaffected sibling (II.3) (Fig. 3B). Her mother had ADA2 enzyme levels in the range of individuals carrying one pathogenic variant in ADA2. ADA2 enzymatic activity for the proband’s unaffected older sister (II.1) was determined in a CLIA-certified outside laboratory to be in the range of carriers (Supplementary table 2). Semiquantitative MLPA analysis identified a homozygous duplication of a region comprising exon 7 of ADA2 in the proband, father, and younger sister (II.2, I.2, II.3) (Fig. 3C). Her mother’s copy number analysis indicated heterozygosity for the duplication. ADA2 exon 7 is an out-of-frame exon and quantitative RT-PCR analysis found a complete loss of RNA expression in the individuals homozygous for the duplication (Fig. 3D). In agreement with the mother’s carrier status, she showed an approximately 50% reduction in ADA2 mRNA. Long-range PCR using a primer set flanking ADA2 exon 7 confirmed a tandem exon duplication in a homozygous state in the proband, father, and her younger sibling (II.2, I.2, II.3) and confirmed her mother’s carrier status (I.1) (Fig. 3E). Long-read sequence analysis was performed on the amplified PCR products and indicated that the potential underlying mechanism for the pathogenic event in this family is a non-homologous meiotic recombination event between two repetitive Alu elements flanking ADA2 exon 7 (Fig. 3F).
Figure 3.
Pedigree, ADA2 enzymatic and molecular analysis of ADA2 in patient 2.
A: Pedigree of the two-generation Pakistani family. Arrow points at index patient. Double lines indicate consanguinity.
B: ADA2 enzymatic activity is strongly reduced/absent in I.2, index patient II.2, and II.3. Mother (I.1) exhibits ADA2 enzyme levels in the range of monoallelic carriers. HC, healthy controls; MC, monoallelic carriers; DADA2, patients with biallelic pathogenic variants in ADA2.
C: Multiplex ligation-dependent probe amplification (MLPA) analysis of the ADA2 gene locus in I.1, I.2, II.2, and II.3. ADA2 probes are depicted on the X axis. The Y axis shows the ratio of relative fluorescence units (RFU) of the specific ADA2 probe and the RFU value of reference probe. Individuals I.2, II.2, and II.3 exhibit a homozygous duplication of exon 7, while I.1 shows a heterozygous duplication. HC, healthy control; Ex, exon; In, intron.
D: Quantitative reverse transcription (qRT) PCR of ADA2 using three distinct sets of primers showing almost complete absence of ADA2 expression in I.2, II.2, and II.3. Mother (1.1) has an approximately 50% reduction in expression compared to healthy controls. HC, healthy control.
E: Long-range PCR using a set of primers flanking ADA2 exon 7 demonstrates a homozygous duplication of approximately 800bp in 1.2, II.2, and II.3, while 1.1 shows a heterozygous duplication. HC, healthy control. Marker, DNA Ladder (GeneRuler 1 kb Plus, Thermo Fisher).
F: Schematic representation of wildtype ADA2 locus with exons 6–8 and Alu-mediated duplication of exon 7. WT, wildtype; MT, mutant; Primer for, Forward primep Primer rev, Reverse primer; AluY/Sc8, Alu-element subfamilies AluY and AluSc8.
The patient’s clinically asymptomatic father and younger sister (II.3) underwent evaluation and had no significant clinical history. On laboratory evaluation, the father had a low IgG 508 mg/dL (normal 700–1600 mg/dL) but otherwise unremarkable values. The younger sister had hypogammaglobulinemia (IgG/IgA/IgM) and a positive anti-dsDNA antibody but was otherwise normal. Anti-TNF therapy was declined by the family and, upon follow-up communication, the proband was still awaiting hematopoietic stem cell transplantation.
DISCUSSION
DADA2 is an autosomal recessive monogenic autoinflammatory disorder where most patients carry LOF variants in the coding region of ADA2. Molecular diagnosis can be complicated as ADA2 is highly polymorphic and there are over 300 SNVs (missense, nonsense, canonical splice site) with a minor allele frequency of <0.001 identified throughout the gene locus [25]. Clinical significance for most of these variants still needs to be determined. DADA2 is considered a rare disease; however, since its initial description in 2014, over 200 affected individuals in multiple populations have been reported in the literature. DADA2 prevalence is higher in founder populations with a high degree of consanguinity.
We report the identification of two pathogenic variants in ADA2 that were missed by standard clinical genetic testing, including chromosomal microarray and exome sequencing. Patient 1 is compound heterozygous for the known pathogenic variant p.Y453C and the canonical donor splice site variant c.−47+2T>C. This splice site variant is located at the boundary of non-coding exon 1 and intron 1 of ADA2 and had been previously identified in a patient who presented with a severe hematological phenotype as result of having two severe LOF variants in ADA2 (c.[144del];[−47+2T>C]) [4]. Splice variants in ADA2 have been reported in patients with vasculitis, bone marrow failure (BMF), and pure red cell aplasia (PRCA), and the phenotypic severity is related to the level of enzymatic activity and protein expression of ADA2 [7, 18, 26–28]. Our patient 1 presented with vasculitis highly responsive to anti-TNF therapy. The crucial role of the 5’-UTR in the regulation of ADA2 function was indicated in the original report on DADA2 and thus this noncoding region of ADA2 should routinely be included in CLIA-approved sequencing assays [1].
Case 1 is also a prime example of how the type of exome sequencing capture kit can influence clinical sensitivity and specificity. During the exome capture process, protein coding regions are captured via hybridization of genomic DNA to biotinylated oligonucleotide probes, called baits [29]. However, the four major commercially used exome capture technologies exhibit significant differences in regards to target choice, bait lengths, and bait density [30–32]. Exome sequencing for the present study was performed using the Roche SeqCap EZ Human Exome Probes v3.0 capture technology, which does not include baits for the untranslated exon 1 of ADA2 transcript NM_001282225.2. This exon was therefore originally not sequenced, and no variants could be identified in the downstream analysis. The described concern also applies to targeted gene panels capturing specific genes of interest. It is therefore crucial for healthcare providers to make an informed decision on what gene panel or exome sequencing technology is most appropriate.
Patient 2 is homozygous for a novel structural variant in ADA2 consisting of an exon 7 tandem duplication (Fig. 3F). To our knowledge, this is the second report on a duplication within the ADA2 gene locus but the first to describe a single exon duplication [33]. Long-read sequencing indicates that the causative mechanism is the non-allelic homologous recombination (NAHR) mediated by two Alu elements flanking exon 7 [34]. Meiotic NAHR is mediated by low-copy repeats, such as Alu elements and can result in genomic loss or gain that can be inherited by the next generation [35]. The two Alu elements flanking ADA2 exon 7 (subfamilies: AluY and AluSc8) are highly homologous (86% identity) and genomic rearrangements involving this specific combination of Alu elements have been previously associated with human disease previously [36, 37]. In line with this, a heterozygous CNV resulting in loss of ADA2 exon 7 was described in three sisters with DADA2, indicating a similar Alu-mediated mechanism [38].
The structural variant in patient 2 was missed by a variety of methods, including Sanger sequencing of ADA2, chromosomal microarray analysis, and exome sequencing. The size of the duplication is approximately 800bp and therefore too large to be confidently identified by Sanger sequencing. The maximum resolution of the chromosomal microarray platform used in this study is stated as approximately 10kb and is therefore not high enough to detect structural variants in this size range. CNV detection using exome sequencing data is mainly based on the identification of statistically significant changes in read depth between samples [39]. However, this method is still challenging, especially for smaller CNVs. The three distinct calling algorithms (XHMM, CNVnator, Atlas-CNV) used in this study failed to detect the ADA2 exon 7 duplication [40–42]. Because of the difficulty in identifying this type of pathogenic variant, the availability of ADA2 enzymatic testing and high-resolution copy number analysis via MLPA are important for a diagnosis of DADA2.
Interestingly, the proband’s father, genetically and enzymatically diagnosed with DADA2, has not shown any clinical symptoms by age 38 years. Asymptomatic adults with deficient ADA2 enzyme activity due to confirmed biallelic pathogenic variants have been reported previously and indicate high variable expressivity and/or incomplete penetrance of DADA2 [10]. The fact that the patient and her father are clinically distinct despite the same homozygous pathogenic variant suggests the presence of additional disease modifiers, such as susceptibility alleles and epigenetic or environmental factors.
Of note, case 2 demonstrates pseudodominant transmission of an autosomal recessive disease and raises the question of the frequency of the ADA2 exon 7 duplication in the Pakistani population. It is surmised that the patient’s paternal grandparents must have inherited the identical-by-descent haplotype encompassing the deleterious variant from a common ancestor. It is not known how many generations ago this common ancestor lived. Considering the high rate of first cousin marriages (≈50%) in the Pakistani population, leading to an increase in the frequency of deleterious alleles, the minor allele frequency of this variant might be unexpectedly high [43, 44]. We suggest that patients of Pakistani ancestry suspected to have DBA-like presentation or severe anemia to be tested for the exon 7 duplication using a simple PCR method described in Figure 3E.
In summary, this study identified and evaluated the effect of two null variants in ADA2 that were missed by a variety of sequencing methods used in routine diagnostics. The importance of molecular diagnosis was highlighted recently by the observation that variants associated with a complete loss of ADA2 enzyme activity are also associated with PRCA and BMF, while hypomorphic variants preserving some residual enzyme activity were often present in patients with vasculitis [18]. Together, these findings indicate that genetic testing, including high resolution CNV detection, should be complemented by enzymatic testing in order to make informed treatment decisions.
Genetic and enzymatic testing should also be performed on asymptomatic family members in order to counsel those individuals on the risk of disease and to optimize their evaluation and medical care. Based on our experience with Family 1, we would like to recommend that asymptomatic DADA2 patients should be prophylactically treated. Patients who carry two null variants and have absent ADA2 activity are unlikely to respond to anti-TNF therapy and they should therefore be considered for transplantation. Additionally, a combination of enzymatic and genetic testing is advisable since reduced penetrance, subclinical family members, and unexpected inheritance patterns can further complicate the diagnosis of DADA2.
Supplementary Material
Supplementary Figure 1:
A: Haplotype analysis of the two-generation family 1. All four children inherited the allele carrying the c.1358A>G; p.Y453C pathogenic variant from their father (depicted in red). The three affected children II.1, II.2, and II.4 inherited the same haplotype (depicted in green) from their mother, which is associated with a second pathogenic variant.
B: Multiplex ligation-dependent probe amplification (MLPA) analysis of the ADA2 gene locus in family 1 did not identify CNVs in individuals I.1, II.1, II.2, II.3, and II.4. ADA2 probes are depicted on the X axis. The Y axis shows the ratio of relative fluorescence units (RFU) of the specific ADA2 probe and the RFU value of reference probe. HC, healthy control; Ex, exon; In, intron.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Programs of the NHGRI and the NIH Clinical Center. The study was additionally supported by a NIDCR, NIH intramural research grant (1ZIADE000695) to JAC and a DDIR Innovation Award to JAC and DGM. Genome sequencing was supported by the Department of Defense (award W81XWH-09-2-0128). This work utilized the computational resources of the NIH HPC Biowulf cluster. OS is supported by the German Research Foundation. We would like to thank the patients, the families, and the healthy controls for their enthusiastic support during this study. We also thank Adelani Adeleye, Camille Alba, Dagmar Bacikova, Daniel N. Hupalo, Elisa McGrath Martinez, Anthony R. Soltis, Gauthaman Sukumar, and Xijun Zhang from the American Genome Center (Uniformed Services University of the Health Sciences, Bethesda, MD, USA) and Harvey B. Pollard and Matthew D. Wilkerson from the Department of Anatomy, Physiology & Genetics (Uniformed Services University of the Health Sciences, Bethesda, MD, USA).
Footnotes
DISCLOSURE OF CONFLICTS OF INTERESTS
The authors declare that they have no conflict of interest
ETHICAL APPROVAL
The study was approved by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) Institutional Review Board.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- 1.Zhou Q, Yang D, Ombrello AK, Zavialov AV, Toro C, Zavialov AV, et al. Early-onset stroke and vasculopathy associated with mutations in ADA2. The New England journal of medicine. 2014; 370(10):911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navon Elkan P, Pierce SB, Segel R, Walsh T, Barash J, Padeh S, et al. Mutant Adenosine Deaminase 2 in a Polyarteritis Nodosa Vasculopathy. New England Journal of Medicine 2014. 2014/03/06; 370(10):921–931. [DOI] [PubMed] [Google Scholar]
- 3.Hashem H, Egler R, Dalai J. Refractory Pure Red Cell Aplasia Manifesting as Deficiency of Adenosine Deaminase 2. Journal of pediatric hematology/oncology 2017. July; 39(5):e293–e296. [DOI] [PubMed] [Google Scholar]
- 4.Hashem H, Kumar AR, Muller I, Babor F, Bredius R, Dalai J, et al. Hematopoietic stem cell transplantation rescues the hematological, immunological, and vascular phenotype in DADA2. Blood. 2017. December 14; 130(24):2682–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Ami T, Revel-Vilk S, Brooks R, Shaag A, Hershfield MS, Kelly SJ, et al. Extending the Clinical Phenotype of Adenosine Deaminase 2 Deficiency. The Journal of Pediatrics. 2016. 2016/10/01/; 177:316–320. [DOI] [PubMed] [Google Scholar]
- 6.Schepp J, Bulashevska A, Mannhardt-Laakmann W, Cao H, Yang F, Seidl M, et al. Deficiency of Adenosine Deaminase 2 Causes Antibody Deficiency. J Clin Immunol. 2016. April; 36(3):179–186. [DOI] [PubMed] [Google Scholar]
- 7.Schepp J, Proietti M, Frede N, Buchta M, Hubscher K, Restrepo JR, et al. Screening of 181 Patients With Antibody Deficiency for Deficiency of Adenosine Deaminase 2 Sheds New Light on the Disease in Adulthood. Arthritis & Rheumatology. 2017; 69(8):1689–1700. [DOI] [PubMed] [Google Scholar]
- 8.Özen S,,Batu ED, Taşkiran EZ, Özkara HA, Ünal Ş, Güleray N, et al. A Monogenic Disease with a Variety of Phenotypes: Deficiency of Adenosine Deaminase 2. The Journal of Rheumatology. 2020; 47(1 ):117–125. [DOI] [PubMed] [Google Scholar]
- 9.Van Nieuwenhove E, Humblet-Baron S, Van Eyck L, De Somer L, Dooley J, Tousseyn T, et al. ADA2 Deficiency Mimicking Idiopathic Multicentric Castleman Disease. Pediatrics. 2018. September; 142(3). [DOI] [PubMed] [Google Scholar]
- 10.Nanthapisal S, Murphy C, Omoyinmi E, Hong Y, Standing A, Berg S, et al. Deficiency of Adenosine Deaminase Type 2: A Description of Phenotype and Genotype in Fifteen Cases. Arthritis & Rheumatology 2016; 68(9):2314–2322. [DOI] [PubMed] [Google Scholar]
- 11.Van Montfrans JM, Hartman EA, Braun KP, Hennekam EA, Hak EA, Nederkoorn PJ, et al. Phenotypic variability in patients with ADA2 deficiency due to identical homozygous R169Q mutations. Rheumatology (Oxford). 2016. May; 55(5):902–910. [DOI] [PubMed] [Google Scholar]
- 12.Springer JM, Gierer SA, Jiang H, Kleiner D,Deuitch N,Ombrello AK, et al. Deficiency of Adenosine Deaminase 2 in Adult Siblings: Many Years of a Misdiagnosed Disease With Severe Consequences. Frontiers in Immunology 2018. 2018-June-14; 9(1361). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batu ED, Karadag O, Taskiran EZ, Kalyoncu U, Aksentijevich I, Alikasifoglu M, et al. A Case Series of Adenosine Deaminase 2-deficient Patients Emphasizing Treatment and Genotype-phenotype Correlations. J Rheumatol. 2015. August; 42(8):1532–1534. [DOI] [PubMed] [Google Scholar]
- 14.Claassen D, Boals M, Bowling KM, Cooper GM, Cox J, Hershfield M, et al. Complexities of genetic diagnosis illustrated by an atypical case of congenital hypoplastic anemia. Molecular Case Studies. 2018; 4(6):a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caorsi R, Penco F, Grossi A, Insalaco A, Omenetti A, Alessio M, et al. ADA2 deficiency (DADA2) as an unrecognised cause of early onset polyarteritis nodosa and stroke: a multicentre national study. Annals of the Rheumatic Diseases. 2017; 76(10):1648–1656. [DOI] [PubMed] [Google Scholar]
- 16.Aksentijevich I, Moura NS, Barron K. Adenosine Deaminase 2 Deficiency. GeneReviews®[Internet]: University of Washington, Seattle; 2019. [PubMed] [Google Scholar]
- 17.Gibson KM, Morishita KA, Dancey P, Moorehead P, Drogemoller B, Han X, et al. Identification of novel Adenosine Deaminase 2 gene variants and varied clinical phenotype in pediatric vasculitis. Arthritis & rheumatology (Hoboken, NJ). 2019. April 22. [DOI] [PubMed] [Google Scholar]
- 18.Lee PY, Kellner ES, Huang Y, Furutani E, Huang Z, Bainter W, et al. Genotype and functional correlates of disease phenotype in deficiency of adenosine deaminase 2 (DADA2). J Allergy Clin Immunol. 2020. January 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashem H, Vatsayan A, Gupta A, Nagle K, Hershfield M, Dalal J. Successful reduced intensity hematopoietic cell transplant in a patient with deficiency of adenosine deaminase 2. Bone Marrow Transplantation 2017. 2017/11/01; 52(11):1575–1576. [DOI] [PubMed] [Google Scholar]
- 20.Van Eyck L, Hershfield MS, Pombal D, Kelly SJ, Ganson NJ, Moens L, et al. Hematopoietic stem cell transplantation rescues the immunologic phenotype and prevents vasculopathy in patients with adenosine deaminase 2 deficiency. The Journal of allergy and clinical immunology. 2015; 135(1):283–287.e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ombrello A, Stone D, Hoffmann P, Jones A, Barham B, Barron K, et al. The deficiency of adenosine deaminase type 2-results of therapeutic intervention. Pediatric Rheumatology Online Journal. 2015. September/28; 13(Suppl 1):O40–O40. [Google Scholar]
- 22.Ombrello AK, Qin J, Hoffmann PM, Kumar P, Stone D, Jones A, et al. Treatment Strategies for Deficiency of Adenosine Deaminase 2. New England Journal of Medicine. 2019 2019/April/18; 380(16):1582–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002. January; 30(1):97–101. [DOI] [PubMed] [Google Scholar]
- 24.Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nature Methods. 2018. 2018/June/01; 15(6):461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019:531210 [Google Scholar]
- 26.Chong-Neto HJ, Segundo GRS, Bandeira M, Chong-Silva DC, Rosário CS, Riedi CA, et al. Homozygous Splice ADA2 Gene Mutation Causing ADA-2 Deficiency. Journal of Clinical Immunology. 2019. 2019/November/01; 39(8):842–845. [DOI] [PubMed] [Google Scholar]
- 27.Trotta L, Martelius T, Siitonen T, Hautala T, Hamalainen S, Juntti H, et al. ADA2 deficiency: Clonal lymphoproliferation in a subset of patients. J Allergy Clin Immunol. 2018. April; 141(4):1534–1537.e1538. [DOI] [PubMed] [Google Scholar]
- 28.Alsultan A, Basher E, Alqanatish J, Mohammed R, Alfadhel M. Deficiency of ADA2 mimicking autoimmune lymphoproliferative syndrome in the absence of livedo reticularis and vasculitis. Pediatric blood & cancer. 2018. April; 65(4). [DOI] [PubMed] [Google Scholar]
- 29.Clark MJ, Chen R, Lam HYK, Karczewski KJ, Chen R, Euskirchen G, et al. Performance comparison of exome DNA sequencing technologies. Nature biotechnology. 2011; 29(10):908–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shigemizu D, Momozawa Y, Abe T, Morizono T, Boroevich KA, Takata S, et al. Performance comparison of four commercial human whole-exome capture platforms. Scientific Reports. 2015 2015/August/03; 5(1):12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chilamakuri CSR, Lorenz S, Madoui M-A, Vodák D,Sun J, Hovig E, et al. Performance comparison of four exome capture systems for deep sequencing. BMC Genomics. 2014; 15(1):449-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asan, Xu Y, Jiang H, Tyler-Smith C, Xue Y, Jiang T, et al. Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol. 2011; 12(9):R95–R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grossi A, Cusano R, Rusmini M, Penco F, Schena F, Podda RA, et al. ADA2 deficiency due to a novel structural variation in 22q11.1. Clinical Genetics. 2019; 95(6):732–733. [DOI] [PubMed] [Google Scholar]
- 34.Song X, Beck CR, Du R, Campbell IM, Coban-Akdemir Z, Gu S, et al. Predicting human genes susceptible to genomic instability associated with Alu/Alu-mediated rearrangements. Genome Res. 2018. August; 28(8):1228–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weckselblatt B, Rudd MK. Fluman Structural Variation: Mechanisms of Chromosome Rearrangements. Trends in genetics: TIG. 2015. October; 31 fl0):587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conceição Pereira M, Loureiro JL, Pinto-Basto J, Brandão E, Margarida Lopes A, Neves G, et al. Alu elements mediate large SPG11 gene rearrangements: further spatacsin mutations. Genetics in Medicine. 2012. 2012/January/01; 14(1 ):143–151. [DOI] [PubMed] [Google Scholar]
- 37.Gu W, Zhang F, Lupski JR. Mechanisms fbjjTiuman genomic rearrangements. Pathogenetics. 2008; 1(1 ):4–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liebowitz J, Hellmann DB, Schnappauf O. Thirty Years of Followup in 3 Patients with Familial Polyarteritis Nodosa due to Adenosine Deaminase 2 Deficiency. The Journal of Rheumatology. 2019:jrheum.180820. [DOI] [PubMed] [Google Scholar]
- 39.Yao R, Zhang C, Yu T, Li N, Hu X, Wang X, et al. Evaluation of three read-depth based CNV detection tools using whole-exome sequencing data. Mol Cytogenet 2017; 10:30–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang T, Wu T-J, Hu J, Sedlazeck FJ, White S, et al. Atlas-CNV: a validated approach to call single-exon CNVs in the eMERGESeq gene panel. Genetics in Medicine. 2019. 2019/September/01; 21 (9):2135–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011. June; 21 (6):974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012. October 5; 91 (4):597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erzurumluoglu AM, Shihab HA, Rodriguez S, Gaunt TR, Day INM. Importance of Genetic Studies in Consanguineous Populations for the Characterization of Novel Human Gene Functions. Annals of human genetics. 2016; 80(3):187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bittles AH, Black ML. Consanguinity, human evolution, and complex diseases. Proceedings of the National Academy of Sciences. 2010; 107(suppl 1):1779–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1:
A: Haplotype analysis of the two-generation family 1. All four children inherited the allele carrying the c.1358A>G; p.Y453C pathogenic variant from their father (depicted in red). The three affected children II.1, II.2, and II.4 inherited the same haplotype (depicted in green) from their mother, which is associated with a second pathogenic variant.
B: Multiplex ligation-dependent probe amplification (MLPA) analysis of the ADA2 gene locus in family 1 did not identify CNVs in individuals I.1, II.1, II.2, II.3, and II.4. ADA2 probes are depicted on the X axis. The Y axis shows the ratio of relative fluorescence units (RFU) of the specific ADA2 probe and the RFU value of reference probe. HC, healthy control; Ex, exon; In, intron.