Abstract
Mature B-cell neoplasms are the fifth most common neoplasm. Due to significant heterogeneity at the clinical and genetic levels, current therapies for these cancers fail to provide long-term cures. The clinical success of proteasome inhibition for the treatment of multiple myeloma and B-cell lymphomas has made the ubiquitin pathway an important emerging therapeutic target. In this study, we assessed the role of the E3 ligase FBXW7 in mature B-cell neoplasms. FBXW7 targeted the frequently inactivated tumor suppressor KMT2D for protein degradation, subsequently regulating gene expression signatures related to oxidative phosphorylation (OxPhos). Loss of FBXW7 inhibited diffuse large B-cell lymphoma cell growth and further sensitized cells to OxPhos inhibition. These data elucidate a novel mechanism of regulation of KMT2D levels by the ubiquitin pathway and uncover a role of FBXW7 in regulating oxidative phosphorylation in B-cell malignancies.
Introduction
The ubiquitin-proteasome system (UPS) specifically promotes degradation of the majority of cellular proteins and regulates a variety of cellular processes. E3 ubiquitin ligases are the UPS effectors that recognize target proteins and promote ubiquitylation and degradation(1,2).
FBXW7 (F-box/WD40 repeat-containing protein 7; gene ID: 55294) is a member of the F-box family of proteins that functions as the substrate-targeting subunit of the Skp1-Cul1-F-box ubiquitin ligase complex SCF-FBXW7(3,4). The F-box domain of FBXW7 allows interaction with Skp1, which links FBXW7 to the rest of the SCF complex(3,4). Through a C-terminal WD40 domain, FBXW7 recognizes a phosphorylated amino acid motif (or phosphodegron) in substrates(3,4). The FBXW7 phospho-degron is conserved from yeast to mammals and follows the general pattern of pS/pT-P-P-X-pS/pT/E/D (that is, a pSer or pThr, two Pro repeats, any amino acid, pSer or pThr or a phosphomimetic glutamic or aspartic acid)(3,4). Amino acid mutation of key WD40 residues or amino acid mutation of a substrate degron can ablate FBXW7-substrate interactions(5–7).
FBXW7 promotes degradation of multiple substrates including CYCLIN E1, c-MYC, JUN, MCL1, and NOTCH1, several of which are known to contribute to carcinogenesis(3,4). Hence, FBXW7 is generally considered to be a tumor suppressor through its negative regulation of these oncogenic proteins. Importantly, mutations in the FBXW7 gene have been described in many solid tumors (mutation rates ranging from 4% to 30% depending on cancer type)(3,8–10). However, FBXW7 mutations are rarely observed in B-cell malignancies (11–14), suggesting a different role in this cell context.
B-cell cancers hijack the regulatory mechanisms controlling physiologic B-cell differentiation for their own advantage of growth and survival. The majority of B-cell non-Hodgkin’s lymphomas (B-NHLs) (including diffuse large B-cell lymphoma (DLBCL)) and multiple myeloma (MM) are neoplastic processes derived from germinal or post-germinal center B-cells. DLBCL and MM display an elevated genetic heterogeneity that limits the efficacy of current cancer therapies(15,16).
Importantly, FBXW7 was highlighted as a pro-survival factor in non-malignant B-cells: multiple myeloma, chronic myeloid leukemia (CML) and B-cell acute lymphoblastic leukemia (B-ALL) (5,17–19). Thus, FBXW7 may be tumor suppressive or pro-survival in different hematologic cancer types depending on the cell context and the relevant substrate(s) targeted for proteolysis.
Based on the clinical success of a proteasome inhibitor, bortezomib, as well as E3 ubiquitin ligase activators, such as the immunomodulatory imide drugs (IMiDs), the sulfonamides, and the proteolysis targeting chimeras (PROTACs)(20–23), it is critical to understand how protein ubiquitylation is achieved and what the downstream consequences of such molecular events are for the development of novel therapeutics. In this study, we focus on expanding the functional understanding of FBXW7 in B-cell cancers and propose the lysine methyl transferase 2D (KMT2D, also known as MLL2, ALR or MLL4; gene ID: 8085) as a novel substrate of FBXW7.
Material and Methods
Cell culture and drug treatment
OPM-1, KMS-11, BJAB, SUDHL10, U2932, HLY1, and HT cells were maintained in RPMI-1640 media containing 10% fetal bovine serum (FBS). HEK293T cells were maintained in Dulbecco’s modified Eagle’s media (DMEM) containing 10% FBS. HEK293T cells were purchased from ATCC. B-cell cancer cell lines were kindly provided by Dr. Laura Pasqualucci, Dr. Michele Pagano and Dr. Yibin Yang. Cell line authentication was not performed as cells were not listed in the commonly misidentified category. All cell lines were free of mycoplasma contamination (MycoAlert kit, Lonza). Cell lines were passaged for less than 6 months after receipt or resuscitation. The following drugs were used: MG132 (Peptides International, #IZL-3175-v; 10 μM final concentration), MLN4924 (EMD Millipore, #505477; 5 μM final concentration), cycloheximide (Sigma-Aldrich, #C7698; 20 mg/mL final concentration), Doxycycline hyclate (Sigma-Aldrich, #D9891; 1 ng/mL or 1 mg/mL final concentration), and IACS-010759 (Selleck Chemicals, #S8731), puromycin (Sigma-Aldrich, 0.5–1 μg/ml final concentration), hygromycin B (EMD Millipore, #400052, 100–200 μg/mL), zeocin (100–250 μg/mL), and blasticidin S HCl (Gibco, #R21001, 3–15 μg/mL). CRISPR-derived HEK293T FBXW7–/– clones were selected using single cell cloning in 96-well plates.
Cell proliferation assay and generation of drug response curve
Cells were cultured in 24-well plates with 1 mL of medium and plated at equal densities on Day 0; cell numbers were counted every 3 days. After each counting, cells were re-plated at the same dilutions in 1 mL until the experimental endpoint. To assess cellular sensitivity to IACS-010759, cells were plated at equal densities for 4 days at the indicated doses of IACS-010759. Cell counting and viability were assessed with the Attune NxT flow cytometer (Thermo Scientific).
Reagents
The following antibodies were used: FLAG (Sigma, #F7425), KMT2D (Bethyl Laboratories, #A300-BL1185), UTX (Bethyl Laboratories, #A302–374A), ASH2L (Bethyl Laboratories, #A300–489A), WDR5 (Bethyl Laboratories, #A302–430A), DPY30 (Bethyl Laboratories, #A304–296A), NCOA6 (Bethyl Laboratories, #A300–411A), CUL1 (Invitrogen, #718700), FBXW7 (Bethyl Laboratories, #A301–720), C-MYC (Santa Crus, #sc-764), c-MYC-pT58 (Applied Biological Materials, #Y011034), HA (Biolegend, #901513), HA (Cell Signaling Technology, #3724), α-Tubulin (Santa Crus, #sc-8035), Skp1 (Santa Cruz, #sc-7163), CyclinE1 (Cell Signaling Technology, #4129), Cdk2 (Santa Cruz, #sc-6248), CyclinE (clone HE12), Vinculin (Santa Cruz, #sc-73614), ubiquitin K48 (EMD Millipore, 05–1307), Rabbit IgG-HRP (GE Healthcare, NA934V), mouse IgG-HRP (GE Healthcare, NA931V) and anti-FLAG M2 Affinity Gel (Sigma, #A2220).
Plasmids
KMT2D deletion mutants were generated using PCR with primers designed with the QuikChange Primer design tool. Domain deletions in FBXW7 and KMT2D were achieved by intramolecular ligation cloning strategies. The Flp-In™ T-Rex™ System manufacturer’s protocol was used to generate BJAB cells expressing exogenous KMT2D. Retroviral expression of FBXW7 was performed using pMSCV-puro plasmid. Lentivirus encoding shRNAs targeting human FBXW7 were described previously (5).
Lentiviruses encoding sgRNAs targeting human FBXW7 and KMT2D were generated from a Lenti-Guide-Puro, Lenti-Guide-GFP, or Lenti-Guide-mCherry vector. Cas9 was delivered using the lentiCas9-Blast vector. The target sequences to knockout human FBXW7 and KMT2D were:
hFBXW7 sgRNA#2: 5’-AGCAAAAGACGACGAACTGG AGG-3’
hFBXW7 sgRNA#4: 5’-TGAAGAACAGATGAATCGTG TGG-3’
hKMT2D sgRNA#1: 5’-GACCTGCCCAACCCACATGT GGG-3’
hKMT2D sgRNA#2: 5’-GACCTCTCCCACATGTGGGT TGG-3’
Luciferase control sgRNA: 5’-CTTCGAAATGTCCGTTCGGT-3’
Gene silencing by siRNA
The following siRNAs were obtained from Dharmacon: siKMT2D (ON-TARGETplus Human KMT2D (8085) siRNA - Individual, 10 nmol, #J-004828–07-0010). Non-targeting Control siRNAs were obtained from Dharmacon (D-001810–01-20). Custom siRNA against all human FBXW7 isoforms was ordered from Dharmacon with the following sequence: 5’ACAGGACAGUGUUUACAAAdTdT3’.
Transfection and retrovirus- or lentivirus-mediated gene transduction
HEK293T cells were transfected using polyethylenimine (PEI) (Polysciences, #24765). For siRNA or DNA transfection in suspension cell lines, the Neon transfection system (Thermo Fisher Scientific, #MPK5000) was used. Adherent cell siRNA transfection was carried out with Lipofectamine™ RNAiMAX Transfection Reagent (ThermoFisher #13778075) according to manufacturer’s protocol. All siRNAs were transfected with the following amounts: 150 pmol siRNA per 1 million suspension cells or 150 pmol siRNA per 10cm tissue culture dish at 60% confluency. All transducing viruses were generated from HEK293T cells. For lentivirus production: psPAX2 and pMD2.G were used. For retrovirus production, GP-293 packaging cells (Clontech) were used. Sixteen hours post-transfection, HEK293T media was changed and viral supernatants were collected at 48 hours and 72 hours and pooled. Target cells were spin-infected in viral supernatant with 8 μg/mL polybrene at 2,400 rpm for 90 mins at 30 °C, after which new media was added. Antibiotic selection or cell sorting (GFP, mCherry, or both) was started 2–7 day post-infection.
Immunoprecipitation and immunoblot
Extract preparation, immunoprecipitation, and immunoblotting were carried out as previously described(24). Normalization and quantification of protein levels were carried out as previously described(24). For λ-phosphatase experiments: α-FLAG immunoprecipitates were incubated for 1 hour at 30°C with or without λ-phosphatase (Lambda PP, NEB, #P0753S). ImageJ was used to quantify bands from immunoblots.
LCMS/MS. Purification and analysis of FBXW7 interactors
HEK293T cells were transfected with FLAG–HA-tagged FBXW7α(WT) or FBXW7α(∆WD40) mutant. Immunopurification, sample preparation, and analyses were carried out as previously described(5). For analysis of KMT2D phosphopeptides, HEK293T cells were transfected with FLAG-KMT2D (a.a. 199–1059) and lysed in 1% SDS. Lysate was diluted to 0.1% SDS in NP-40 IP buffer and FLAG-immunopurified. Phosphoproteomic analysis of immunopurified FLAGKMT2D (a.a. 199–1059) was carried out as previously described(25).
RNA-Seq
Total RNA was extracted from OPM-1 cells in low serum (with three biological replicates) using RNeasy Mini Kit (QIAGEN, #74104) and polyA+ transcripts were isolated with NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB, #7490S). RNA-Seq libraries were prepared with NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina® (NEB, #E7760S). Libraries were quantified using NEBNext® Library Quant Kit for Illumina (NEB, #E7630S) and then sequenced using a NextSeq500 platform (75-base-pair [bp] single-end reads) (Illumina). Differentially expressed genes (DEGs) were identified through the in-house pipeline. Raw reads (fastq) were mapped to the human reference genome (hg38) using “STAR” (Spliced Transcripts Alignment to a Reference) aligner followed by read trimming using “Trimmomatic” to remove adaptors and low-quality reads. “FeatureCounts” calculated how many reads map to each gene/feature with the parameters ‘-C’ (without reads whose pairs mapped to different chromosomes), ‘-M’ (with multi-mapped reads) and ‘-t exon’ (within exon region). Detailed information of featureCounts can be found in the user guide, https://bioconductor.org/packages/release/bioc/vignettes/Rsubread/inst/doc/SubreadUsersGuide.pdf. Finally, “DESeq2” identified DEGs by comparing the gene expression values of genes across differentially treated samples. GO analyses were performed using the BiNGO Cytoscape tool(26). RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE148131.
Xenotransplantation experiments
All mouse work was performed under our laboratory’s ethical guidelines and protocols approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Female NSG mice were purchased from The Jackson Laboratory. Eight- to twelve-week-old NSG mice received subcutaneous flank injections of 1×107 U2932 CRISPR-Cas9 cells (in 100 μL sterile PBS). Tumor burden was monitored daily by visual checks and palpations. Tumor volume was calculated by caliper measure and tumor weights were determined for excised tumors at the experimental endpoint.
In-vitro ubiquitylation assay
The in-vitro ubiquitylation of KMT2D by FBXW7 was performed using anti-FLAG immunopurified fragments (A, B, C, D, and E) of KMT2D. KMT2D fragments were incubated for 60 minutes at 30 °C in a ubiquitylation reaction mix containing: recombinant SCF-FBXW7, E1 enzyme (100 nM), UbcH5c (15 ng/μl), ubiquitin (2.5 μg/μl) and ATP (2 mM). After ubiquitylation reactions, denatured FLAG-KMT2D fragments were FLAG-immunopurified and probed for ubiquitin signals by SDS-PAGE and immunoblotting.
Measurements of NAD+/NADH and ATP levels
Total NAD+/NADH and NADH levels were determined using the NAD+/NADH Quantification kit (BioVision, #K337–100) according to the manufacturer’s protocol. Briefly, 1.5×106 cells were used per sample in a 400 μL lysis buffer volume. This volume was equally split for analysis of (1) total NAD+/NADH levels and (2) NADH levels only, which were measured after decomposition of NAD+ at 60 °C for 30 mins. The “total” lysate was diluted 1:10 before measurement. For ATP measurement, cells were plated at equal densities in medium and left overnight without or with 5 nM IACS-010759. The next day, cells were plated in white 96-well plates at 100 μL medium per well and ATP levels were measured with the CellTiter-Glo® Luminescent Cell Viability Assay (Promega, G7570).
qPCR gene expression or DNA analysis
Total RNA was extracted using the RNeasy Kit (Qiagen). cDNA synthesis was performed using the RNA to cDNA Ecodry Premix oligo dT kit (Takara, #639542). Total DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, #69504). Quantitative PCR analysis using SYBRGreen PCR Master Mix (Thermo Fisher Scientific, #4309155) was performed according to standard procedures. Primer sequences (5’ to 3’) used:
ND4_qFOR: CTCGCTAACCTCGCCTTACC
ND4_qREV: AGTGAGCCCCATTGTGTTGT
ND4L_qFOR: TAACCCTCAACACCCACTCC
ND4L_qREV: GGCCATATGTGTTGGAGATTG
ATP6_qFOR: CGCCACCCTAGCAATATCAA
ATP6_qREV: TTAAGGCGACAGCGATTTCT
COX1_qFOR: CGATGCATACACCACATGAA
COX1_qREV: AGCGAAGGCTTCTCAAATCA
COX3_qFOR: GGCATCTACGGCTCAACATT
COX3_qREV: CGAAGCCAAAGTGATGTTTG
COX2_qFOR: TGAAGCCCCCATTCGTATAA
COX2_qREV: ACGGGCCCTATTTCAAAGAT
CYTB_qFOR: AATTCTCCGATCCGTCCCTA
CYTB_qREV: GGAGGATGGGGATTATTGCT
16SRNA_qFOR: GGGATAACAGCGCAATCCTA
16SRNA_qREV:CCTGGATTACTCCGGTCTGA
ND1_qFOR: ATACCCCCGATTCCGCTACGAC
ND1_qREV: GTTTGAGGGGGAATGCTGGAGA
hGAPDH_qFOR : GGAGCGAGATCCCTCCAAAAT
hGAPDH_qREV: GGCTGTTGTCATACTTCTCATGG
β-globin_qFOR: CAACTTCATCCACGTTCACC
β-globin_qREV: GAAGAGCCAAGGACAGGTAC
Seahorse XF Oxygen Consumption Rate Analysis
Seahorse XFe96 sensor cartridge was hydrated in a utility plate with 200 μL of Seahorse XF Calibrant solution overnight at 37 °C with 0% CO2. One hour prior to assay, U2932 cells were plated at a density of 1×105 per 180 μL per well in a poly-D-lysine coated XF96 cell culture microplate with 6 replicates. Cells were plated in Seahorse XF RPMI medium containing 2.05 mM L-glutamine and 11.11 mM glucose. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) was measured by the Seahorse XFe96 Analyzer.
MitoTracker Staining
Cells were plated at equal densities and incubated with 50 nM MitoTracker DeepRed FM (Thermo Fisher Scientific, #M22426) at 37 °C for 30 minutes. After staining, cells were washed with and resuspended in PBS before analysis by flow cytometry using the Attune NxT flow cytometer. Data analysis was carried out in FloJo.
Statistical analyses
With the exception of RNA-seq, statistical analyses were performed with Prism 6 (GraphPad). Statistical testing, sample sizes, and reproducibility for each figure are indicated in the corresponding legends.
Patient expression data
Differential expression data for FBXW7 in normal and different cancer types was obtained from Gene Expression across Normal and Tumor tissue (GENT) database. Publicly available microarray data for multiple myeloma (GSE5900) were obtained from Oncomine (www.oncomine.org). Intensities from the FBXW7 probe (229419_at) were plotted. Mitochondrial gene signature analysis was carried out using the publicly available dataset (GSE10846). The following probes were used: PDHB (211023_at), NDUFA5 (201304_at), NDUFS3 (201740_at), ACAT1 (205412_at), ETFA (201931_at), MTHFD2 (201761_at), and SOD2 (216841_s_at).
Data availability
The authors declare that the data supporting the findings of this study are available upon reasonable requests.
Results
Proteomics-based approach to identify putative substrates of FBXW7
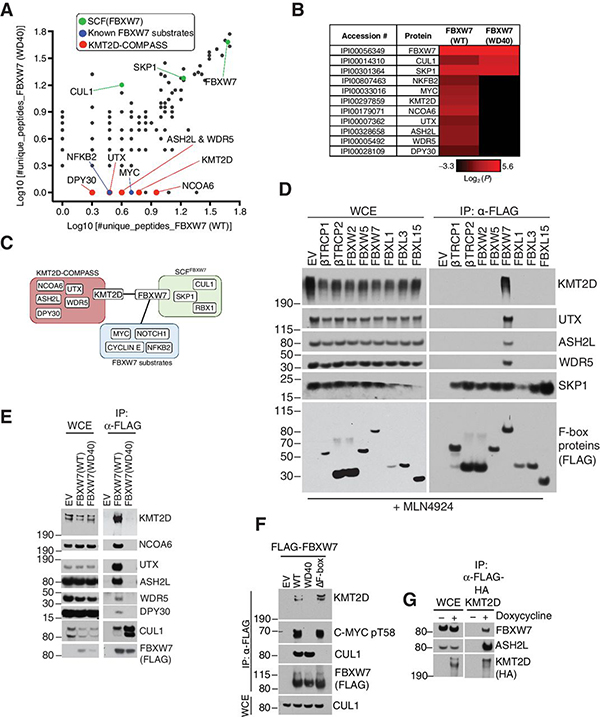
To identify substrates of FBXW7, we immunopurified FLAG-HA-tagged FBXW7 and coupled to mass spectrometry (Supplementary Fig. S1a)(5). The strategy involved purification of FBXW7 wild-type (WT) or FBXW7(WD40). Substrates were defined by the peptides identified in the FBXW7(WT) but not the FBXW7(WD40) immunoprecipitates (Supplementary Fig. S1a). We identified common components of the SCF complex such as CUL1 and SKP1 in both FBXW7(WT) and FBXW7(WD40) immunoprecipitates (Fig. 1a,b), while known substrates, such as p100 (NFKB2) and MYC, were specifically identified in FBXW7(WT) but not FBXW7(WD40) purifications (Fig. 1a,b). Notably, peptides of KMT2D and associated COMPASS complex members (NCOA6, UTX, ASH2L, WDR5, and DPY30(27)) were identified in the FBXW7(WT) but not FBXW7(WD40) immunoprecipitates (Fig. 1a,b), suggesting that KMT2D-COMPASS proteins may be targets of FBXW7 (Fig. 1c).
Figure 1. Proteomics-based approach identifies KMT2D as a substrate-like interactor of FBXW7.
A, Scatter Plot of log10(#unique peptides+1) identified in the mass spectrometry analysis of FBXW7 immunoprecipitates. Known SCF interactors (green), known substrates (blue), and putative substrates (red) are highlighted. B, Heatmap of log2(#unique peptides+0.1) for the proteomic in (a.) C, Schematic representation of SCF-FBXW7 substrate interaction network. D, Immunoblot of indicated proteins from whole cell extracts (WCE) or α-FLAG immunoprecipitates (FLAG-IP) from HEK293T cells overexpressing the indicated FLAG-F-box proteins. Treatment with MLN4924 was for 6 hours. E, Immunoblot of indicated proteins from WCE or FLAG-IP samples from HEK293T cells overexpressing the indicated FLAG-FBXW7 constructs. F, Immunoblot of indicated proteins from FLAG-IP samples from HEK293T cells overexpressing the indicated FLAG-sFBXW7 constructs. G, Immunoblot of the indicated proteins from WCE or FLAG-IP samples in BJAB cells. FLAG-HA-KMT2D was stably expressed in BJAB DLBCL cells using the doxycycline-inducible Flp-In™ T-Rex™ System (doxycycline treatment: 1 μg/mL for 16 hours).
KMT2D methylates histone H3 lysine K4 (H3K4) at enhancer sites in mammalian cells to mark transcriptionally active chromatin(28–30). To investigate whether the interaction between FBXW7 and KMT2D-COMPASS proteins is specific, we screened a panel of FLAG-tagged WD40-containing human F-box proteins. Only FBXW7 out of the eight screened F-box proteins co-immunoprecipitated KMT2D-COMPASS complex members (Fig. 1d). To further validate our proteomic findings, we performed western blot analysis of anti-FLAG co-immunoprecipitates from HEK293T cells expressing FLAG-FBXW7 (WT), FLAG-FBXW7 (WD40), or an empty vector (EV). KMT2D and its associated complex members (NCOA6, UTX, ASH2L, WDR5, and DPY30) were confirmed to interact with FBXW7 in a substrate-like manner, i.e. co-immunoprecipitating with FBXW7(WT) but not with the FBXW7(WD40) substrate-binding mutant (Fig. 1e, f). In a reverse co-immunoprecipitation assay, pull-down of FLAG-HA tagged KMT2D revealed interaction with endogenous FBXW7 (Fig. 1g).
All together, these data identified KMT2D and the members of the COMPASS complex as specific interactors of FBXW7 at the WD40 substrate interface.
KMT2D interacts with FBXW7 in a proteasomal- and neddylation-dependent manner
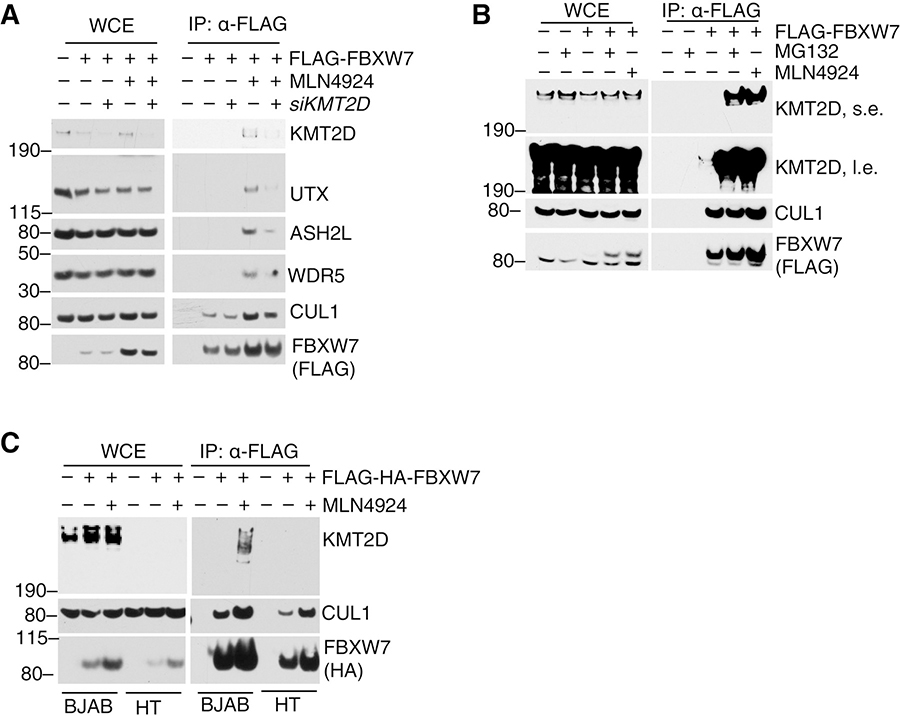
Next, we assessed whether KMT2D was the main FBXW7 interactor. Downregulation of KMT2D reduced interaction between FBXW7 and UTX, ASH2L and WDR5 (Fig. 2a), suggesting that KMT2D mediates COMPASS sub-members interaction with FBXW7. Importantly, KMT2D knockdown did not reduce the total cellular levels of UTX, ASH2L, or WDR5 (Fig. 2a).
Figure 2. KMT2D interacts with FBXW7 in a proteasomal- and neddylation-dependent manner.
A, Immunoblot for the indicated proteins from WCE or FLAG-IP samples from HEK293T cells overexpressing FLAG-FBXW7. Cells were treated with DMSO or MLN4924 (6 hours) and siCTRL or siKMT2D (12 hours). B, Immunoblot for the indicated proteins from WCE or FLAG-IP samples from HEK293T cells overexpressing FLAG-FBXW7 and treated with DMSO or the proteasome inhibitor MG132 for 6 hours. C, Immunoblot for indicated proteins from WCE or FLAG-IP samples from DLBCL cell lines BJAB (KMT2D-positive) or HT (KMT2D-negative) that were infected with retroviruses expressing FLAG-FBXW7 and treated with the neddylation inhibitor MLN4924 for 6 h.
It was recently reported that the lysine-specific histone demethylase 1 (LSD1) interacts with and regulates the function of FBXW7(31). To assess whether KMT2D regulates FBXW7 function, we investigated the effect of KMT2D gain- and loss-of-function on FBXW7 substrate levels. Importantly, over-expression or knockout of KMT2D did not alter the half-life of known FBXW7 substrates such as c-MYC or CYCLIN E1 (Supplementary Fig. S1b, c).
In contrast, we observed that KMT2D-FBXW7 interaction was drastically increased upon proteasomal blockage or inhibition of the NEDD8-activating enzyme (NAE) with MG132 and MLN4924, respectively (Fig. 2b). MG132 blocks the proteasome, abolishing degradation of ubiquitylated proteins(32), while MLN4924 decreases NEDD8–Cullin conjugates and inhibits protein ubiquitylation and degradation mediated by Cullin-based ligases (33). Similar data were obtained in the BJAB DLBCL cell line treated with MLN4924 (Fig. 2c). As a negative control, HT cells carrying KMT2D truncating mutations(34) displayed no interaction between FBXW7 and KMT2D even upon MLN4924 treatment (Fig. 2c).
Altogether, these data demonstrate that interaction of KMT2D with FBXW7 is regulated by the proteasome and by Cullin-neddylation.
The FBXW7-KMT2D interaction is dictated by multiple N-terminal phosphodegrons
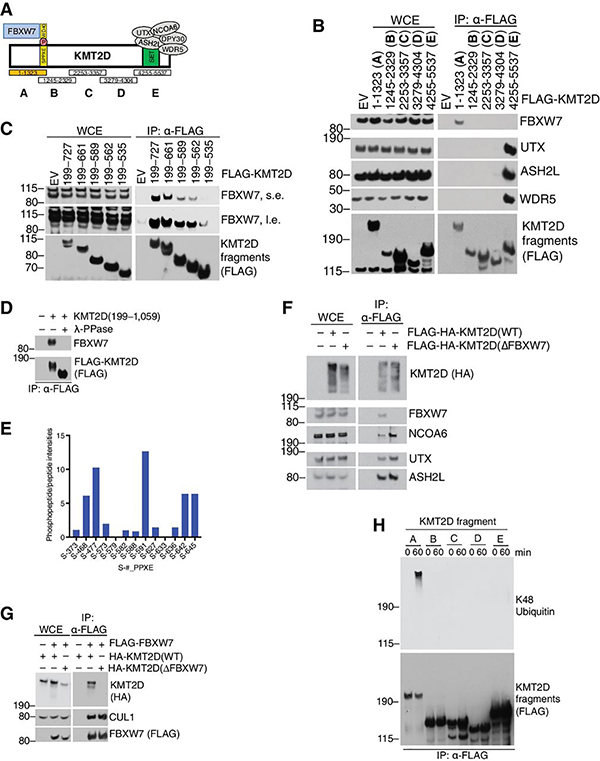
KMT2D is a 5,537 amino acid long protein (~593 kDa) containing a SET domain, seven PHD fingers and an HMG-I binding motif(27). Through the C-Terminal SET domain, KMT2D recruits the COMPASS complex members to promote H3K4 methylation(35) (Fig. 3a). Since the FBXW7 WD40-domain interacts with substrates via a phosphodegron, we decided to map the binding site within KMT2D. We cloned and expressed overlapping FLAG-tagged fragments spanning the large KMT2D protein (Fig. 3a, b). An N-terminal region (amino acids 1 to 1,323, Fragment A) specifically mediated KMT2D interaction with FBXW7. As expected, KMT2D-COMPASS sub-members (ASH2L, WDR5) interacted with KMT2D at the C-terminal SET-domain containing region (Fig. 3a,b) (36), further supporting the evidence that FBXW7 interacts with the COMPASS sub-members via KMT2D.
Figure 3. The FBXW7-KMT2D interaction is dictated by multiple N-terminal phosphodegrons.
A, Schematic of KMT2D interaction with FBXW7 and COMPASS-complex members. KMT2D fragments A-E are indicated. B, Immunoblot for the indicated proteins from WCE or FLAG-IP samples from HEK293T cells overexpressing the indicated FLAG-tagged KMT2D fragments. C, same as in (b), except the indicated KMT2D mutants were tested. D, Immunoblot for indicated proteins from α-FLAG immunoprecipitates of KMT2D (a.a. 199–1,059). α-FLAG immunoprecipitates were treated with or without λ-phosphatase. E, Graph showing relative quantification of KMT2D phosphopeptide to peptide intensities. Peptides were quantified by LC-MS/MS of FLAG-immunoprecipitates from HEK293T cells overexpressing a KMT2D fragment (a.a. 199–1059) and treated with proteasome inhibitor MG132 for 6 hours. F, Immunoblots for the indicated proteins from WCE or FLAG-IP samples from BJAB cells stably expressing doxycycline-inducible FLAG-HA-KMT2D (WT) or (∆FBXW7). G, Immunoblot for indicated proteins from WCE or α-FLAG immunoprecipitates from HEK293T cells overexpressing the indicated proteins. H, Immunoblot for indicated proteins. FLAG-immunopurified KMT2D fragments were subjected to in-vitro ubiquitylation reactions for 0 or 60 mins. Upon reactions, KMT2D fragments were FLAG-immunopurified in denaturing conditions and immunoblotted as indicated.
Closer inspection of the amino acid sequence of the FBXW7-interacting region in KMT2D revealed 25 SPPXE motifs spanning amino acids 450 to 987 (Supplementary Fig. S2a); an unprecedented feature for any FBXW7 substrate(3). Canonical FBXW7 degrons (S/TPPXS/E) require two essential negatively charged amino acids for WD40 domain binding(3). In KMT2D, the distal phospho-site is achieved by a negatively charged Glutamate (Glu, E) (Supplementary Fig. S2a). In the search for a critical SPPXE motif important for interaction with FBXW7, we utilized sub-fragments containing varying numbers of SPPXE motifs and found a proportionally decreased interaction as the number of SPPXE sites decreased (Fig. 3c). Hence, multiple SPPXE motifs are critical for recruiting FBXW7 and might function in a redundant manner.
To assess whether the KMT2D-FBXW7 interaction is phosphorylation dependent, we treated FLAG-KMT2D immunoprecipitates with l-phosphatase and found that interaction with FBXW7 was ablated (Fig. 3d). Strikingly, the KMT2D fragment (containing the SPPXE repeats) displayed a faster migration on gel upon l-phosphatase treatment, suggesting that the region between amino acids 199–1,059 is phosphorylated (Fig. 3d). Mass spectrometry analysis of immunopurified FLAG-KMT2D(aa 199–1,059) revealed that multiple serine residues within the SPPXE degron region were indeed phosphorylated in cells (Fig. 3e).
We cloned a KMT2D protein lacking interaction with FBXW7 (DFBXW7) by deleting the amino acid region 450 to 987 (containing 25 SPPXE degrons). In line with our previous data, immunoprecipitation experiments via FLAG-tagged KMT2D (Fig. 3f) or FLAG-tagged-FBXW7 (Fig. 3g) demonstrated the requirement of these degrons for interaction with FBXW7. Notably, FLAG-KMT2D(DFBXW7) maintained an intact interaction with COMPASS-complex proteins such as NCOA6, UTX, ASH2L, WDR5 (Fig. 3f), suggesting that KMT2D(DFBXW7) is correctly folded.
Finally, in vitro ubiquitylation assay was carried out by incubating the KMT2D fragments (Fig. 3a) with recombinant SCF-FBXW7 (Fig. 3h). In agreement with the mapping data (Fig. 3a, b), Fragment A was specifically ubiquitylated by FBXW7.
Overall, we identified multiple N-terminal phospho-degrons in KMT2D that mediate interaction with the E3 ligase FBXW7.
FBXW7 promotes degradation of KMT2D
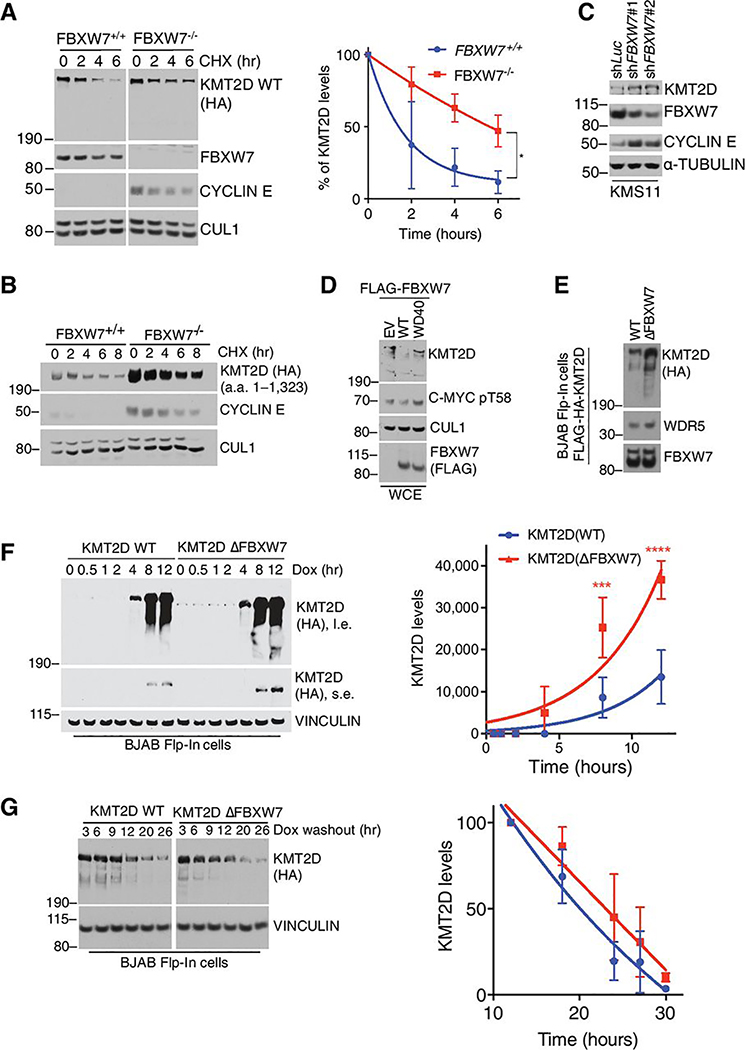
With the observation that KMT2D interacts with FBXW7, we investigated whether loss of FBXW7 had an effect on KMT2D protein turnover. We utilized cycloheximide-chase analyses in HEK293T FBXW7+/+ or FBXW7−/− cells or cells where depletion of FBXW7 was achieved by siRNA. Both experiments revealed that downregulation of FBXW7 led to an increased half-life of KMT2D (Fig. 4a and Supplementary Fig. S2b). Similar data were obtained when we expressed an N-Terminal fragment of KMT2D containing the FBXW7 degrons (Fig. 4b and Supplementary Fig. S2c). These data were also validated in multiple myeloma B-cell cancer cells, KMS11, where knockdown of FBXW7 with two different shRNAs led to an increase in endogenous KMT2D protein levels (Fig. 4c). In addition to loss of function experiments, gain of function via overexpression of FBXW7(WT) reduced KMT2D protein levels, which was not observed when FBXW7(WD40) mutant was overexpressed (Fig. 4d).
Figure 4. FBXW7 promotes degradation of KMT2D.
A, Immunoblot for the indicated proteins from HEK293T cells (FBXW7+/+ or FBXW7−/− CRISPR clones) overexpressing HA-KMT2D WT full-length and treated with cycloheximide (CHX) for the indicated times. ImageJ quantification of band intensity is plotted in the graph (n=3; mean ± SD; one-tailed Student’s t-test (*p<0.05)). KMT2D half-life is 1.9h and 5.5h in the FBXW7+/+ and FBXW7−/− cells, respectively B, Same as in (a), except that indicated HA-KMT2D fragment was expressed. C, Immunoblot for the indicated proteins from KMS11 multiple myeloma cells infected with lentiviruses encoding the indicated shRNAs. D, Immunoblot for the indicated proteins in HEK293T cells overexpressing the indicated FLAG-FBXW7 constructs. E, Immunoblot analysis for the indicated proteins from Flp-In™ T-Rex™ BJAB DLCBCL cells stably expressing KMT2D(WT) or KMT2D(∆FBXW7) upon doxycycline treatment (1 μg/mL). F, Immunoblot for indicated proteins of Flp-In™ T-Rex™ BJAB cells. Cells were treated with doxycycline (1 ng/mL) for the indicated times. Quantification of HA-KMT2D signal was done using ImageJ software (n=3, mean ± SD, two-way ANOVA (***p<0.0002, ****p<0.0001)). G, Immunoblot for indicated proteins of Flp-In™ T-Rex™ BJAB cells. Cells were treated with doxycycline (1 ng/mL) for 4 h. Doxycycline was washed-out and cells harvested at the indicated times. Quantification of HA-KMT2D signal was done using ImageJ software (n=3, data are mean ± SD, two-way ANOVA).
The KMT2D(DFBXW7) mutant, which fails to interact with FBXW7, was expressed at higher levels than KMT2D(WT) (Fig. 4e). Taking advantage of our dox-inducible system, we assessed whether FBXW7 targets neo-synthesized KMT2D. To this end, BJAB cells were treated with doxycycline and KMT2D levels were monitored over time. Interestingly, KMT2D(DFBXW7) accumulated quicker than KMT2D(WT) (Fig. 4f); however, withdrawal of doxycycline showed similar decay kinetics for KMT2D(WT) and KMT2D(DFBXW7) (Fig. 4g). These data suggest that actively translated KMT2D protein could be targeted for proteolysis by FBXW7.
Collectively, our observations suggest that FBXW7 controls KMT2D levels by regulating its protein stability.
FBXW7 promotes proliferation of mature B-cell neoplasms
KMT2D is a tumor suppressor gene (34,37) mutated in 30% of de novo cases of diffuse large B-cell lymphomas and ~90% cases of follicular lymphomas (15,38–41). The spectrum of mutations is largely represented by premature stop codons, frameshift indels, and splice-site mutations (12,15,40,41). Hence, we hypothesized that FBXW7-mediated degradation of KMT2D could favor tumor growth by reducing KMT2D levels.
First, we investigated the expression status of FBXW7 in multiple myeloma and DLBCL patient samples. Expression of FBXW7 was significantly upregulated in multiple myeloma and DLBCL samples compared to normal tissue (Supplementary Fig. S3a). Stage-wise assessment in multiple myeloma samples also revealed a significant upregulation of FBXW7 with disease progression (Supplementary Fig. S3b). Additionally, CRISPR-screens revealed that FBXW7 negatively impacted cell proliferation in DLBCL cells (Supplementary Fig. S3c)(42). These observations suggest that FBXW7 may play a pro-tumoral role in mature B-cell cancers.
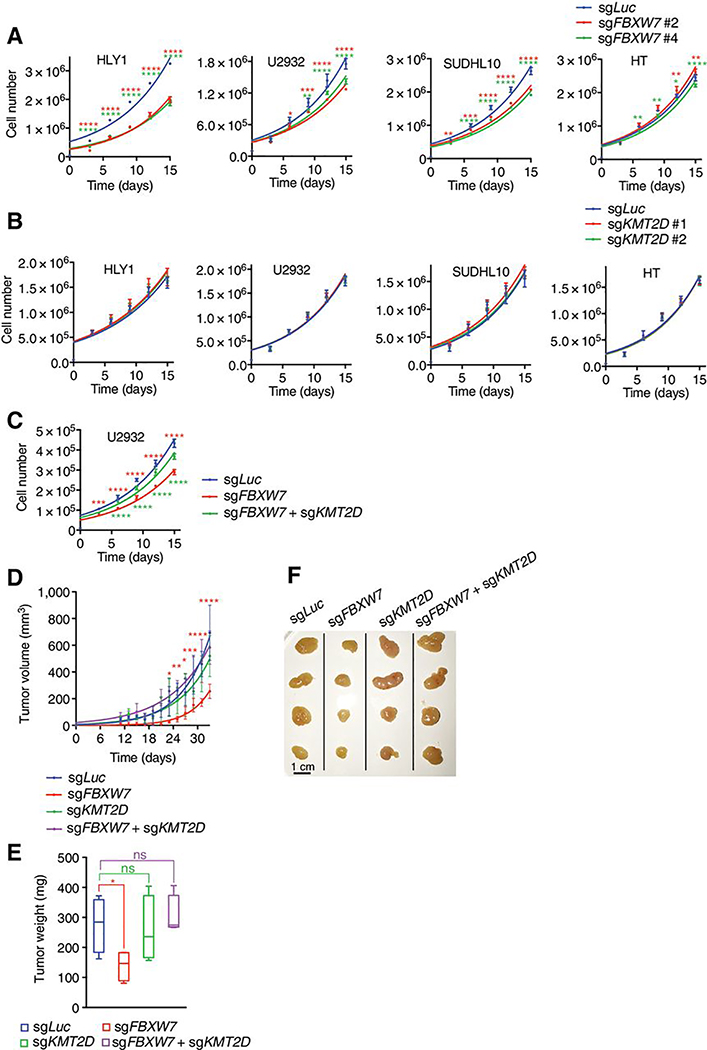
To functionally assess whether loss of FBXW7 inhibits cell proliferation, we performed bulk FBXW7 knockout by CRISPR-mediated gene editing in DLBCL cells (Fig. 5a and Supplementary Fig. S4a). We found that ablation of FBXW7 with 2 sgRNAs resulted in reduced proliferation of HLY1, U2932, and SUDHL10. Interestingly, HT cells carrying a loss of KMT2D protein (Fig. 2c and (34)), were insensitive to FBXW7 deletion. Taken together, these results suggest a pro-survival role of FBXW7 in DLBCL.
Figure 5. FBXW7 promotes proliferation of mature B-cell neoplasms.
A,B,C, Plots of cell counts over time. Indicated cells were infected with lentiviruses encoding sgRNAs targeting human FBXW7, KMT2D or combination (n=3; mean ± SD; two-way ANOVA (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001)). D, Tumor volume measurement over time from xenografts of U2932 cells that were subcutaneously injected into NSG mice flanks (n=4; mean ± SD; two-way ANOVA (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001)). E, Weights of tumors excised from (d) at end-point (n=4, mean ± SD, two-tailed Student’s t-test*p<0.05). F, Pictures of tumors from (d).
To assess the effect of KMT2D loss on cell proliferation, we used 2 KMT2D-targeting sgRNAs in DLBCL cells (Fig. 5b and Supplementary Fig. S4b). Loss of KMT2D had no effect on cell proliferation, suggesting that ablation of this tumor suppressor protein was incapable of further promoting proliferation of DLBCL cells. However, loss of KMT2D rescued the proliferation defect induced by FBXW7 deletion (Fig. 5c and Supplementary Fig. S4c), suggesting that stabilization of KMT2D is limiting for DLBCL proliferation.
The in vitro data were confirmed in a xenotransplantation model. Here, U2932 cells expressing sgRNAs targeting FBXW7, KMT2D or both were injected in the flack of NSG mice. Tumor volume (Fig. 5d) and tumor weight (Fig. 5e,f) analysis demonstrated that loss of FBXW7 blunted tumor growth, a phenotype rescued by concomitant loss of KMT2D.
Altogether, these findings suggest that FBXW7 is a positive regulator of DLBCL cells via downregulation of KMT2D protein levels.
The FBXW7-KMT2D axis regulates transcription of genes in the oxidative phosphorylation pathway
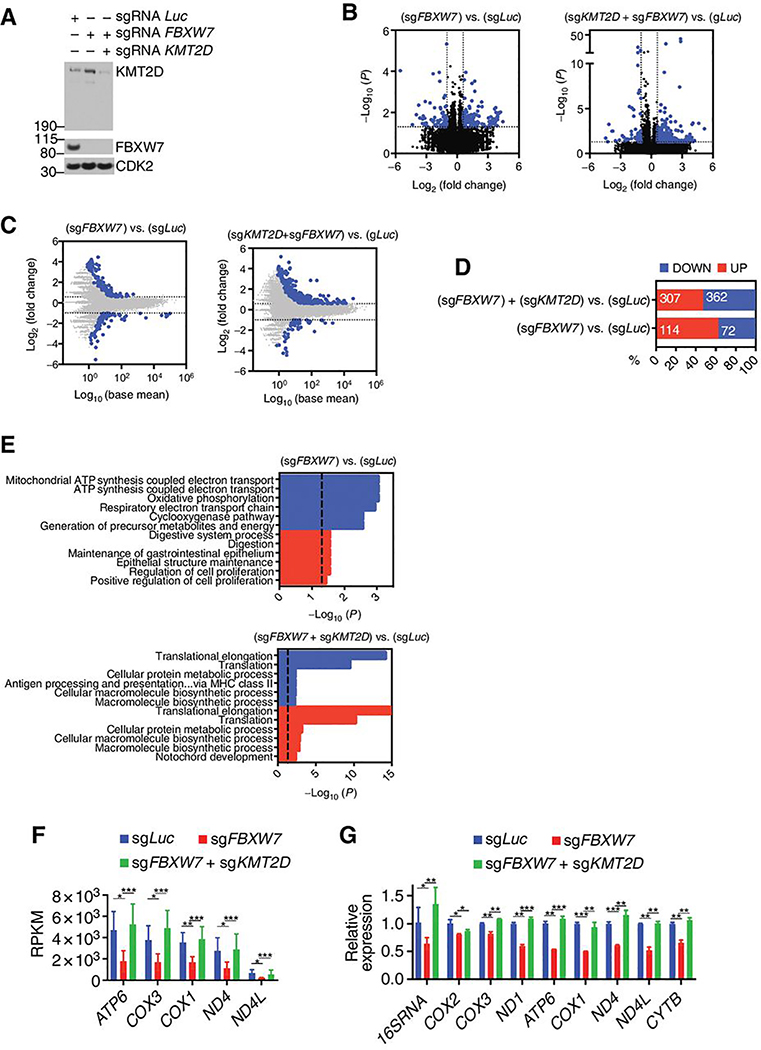
Given that KMT2D can alter chromatin state to regulate gene expression(28), we sought to investigate gene signatures regulated by the FBXW7-KMT2D axis. To this purpose, we generated bulk CRISPR-knockout OPM-1 cells expressing sgRNAs against luciferase (sgLuc), FBXW7 (sgFBXW7), or FBXW7 together with KMT2D (sgFBXW7+sgKMT2D) (Fig. 6a). Similarly to what we have shown so far, loss of FBXW7 led to upregulation of endogenous KMT2D protein levels (Fig. 6a) but did not alter gene expression of KMT2D (Supplementary Fig. S5a).
Figure 6. The FBXW7-KMT2D degradation axis regulates genes of the oxidative phosphorylation pathway.
A, Immunoblot for the indicated proteins from OPM-1 cells expressing the indicated sgRNAs. B, Volcano plot showing differentially expressed genes (DEGs) in for cells showed in (a) (n = 3, DeSeq2). Shown in blue are the mRNAs with at least 50% fold change increases or decreases with a p-value ≤ 0.05. C, Fold-change plot of gene expression levels for the cells shown in (a) (n = 3, DeSeq2). Shown in blue are mRNAs with at least 50% fold change increase or decrease. D, Proportion of significant upregulated or downregulated genes from (b). E, Gene ontology (GO) analysis of the upregulated or downregulated genes from (d). Dashed line indicates −log10(P)= 1.301, corresponding to p-value=0.05. F, Reads Per Kilobase per Million mapped reads (RPKM) values for indicated genes (n=3; mean ± SD, Deseq2 test; *p<0.05, **p<0.01, ***p<0.001).. G, Relative mRNA expression levels for indicated genes analyzed by qPCR, (n=3; mean ± SD, two-tailed Student’s t-test; *p<0.05, **p<0.01, ***p<0.001).
Analyses of differentially expressed genes (DEGs) upon loss of FBXW7 alone or in combination with KMT2D were compared to sgLuc expressing cells (Fig. 6b,c). Loss of FBXW7 displayed a higher proportion of upregulated genes than downregulated genes (Fig. 6d). However, this higher proportion of upregulated genes was reduced by concomitant loss of KMT2D (Fig. 6d). To identify significant gene signature changes, Gene ontology (GO) analysis was carried out on the [sgFBXW7 vs. sgLuc] and [sgFBXW7+sgKMT2D vs. sgLuc] DEGs. We considered the pathways deregulated by FBXW7 loss and rescued by concomitant KMT2D ablation as the ones specifically regulated by the FBXW7-KMT2D axis. GO analysis revealed that loss of FBXW7 deregulated processes related to oxidative phosphorylation and ATP synthesis (e.g. “mitochondrial ATP synthesis coupled electron transport” and “respiratory electron transport chain”) (Fig. 6e). Deregulation of these oxidative phosphorylation and ATP synthesis processes was, however, not observed with concurrent loss of KMT2D (Fig. 6e). Notably, the genes deregulated were mitochondrial-encoded genes (Fig. 6f) that were further validated in secondary qPCR analysis of gene expression in OPM-1 (Fig. 6g) as well as mtDNA content in U2932 cells (Supplementary Fig. S5b).
Hence, ablation of FBXW7 promotes deregulation of gene sets related to oxidative phosphorylation (OxPhos) in a KMT2D-dependent manner.
FBXW7 promotes OxPhos in DLBLC
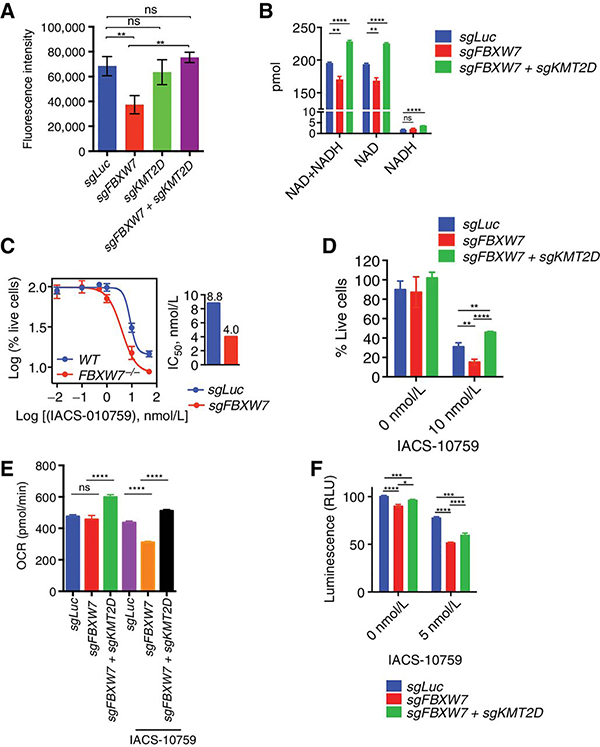
Since the FBXW7-KMT2D axis controls mitochondrial-encoded genes, we hypothesized that loss of FBXW7 would impact mitochondrial mass. We analyzed MitoTracker Deep Red FM uptake in U2932 cells expressing sgLuc, sgFBXW7, sgKMT2D or sgFBXW7+sgKMT2D (Fig. 7a). FBXW7 knockout cells displayed a decreased staining, which was rescued by concomitant KMT2D deletion. This mitochondrial defect was also confirmed by measurement of NAD, the by-product of complex I(43) (Fig. 7b). Levels of NADH were not changed in the sgFBXW7 expressing cells. Interestingly, concomitant knockout of KMT2D rescued the NAD levels.
Figure 7. FBXW7 promotes OxPhos in DLBLC.
A, Fluorescence intensity for U2932 cells expressing the indicated sgRNAs and stained with 50 nM MitoTracker Deep Red FM dye for the 30min (n=3, mean ± SD, one-way ANOVA; (**p<0.001, ns=not-significant). B, Total NAD+ and NADH levels were determined in U2932 cells expressing the indicated sgRNAs. (n=3; mean ± SD, two-tailed Student’s t-test; **p<0.01, ****p<0.0001). C, U2932 cells expressing the indicated sgRNAs were treated for 4 days with the indicated concentrations of IACS-010759. Live cell numbers were plotted (n=3, mean ± SD). IC50 is shown in the bar graph. D, U2932 cells expressing the indicated sgRNAs were treated for 4 days with DMSO or 10 nM IACS-010759. Live cell numbers were plotted (n=3; mean ± SD,two-tailed Student’s t-test; **p<0.01, ****p<0.0001 ). E, Oxygen consumption rates (OCR) of U2932 expressing the indicated sgRNAs pretreated with DMSO or with 15 nM IACS-010759. Measurements were taken using the Seahorse XFe96 Analyzer (n=6; mean ± SD; two-tailed Student’s t-test; ****p<0.0001, ns=not-significant). F, Quantification of ATP levels in U2932 cells expressing the indicated sgRNAs. Cells were treated with DMSO or 5 nM IACS-010759 for 16h (n=3; mean ± SD; two-tailed Student’s t-test; *p<0.05, ***p<0.001, ****p<0.0001).
As our data pointed at a mitochondrial defect in the sgFBXW7 cells, we hypothesized that these cells were more sensitive to the OxPhos inhibitor, IACS-010759(44). IACS-010759 is a clinical-grade small-molecule inhibitor of complex I of the mitochondrial electron transport chain(44) that is potent in inhibiting cell proliferation in lymphoma, leukemia, and solid tumors(44–47). U2932 sgFBXW7 cells displayed an increased sensitivity to OxPhos inhibition as measured by cell viability (Fig. 7c). Co-ablation of KMT2D rescued this phenotype (Fig. 7d). Furthermore, measurement of oxygen consumption rates (OCR) (Fig. 7e) and ATP levels (Fig. 7f) confirmed the mitochondrial/OxPhos defect of sgFBXW7 cells, particularly in the presence of IACS-010759; these defects were rescued by concomitant loss of KMT2D.
To query the relevance of KMT2D in primary DLBCLs, we assessed its expression with components of the mitochondrial signature in public dataset(48). We found a significant inverse correlation between KMT2D levels and OxPhos genes expression (Supplementary Fig. S6a, b) (i.e.; low KMT2D correlated with high OxPhos signature).
Altogether, our findings suggest that downregulation of KMT2D levels positively correlate with the OxPhos pathway.
Discussion
We report that FBXW7 is the E3 ubiquitin ligase for the lysine methyltransferase KMT2D, a gene highly mutated across several cancer types and particularly in B-cell malignancies such as B-cell lymphomas. Since FBXW7 was previously highlighted as a pro-survival factor in normal and cancerous B-cells (17–19), its identification as an E3 ligase for KMT2D could provide a mechanistic explanation for this function.
KMT2D is a tumor suppressor gene in DLBCL and Follicular lymphoma (FL). Genetic ablation of KMT2D in a BCL2-overexpression driven model of B-cell lymphoma promotes a higher penetrance of DLBCL and FL(34,37). While genetic mutations of KMT2D in human lymphoma have been explored as the mechanism that promote KMT2D loss-of-function, the regulation of KMT2D at post-translational levels by the UPS has not been well defined.
We show that KMT2D interacts with FBXW7 in a substrate-like manner. KMT2D contains multiple N-terminal phosphodegrons required to promote interaction with FBXW7. Phosphorylation of these degrons is observed in vivo and FBXW7-KMT2D interaction is dependent on phosphorylation. KMT2D is the first known FBXW7 substrate that contains such unique degron arrangement. For instance, CYCLIN E1 has two degrons while c-MYC, p100, and NOTCH1 have only one known degron(5,6,49,50). The 25 consecutive KMT2D degrons follow the general consensus of “SPPXE”. The negative glutamic acid residue serves as one of the two required negative charges for the FBXW7-WD40 domain to bind KMT2D. The serine residue must therefore be phosphorylated by one or several kinases to facilitate FBXW7 interaction. Analyses of the post-translational modifications (PTM) of the KMT2D protein and the PTM effects on KMT2D protein levels/function have only now begun to be explored(51–54). Since we observe interaction between FBXW7 and KMT2D as well as serine phosphorylation at steady state, we suggest that one or multiple kinases constitutively mediate this interaction. However, our proteomic data shows that not all of the SPPXE serine residues were phosphorylated, and it is thus likely that a complex regulation of signaling orchestrates the interaction to FBXW7. It is worth mentioning that identification of the kinase(s) responsible for mediating the phosphorylation-dependent KMT2D-FBXW7 interaction would allow for utilization of a kinase inhibitor to specifically disrupt FBXW7-mediated KMT2D degradation.
Notably, it was previously reported that KMT2D protein is destabilized when (i) its SET-domain is inactivated by mutations or (ii) upon expression of an H3.3K4 methylation-defective mutant histone(53,54). However, not all SET-impaired KMT2D mutants are unstable(55,56), hence further assessment of SET-impaired KMT2D mutants’ degradation and dependency on FBXW7 will be needed to understand the crosstalk between KMT2D catalytic activity and protein stability.
FBXW7 regulates different biological processes through degradation of specific substrates. For example, FBXW7 controls gene transcription through degradation of transcription factors such as c-MYC. c-MYC is a potent oncogene in myeloma and lymphoma(57,58) although the mechanism of c-MYC upregulation in these type of B-cell cancer is associated with overexpression of the mRNA(59–61), which could disengage c-MYC from the control by FBXW7. CRIPSR screens (42), previous studies (17–19) and the current work points at FBXW7 as a pro-survival gene in B-type cancer. This is also highlighted by the low frequency of FBXW7 mutation in myeloma and B-cell lymphoma as opposed to the high mutations frequency in T-ALL, where loss of FBXW7 function stabilizes oncogenic NOTCH1 and results in an increase of NOTCH1 target gene transcription(62,63). Interestingly, we observed a greater effect of FBXW7 loss on DLBCL growth in vivo versus in vitro. This could be due to the longer experimental time-course in vivo as well as the presence of additional in vivo biological factors from the host microenviroment that may feed into cellular processes regulated by the FBXW7-KMT2D axis.
Using unbiased RNA-seq studies, we show that loss of FBXW7 deregulated OxPhos transcriptional gene programs in a manner that is dependent on KMT2D loss. Our data add to previous observations that FBXW7 mutation alters mitochondrial signature genes and OxPhos metabolism(64) as well as recent data suggesting that KMT2D is implicated in metabolic gene pathways(65). We also showed that FBXW7-loss further sensitizes cells to OxPhos inhibition via a complex I inhibitor, IACS-010759(44), likely due to defects in mitochondrial function. Hence, coupling OxPhos inhibition with loss of FBXW7 function confers greater toxicity in DLBCL cells.
Supplementary Material
Significance.
Findings characterize FBXW7 as a pro-survival factor in B-cell lymphoma via degradation of the chromatin modifier KMT2D.
Acknowledgements
The authors thank Grant Grothusen for critically reading the manuscript, Taehyong Kim for help with the RNA-seq analysis, Laura Pasqualucci and Jiyuan Zhang for providing the full-length KMT2D cDNA, Laura Pasqualucci, Yibin Yang, and Michele Pagano for providing DLBCL cell lines. This work was supported in part by grant R01-CA207513–01 and 3R01CA207513–03S1 from the National Cancer Institute, the MMRF Research Fellow Award and The Research Scholar Grant American Cancer society to LB.
Footnotes
The authors declare no competing financial interests.
References
- 1.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 2013;14:369–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 2004;5:739–51 [DOI] [PubMed] [Google Scholar]
- 3.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 2008;8:83–93 [DOI] [PubMed] [Google Scholar]
- 4.Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell 2014;26:455–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busino L, Millman SE, Scotto L, Kyratsous CA, Basrur V, O’Connor O, et al. Fbxw7alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat Cell Biol 2012;14:375–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 2004;23:2116–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell 2007;26:131–43 [DOI] [PubMed] [Google Scholar]
- 8.Yeh CH, Bellon M, Pancewicz-Wojtkiewicz J, Nicot C. Oncogenic mutations in the FBXW7 gene of adult T-cell leukemia patients. Proc Natl Acad Sci U S A 2016;113:6731–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015;163:506–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 2014;46:467–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 2018;24:679–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A 2012;109:3879–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 2017;171:481–94 e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vikova V, Jourdan M, Robert N, Requirand G, Boireau S, Bruyer A, et al. Comprehensive characterization of the mutational landscape in multiple myeloma cell lines reveals potential drivers and pathways associated with tumor progression and drug resistance. Theranostics 2019;9:540–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011;476:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 2014;25:91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramezani-Rad P, Leung CR, Apgar JR, Rickert RC. E3 Ubiquitin Ligase Fbw7 Regulates the Survival of Mature B Cells. J Immunol 2020;204:1535–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeishi S, Matsumoto A, Onoyama I, Naka K, Hirao A, Nakayama KI. Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer Cell 2013;23:347–61 [DOI] [PubMed] [Google Scholar]
- 19.Reavie L, Buckley SM, Loizou E, Takeishi S, Aranda-Orgilles B, Ndiaye-Lobry D, et al. Regulation of c-Myc ubiquitination controls chronic myelogenous leukemia initiation and progression. Cancer Cell 2013;23:362–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 2014;343:305–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou N, Gutierrez-Uzquiza A, Zheng XY, Chang R, Vogl DT, Garfall AL, et al. RUNX proteins desensitize multiple myeloma to lenalidomide via protecting IKZFs from degradation. Leukemia 2019;33:2006–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Shaffer AL 3rd, Emre NC, Ceribelli M, Zhang M, Wright G, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell 2012;21:723–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han T, Goralski M, Gaskill N, Capota E, Kim J, Ting TC, et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 2017;356. [DOI] [PubMed] [Google Scholar]
- 24.Choi J, Lee K, Ingvarsdottir K, Bonasio R, Saraf A, Florens L, et al. Loss of KLHL6 promotes diffuse large B-cell lymphoma growth and survival by stabilizing the mRNA decay factor roquin2. Nat Cell Biol 2018;20:586–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006;127:635–48 [DOI] [PubMed] [Google Scholar]
- 26.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005;21:3448–9 [DOI] [PubMed] [Google Scholar]
- 27.Froimchuk E, Jang Y, Ge K. Histone H3 lysine 4 methyltransferase KMT2D. Gene 2017;627:337–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shilatifard A The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 2012;81:65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol 2013;33:4745–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Mittal A, Reid J, Reich S, Gamblin SJ, Wilson JR. Evolving Catalytic Properties of the MLL Family SET Domain. Structure 2015;23:1921–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan H, Tan M, Zhang Q, Yang F, Wang S, Li H, et al. LSD1 destabilizes FBXW7 and abrogates FBXW7 functions independent of its demethylase activity. Proc Natl Acad Sci U S A 2019;116:12311–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 1994;78:761–71 [DOI] [PubMed] [Google Scholar]
- 33.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009;458:732–6 [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med 2015;21:1190–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer 2015;15:334–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhar SS, Lee SH, Kan PY, Voigt P, Ma L, Shi X, et al. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev 2012;26:2749–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortega-Molina A, Boss IW, Canela A, Pan H, Jiang Y, Zhao C, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med 2015;21:1199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green MR, Gentles AJ, Nair RV, Irish JM, Kihira S, Liu CL, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood 2013;121:1604–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A 2015;112:E1116–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, et al. Genetics of follicular lymphoma transformation. Cell Rep 2014;6:130–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet 2011;43:830–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phelan JD, Young RM, Webster DE, Roulland S, Wright GW, Kasbekar M, et al. A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature 2018;560:387–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Canto C, Menzies KJ, Auwerx J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab 2015;22:31–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molina JR, Sun Y, Protopopova M, Gera S, Bandi M, Bristow C, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med 2018;24:1036–46 [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Yao Y, Zhang S, Liu Y, Guo H, Ahmed M, et al. Metabolic reprogramming toward oxidative phosphorylation identifies a therapeutic target for mantle cell lymphoma. Sci Transl Med 2019;11. [DOI] [PubMed] [Google Scholar]
- 46.Vangapandu HV, Alston B, Morse J, Ayres ML, Wierda WG, Keating MJ, et al. Biological and metabolic effects of IACS-010759, an OxPhos inhibitor, on chronic lymphocytic leukemia cells. Oncotarget 2018;9:24980–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lissanu Deribe Y, Sun Y, Terranova C, Khan F, Martinez-Ledesma J, Gay J, et al. Mutations in the SWI/SNF complex induce a targetable dependence on oxidative phosphorylation in lung cancer. Nat Med 2018;24:1047–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 2008;359:2313–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye X, Nalepa G, Welcker M, Kessler BM, Spooner E, Qin J, et al. Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J Biol Chem 2004;279:50110–9 [DOI] [PubMed] [Google Scholar]
- 50.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004;306:269–71 [DOI] [PubMed] [Google Scholar]
- 51.Toska E, Osmanbeyoglu HU, Castel P, Chan C, Hendrickson RC, Elkabets M, et al. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science 2017;355:1324–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toska E, Castel P, Chhangawala S, Arruabarrena-Aristorena A, Chan C, Hristidis VC, et al. PI3K Inhibition Activates SGK1 via a Feedback Loop to Promote Chromatin-Based Regulation of ER-Dependent Gene Expression. Cell Rep 2019;27:294–306 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jang Y, Wang C, Zhuang L, Liu C, Ge K. H3K4 Methyltransferase Activity Is Required for MLL4 Protein Stability. J Mol Biol 2017;429:2046–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jang Y, Broun A, Wang C, Park YK, Zhuang L, Lee JE, et al. H3.3K4M destabilizes enhancer H3K4 methyltransferases MLL3/MLL4 and impairs adipose tissue development. Nucleic Acids Res 2019;47:607–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dorighi KM, Swigut T, Henriques T, Bhanu NV, Scruggs BS, Nady N, et al. Mll3 and Mll4 Facilitate Enhancer RNA Synthesis and Transcription from Promoters Independently of H3K4 Monomethylation. Mol Cell 2017;66:568–76 e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rickels R, Herz HM, Sze CC, Cao K, Morgan MA, Collings CK, et al. Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat Genet 2017;49:1647–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spevak CC, Park CY. Novel Mechanism Regulates c-Myc Expression in Diffuse Large B-Cell Lymphoma. J Natl Cancer Inst 2020;112:7–9 [DOI] [PubMed] [Google Scholar]
- 58.Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia 2011;25:1026–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stasik CJ, Nitta H, Zhang W, Mosher CH, Cook JR, Tubbs RR, et al. Increased MYC gene copy number correlates with increased mRNA levels in diffuse large B-cell lymphoma. Haematologica 2010;95:597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ott G, Rosenwald A, Campo E. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Blood 2013;122:3884–91 [DOI] [PubMed] [Google Scholar]
- 61.Jovanovic KK, Roche-Lestienne C, Ghobrial IM, Facon T, Quesnel B, Manier S. Targeting MYC in multiple myeloma. Leukemia 2018;32:1295–306 [DOI] [PubMed] [Google Scholar]
- 62.Close V, Close W, Kugler SJ, Reichenzeller M, Yosifov DY, Bloehdorn J, et al. FBXW7 mutations reduce binding of NOTCH1, leading to cleaved NOTCH1 accumulation and target gene activation in CLL. Blood 2019;133:830–9 [DOI] [PubMed] [Google Scholar]
- 63.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med 2007;204:1825–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis RJ, Gonen M, Margineantu DH, Handeli S, Swanger J, Hoellerbauer P, et al. Pan-cancer transcriptional signatures predictive of oncogenic mutations reveal that Fbw7 regulates cancer cell oxidative metabolism. Proc Natl Acad Sci U S A 2018;115:5462–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carosso GA, Boukas L, Augustin JJ, Nguyen HN, Winer BL, Cannon GH, et al. Precocious neuronal differentiation and disrupted oxygen responses in Kabuki syndrome. JCI Insight 2019;4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available upon reasonable requests.