Abstract
Madagascar's ring‐tailed lemurs (Lemur catta) are experiencing rapid population declines due to ongoing habitat loss and fragmentation, as well as increasing exploitation for bushmeat and the illegal pet trade. Despite being the focus of extensive and ongoing behavioral studies, there is comparatively little known about the genetic population structuring of the species. Here, we present the most comprehensive population genetic analysis of ring‐tailed lemurs to date from across their likely remaining geographic range. We assessed levels of genetic diversity and population genetic structure using multilocus genotypes for 106 adult individuals from nine geographically representative localities. Population structure and F ST analyses revealed moderate genetic differentiation with localities being geographically partitioned into northern, southern, western and also potentially central clusters. Overall genetic diversity, in terms of allelic richness and observed heterozygosity, was high in the species (AR = 4.74, H O = 0.811). In fact, it is the highest among all published lemur estimates to date. While these results are encouraging, ring‐tailed lemurs are currently affected by ongoing habitat fragmentation and occur at lower densities in poorer quality habitats. The effects of continued isolation and fragmentation, coupled with climate‐driven environmental instability, will therefore likely impede the long‐term viability of the species.
Keywords: conservation genetics, Madagascar, microsatellites, strepsirrhines
Here, we present the most comprehensive population genetic analysis of ring‐tailed lemurs to date from across their likely remaining geographic range. Our study uses multilocus genotypes for 106 adult individuals from nine geographically representative localities to evaluate population genetic diversity and structure on a range‐wide scale. The results described herein demonstrate that, while Madagascar's remaining ring‐tailed lemur populations are geographically isolated, they have retained high levels of genetic diversity, with moderate genetic differentiation among populations despite a relatively large geographic distribution.
1. INTRODUCTION
Sustaining natural levels of genetic diversity within wildlife populations is a key concern for conservation biologists (Frankham, 1995, 2003, 2005). Pressures from climate change, anthropogenic habitat modification, overexploitation, and the introduction of novel competitors and infectious diseases are producing rapidly and ever‐changing environments, forcing species to adapt and evolve or go extinct (Di Marco, Venter, Possingham, & Watson, 2018; Frankham, Ballou, & Briscoe, 2010). Genetic diversity, the variation of alleles and genotypes present within a population, is the foundation on which natural selection acts and is therefore necessary for adaptive evolutionary change to occur (Frankham, 1995, 2003, 2005; Frankham et al., 2010). Populations with low levels of genetic diversity struggle to evolve in modified environments. For instance, the Tasmanian devil (Sarcophilus harrisii) has become vulnerable to the spread of devil facial tumor disease due to the lack of diversity across immune genes following human‐induced population crashes from introduced diseases (Guiler, 1970; Morris, Wright, Grueber, Hogg, & Belov, 2015).
Habitats modified by human activity hold less genetic diversity than pristine environments, thus putting their inhabitants at high risk (Miraldo et al., 2016). That is because deforestation, fragmentation, and habitat degradation—all critical threats to biodiversity—interact to restrict the amount of viable habitat available to species, reduce carrying capacity and consequently maximum population size, and create isolated patches separated by matrix (i.e., inhospitable habitat) that impedes gene flow among remaining species' populations (Baden et al., 2019; Holmes et al., 2013; Stangel, Lennartz, & Smith, 1992). Combined, these processes lead to greater inbreeding, reduced genetic diversity, and ultimately an increased extinction risk (Frankham et al., 2010; Lino, Fonseca, Rojas, Fischer, & Pereira, 2019; Struebig et al., 2011).
Though not alone in its vulnerability to habitat loss, Madagascar's biodiversity is considered to be a top concern, in part because of its incredible concentration of species endemism (Myers, Mittermeier, Mittermeier, da Fonseca, & Kent, 2000). Since its colonization by humans as recently as 4,000 years ago, the island has undergone extensive forest cover loss and with it more than 17 species of large‐bodied lemurs (Dewar et al., 2013; Godfrey & Irwin, 2007; Kistler et al., 2015; Myers et al., 2000). Unfortunately, deforestation in Madagascar continues unabated (Vieilledent et al., 2018) and scientists anticipate that remaining rainforest habitat will be lost before the end of this century (Morelli et al., 2020). When considered alongside the impacts of climate change, this threat poses significant risk to the persistence of remaining extant lemur species (Brown & Yoder, 2015; Morelli et al., 2020). It is therefore an urgent conservation priority to quantify the genetic variability present within Madagascar's only endemic primate radiation to assess whether and to what extent lemur species can cope with intensifying environmental pressures.
Of particular concern is Madagascar's most charismatic species, the ring‐tailed lemur (Lemur catta, Figure 1). Ring‐tailed lemurs are medium‐sized (average 2.2 kg) terrestrial strepsirrhines that can be found throughout southern Madagascar (Cameron & Gould, 2013; Fardi, Sauther, Cuozzo, Youssouf, & Bernstein, 2018; Gould, Sussman, & Sauther, 2003; Sauther, Gould, Cuozzo, O'Mara, 2015; Sussman, 1991). They are considered a generalist taxon, maintaining a diverse frugivorous–folivorous diet (Cameron & Gould, 2013; Sauther, 1998; Sauther, Sussman, & Gould, 1999) and exhibiting considerable ecological flexibility (Cameron & Gould, 2013; Fardi et al., 2018; Gould et al., 2003; Sussman, 1991). The species occupies diverse habitat types ranging from rainforests to subalpine, deciduous, gallery, and spiny bush forests to anthropogenic savannah (Cameron & Gould, 2013; Gabriel, 2013; Goodman & Langrand, 1996; Goodman et al., 2002; Goodman, Rakotoarisoa, & Wilme, 2006; LaFleur & Gould, 2009; Sauther et al., 2006); however, much of their habitat has been altered by human activities, including clearing for agriculture, burning for charcoal production, and deforesting areas to create settlements (Sussman, Green, Porton, Andrianasolondraibe, & Ratsirarson, 2003). In the past 40 years alone, ring‐tailed lemurs have lost over 45% of their habitat (Brinkmann, Noromiarilanto, Ratovonamana, & Buerkert, 2014; LaFleur, Clarke, Ratzimbazafy, & Reuter, 2017a); and by 2080, it is estimated that 63% of their remaining range will shift due to climate change alone (Brown & Yoder, 2015). Furthermore, there has been a recent uptick in exploitation for the illegal pet trade, causing severe population declines, and in some cases local extinctions, throughout their remaining geographic range (Gardner & Davies, 2014; Gould & Sauther, 2016; LaFleur, Clarke, Reuter, Schaefer, & terHorst, 2019; LaFleur, Clarke, Reuter, & Schaeffer, 2017b; LaFleur & Gould, 2009; Reuter et al., 2019; Reuter & Schaefer, 2016). At present, there are estimated to be fewer than 2,400 individuals within sampled locations (Gould & Sauther, 2016; LaFleur et al., 2017b), though population estimates are still lacking throughout much of their range (e.g., Murphy, Ferguson, & Gardner, 2017).
FIGURE 1.

Photograph of ring‐tailed lemur (Lemur catta) by M LaFleur
Despite being one of the most‐studied lemur species, there is relatively little known about the genetic diversity and population structure of remaining wild ring‐tailed lemur individuals. Existing studies suggest that northern (Anja Community Reserve, Sakaviro, and Tsaranoro Valley Fragments; Clarke, Gray, Gould, & Burrell, 2015) and western localities (Bezà Mahafaly Special Reserve and Tsimanampetsotsa National Park; Parga, Sauther, Cuozzo, Jacky, & Lawler, 2012) maintain moderate levels of genetic diversity, with the smallest fragments (e.g., Sakaviro), isolated by roads and anthropogenic savannah, containing relatively lower levels of allelic richness and “mean number of alleles” than the larger western localities (Clarke et al., 2015; Parga et al., 2012). Moreover, low F ST values among sites in the north (Clarke et al., 2015) and among those in the west (Parga et al., 2015) indicate minimal genetic differentiation, suggesting the presence of historical gene flow. While encouraging, there is also evidence that genetic erosion within the species has already begun to negatively impact their health and fitness (e.g., Charpentier, Williams, & Drea, 2008; Grogan, Sauther, Cuozzo, & Drea, 2017). It is therefore likely that at least some of Madagascar's remaining ring‐tailed lemur populations are already experiencing a time‐delayed response (i.e., extinction debt), as extinctions do not typically occur until several generations after a fragmentation event (Jackson & Sax, 2010; Tilman, May, Lehman, & Nowak, 1994).
In an effort to characterize the remaining genetic diversity present within the species and identify how this diversity is apportioned among remnant populations, we provide a preliminary population genetic assessment of ring‐tailed lemurs across their remaining geographic range. We evaluate within‐ and across‐site levels of genetic diversity and infer population genetic structure to better understand this species' adaptive potential and diagnose possible conservation priorities.
2. METHODS
2.1. Sample collection
Our sample included 106 adult ring‐tailed lemurs from nine geographically representative localities from across their existing range (Table 1, Figure 2). This dataset includes previously published genetic data from 75 adult ring‐tailed lemurs from five localities (Clarke et al., 2015; Parga et al., 2012), as well as 31 new individuals from an additional four sites (Table 1). Published genetic data were collected in May through August 2006 (Parga et al., 2012) and August to October 2012 (Clarke et al., 2015). Data for new individuals were generated from fecal samples collected from Isalo in July 2016 and from Ambirary (AMB), Beoloke (BLK), and Berenty (BER) in June and July 2017. Multiple individuals and groups were sampled at each locality (Table 1). Fecal samples were immediately stored in RNAlater (Ambion) to prevent DNA degradation and were banked within 1 month of collection at −80°C for long‐term storage. Sample collection and export/import protocols adhered to Malagasy and International laws and were approved by Malagasy wildlife authorities and the US Fish and Wildlife Service.
TABLE 1.
Sampling localities, geographic coordinates, and sample sizes of ring‐tailed lemur populations used in this study
| Site name | Site code | Geographic region | Habitat type | Protection status | Latitude | Longitude | n ind. | n groups | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Ambirary P.R. | AMB | SE | Gallery, scrub, and spiny forest | Private Reserve | 24°59′53″ S | 46°18′07″ E | 5 | 3 | This study |
| Anja C.R. | ANJA | NE | Mixed xerophytic, deciduous vegetation, large granite outcrops | Community‐level association | 21°51′12″ S | 46°50′40″ E | 10 | 3 | Clarke et al. (2015) |
| Beoloke P.R. | BLK | SE | Gallery forest | Private Reserve | 25°00′59″ S | 46°18′51″ E | 5 | 4 | This study |
| Berenty P.R. | BER | SE | Gallery, scrub, and spiny forest | Private Reserve | 25°00′00″ S | 46°18′00″ E | 13 | 8 | This study |
| Beza Mahafaly S.R. | BEZA | W | Gallery forest | Special Reserve | 23°30′00″ S | 44°40′00″ E | 20 | 6 | Parga et al. (2012) |
| Isalo N.P. | ISALO | C | Grass savannah, large rock formations | National Park | 22°29′26″ S | 45°22′44″ E | 8 | 2 | This study |
| Sakaviro | SAKA | NE | Mixed xerophytic, deciduous vegetation, large granite outcrops | Community‐level association | 21°47′03″ S | 46°52′02″ E | 10 | During census | Clarke et al. (2015) |
| Tsaranoro V.F. | TSARA | NE | Mixed xerophytic, deciduous vegetation, large granite outcrops | Community‐level association | 22°05′11″ S | 46°46′14″ E | 10 | 4 | Clarke et al. (2015) |
| Tsimanampetsotsa N.P. | TNP | W | Spiny, succulent, xerophilic vegetation | National Park | 24°06′00″ S | 43°50′00″ E | 25 | 4 | Parga et al. (2012) |
| Total sample | 106 | 34 |
Abbreviations: C, central; CR, Community Reserve; NE, northeast; NP, National Park; PR, Private Reserve; SE, southeast; SR, Special Reserve; VF, Valley Forest; W, west.
FIGURE 2.
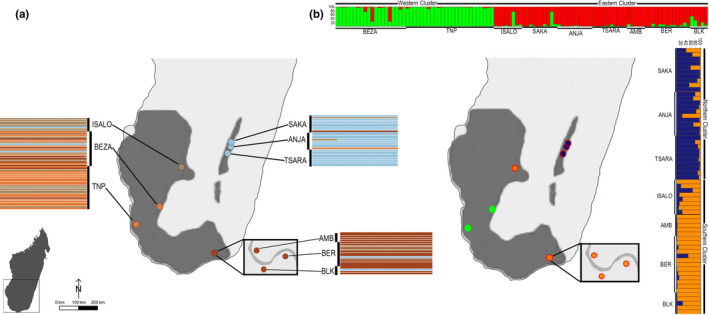
Map illustrating the nine ring‐tailed lemur localities sampled and population genetic structure results from DAPC (a) and Structure (b) analyses. Each bar illustrates the proportional membership (Q) of each individual lemur belonging to the clusters identified. DAPC (a) identified four genetic clusters (orange: west; beige: central; blue: north; brown: south). Structure (b) identified three genetic clusters. The horizontal bar (b) illustrates results from primary Structure analysis (n = 9 sites), showing that localities partitioned into eastern and western genetic clusters, as indicated by red and green point outlines, respectively. Vertical bar (b) illustrates results from secondary Structure analysis of eastern sites (n = 7 sites), indicating further subdivision into northern and southern clusters as indicated by blue and orange point fills, respectively. For full site names see Table 1. Gray shading illustrates historic ring‐tailed lemur distribution across southern Madagascar retrieved from IUCN website
2.2. DNA extraction
Total genomic DNA (gDNA) was extracted from new samples (n = 31) using QIAamp DNA Stool Mini Kits (QIAGEN) following Clarke et al. (2015). Samples were amplified at six microsatellite markers that have been shown to reliably amplify fecal DNA: L‐2 (Merenlender, 1993), Lc5, Lc6, Lc7 (Pastorini, Fernando, Forstner, & Melnick, 2005), 69HDZ267, and 69HDZ299 (Zaonarivelo et al., 2007) (Appendix S1).
2.3. Microsatellite genotyping
Extraction products were amplified via PCR in a 13 μl reaction volume using 6.25 μl HotStarTaq DNA polymerase Master Mix, 20 mg/ml BSA, 10 μM primer pairs, and 3 μl (0.25–1 ng) gDNA using annealing temperatures outlined in the Appendix S1. The 5′ end of the forward primer was fluorescently labeled. PCR products were separated by capillary electrophoresis (ABI 3730xl Genetic Analyzer), and alleles were sized to an internal size standard (Rox‐500) using GeneMarker software v.2.6.7 (SoftGenetics). Genotype assignment was based on multiple independent reactions, where heterozygotes were confirmed with at least two independent reactions and homozygotes were confirmed with five independent reactions (Morin, Chambers, Boesch, & Vigilant, 2001; Taberlet et al., 1996). Individuals from earlier studies (n = 45, Parga et al., 2012; n = 30, Clarke et al., 2015) were regenotyped and scaled to ensure datasets were comparable. CERVUS v.3.0 (Kalinowski, Taper, & Marshall, 2007) was used to calculate probability of identity (P ID), that is, the probability that two randomly drawn individuals from a population will show identical multilocus genotypes.
2.4. Population genetic analysis
2.4.1. Genetic diversity
Using Micro‐Checker (van Oosterhout, Hutchinson, Wills, & Shipley, 2004), loci were checked for the presence of null alleles and were tested for deviations from Hardy–Weinberg equilibrium and linkage disequilibrium using the program Genepop v.4.2 (Raymond & Rousset, 1995). They were evaluated using a 10,000 iteration dememorization phase, followed by 100 batches of 10,000 iterations. Measures of genetic diversity, including number of alleles per locus (nA), mean number of alleles per locus (MNA), allelic richness (AR), observed (H O), and expected (H E) heterozygosities, and Wright's F IS for each sampling location were calculated using GenoDive (Meirmans & Van Tienderen, 2004). We standardized allelic richness (AR) to the smallest sample size in the dataset to account for uneven sampling between populations using HP‐Rare 1.1 (Kalinowski, 2005).
2.4.2. Population genetic structure
To assess the genetic distances between sampling localities, Wright's F ST (Weir & Cockerham, 1984) was calculated for all pairs of populations using GenoDive (Meirmans & Van Tienderen, 2004). F ST is a measure of genetic differentiation among subpopulations and illustrates whether and to what extent populations are considered genetically distinct (Frankham et al., 2010). Significance was calculated using 10,000 permutations corrected for multiple comparisons (Bonferroni adjusted p = 0.001).
The presence of isolation‐by‐distance (IBD) was evaluated using the program GenAlEx v.6.5 (Peakall & Smouse, 2012) and significance estimated with Mantel's test using 10,000 permutations (Mantel, 1967). Genetic distances between populations were estimated using F ST/(1 − F ST).
To identify genetic clusters, we used three different methods. First, we used a model‐based Bayesian clustering method implemented in Structure v2.3.4 (Pritchard, Stephens, & Donnelly, 2000). This method is used to estimate the number of genetically distinct clusters (K) with no a priori information regarding the individuals' geographic sampling locations provided, so clusters were formed solely on genetic information. We evaluated the hypothesis K = 1–12, three more than the number of wild populations (Evanno, Regnaut, & Goudet, 2005), using 100,000 iterations of MCMC following a burn‐in of 50,000 iterations, as longer burn‐in or MCMC did not significantly change our results. We implemented 20 runs for each value of K and assumed admixture and correlated allele frequencies. The admixture model allows individuals to have mixed ancestry, assuming that a portion of an individual's genome, q, comes from a subpopulation, k (where ; Falush, Stephens, & Pritchard, 2003). To account for unbalanced sampling, the ALPHA value was changed from the default value (1.0) to 0.5 (Wang, 2016). The most likely number of genetic clusters (K) was assessed using the highest value of ∆K (Evanno et al., 2005) using the program Structure Harvester v0.6.94 (Earl & vonHoldt, 2012). Structure Harvester calculates the second‐order rate of change of the likelihood distribution (ΔK), which indicates the most pronounced subdivision within the data and the optimal number of genetic clusters. We implemented a two‐step approach to evaluate further substructure in the dataset. We first identified the most likely number of clusters within the overall sample (n = 106) and then ran subsequent analyses within each of the K clusters (Evanno et al., 2005).
To corroborate the Structure analysis, a discriminant analysis of principal components (DAPC) was performed in R, using the adegenet package (Jombart, 2008; R Core Team, 2017). This multivariate method identifies clusters of genetically related individuals that maximize between‐group variability and minimize within‐group variability by using a set of retained principal components (determined by the user to optimize variance explained; Jombart, Devillard, & Balloux, 2010). The optimal number of clusters is determined by the number of clusters with the smallest Bayesian information criterion (BIC) value.
To further substantiate both the Structure and DAPC results, a principal coordinates analysis (PCoA) was performed with a standard genetic distance matrix (Nei, 1978) using GenAlEx v.6.5 (Peakall & Smouse, 2012).
3. RESULTS
3.1. Genetic diversity
The nonexclusion combined probability of identity (P ID; Paetkau & Strobeck, 1994) of the six markers used in this study was 5.22 × 10−10 and for P ID‐sibs was 1.13 × 10−3. These values demonstrate a low probability that two individuals would share the same multilocus genotype. The six loci were highly polymorphic, with the number of alleles ranging between 12 and 15 alleles (Table 2). Individuals were pooled across sampling localities, and there was no evidence of linkage disequilibrium. One locus was found to significantly deviate from Hardy–Weinberg equilibrium (L‐2; Table 2); however, it did not deviate at any one site specifically and was therefore included in further analyses.
TABLE 2.
Characteristics of 6 microsatellite markers amplified in 106 ring‐tailed lemur samples, including the number of alleles per locus (n A), observed (H o) and expected (H e) heterozygosity, deviations from Hardy–Weinberg equilibrium (HWE), and polymorphic information content (PIC)
| Marker | Size range (bp) | Annealing temp (°C) | n A | H o | H e | HWE | PIC | GenBank |
|---|---|---|---|---|---|---|---|---|
| L‐2 | 179–203 | 48 | 15 | 0.812 | 0.881 | 0.0294 | 0.853 | – |
| Lc5 | 127–151 | 60 | 12 | 0.680 | 0.847 | 0.5353 | 0.835 | AY366441 |
| Lc6 | 248–270 | 60 | 12 | 0.788 | 0.809 | 0.3804 | 0.791 | AY366442 |
| Lc7 | 172–198 | 60 | 14 | 0.805 | 0.862 | 0.2387 | 0.857 | AY366443 |
| 69HDZ267 | 156–178 | 55 | 15 | 0.902 | 0.916 | 0.1641 | 0.901 | EF093488 |
| 69HDZ299 | 238–262 | 58 | 15 | 0.881 | 0.915 | 0.8349 | 0.896 | EF093489 |
Significant p values (p < 0.05) are shown in bold.
As a species, ring‐tailed lemurs maintain high levels of genetic diversity despite severe habitat fragmentation across their range. Mean number of alleles (MNA) ranged from 4.33 to 8.67 (Table 3). The mean observed heterozygosity across sampling sites was 0.811 ± 0.044, while mean expected heterozygosity was 0.775 ± 0.054. Overall F IS was −0.052, and values ranged from −0.194 at ISALO to 0.042 at BER (Table 3).
TABLE 3.
Allelic diversity within each of the nine ring‐tailed lemur sampling localities, including mean number of alleles (MNA), allelic richness (AR), observed (H o) and expected (H e) heterozygosity, inbreeding coefficient (F IS), and significant deviations from Hardy–Weinberg Equilibrium (HWE) calculated using 10,000 iterations
| Site | N | MNA | AR(SE) | H o | H e | F IS | HWE |
|---|---|---|---|---|---|---|---|
| AMB | 5 | 4.667 | 4.67 | 0.833 | 0.779 | −0.075 | 0.7111 |
| BLK | 5 | 4.333 | 4.33 | 0.836 | 0.803 | −0.042 | 0.9666 |
| BER | 13 | 8.000 | 5.88 | 0.828 | 0.865 | 0.042 | 0.2995 |
| ANJ | 10 | 4.833 | 4.19 | 0.771 | 0.716 | −0.077 | 0.3493 |
| SAKA | 10 | 4.667 | 4.09 | 0.857 | 0.727 | −0.179 | 0.0039 |
| TSARA | 10 | 6.167 | 4.91 | 0.722 | 0.730 | 0.011 | 0.7723 |
| ISALO | 8 | 4.667 | 4.19 | 0.854 | 0.719 | −0.194 | 0.1452 |
| BEZA | 20 | 8.500 | 5.15 | 0.792 | 0.821 | 0.035 | 0.3109 |
| TNP | 25 | 8.667 | 5.23 | 0.807 | 0.816 | 0.011 | 0.2090 |
| Overall | 106 | 6.056 | 4.74 | 0.811 | 0.775 | −0.052 | ‐ |
Significant values (p < 0.05) are shown in bold.
3.2. Population genetic structure
Pairwise values of F ST among sampling localities ranged from 0.034 to 0.183, with a mean of 0.129. Genetic differentiation among sampling localities was significant in 31 out of 36 cases with nonsignificant F ST comparisons between eastern localities AMB‐BLK‐BER, AMB‐ANJA, and ANJA‐SAKA (Table 4). Wright (1978), perhaps somewhat subjectively, considered F ST values between 0.05 and 0.15 to indicate moderate genetic differentiation, whereas values of 0.15–0.25 to indicate great genetic differentiation (though Wright's recommendations were made at a time when highly mutable genetic markers, such as microsatellites, were not used). Pairwise distances (F ST) between BER and BLK (0.053), BER and AMB (0.071), and between BEZA and TNP (0.034) indicate minimal to moderate genetic differentiation between these localities and are consistent with the clustering seen in the subsequent Structure analyses. Larger values of F ST (>0.15), suggesting great differentiation, were observed between ISALO and sampling locales in the north (SAKA and TSARA) and south (AMB and BLK). Despite great geographic distance, there was only moderate F ST between BEZA and BER (0.077), BEZA and BLK (0.083), and TNP and BER (0.091). The results of Mantel's test (Figure 3) revealed a significant pattern of isolation‐by‐distance (IBD; R = 0.418, p = 0.007 based on 1,000 permutations) with geographical distance explaining over 17% of the variation in genetic distance. However, there was a considerable amount of unexplained variation in the IBD data. For example, in the eastern cluster there is relatively little geographic distance between the three populations comprising the northern (ANJA‐SAKA‐TSARA) and southern groups (AMB‐BER‐BLK), yet some sampling sites within each of these triads reflect moderate genetic differentiation (Figure 3).
TABLE 4.
Pairwise F ST values (above diagonal) and indication of significant F ST values (below diagonal) among sampling localities of ring‐tailed lemurs
| AMB | BLK | BER | ANJA | SAKA | TSARA | ISALO | BEZA | TNP | |
|---|---|---|---|---|---|---|---|---|---|
| AMB | – | 0.125 | 0.071 | 0.124 | 0.129 | 0.185 | 0.146 | 0.111 | 0.139 |
| BLK | NS | – | 0.053 | 0.141 | 0.136 | 0.164 | 0.183 | 0.083 | 0.107 |
| BER | NS | NS | – | 0.117 | 0.107 | 0.129 | 0.101 | 0.077 | 0.091 |
| ANJA | NS | * | * | – | 0.072 | 0.109 | 0.150 | 0.122 | 0.155 |
| SAKA | * | * | * | NS | – | 0.131 | 0.178 | 0.121 | 0.124 |
| TSARA | * | * | * | * | * | – | 0.181 | 0.139 | 0.136 |
| ISALO | * | * | * | * | * | * | – | 0.106 | 0.125 |
| BEZA | * | * | * | * | * | * | * | – | 0.034 |
| TNP | * | * | * | * | * | * | * | * | – |
Significant values indicated with * (p < 0.001 after Bonferroni corrections).
FIGURE 3.
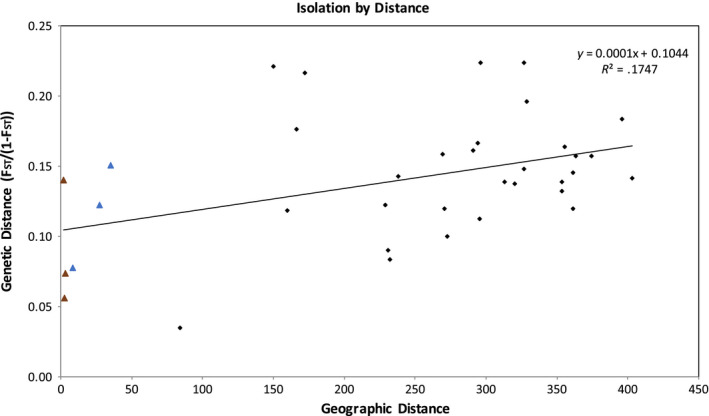
Results from Mantel's test for isolation‐by‐distance (IBD) in ring‐tailed lemurs from the nine localities sampled. The closest geographic localities (AMB‐BER‐BLK, brown triangles; and SAKA‐ANJA‐TSARA, blue triangles) did not reflect IBD
Further analyses demonstrate that eight of nine wild populations of ring‐tailed lemurs can be geographically grouped into two structured genetic clusters: a western cluster (BEZA, TNP) and an eastern cluster (ANJA, SAKA, TSARA, BER, AMB, and BLK), as indicated by the highest value of ΔK (Figure 2b). Cluster 1 comprised individuals from the six eastern localities and one central locality (61 of 106 individuals), and Cluster 2 comprised of individuals from the two western localities (45 of 106 individuals). The analysis was repeated with each of the K = 2 clusters separately following Evanno et al. (2005) and found that Cluster 1 (eastern localities) could be further subdivided into K = 2 geographically structured clusters (southern, subcluster 1: AMB, BLK, BER, and ISALO; northern, subcluster 2: ANJA, SAKA, and TSARA; Figure 2b). We found that Cluster 2 (western localities) was comprised of one genetic cluster, as the mean L(K) could not confidently exclude K = 1. Mean L(K) and ΔK plots of all Structure runs are provided in supplementary information (Figure S1).
We detected similar patterns of structuring in the discriminant analysis of principal components (DAPC, Figure 2a) between the northern, southern, and western localities; however, according to the smallest BIC value, K‐means clustering estimated four genetic clusters. The majority of individuals clustered geographically with southern localities (AMB, BLK, BER) showing a higher membership probability to Cluster 1, northern localities (ANJA, SAKA, TSARA) to Cluster 2, and the central locality (ISALO) to Cluster 3. Individuals in the western localities (BEZA and TNP) showed higher membership probabilities to Cluster 4. From our initial analyses, we could not confidently group ISALO, as our Structure results indicated that ISALO clustered with southern localities (BER‐AMB‐BLK) despite the distance (301 km) and our DAPC results grouped the site into its own central cluster. Upon further investigation, we did find support in our Structure analysis for higher K‐values (K = 3–4, Figure S2), supporting the DAPC results and grouping ISALO into a separate central cluster.
The principal coordinate analysis (PCoA; Figure 4) shows loose clustering between sampling localities according to geographic location. Western localities (BEZA and TNP) clustered together along axis one, while axis 2 separated sampling localities between north (ANJ, SAKA, TSARA) and south (AMB, BLK, BER). Though these results indicate geographic separation, the PCoA does show overlap between sampling localities in west, central, and southern Madagascar.
FIGURE 4.
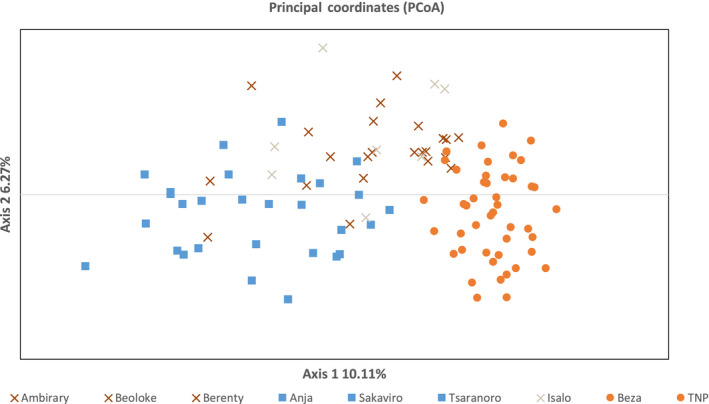
Principal coordinate analysis (PCoA). Data points are color coded according to DAPC analysis (orange: west; beige: central; blue: North; brown: South) while marker shapes correspond to STRUCTURE analysis (•: Western; □: North; x: South)
4. DISCUSSION
4.1. Genetic diversity and population structure
Although most of Madagascar's ring‐tailed lemur populations are geographically isolated, evidence described herein demonstrate that they have retained high levels of genetic diversity, with moderate genetic differentiation among populations despite being separated by a relatively large geographic distance. We describe levels of genetic diversity (MNA = 6.056, AR = 4.74, global H O = 0.811; Table 3) that are higher than those found in any other strepsirrhine, including mouse lemurs (MNA = 2.75–4.38, H O = 0.557–0.695; Olivieri, Sousa, Chikhi, & Radespiel, 2008; Radespiel, Rakotondravony, & Chikhi, 2008; Table 5). Though differing in size, habitat type, and protection status, the nine localities sampled in our study showed similar levels of genetic diversity. Interestingly, ISALO, the largest forest included in this analysis (81,500 ha), showed one of the lowest levels of allelic diversity (AR = 4.19) and some of the highest levels of heterozygosity (H O = 0.85). In fact, the genetic diversity within ISALO was most similar to the smallest fragment included in our study, SAKA (14 ha, AR = 4.09, H O = 0.86; Table 3). This is likely due to the relatively small number of markers used and our limited sample size (n = 8 individuals) at the time of this analysis.
TABLE 5.
Indices of allelic diversity across twelve lemur taxa, including minimum, maximum, and mean number of alleles (MNA), observed (H o) and expected (H e) heterozygosity, and population differentiation (F ST)
| Family | Species | Sample | Marker | n ind. | n sites | MNA | H o | H e | F ST | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Min | Max | Mean | ||||||||
| Daubentoniidae | Daubentonia madagascariensis 1 | Tissue | SNPs | 12 | 8 | – | – | – | – | – | (0.129) | (0.194) | (0.164 ± 0.033) |
| Cheirogaleidae | Microcebus bongolavensis 2 | Tissue | 8 microsats | 45 | 3 | 3.63 | 5.00 | 4.17 | 0.557 | 0.582 | 0.057 | 0.102 | 0.076 ± 0.023 |
| Microcebus danfossi 2 | Tissue | 8 microsats | 78 | 7 | 2.75 | 6.63 | 4.91 | 0.628 | 0.654 | 0.025 | 0.195 | 0.096 ± 0.049 | |
| Microcebus murinus 3 | Tissue | 10 microsats | 167 | 3 | – | – | – | – | – | 0.004 | 0.016 | – | |
| Microcebus ravelobensis 4 | Tissue | 8 microsats | 187 | 12 | – | – | – | 0.695 | 0.692 | −0.002 | 0.122 | 0.052 ± 0.027 | |
| Microcebus ravelobensis 2 | Tissue | 8 microsats | 205 | 8 | 4.38 | 6.50 | 5.76 | 0.615 | 0.605 | 0.006 | 0.156 | 0.072 ± 0.035 | |
| Lepilemuridae | Lepilemur edwardsi 5 | Tissue | 14 microsats | 20 | 2 | 3.86 | 4.00 | – | – | – | −0.09 | 1.00 | – |
| Lemuridae | Eulemur cinereiceps 6 | Tissue | 26 microsats | 53 | 4 | 2.71* | 3.36* | 3.06* | 0.520 | 0.527 | 0.020 | 0.076 | 0.054 ± 0.019 |
| Varecia rubra 7 | Tissue | 15 microsats | 32 | 2 | 1.0 | 8.0 | 5.03 | 0.666 | 0.643 | – | – | 0.077 | |
| Varecia variegata 8 | Blood, feces | 16 microsats | 55 | 5 | 2.47* | 3.23* | 2.84* | – | – | 0.039 | 0.291 | 0.197 ± 0.084 | |
| Varecia variegata 9 | Blood, feces | 10 microsats | 209 | 19 | 2.20 | 4.70 | 3.42 | 0.519 | 0.573 | 0.002 | 0.441 | 0.247 ± 0.094 | |
| Lemur catta 10 | Feces | 6 microsats | 30 | 3 | 4.83 | 5.83 | 5.27 | 0.797 | 0.703 | 0.05 | 0.11 | – | |
| Lemur catta 11 | Feces | 6 microsats | 106 | 9 | 4.33 | 8.67 | 6.06 | 0.811 | 0.775 | 0.034 | 0.183 | 0.129 ± 0.032 | |
| Indriidae | Propithecus tattersali 12 | Feces | 13 microsats | 82 | 3 | (3.43)* | (3.99)* | – | (0.707) | (0.621) | (0.136) | (0.160) | (0.147) |
| Propithecus tattersali 13 | Feces | 13 microsats | 224 | 9 | 3.00 | 6.00 | 4.92 | 0.690 | 0.660 | 0.010 | 0.300 | 0.119 ± 0.067 | |
| Propithecus verreauxi 14 | Tissue | 7 microsats | 77–131 | 10–28 | – | – | – | – | – | 0.024 | 0.075 | 0.052 | |
| Propithecus perrieri 15 | Feces | 24 microsats | 42 | 3 | 3.58 | 4.25 | 3.83 | 0.610 | 0.640 | 0.023 | 0.061 | 0.039 | |
| Indri indri 16 | Blood, tissue | 17 microsats | 43 | 3 | 4.81* | 5.36* | – | – | 0.850 | 0.0746 | 0.129 | 0.105 | |
F ST comparisons are among sampling localities unless otherwise noted. Values in parentheses denote comparisons among inferred clusters. Asterisk values denote allelic richness of sampling localities.
Perry et al. (2013).
Olivieri et al. (2008).
Fredsted, Pertoldi, Schierup, and Kappeler (2005).
Radespiel et al. (2008).
Craul et al. (2009).
Brenneman et al. (2012).
Razakamaharavo, McGuire, Vasey, Louis, and Brenneman (2010).
Holmes et al. (2013).
Baden et al. (2014).
Clarke et al. (2015).
This study.
Quéméré, Louis, Louis, Ribéron, Chikhi, and Crouau‐Roy (2010).
Quéméré, Crouau‐Roy, Crouau‐Roy, Rabarivola, Louis, and Chikhi (2010).
Lawler, Richard, and Riley (2003).
Salmona et al. (2015).
Nunziata et al. (2016).
Nevertheless, despite the limited number of markers used in this study, we found continuous patterns of structure across this species' range, including subdivision of eastern localities into northern and southern groups. Though there are large geographic distances between localities, our F ST results indicated only moderate differentiation between sites. This may be attributed to the dispersal ability of this species. Being the most terrestrial of living lemurs (Jolly, 1966; Sussman, 1972, 1974), ring‐tailed lemurs can disperse more easily across nonforested areas than forest‐dependent arboreal species. Furthermore, the largest river drainage systems in southern Madagascar are seasonal and therefore do not pose permanent dispersal barriers to this species; in fact, they may actually be used as dispersal corridors (Goodman et al., 2006).
There was strong evidence for isolation‐by‐distance (IBD), meaning there was a positive correlation between genetic and geographic distances among populations. There are currently no records of subfossil ring‐tailed lemurs outside of their current distribution (Godfrey, Jungers, Simons, Chatrath, & Rakotosamimanana, 1999), suggesting that this broad geographical range has been stable through geological time (Goodman et al., 2006). If localities had been isolated for a long time, we would expect genetic drift to erase any pattern of IBD (Baden et al., 2014; Olivieri et al., 2008). Therefore, there is potentially movement and relatively recent interconnectivity between ring‐tailed populations via river basins (e.g., Mandrare River; Goodman et al., 2006). Genetic structure can result from limited gene flow or from historical events such as fragmentation; however, distinguishing between these processes can be challenging, especially when demography is unknown and forest fragmentation is recent.
Measures of genetic diversity, gene flow, and population structure are subjected to time‐lag effects (Epps & Keyghobadi, 2015). For instance, F ST values typically reflect historic rather than current population structure if populations have not yet reached migration–drift equilibrium (Whitlock & McCauley, 1999). Moreover, heterozygosity is slow to decline in previously large populations following a genetic health bottleneck (Cornuet & Luikart, 1996), a pattern which has been documented in the western localities of this species range (BEZA and TNP; Parga et al., 2012). Because deforestation has occurred within the last few decades (Brinkmann et al., 2014; Clarke et al., 2015; Gardner & Davies, 2014), it may therefore be too recent to gauge whether habitat loss has negatively impacted genetic diversity and gene flow in this species (Keyghobadi, Roland, Matter, & Strobeck, 2005; Nei, Maruyama, & Chakraborty, 1975).
4.2. Conservation implications
Genetic diversity is lost more rapidly within fragmented and isolated habitats, elevating a species' extinction risk (Frankham, 1995, 2003, 2005). Our results indicate that ring‐tailed lemur populations have high levels of genetic diversity. While this is encouraging for the conservation of the species, this may reflect past, not current population processes. Though this species is considered the most ecologically flexible lemur, exploiting anthropogenic landscapes and persisting in small fragments (Cameron & Gould, 2013; Gabriel, 2013; LaFleur & Gould, 2009; Sauther et al., 2006), they are significantly affected by fragmentation and occur at lower densities in poorer habitats (Eppley, Santini, Tinsman, & Donati, 2020; Gabriel, 2013; Kelley, 2011; Sussman et al., 2003). In addition, continued fragmentation and further isolation, coupled with climate change, may prove too much for this historically abundant lemur species. Climatic cycles have been shown to strongly affect mortality rate within this species; a 2‐year drought period in southwestern Madagascar resulted in a tenfold increase (3%–27%) in mortality among adult populations in this region (Gould et al., 2003).
Our future work aims to increase sampling efforts in underrepresented and unprotected regions, to reflect the full geographic range of this species and provide a species‐wide genetic health assessment (e.g., Calkins & Baden, in review). Moreover, this dataset forms the basis for future landscape genetics analyses which will be used to infer migration and gene flow across the species' remaining range. Because ring‐tailed lemurs are highly terrestrial and are suspected to utilize river basins as dispersal corridors (Goodman et al., 2006), they may have been able to disperse across Madagascar more easily than the more restricted arboreal lemur species. We can use landscape genetic analyses to test this hypothesis to not only better understand what facilitates and impedes gene flow, but also to develop targeted management plans moving forward.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTION
Aparna Chandrashekar: Data curation (lead); Formal analysis (lead); Investigation (equal); Visualization (lead); Writing‐original draft (lead); Writing‐review & editing (equal). Jessica A. Knierim: Data curation (equal); Formal analysis (equal); Funding acquisition (equal); Investigation (equal); Writing‐review & editing (equal). Sohail Khan: Formal analysis (supporting); Investigation (supporting); Writing‐review & editing (equal). Dominique L. Raboin: Formal analysis (supporting); Investigation (supporting); Writing‐review & editing (equal). Sateesh Venkatesh: Formal analysis (supporting); Investigation (supporting); Writing‐review & editing (equal). Tara A. Clarke: Data curation (equal); Formal analysis (supporting); Funding acquisition (equal); Investigation (equal); Resources (equal); Writing‐review & editing (equal). Frank P. Cuozzo: Formal analysis (supporting); Funding acquisition (equal); Investigation (equal); Writing‐review & editing (equal). Marni LaFleur: Data curation (equal); Funding acquisition (equal); Investigation (equal); Resources (equal); Writing‐review & editing (equal). Richard R. Lawler: Formal analysis (equal); Visualization (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Joyce A. Parga: Data curation (equal); Formal analysis (supporting); Funding acquisition (equal); Investigation (equal); Resources (equal); Writing‐review & editing (equal). Hantanirina R. Rasamimanana: Formal analysis (supporting); Investigation (equal); Writing‐review & editing (equal). Kim E. Reuter: Formal analysis (supporting); Writing‐review & editing (equal). Michelle L. Sauther: Formal analysis (supporting); Funding acquisition (equal); Investigation (equal); Writing‐review & editing (equal). Andrea L. Baden: Conceptualization (lead); Data curation (equal); Formal analysis (equal); Funding acquisition (equal); Investigation (equal); Resources (equal); Supervision (lead); Visualization (equal); Writing‐original draft (lead); Writing‐review & editing (equal).
Supporting information
Fig S1‐S2
Appendix S1
ACKNOWLEDGMENTS
Logistical support in Madagascar was provided by Madagascar Biodiversity Partnership, MICET, ANGAP/MNP. Laboratory support was provided by the Hunter Primate Molecular Ecology Lab. Funding was generously provided by Lemur Love (MLF, TAC), The Animal Behavior and Conservation Program of Hunter College (JAK), The University of Colorado Boulder, the National Geographic Society (MLS, FPC), the St. Louis Zoological Park FRC 06‐1 (MLS, FPC), the National Science Foundation Minority Postdoctoral Fellowship (JAP), Duke University (TAC), Margot Marsh Biodiversity Foundation (MLF), University of San Diego (MLF), and The City University of New York (ALB).
Chandrashekar A, Kneirim JA, Khan S, et al. Genetic population structure of endangered ring‐tailed lemurs (Lemur catta) from nine sites in southern Madagascar. Ecol Evol. 2020;10:8030–8043. 10.1002/ece3.6337
DATA AVAILABILITY STATEMENT
Raw data are publicly available on Zenodo (https://doi.org/10.5281/zenodo.3750377, https://doi.org/10.5281/zenodo.3750382).
REFERENCES
- Baden, A. L. , Holmes, S. M. , Johnson, S. E. , Engberg, S. E. , Louis, E. E. Jr , & Bradley, B. J. (2014). Species‐level view of population structure and gene flow for a critically endangered primate (Varecia variegata). Ecology and Evolution, 4:2675‐2692. 10.1002/ece3.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden, A. L. , Mancini, A. N. , Federman, S. , Holmes, S. M. , Johnson, S. E. , Kamilar, J. , … Bradley, B. J. (2019). Anthropogenic pressures drive population genetic structuring across a Critically Endangered lemur species range. Scientific Reports, 9(1), 1–11. 10.1038/s41598-019-52689-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenneman, R. A. , Johnson, S. E. , Bailey, C. A. , Ingraldi, C. , Delmore, K. E. , Wyman, T. M. , … Louis, E. E. (2012). Population genetics and abundance of the Endangered grey‐headed lemur Eulemur cinereiceps in south‐east Madagascar: Assessing risks for fragmented and continuous populations. Oryx, 46(2), 298–307. [Google Scholar]
- Brinkmann, K. , Noromiarilanto, F. , Ratovonamana, R. Y. , & Buerkert, A. (2014). Deforestation processes in south‐western Madagascar over the past 40 years: What can we learn from settlement characteristics? Agriculture, Ecosystems and Environment, 195, 231–243. 10.1016/j.agee.2014.06.008 [DOI] [Google Scholar]
- Brown, J. L. , & Yoder, A. D. (2015). Shifting ranges and conservation challenges for lemurs in the face of climate change. Ecology and Evolution, 5(6), 1131–1142. 10.1002/ece3.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, A. , & Gould, L. (2013). Fragment‐adaptive behavioural strategies and intersite variation in the ring‐tailed lemur (Lemur catta) in south‐central Madagascar In Marsh L. K., & Chapman C. A. (Eds.), Primates in fragments: Complexity and resilience (pp. 227–243). New York, NY: Springer New York; 10.1007/978-1-4614-8839-2_16 [DOI] [Google Scholar]
- Charpentier, M. J. , Williams, C. V. , & Drea, C. M. (2008). Inbreeding depression in ring‐tailed lemurs (Lemur catta): Genetic diversity predicts parasitism, immunocompetence, and survivorship. Conservation Genetics, 9(6), 1605–1615. 10.1007/s10592-007-9499-4 [DOI] [Google Scholar]
- Clarke, T. A. , Gray, O. , Gould, L. , & Burrell, A. S. (2015). Genetic diversity of the ring‐tailed lemur (Lemur catta) in South‐Central Madagascar. Folia Primatologica, 86(1–2), 76–84. 10.1159/000368668 [DOI] [PubMed] [Google Scholar]
- Cornuet, J. M. , & Luikart, G. (1996). Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics, 144(4), 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craul, M. , Chikhi, L. , Sousa, V. , Olivieri, G. L. , Rabesandratana, A. , Zimmermann, E. , & Radespiel, U. (2009). Influence of forest fragmentation on an endangered large‐bodied lemur in northwestern Madagascar. Biological Conservation, 142(12), 2862–2871. 10.1016/j.biocon.2009.05.026 [DOI] [Google Scholar]
- Dewar, R. E. , Radimilahy, C. , Wright, H. T. , Jacobs, Z. , Kelly, G. O. , & Berna, F. (2013). Stone tools and foraging in northern Madagascar challenge Holocene extinction models. Proceedings of the National Academy of Sciences, 110(31), 12583–12588. 10.1073/pnas.1306100110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marco, M. , Venter, O. , Possingham, H. P. , & Watson, J. E. (2018). Changes in human footprint drive changes in species extinction risk. Nature Communications, 9(1), 1–9. 10.1038/s41467-018-07049-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl, D. A. , & vonHoldt, B. M. (2012). Structure Harvester: A website and program for visualizing Structure output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Eppley, T. M. , Santini, L. , Tinsman, J. C. , & Donati, G. (2020). Do functional traits offset the effects of fragmentation? The case of large‐bodied diurnal lemur species. American Journal of Primatology, 82(4), e23104. [DOI] [PubMed] [Google Scholar]
- Epps, C. W. , & Keyghobadi, N. (2015). Landscape genetics in a changing world: Disentangling historical and contemporary influences and inferring change. Molecular Ecology, 24(24), 6021–6040. 10.1111/mec.13454 [DOI] [PubMed] [Google Scholar]
- Evanno, G. , Regnaut, S. , & Goudet, J. (2005). Detecting the number of clusters of individuals using the software Structure: A simulation study. Molecular Ecology, 14(8), 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Falush, D. , Stephens, M. , & Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics, 164(4), 1567–1587. 10.3410/f.1015548.197423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardi, S. , Sauther, M. L. , Cuozzo, F. P. , Youssouf, J. I. A. , & Bernstein, R. (2018). The effect of extreme weather events on hair cortisol and body weight in a wild ring‐tailed lemur population (Lemur catta) in southwestern Madagascar. American Journal of Primatology, 80(2), e22731 10.1002/ajp.22731/full [DOI] [PubMed] [Google Scholar]
- Frankham, R. (1995). Conservation genetics. Annual Review of Genetics, 29(1), 305–327. 10.1146/annurev.ge.29.120195.001513 [DOI] [PubMed] [Google Scholar]
- Frankham, R. (2003). Genetics and conservation biology. Comptes Rendus Biologies, 326, 22–29. 10.1016/S1631-0691(03)00023-4 [DOI] [PubMed] [Google Scholar]
- Frankham, R. (2005). Genetics and extinction. Biological Conservation, 126(2), 131–140. 10.1016/j.biocon.2005.05.002 [DOI] [Google Scholar]
- Frankham, R. , Ballou, J. D. , & Briscoe, D. A. (2010). Introduction to conservation genetics. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Fredsted, T. , Pertoldi, C. , Schierup, M. H. , & Kappeler, P. M. (2005). Microsatellite analyses reveal fine‐scale genetic structure in grey mouse lemurs (Microcebus murinus). Molecular Ecology, 14, 2363–2372. [DOI] [PubMed] [Google Scholar]
- Gabriel, D. N. (2013). Habitat use and activity patterns as an indication of fragment quality in a strepsirrhine primate. International Journal of Primatology, 34, 388–406. 10.1007/s10764-013-9668-x [DOI] [Google Scholar]
- Gardner, C. J. , & Davies, Z. G. (2014). Rural bushmeat consumption within multiple‐use protected areas: Qualitative evidence from southwest Madagascar. Human Ecology, 42, 21–34. [Google Scholar]
- Godfrey, L. R. , & Irwin, M. T. (2007). The evolution of extinction risk: Past and present anthropogenic impacts on the primate communities of Madagascar. Folia Primatologica, 78(5–6), 405–419. 10.1159/000105152 [DOI] [PubMed] [Google Scholar]
- Godfrey, L. R. , Jungers, W. L. , Simons, E. L. , Chatrath, P. S. , & Rakotosamimanana, B. (1999). Past and present distributions of lemurs in Madagascar In New directions in lemur studies (pp. 19–53). Boston, MA:Springer. [Google Scholar]
- Goodman, S. M. , & Langrand, O. (1996). A high mountain population of the ring‐tailed lemur Lemur catta on the Andringitra Massif, Madagascar. Oryx, 30, 259–268. [Google Scholar]
- Goodman, S. M. , Raherilalao, M. J. , Rakotomalala, D. , Rakotondravony, D. , Raselimanana, A. P. , Razakarivony, H. V. , & Soarimalala, V. (2002). Inventaire des vertébrés du Parc National de Tsimanampetsotsa (Toliara). Akon'ny Ala, 28, 1–36. [Google Scholar]
- Goodman, S. M. , Rakotoarisoa, S. V. , & Wilme, L. (2006). The distribution and biogeography of the Ringtailed Lemur (Lemur catta) in Madagascar In Ringtailed Lemur biology (pp. 3–15). Boston, MA:Springer; 10.1007/978-0-387-34126-2_1 [DOI] [Google Scholar]
- Gould, L. , & Sauther, M. L. (2016). Going, going, gone… is the iconic ring‐tailed lemur (Lemur catta) headed for imminent extirpation? Primate Conservation, 2016(30), 89–101. [Google Scholar]
- Gould, L. , Sussman, R. W. , & Sauther, M. L. (2003). Demographic and life‐history patterns in a population of ring‐tailed lemurs (Lemur catta) at Beza Mahafaly Reserve, Madagascar: A 15‐year perspective. American Journal of Physical Anthropology, 120(2), 182–194. 10.1002/ajpa.10151 [DOI] [PubMed] [Google Scholar]
- Grogan, K. E. , Sauther, M. L. , Cuozzo, F. P. , & Drea, C. M. (2017). Genetic wealth, population health: Major histocompatibility complex variation in captive and wild ring‐tailed (Lemur catta). Ecology and Evolution, 7, 7638–7649. 10.1002/ece3.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiler, E. R. (1970). Tasmanian devils and agriculture. Tasmanian Journal of Agriculture, 41, 134–137. [Google Scholar]
- Holmes, S. M. , Baden, A. L. , Brenneman, R. A. , Engberg, S. E. , Louis, E. E. , & Johnson, S. E. (2013). Patch size and isolation influence genetic patterns in black‐and‐white ruffed lemur (Varecia variegata) populations. Conservation Genetics, 14(3), 615–624. 10.1007/s10592-013-0455-1 [DOI] [Google Scholar]
- Jackson, S. T. , & Sax, D. F. (2010). Balancing biodiversity in a changing environment: Extinction debt, immigration credit and species turnover. Trends in Ecology & Evolution, 25(3), 153–160. 10.1016/j.tree.2009.10.001 [DOI] [PubMed] [Google Scholar]
- Jolly, A. (1966). Lemur behavior: A Madagascar field study (p. 187, xiv). Chicago, IL: University of Chicago Press. [Google Scholar]
- Jombart, T. (2008). adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics, 24(11), 1403–1405. 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- Jombart, T. , Devillard, S. , & Balloux, F. (2010). Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genetics, 11(1), 94 10.1186/1471-2156-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski, S. T. (2005). HP‐RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes, 5(1), 187–189. 10.1111/j.1471-8286.2004.00845.x [DOI] [Google Scholar]
- Kalinowski, S. T. , Taper, M. L. , & Marshall, T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology, 16, 1099–1106. 10.1111/j.1365-294x.2007.03089.x [DOI] [PubMed] [Google Scholar]
- Kelley, E. A. (2011). Lemur catta in the region of Cap Sainte‐Marie, Madagascar: Introduced cacti, xerophytic Didiereaceae‐Euphorbia bush, and tombs (Unpublished doctoral dissertation). Washington University, St. Louis. [Google Scholar]
- Keyghobadi, N. , Roland, J. , Matter, S. F. , & Strobeck, C. (2005). Among‐and within‐patch components of genetic diversity respond at different rates to habitat fragmentation: An empirical demonstration. Proceedings of the Royal Society B: Biological Sciences, 272(1562), 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler, L. , Ratan, A. , Godfrey, L. R. , Crowley, B. E. , Hughes, C. E. , Lei, R. , … Perry, G. H. (2015). Comparative and population mitogenomic analyses of Madagascar's extinct, giant ‘subfossil’ lemurs. Journal of Human Evolution, 79, 45–54. 10.1016/j.jhevol.2014.06.016 [DOI] [PubMed] [Google Scholar]
- LaFleur, M. , Clarke, T. A. , Ratzimbazafy, J. , & Reuter, K. E. (2017a). Lemur catta (Linnaeus, 1758) In Schwitzer C., Mittermeier R. A., Rylands A. B., Chiozza F., Williamson E. A., Macfie E. J., Wallis J., & Cotton A. (Eds.), Primates in peril: The world's 25 most endangered primates 2016–2018 (pp. 35–37). Arlington, VA: IUCN SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), and Bristol Zoological Society. [Google Scholar]
- LaFleur, M. , Clarke, T. , Reuter, K. , Schaefer, M. , & terHorst, C. (2019). Illegal trade of wild‐captured Lemur catta in Madagascar. Folia Primatologica, 90, 199–214. 10.1159/000496970 [DOI] [PubMed] [Google Scholar]
- LaFleur, M. , Clarke, T. A. , Reuter, K. E. , & Schaeffer, T. (2017b). Rapid decrease in populations of wild ring‐tailed lemurs (Lemur catta) in Madagascar. Folia Primatologica, 87(5), 320–330. 10.1159/000455121 [DOI] [PubMed] [Google Scholar]
- LaFleur, M. , & Gould, L. (2009). Feeding outside the forest: The importance of crop raiding and an invasive weed in the diet of gallery forest ring‐tailed lemurs (Lemur catta) following a cyclone at the Beza Mahafaly special reserve, Madagascar. Folia Primatologica, 80(3), 233–246. 10.1159/000240968 [DOI] [PubMed] [Google Scholar]
- Lawler, R. R. , Richard, A. F. , & Riley, M. A. (2003). Genetic population structure of the white sifaka (Propithecus verreauxi verreauxi) at Beza Mahafaly Special Reserve, southwest Madagascar (1992–2001). Molecular Ecology, 12, 2307–2317. [DOI] [PubMed] [Google Scholar]
- Lino, A. , Fonseca, C. , Rojas, D. , Fischer, E. , & Pereira, M. J. R. (2019). A meta‐analysis of the effects of habitat loss and fragmentation on genetic diversity in mammals. Mammalian Biology, 94(1), 69–76. 10.1016/j.mambio.2018.09.006 [DOI] [Google Scholar]
- Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Research, 27(2 Part 1), 209–220. [PubMed] [Google Scholar]
- Meirmans, P. G. , & Van Tienderen, P. H. (2004). GENOTYPE and GENODIVE: Two programs for the analysis of genetic diversity of asexual organisms. Molecular Ecology Notes, 4(4), 792–794. 10.1111/j.1471-8286.2004.00770.x [DOI] [Google Scholar]
- Merenlender, A. (1993). The effects of sociality on the demography and genetic structure of Lemur fulvus rufus (polygamous) and Lemur rubriventer (monogamous) and the conservation implications (Doctoral dissertation). University of Rochester. [Google Scholar]
- Miraldo, A. , Li, S. , Borregaard, M. K. , Flórez‐Rodríguez, A. , Gopalakrishnan, S. , Rizvanovic, M. , … Nogués‐Bravo, D. (2016). An Anthropocene map of genetic diversity. Science, 353(6307), 1532–1535. [DOI] [PubMed] [Google Scholar]
- Morelli, T. L. , Smith, A. B. , Mancini, A. N. , Balko, E. A. , Borgerson, C. , Dolch, R. , … Baden, A. L. (2020). The fate of Madagascar's rainforest habitat. Nature Climate Change, 10(1), 89–96. 10.1038/s41558-019-0647-x [DOI] [Google Scholar]
- Morin, P. A. , Chambers, K. E. , Boesch, C. , & Vigilant, L. (2001). Quantitative polymerase chain reaction analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzees (Pan troglodytes verus). Molecular Ecology, 10(7), 1835–1844. 10.1046/j.0962-1083.2001.01308.x [DOI] [PubMed] [Google Scholar]
- Morris, K. M. , Wright, B. , Grueber, C. E. , Hogg, C. , & Belov, K. (2015). Lack of genetic diversity across diverse immune genes in an endangered mammal, the Tasmanian devil (Sarcophilus harrisii). Molecular Ecology, 24(15), 3860–3872. [DOI] [PubMed] [Google Scholar]
- Murphy, A. J. , Ferguson, B. , & Gardner, C. J. (2017). Recent estimates of ring‐tailed lemur (Lemur catta) population declines are methodologically flawed and misleading. International Journal of Primatology, 38, 623–628. [Google Scholar]
- Myers, N. , Mittermeier, R. A. , Mittermeier, C. G. , da Fonseca, G. A. B. , & Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature, 403, 853–858. [DOI] [PubMed] [Google Scholar]
- Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89(3), 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M. , Maruyama, T. , & Chakraborty, R. (1975). The bottleneck effect and genetic variability in populations. Evolution, 29, 1–10. 10.1111/j.1558-5646.1975.tb00807.x [DOI] [PubMed] [Google Scholar]
- Nunziata, S. O. , Wallenhorst, P. , Barrett, M. A. , Junge, R. E. , Yoder, A. D. , & Weisrock, D. W. (2016). Population and conservation genetics in an Endangered lemur, Indri indri, across three forest reserves in Madagascar. International Journal of Primatology, 37, 688–702. 10.1007/s10764-016-9932-y [DOI] [Google Scholar]
- Olivieri, G. L. , Sousa, V. , Chikhi, L. , & Radespiel, U. (2008). From genetic diversity and structure to conservation: Genetic signature of recent population declines in three mouse lemur species (Microcebus spp.). Biological Conservation, 141(5), 1257–1271. 10.1016/j.biocon.2008.02.025 [DOI] [Google Scholar]
- Paetkau, D. , & Strobeck, C. (1994). Microsatellite analysis of genetic variation in black bear populations. Molecular Ecology, 3, 489–495. 10.1111/j.1365-294X.1994.tb00127.x [DOI] [PubMed] [Google Scholar]
- Parga, J. A. , Sauther, M. L. , Cuozzo, F. P. , Jacky, I. A. Y. , Gould, L. , Sussman, R. W. , … Pastorini, J. (2015). Genetic evidence for male and female dispersal in wild Lemur catta . Folia Primatologica, 86(1–2), 66–75. [DOI] [PubMed] [Google Scholar]
- Parga, J. A. , Sauther, M. L. , Cuozzo, F. P. , Jacky, I. A. Y. , & Lawler, R. R. (2012). Evaluating ring‐tailed lemurs (Lemur catta) from southwestern Madagascar for a genetic population bottleneck. American Journal of Physical Anthropology, 147(1), 21–29. 10.1002/ajpa.21603 [DOI] [PubMed] [Google Scholar]
- Pastorini, J. , Fernando, P. , Forstner, M. R. J. , & Melnick, D. J. (2005). Characterization of new microsatellite loci for the ring‐tailed lemur (Lemur catta). Molecular Ecology Notes, 5(1), 149–151. 10.1111/j.1471-8286.2004.00865.x [DOI] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2012). GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research‐an update. Bioinformatics, 28(19), 2537–2539. 10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, G. H. , Melsted, P. , Marioni, J. C. , Wang, Y. , Bainer, R. , Pickrell, J. K. , … Gilad, Y. (2013). Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Resources, 22, 602–610. 10.1101/gr.130468.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. 10.1111/j.1471-8286.2007.01758.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quéméré, E. , Crouau‐Roy, B. , Rabarivola, C. , Louis, E. E. Jr , & Chikhi, L. (2010). Landscape genetics of an endangered lemur (Propithecus tattersalli) within its entire fragmented range. Molecular Ecology, 19, 1606–1621. 10.1111/j.1365-294X.2010.04581.x [DOI] [PubMed] [Google Scholar]
- Quéméré, E. , Louis, E. E. , Ribéron, A. , Chikhi, L. , & Crouau‐Roy, B. (2010). Non‐invasive conservation genetics of the critically endangered golden‐crowned sifaka (Propithecus tattersalli): High diversity and significant genetic differentiation over a small range. Conservation Genetics, 11(3), 675–687. 10.1007/s10592-009-9837-9 [DOI] [Google Scholar]
- R Core Team (2017). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; Retrieved from http://www.Rproject.org [Google Scholar]
- Radespiel, U. , Rakotondravony, R. , & Chikhi, L. (2008). Natural and anthropogenic determinants of genetic structure in the largest remaining population of the endangered golden‐brown mouse lemur (Microcebus ravelobensis). American Journal of Primatology, 70(9), 860–870. 10.1002/ajp.20574 [DOI] [PubMed] [Google Scholar]
- Raymond, M. , & Rousset, F. (1995). GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. Journal of Heredity, 86(3), 248–249. [Google Scholar]
- Razakamaharavo, V. R. , McGuire, S. M. , Vasey, N. , Louis, E. E. , & Brenneman, R. A. (2010). Genetic architecture of two red ruffed lemur (Varecia rubra) populations of Masoala National Park. Primates, 51, 53–61. [DOI] [PubMed] [Google Scholar]
- Reuter, K. E. , LaFleur, M. , Clarke, T. A. , Holiniaina Kjeldgaard, F. , Ramanantenasoa, I. , Ratolojanahary, T. , … Schaefer, M. S. (2019). A national survey of household pet lemur ownership in Madagascar. PLoS ONE, 14(5), e0216593 10.1371/journal.pone.0216593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, K. E. , & Schaefer, M. S. (2016). Illegal captive lemurs in Madagascar: Comparing the use of online and in‐person data collection methods. American Journal of Primatology, 79(11), e22541 10.1002/ajp.22541 [DOI] [PubMed] [Google Scholar]
- Salmona, J. , Teixeira, H. , Rasolondraibe, E. , Aleixo‐Pais, I. , Kun‐Rodrigues, C. , Rakotonanahary, A. N. , … Chikhi, L. (2015). Genetic diversity, population size, and conservation of the Critically Endangered Perrier's sifaka (Propithecus perrieri). International Journal of Primatology, 36, 1132–1153. 10.1007/s10764-015-9881-x [DOI] [Google Scholar]
- Sauther, M. L. (1998). The interplay of phenology and reproduction in ring‐tailed lemurs: Implications for ring‐tailed lemur conservation. Folia Primatologica, 69, 309–320. 10.1159/000052719 [DOI] [Google Scholar]
- Sauther, M. L. , Fish, K. D. , Cuozzo, F. P. , Miller, D. S. , Hunter‐Ishikawa, M. , & Culbertson, H. (2006). Patterns of health, disease, and behavior among wild ringtailed lemurs, Lemur catta: Effects of habitat and sex In Jolly A., Sussman R. W., Koyama N., & Rasamimanana H. (Eds.), Ringtailed lemur biology: Lemur catta in Madagascar (pp. 313–331). New York, NY: Springer. [Google Scholar]
- Sauther, M. L. , Gould, L. , Cuozzo, F. P. , O'Mara, M. T. (2015). Ring‐Tailed Lemurs: A Species Re‐Imagined. Folia Primatologica. 86(1‐2),5‐13. [DOI] [PubMed] [Google Scholar]
- Sauther, M. L. , Sussman, R. W. , & Gould, L. (1999). The socioecology of the ring‐tailed lemur: Thirty‐five years of research. Evolutionary Anthropology, 8, 120–132. [DOI] [Google Scholar]
- Stangel, P. W. , Lennartz, M. R. , & Smith, M. H. (1992). Genetic variation and population structure of Red‐cockaded Woodpeckers. Conservation Biology, 6(2), 283–292. 10.1046/j.1523-1739.1992.620283.x [DOI] [Google Scholar]
- Struebig, M. J. , Kingston, T. , Petit, E. J. , Le Comber, S. C. , Zubaid, A. , Mohd‐Adnan, A. , & Rossiter, S. J. (2011). Parallel declines in species and genetic diversity in tropical forest fragments. Ecology Letters, 14(6), 582–590. 10.1111/j.1461-0248.2011.01623.x [DOI] [PubMed] [Google Scholar]
- Sussman, R. W. (1972). An ecological study of two Madagascan primates: Lemur fulvus rufus Audebert and Lemur catta Linnaeus. Ph.D. dissertation. Duke University, Raleigh, NC. [Google Scholar]
- Sussman, R. W. (1974). Ecological distinctions between two species of lemur In Martin R. D., Doyle D. A., & Walker C. (Eds.), Prosimian biology (pp. 75–108). London, UK: Duckworth. [Google Scholar]
- Sussman, R. W. (1991). Demography and social organization of free‐ranging Lemur catta in the Beza Mahafaly Reserve, Madgascar. American Journal of Physical Anthropology, 58, 43–58. 10.1002/ajpa.1330840105 [DOI] [Google Scholar]
- Sussman, R. W. , Green, G. M. , Porton, I. , Andrianasolondraibe, O. , & Ratsirarson, J. (2003). A survey of the habitat of Lemur catta in Southwestern and Southern Madagascar. Primate Conservation, 19(19), 32–57. [Google Scholar]
- Taberlet, P. , Griffin, S. , Goossens, B. , Questiau, S. , Manceau, V. , Escaravage, N. , … Bouvet, J. (1996). Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Research, 24(16), 3189–3194. 10.1093/nar/24.16.3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman, D. , May, R. M. , Lehman, C. L. , & Nowak, M. A. (1994). Habitat destruction and the extinction debt. Nature, 371, 65–66. 10.1038/371065a0 [DOI] [Google Scholar]
- van Oosterhout, C. , Hutchinson, W. , Wills, D. , & Shipley, P. (2004). Micro‐checker: Software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4(3), 535–538. 10.1111/j.1471-8286.2004.00684.x [DOI] [Google Scholar]
- Vieilledent, G. , Grinand, C. , Rakotomalala, F. A. , Ranaivosoa, R. , Rakotoarijaona, J. R. , Allnut, T. F. , & Achard, F. (2018). Combining global tree cover loss data with historical national forest cover maps to look at six decades of deforestation and forest fragmentation in Madagascar. Conservation Biology, 222, 189–197. 10.1016/j.biocon.2018.04.008 [DOI] [Google Scholar]
- Wang, J. (2016). The computer program STRUCTURE for assigning individuals to populations: Easy to use but easier to misuse. Molecular Ecology Resources, 17(5), 981–990. 10.1111/1755-0998.12650 [DOI] [PubMed] [Google Scholar]
- Weir, B. S. , & Cockerham, C. C. (1984). Estimating F‐statistics for the analysis of population structure. Evolution, 38(6), 1358 10.1111/j.1558-5646.1984.tb05657.x [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C. , & McCauley, D. E. (1999). Indirect measures of gene flow and migration: FST≠1/(4Nm+1). Heredity, 82, 117–125. 10.1038/sj.hdy.6884960 [DOI] [PubMed] [Google Scholar]
- Wright, S. (1978). Evolution and the genetics of populations: Variability within and among natural populations (Vol. 4). Chicago, IL: University of Chicago Press. [Google Scholar]
- Zaonarivelo, J. R. , Andriantompohavana, R. , Shore, G. E. , Engberg, S. E. , McGuire, S. M. , Louis, E. E. , & Brenneman, R. A. (2007). Characterization of 21 microsatellite marker loci in the ring‐tailed lemur (Lemur catta). Conservation Genetics, 8(5), 1209–1212. 10.1007/s10592-006-9259-x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S2
Appendix S1
Data Availability Statement
Raw data are publicly available on Zenodo (https://doi.org/10.5281/zenodo.3750377, https://doi.org/10.5281/zenodo.3750382).


