Abstract
Key Message
Distinct catalytic features of the Poaceae TPS-a subfamily arose early in grass evolution and the reactions catalyzed have become more complex with time.
Abstract
The structural diversity of terpenes found in nature is mainly determined by terpene synthases (TPS). TPS enzymes accept ubiquitous prenyl diphosphates as substrates and convert them into the various terpene skeletons by catalyzing a carbocation-driven reaction. Based on their sequence similarity, terpene synthases from land plants can be divided into different subfamilies, TPS-a to TPS-h. In this study, we aimed to understand the evolution and functional diversification of the TPS-a subfamily in the Poaceae (the grass family), a plant family that contains important crops such as maize, wheat, rice, and sorghum. Sequence comparisons showed that aside from one clade shared with other monocot plants, the Poaceae TPS-a subfamily consists of five well-defined clades I–V, the common ancestor of which probably originated very early in the evolution of the grasses. A survey of the TPS literature and the characterization of representative TPS enzymes from clades I–III revealed clade-specific substrate and product specificities. The enzymes in both clade I and II function as sesquiterpene synthases with clade I enzymes catalyzing initial C10-C1 or C11-C1 ring closures and clade II enzymes catalyzing C6-C1 closures. The enzymes of clade III mainly act as monoterpene synthases, forming cyclic and acyclic monoterpenes. The reconstruction and characterization of clade ancestors demonstrated that the differences among clades I–III were already present in their ancestors. However, the ancestors generally catalyzed simpler reactions with less double-bond isomerization and fewer cyclization steps. Overall, our data indicate an early origin of key enzymatic features of TPS-a enzymes in the Poaceae, and the development of more complex reactions over the course of evolution.
Electronic supplementary material
The online version of this article (10.1007/s11103-020-01037-4) contains supplementary material, which is available to authorized users.
Keywords: Terpene synthase, Evolution, Maize, Grasses, Ancestral sequence reconstruction
Introduction
Terpenes are a structurally diverse group of natural products that are ubiquitous in plants and many other organisms. They are involved in basic physiological processes, but also play important roles in plant–plant and plant–insect interactions. Many plants, for example, produce and release volatile terpenes in response to insect herbivory to attract natural enemies of their herbivores (Unsicker et al. 2009). Infection with plant pathogens can also induce the formation of terpenes, which in turn act as phytoalexins (Block et al. 2019). Until now, more than 40,000 terpenes have been described (Tholl 2015). This structural diversity can mainly be attributed to terpene synthases (TPS), a class of enzymes that catalyze the conversion of prenyl diphosphates into the different terpene skeletons (reviewed in Degenhardt et al. 2009; Karunanithi and Zerbe 2019). Most monoterpene synthases accept the precursor geranyl diphosphate (GPP) as substrate and produce monoterpenes (C10), while most sesquiterpene synthases convert (E,E)-farnesyl diphosphate (FPP) into sesquiterpenes (C15), and diterpene synthases catalyze the conversion of geranylgeranyl diphosphate (GGPP) into the different diterpenes (C20). Interestingly, a number of terpene synthases have been described to act also on cis-prenyl diphosphates such as neryl diphosphate, (Z,Z)-FPP, and nerylneryl diphosphate (Schilmiller et al. 2009; Sallaud et al. 2009; Zi et al. 2014). The terpenes formed may exert a variety of biological functions (Unsicker et al. 2009; Block et al. 2019). They may also act as substrates for modifying enzymes such as cytochrome P450 monooxygenases, O-methyltransferases, and acyltransferases (Dudareva et al. 2004; Degenhardt et al. 2009; Bathe and Tissier 2019).
Although terpene synthases often have broad substrate specificity and accept GPP, FPP, and GGPP in vitro, their in planta function may be narrower due to their subcellular localization (Pazouki and Niinemetz 2016). Monoterpene synthases and diterpene synthases typically contain N-terminal signal peptides and are transported into plastids, the site for GPP and GGPP, but not FPP production. Sesquiterpene synthases, however, are usually found in the cytosol, a site of FPP, but not GPP nor GGPP production (Degenhardt et al. 2009). There is increasing evidence for an exchange of prenyl diphosphates between the different compartments of a plant cell, especially under stress conditions (e.g. Gutensohn et al. 2013; Dong et al. 2016). Thus, the subcellular localization of TPS enzymes as well as the exchange of TPS substrates between compartments can determine the in planta function of terpene synthases with broad substrate specificity (Pazouki and Niinemetz 2016).
Based on sequence similarity and phylogenetic relationships, terpene synthases can be divided into eight subfamilies, TPS-a to TPS-h (Bohlmann et al. 1998; Chen et al. 2011). The subfamilies TPS-a, TPS-b, and TPS-g comprise predominantly monoterpene synthases and sesquiterpene synthases of angiosperms, while the gymnosperm monoterpene and sesquiterpene synthases belong to the TPS-d subfamily. The subfamilies TPS-c and TPS-e/f mainly comprise diterpene synthases from angiosperms and gymnosperms, and the subfamily TPS-h contains diterpene synthases specific for the Lycopodiophyta (clubmosses) (Bohlmann et al. 1998; Chen et al. 2011; Karunanithi and Zerbe 2019). Angiosperm plants typically possess 20–60 TPS genes, most of which belong to the TPS-a subfamily (Chen et al. 2011).
The reaction mechanism catalyzed by terpene synthases starts with the formation of a carbocation intermediate that is either derived by a metal ion-dependent ionization of the prenyl diphosphate substrate (class I TPS) or by protonation of a double bond in the prenyl diphosphate tail (class II TPS) (Degenhardt et al. 2009). While most if not all monoterpene synthases and sesquiterpene synthases belong to class I TPS, diterpene synthases can be found in both classes. After its formation, the carbocation intermediate can undergo a complex cascade of different cyclizations, hydride shifts, and skeleton rearrangements until the reaction is terminated by a deprotonation or addition of a nucleophile such as water (Degenhardt et al. 2009). Many terpene synthases are multiproduct enzymes that produce complex mixtures of compounds (Steele et al. 1998; Garms et al. 2010; Irmisch et al. 2014). However, the individual components of the mixture often share the same terpene skeleton or at least belong to structurally related skeleton types, the formation of which is mainly determined by the first cyclization of the reaction cascade (e.g. Köllner et al. 2004; Garms et al. 2010; Irmisch et al. 2012). The maize sesquiterpene synthase TPS4, for example, produces a mixture of more than 22 compounds that are almost all derived by an initial C6-C1 ring closure (numbering as for FPP) of the (Z,E)-farnesyl carbocation (Köllner et al. 2004). In contrast, all products formed by MrTPS2, a terpene synthase found in Matricaria recutita, are derived by an initial C11-C1 closure of the (E,E)-farnesyl carbocation (Irmisch et al. 2012; Hong et al. 2013). Although the basic phylogeny of the TPS family, including division into subfamilies, is well accepted, evolutionary relationships among many of the reaction types are still not well understood.
As part of our ongoing effort to investigate the formation of terpenes in the grass family (Poaceae), we aimed to understand the evolution of catalytic function in the TPS-a enzymes of this plant family. In the Poaceae, the TPS-a subfamily dominates the TPS gene family in terms of absolute numbers of genes (Chen et al. 2011) and includes a large number of previously characterized TPS enzymes. Ancestral sequences of the Poaceae TPS-a subfamily were reconstructed, synthesized, and heterologously expressed in Escherichia coli. Recombinant enzymes were incubated with the potential substrates GPP and (E,E)-FPP, and reaction products were analyzed using gas chromatography-mass spectrometry (GC–MS). Our data showed that many of the distinct features of specific clades of the Poaceae TPS-a subfamily arose early in grass evolution, but the reactions catalyzed have become more complex with time.
Results
Most of the TPS-a genes in the Poaceae belong to five well-defined clades
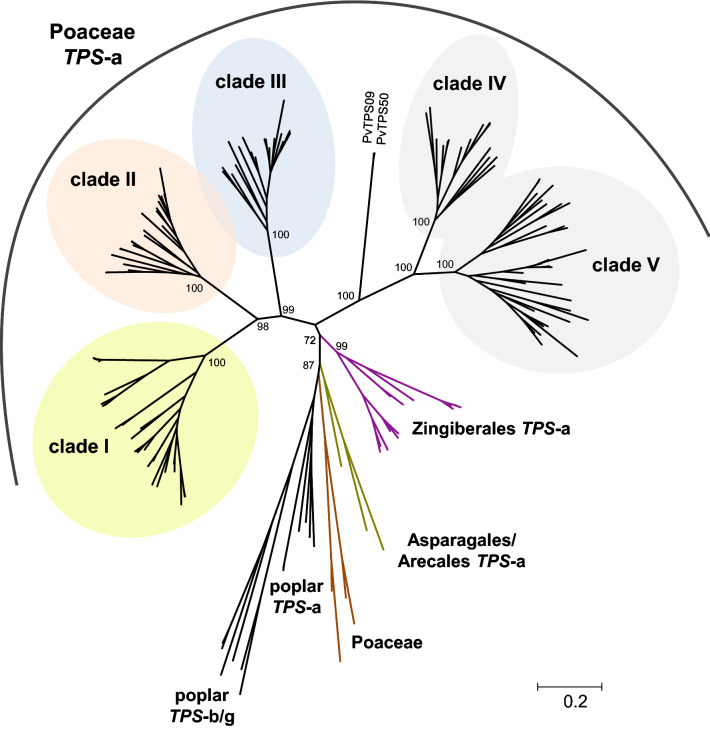
In order to identify TPS-a genes from the Poaceae, we constructed a phylogenetic tree of terpene synthase genes that were extracted using BLASTP analysis from six grass species, including Zea mays, Sorghum bicolor, Setaria italica, Panicum virgatum, Oryza sativa, and Brachypodium distachyon, and a number of other monocotyledonous and dicotyledonous species available in the NCBI database and the Phytozome 9.1 database (Supplemental Figure S1). In accordance with the literature (Chen et al. 2011), all obtained TPS genes clustered into one of the established TPS subfamilies TPS-a, TPS-b, TPS-c, TPS-e, TPS-f, and TPS-g. The TPS-a genes formed two groups, one containing genes from dicotyledonous species and the other containing genes of monocotyledonous species (Supplemental Figure S1, Chen et al. 2011). A deeper dendrogram analysis of the identified monocotyledonous TPS-a genes revealed five well-defined monophyletic clades, I–V, each of which possessed exclusively TPS-a genes from the Poaceae (Fig. 1). Two genes from P. virgatum were similar to clade IV and clade V sequences, but clustered separately. Four other Poaceae TPS-a genes, from rice, sorghum, and bamboo, were found to form a very basal clade among the monocotyledonous TPS-a subfamily, while TPS genes from families outside the Poaceae clustered separately from the five Poaceae clades (Fig. 1).
Fig. 1.
Dendrogram analysis of terpene synthase genes from six Poaceae species (Zea mays, Sorghum bicolor, Setaria italica, Panicum virgatum, Oryza sativa, Brachypodium distachyon) and from diverse species of Arecales, Asparagales, and Zingiberales. The tree was inferred by using the Maximum Likelihood method based on the General Time Reversible model (Rates among sites, G + I). Bootstrap values (n = 1000 replicates) are shown next to each node. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Terpene synthases from Populus trichocarpa were included as an outgroup
The Poaceae TPS-a genes in clades I–V encode enzymes with distinct catalytic features
A literature survey of the properties of the Poaceae TPS-a encoded enzymes characterized to date revealed that clades I–V have distinctive catalytic features (Table 1). All previously characterized clade I enzymes are sesquiterpene synthases that accept (E,E)-FPP as substrate and, with two exceptions, catalyze an initial C10-C1 or C11-C1 closure (Fig. 2). All characterized clade II enzymes are also sesquiterpene synthases, but produce either acyclic terpenes or cyclic terpenes derived from an initial C6-C1 closure. In contrast to the sesquiterpene synthases of clades I and II, characterized clade III enzymes mainly accept GPP as substrate and produce monoterpenes derived from an initial C6-C1 closure (Fig. 2). Moreover, most of the clade III sequences are predicted to contain a transit peptide for plastid localization, indicating that these enzymes likely function as monoterpene synthases in planta. The terpene synthases in clades IV and V, however, are diverse in terms of substrate specificity and initial cyclization reaction. The characterized enzymes in these clades form monoterpenes or sesquiterpenes, and catalyze initial C6-C1, C10-C1, or C11-C1 closures (Table 1).
Table 1.
Characterized TPS-a enzymes in the Poaceae
| Clade | TPS Name | Species | Grass lineage | Main product(s) | Initial cyclization | References |
|---|---|---|---|---|---|---|
| I | Bradi3g14710 | Brachypodium distachyon | BEP | (E)-β-Caryophyllene | 11–1 | This study |
| I | ZS | Chrysopogon zizanoides | PACMAD | (+)-Zizaene | 6–1 | Hartwig et al. (2015) |
| I | ObTPS1 | Oryza barthii | BEP | (E)-β-Caryophyllene | 11–1 | Chen et al. (2014) |
| I | OgTPS1 | Oryza glaberrima | BEP | (E)-β-Caryophyllene/Germacrene A | 11–1/10–1 | Chen et al. (2014) |
| I | OgluTPS1 | Oryza glumaepatula | BEP | (E)-β-Caryophyllene | 11–1 | Chen et al. (2014) |
| I | OnTPS1 | Oryza nivara | BEP | (E)-β-Caryophyllene | 11–1 | Chen et al. (2014) |
| I | OoTPS1 | Oryza officinalis | BEP | (E)-β-Caryophyllene/Germacrene A | 11–1/10–1 | Chen et al. (2014) |
| I | OrTPS1 | Oryza rufipogon | BEP | Germacrene A/Germacrene D | 10–1 | Chen et al. (2014) |
| I | OsTPS1 | Oryza sativa | BEP | (E)-β-Caryophyllene/Germacrene A | 11–1 | Yuan et al. (2008) |
| I | PvTPS1 | Panicum virgatum | PACMAD | Cycloisosativene | 10–1 | Muchlinski et al. (2019) |
| I | PvTPS11 | Panicum virgatum | PACMAD | (E)-β-Caryophyllene | 11–1 | Muchlinski et al. (2019) |
| I | PvTPS14 | Panicum virgatum | PACMAD | (E)-β-Caryophyllene | 11–1 | Muchlinski et al. (2019) |
| I | PvTPS19 | Panicum virgatum | PACMAD | (E)-β-Caryophyllene | 11–1 | Muchlinski et al. (2019) |
| I | SbTPS4 | Sorghum bicolor | PACMAD | (E)-β-Caryophyllene | 11–1 | Zhuang et al. (2012) |
| I | SbTPS5 | Sorghum bicolor | PACMAD | (Z)-γ-Bisabolene/(E)-α-Bergamotene | 6–1 | Zhuang et al. (2012) |
| I | ZmTPS23-Del | Zea mays | PACMAD | (E)-β-Caryophyllene | 11–1 | Köllner et al. (2008a) |
| I | ZmTPS20-Del | Zea mays | PACMAD | Germacrene A | 10–1 | This study |
| I | ZmTPS22-Del | Zea mays | PACMAD | β-Copaene | 10–1 | This study |
| II | Bradi3g15956 | Brachypodium distachyon | BEP | β-Macrocarpene/γ-Macrocarpene | 6–1 | This study |
| II | Oba080 | Oryza barthii | BEP | (E)-β-Farnesene/Zingiberene | no/6–1 | Chen et al. (2020) |
| II | Og100 | Oryza glaberrima | BEP | Zingiberene | 6–1 | Chen et al. (2020) |
| II | On080 | Oryza nivara | BEP | (E)-β-Farnesene | no | Chen et al. (2020) |
| II | Or100 | Oryza rufipogon | BEP | β-Bisabolene/ β-Sesquiphellandrene | 6–1 | Chen et al. (2020) |
| II | Os100 | Oryza sativa | BEP | β-Bisabolene/ β-Sesquiphellandrene | 6–1 | Yuan et al. (2008) |
| II | Os120 | Oryza sativa | BEP | β-Bisabolene | 6–1 | Chen et al. (2020) |
| II | PvTPS16 | Panicum virgatum | PACMAD | (E)-β-Farnesene | no | Muchlinski et al. (2019) |
| II | PvTPS17 | Panicum virgatum | PACMAD | β-Bisabolene | 6–1 | Muchlinski et al. (2019) |
| II | PvTPS20 | Panicum virgatum | PACMAD | β-Bisabolene | 6–1 | Muchlinski et al. (2019) |
| II | PvTPS69 | Panicum virgatum | PACMAD | (E)-β-Farnesene | no | Muchlinski et al. (2019) |
| II | PvTPS94 | Panicum virgatum | PACMAD | α-Santalene | 6–1 | Muchlinski et al. (2019) |
| II | PvTPS109 | Panicum virgatum | PACMAD | (E)-β-Farnesene | no | Muchlinski et al. (2019) |
| II | SbTPS1 | Sorghum bicolor | PACMAD | Zingiberene | 6–1 | Zhuang et al. (2012) |
| II | SbTPS2 | Sorghum bicolor | PACMAD | β-Sesquiphellandrene | 6–1 | Zhuang et al. (2012) |
| II | SbTPS3 | Sorghum bicolor | PACMAD | (E)-β-Farnesene/(E)-α-Bergamotene | no/6–1 | Zhuang et al. (2012) |
| II | ZmTPS4-B73 | Zea mays | PACMAD | 7-epi-Sesquithujene/β-Bisabolene | 6–1 | Köllner et al. (2004) |
| II | ZmTPS5-Del1 | Zea mays | PACMAD | Sesquithujene/β-Bisabolene | 6–1 | Köllner et al. (2004) |
| II | ZmTPS6-B73 | Zea mays | PACMAD | β-Macrocarpene | 6–1 | Köllner et al. (2008b) |
| II | ZmTPS11-B73 | Zea mays | PACMAD | β-Macrocarpene | 6–1 | Köllner et al. (2008b) |
| II | ZmTPS10-B73 | Zea mays | PACMAD | (E)-β-Farnesene/(E)-α-Bergamotene | no/6–1 | Schnee et al. (2006) |
| II | ZmTPS10-dip | Zea diploperennis | PACMAD | (E)-β-Farnesene/(E)-α-Bergamotene | no/6–1 | Köllner et al. (2009) |
| II | ZmTPS10-hue | Zea huehuetenangensis | PACMAD | (E)-β-Farnesene/(E)-α-Bergamotene | no/6–1 | Köllner et al. (2009) |
| II | ZmTPS10-mex | Zea mexicana | PACMAD | (E)-β-Farnesene/(E)-α-Bergamotene | no/6–1 | Köllner et al. (2009) |
| II | ZmTPS10-per | Zea perennis | PACMAD | (E)-β-Farnesene/(E)-α-Bergamotene | no/6–1 | Köllner et al. (2009) |
| III | PvTPS03 | Panicum virgatum | PACMAD | (E)-γ-Bisabolene | 6–1 | Muchlinski et al. (2019) |
| III | PvTPS04 | Panicum virgatum | PACMAD | α-Terpinolene | 6–1MT | Muchlinski et al. (2019) |
| III | PvTPS08 | Panicum virgatum | PACMAD | 1,8-Cineole | 6–1MT | Muchlinski et al. (2019) |
| III | PvTPS36 | Panicum virgatum | PACMAD | Limonene | 6–1MT | Muchlinski et al. (2019) |
| III | PvTPS83 | Panicum virgatum | PACMAD | (E)-γ-Bisabolene | 6–1 | Muchlinski et al. (2019) |
| III | ZmTPS15-B73 | Zea mays | PACMAD | 1,8-Cineole | 6–1MT | This study |
| III | ZmTPS19-B73 | Zea mays | PACMAD | α-Terpineol/γ-Terpinene | 6–1MT | Lin et al. (2008) |
| III | ZmTPS26-B73 | Zea mays | PACMAD | α-Terpineol/γ-Terpinene | 6–1MT | Lin et al. (2008) |
| IV | PvTPS05 | Panicum virgatum | PACMAD | α-Selinene | 10–1 | Muchlinski et al. (2019) |
| IV | PvTPS56 | Panicum virgatum | PACMAD | α-Terpineol | 6–1MT | Muchlinski et al. (2019) |
| IV | ZmEDS | Zea mays | PACMAD | Eudesmane-2,11-diol | 10–1 | Liang et al. (2018) |
| IV | ZmTPS21 | Zea mays | PACMAD | β-Selinene | 10–1 | Ding et al. (2017) |
| V | OsTPS3 | Oryza sativa | BEP | (E,E)-Farnesol | no | Cheng et al. (2007) |
| V | OsTPS19 | Oryza sativa | BEP | Limonene | 6–1MT | Chen et al. (2018) |
| V | OsTPS20 | Oryza sativa | BEP | Limonene | 6–1MT | Chen et al. (2018) |
| V | PvTPS06 | Panicum virgatum | PACMAD | δ-Cadinene | 10–1 | Muchlinski et al. (2019) |
| V | PvTPS53 | Panicum virgatum | PACMAD | Geraniol | noMT | Muchlinski et al. (2019) |
| V | PvTPS54 | Panicum virgatum | PACMAD | Not active | – | Muchlinski et al. (2019) |
| V | PvTPS55 | Panicum virgatum | PACMAD | Germacrene D | 10–1 | Muchlinski et al. (2019) |
| V | PvTPS101 | Panicum virgatum | PACMAD | Geraniol | noMT | Muchlinski et al. (2019) |
| V | TaPS | Triticum aestivum | PACMAD | β-Patchoulene | 11–1 | Pu et al. (2019) |
| V | ZmTPS7 | Zea mays | PACMAD | τ-Cadinol | 10–1 | Ren et al. (2016) |
| V | ZmTPS8 | Zea mays | PACMAD | Germacrene D | 10–1 | Fontana et al. (2011) |
MT monoterpene synthase activity
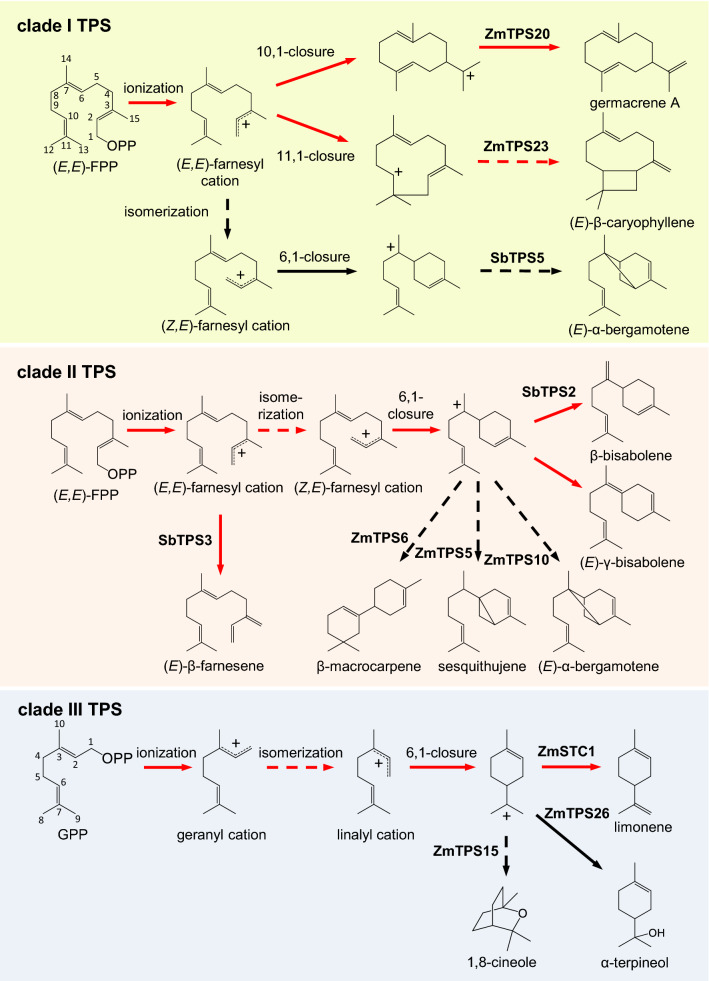
Fig. 2.
Reactions catalyzed by representative TPS enzymes from clades I–III and their respective ancestors including major carbocationic intermediates. Reactions catalyzed by the putative ancestors are marked with red arrows. Dashed arrows indicate multiple reaction steps
Characterization of TPS-a enzymes from maize and Brachypodium distachyon
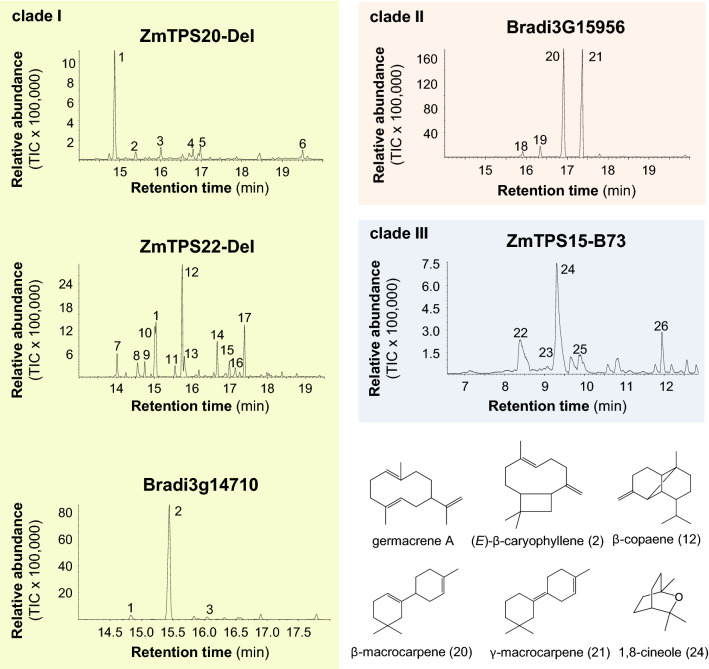
To provide more information about the differences among these Poaceae TPS-a clades, we characterized three previously unstudied TPS-a genes from maize and two TPS-a genes from B. distachyon, a species from which no TPS has been characterized so far. The selected terpene synthases belong to clade I (maize ZmTPS20-Del and ZmTPS22-Del, B. distachyon Bradi3g14710), clade II (B. distachyon Bradi3g15956), and clade III (maize ZmTPS15-B73). Genes were cloned and heterologously expressed in E. coli, and partially purified proteins were incubated with the potential substrates GPP and (E,E)-FPP in the presence of 10 mM Mg2+ as cofactor. Enzyme products were analyzed using GC–MS. The three clade I enzymes ZmTPS20-Del, ZmTPS22-Del, and Bradi3g14710 showed exclusively sesquiterpene synthase activity and produced sesquiterpenes derived by either an initial C10-C1 closure or an initial C11-C1 closure, corresponding to the properties of clade I enzymes in the literature. ZmTPS20-Del produced mainly germacrene A (C10-C1 closure), which appeared as the thermal rearrangement product β-elemene in the traces of the GC chromatograms (Fig. 3; Supplemental Figure S2). ZmTPS22-Del produced a complex mixture of sesquiterpenes with the bicyclic β-copaene (C10-C1 closure) as the major component (Fig. 3). In contrast to ZmTPS20-Del and ZmTPS22-Del, Bradi3g14710 had higher product specificity and formed (E)-β-caryophyllene (C11-C1 closure) with trace amounts of α-humulene and germacrene A (Fig. 3). Monoterpene synthase activity with GPP was not observed for any of the three enzymes. The clade II enzyme Bradi3g15956 converted (E,E)-FPP into β-macrocarpene and γ-macrocarpene, two bicyclic sesquiterpenes both derived by an initial C6-C1-closure (Fig. 3). GPP, however, was not accepted by Bradi3g15956 as substrate. Maize ZmTPS15-B73 showed no activity when provided with (E,E)-FPP. However, it converted GPP into a mixture of monoterpenes dominated by the bicyclic alcohol 1,8-cineole (C6-C1 closure) (Fig. 3). Subcellular localization prediction using three different algorithms revealed the presence of an N-terminal transit peptide for plastid localization (Supplemental Figure S3), indicating that ZmTPS15-B73 likely acts as a monoterpene synthase in maize plastids.
Fig. 3.
Characterization of selected terpene synthases from the Poaceae TPS-a clades I–III. Genes were heterologously expressed in Escherichia coli and partially purified proteins were incubated with (E,E)-FPP (TPS20-Del, TPS22-Del, Bradi3g14710, Bradi3g15956) or GPP (TPS15-B73). TPS reaction products were collected from the headspace of the enzyme assays using a solid phase microextraction (SPME) fiber and analyzed by gas chromatography-mass spectrometry. Total ion current chromatograms are shown. 1, β-elemene; 2, (E)-β-caryophyllene; 3, α-humulene; 4, unidentified sesquiterpene hydrocarbon1; 5, unidentified sesquiterpene hydrocarbon2; 6, unidentified sesquiterpene alcohol1; 7, δ-elemene; 8, unidentified sesquiterpene hydrocarbon3; 9, α-copaene; 10, β-cubebene; 11, β-ylangene; 12, β-copaene; 13, γ-elemene; 14, germacrene D; 15, α-muurolene; 16, cubebol; 17, δ-cadinene; 18, (E)-β-farnesene; 19, unidentified sesquiterpene hydrocarbon4; 20, β-macrocarpene; 21, γ-macrocarpene; 22, myrcene; 23, limonene; 24, 1,8-cineole; 25, (E)-β-ocimene; 26, linalool. Structures of major TPS products are shown
The reconstruction and characterization of TPS-a ancestor enzymes
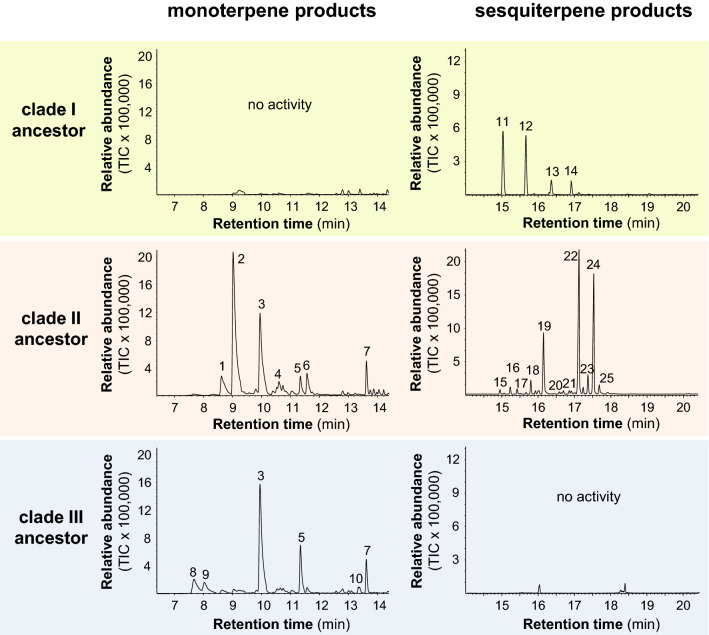
Most of the enzymes in clades I–III share clade-specific catalytic features, catalyzing either a common initial cyclization or having the same substrate specificity (Table 1; Fig. 2). To test whether these features were already pronounced in the ancestors of the different clades, we reconstructed the respective ancestor sequences, expressed the synthesized genes in E. coli, and tested the recombinant enzymes obtained with GPP and (E,E)-FPP. The putative clade I ancestor showed only sesquiterpene synthase activity and produced mainly germacrene A (C10-C1) and (E)-β-caryophyllene (C11-C1) (Figs. 2 and 4, Supplemental Figure S2). The putative clade II ancestor accepted both GPP and (E,E)-FPP as substrate and produced mixtures of monoterpenes and sesquiterpenes. While the monoterpene blend was dominated by the acyclic myrcene, the major sesquiterpenes were identified as β-bisabolene and (E)-γ-bisabolene, which are both formed via an initial C6-C1 closure (Figs. 2 and 4). In contrast to the putative ancestors of clade I and clade II, the clade III ancestor, which was expressed without the N-terminal signal peptide, possessed exclusively monoterpene synthase activity and produced the cyclic limonene (C6-C1) as its major product (Figs. 2 and 4).
Fig. 4.
Proteins encoded by putative ancestors to Poaceae TPS-a clades have terpene synthase activity in vitro. Reconstructed TPS ancestor sequences of clades I, II, and III of the Poaceae TPS-a genes were synthesized, cloned, and heterologously expressed in Escherichia coli. Partially purified proteins were incubated with the potential substrates GPP and (E,E)-FPP. TPS reaction products were collected from the headspace of the enzyme assays using a solid phase microextraction (SPME) fiber and analyzed by gas chromatography-mass spectrometry. Total ion current chromatograms are shown. 1, β-phellandrene; 2, myrcene; 3, limonene; 4, (E)-β-ocimene; 5, terpinolene; 6, linalool; 7, α-terpineol; 8, α-pinene; 9, camphene; 10, terpinen-4-ol; 11, β-elemene; 12; (E)-β-caryophyllene; 13, α-humulene; 14, germacrene D; 15, 7-epi-sesquithujene; 16, sesquithujene; 17, (Z)-α-bergamotene; 18, (E)-α-bergamotene; 19, (E)-β-farnesene; 20, sesquisabinene; 21, zingiberene; 22, β-bisabolene; 23, β-sesquiphellandrene; 24, (E)-γ-bisabolene; 25, (Z)-α-bisabolene
Discussion
Many important crops such as maize, wheat, rice, and sorghum belong to the Poaceae, a plant family known to be rich in terpenes. Terpenes play important roles in plant defense and their biosynthesis has been intensively investigated during the last three decades (Degenhardt et al. 2009). In this study, we aimed to understand the functional evolution of the TPS-a subfamily in the grasses by reconstructing and characterizing ancestral TPS enzymes and comparing their catalytic activities to those of modern terpene synthases.
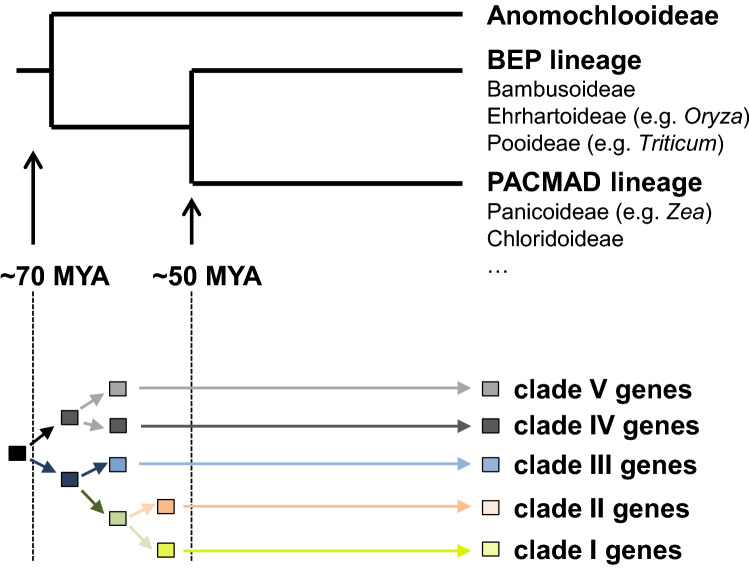
The Poaceae family consists of three major lineages, the PACMAD (Panicoideae, Arundinoideae, Chloridoideae, Micrairoideae, Aristidoideae, Danthonioideae) lineage, the BEP (Bambusoideae, Ehrhartoideae, Pooideae) lineage, and the basal Anomochlooideae (Strömberg 2011, Fig. 5). Fossil records indicate that the common ancestor of the grasses existed more than 70 Ma in the Late Cretaceous, and that the PACMAD and BEP lineages evolved between 70 and 50 Ma (Strömberg 2011). A phylogenetic analysis of TPS-a genes from different monocotyledonous species revealed five monophyletic clades I–V that exclusively contain genes from the Poaceae (Fig. 1). Since all of the five TPS-a clades contain both PACMAD and BEP genes (Table 1; Supplemental Figure S4), it is likely that the direct ancestors of the five clades evolved by at least four consecutive gene duplication events from a common TPS-a progenitor prior to the separation of the two lineages (Fig. 5).
Fig. 5.
Model for the evolution of the five different TPS-a clades in the Poaceae. The phylogenetic relationships of the clades from Fig. 1 are shown together with their alignment to a simplified grass phylogeny. The ancestors of the TPS-a clades I–V all evolved through gene duplications before the split of the PACMAD and BEP lineages
A survey of the TPS literature and characterization of representative TPS in this study revealed that most of the enzymes in clades I, II, IV, and V are sesquiterpene synthases, while clade III possesses predominantly monoterpene synthases (Table 1; Fig. 3). It is thus tempting to speculate that the TPS-a progenitor in the early grasses was a sesquiterpene synthase. In this scenario, the ancestor of clade III had to evolve monoterpene synthase activity, presumably by the accumulation of mutations leading to a decreased active site pocket that no longer allowed binding of the FPP substrate, and gained an N-terminal signal peptide for transport to the plastid, where GPP is produced. Indeed, the reconstruction of this ancestor based on the modern clade III sequences and its functional characterization showed that the enzyme was able to convert GPP into monoterpenes but not (E,E)-FPP into sesquiterpenes (Fig. 4). Interestingly, a few enzymes in clades IV and V have also been described as monoterpene synthases (Table 1), indicating multiple evolution of this enzymatic function within the TPS-a subfamily. OsTPS19 and OsTPS20 from rice, for example, belong to clade V and were found to be localized in plastids. The manipulation of OsTPS19 gene expression in transgenic rice clearly confirmed its function as a monoterpene synthase in planta (Chen et al. 2018). However, the subcellular localization of other putative monoterpene synthases in clades IV and V is rather unclear. Some of them do not appear to contain signal peptides (Muchlinski et al. 2019) and, in contrast to clade III monoterpene synthases that only accept GPP as substrate (Lin et al. 2008; Muchlinski et al. 2019; Fig. 3), they often display both monoterpene and sesquiterpene synthase activity in vitro. Thus, a reliable prediction of their function in vivo is still not possible.
The Poaceae Tps-a clades differ among each other in the type of reactions that they catalyze. Nearly all monoterpene and sesquiterpene synthases are class I TPS enzymes, which catalyze a metal ion-dependent ionization of the substrate by abstracting the pyrophosphate moiety (Degenhardt et al. 2009; Fig. 2). The first reaction intermediate formed from GPP by monoterpene synthases is the geranyl cation. It can be deprotonated or captured by water addition to form acyclic monoterpenes, but due to conformational constraints can only be cyclized after isomerization to the linalyl cation, C1 of which is located near to the C6-C7 double bond (Fig. 2). The ionization of (E,E)-FPP by sesquiterpene synthases results in the formation of the (E,E)-farnesyl cation, which can undergo direct C10-C1 or C11-C1 closures to yield 10-membered or 11-membered rings, respectively. Alternatively, the isomerization of the C2–C3 double bond of the (E,E)-farnesyl cation leads to the tertiary (Z,E)-farnesyl cation, in direct analogy with the isomerization of GPP to the linalyl cation. The (Z,E)-farnesyl cation can then undergo cyclization involving either the central or distal double bond forming 1,6-, 1,7-, 1,10-, or 1,11- products (Degenhardt et al. 2009). Although the cyclic mono- and sesquiterpene cations formed in this way can be modified by further ring closures, ring contractions, and other rearrangements, the basic skeleton type is determined by the initial cyclization and isomerization reactions (Degenhardt et al. 2009). Interestingly, the different TPS-a enzymes in clades I, II, and III catalyze distinct and clade-specific initial reaction steps. With one exception, all clade I enzymes characterized to date produce sesquiterpenes derived from an initial C10-C1 or C11-C1 closure that does not require isomerization of the farnesyl cation (Fig. 2; Table 1). In contrast, clade II and clade III enzymes catalyze the isomerization of the farnesyl cation and the geranyl cation, respectively, prior to a C6-C1 ring closure. The reconstructed ancestors of clades I, II, and III were shown to catalyze the same basic initial reactions as the present day members of the clade (Figs. 2 and 4), indicating that the key enzymatic features of the modern TPS-a enzymes evolved early in the evolution of the grasses. However, in contrast to the modern enzymes, the reconstructed TPS ancestors formed simpler structures that are mainly derived by a deprotonation of the first cyclic cation (Fig. 2). Present day enzymes typically catalyze reaction cascades with additional steps, such as the isomerization of carbon–carbon double bond in the initial cation to allow alternate ring closures or additional cyclizations. The structural diversity of terpenes formed in today’s grasses likely evolved later in the grass evolution within single subfamilies, genera, or even species. Successive gene duplications of the respective clade ancestors and the subsequent accumulation of mutations led to the multitude of modern TPS-a enzymes, many of which catalyze more complex reactions than the ancestor. Although it is universally accepted that evolution of natural product biosynthesis has led to the formation of more and more complex structures, this process has rarely been documented at the level of a specific enzyme and plant group.
Materials and methods
Plant material
Seeds of the maize (Zea mays L.) inbred line B73 were provided by KWS seeds (Einbeck, Germany), and seeds of the maize hybrid variety Delprim were obtained from Delley Samen und Pflanzen (Delley, Switzerland). Plants were grown in commercially available potting soil in a climate-controlled chamber with a 16 h photoperiod, 1 mmol (m2)−1 s−1 of photosynthetically-active radiation, a temperature cycle of 22 ºC/18 ºC (day/night), and 65% relative humidity. Ten day old seedlings were harvested and immediately frozen in liquid nitrogen. Seeds of the Brachypodium distachyon inbred diploid accession Bd21.3 were obtained from the National Plant Germplasm System, US Department of Agriculture. Seeds were sowed into pots and placed at 4 °C in the dark for 2 days, and then the pots were transferred into a Percival growth chamber. Plants were grown under 16 h light/8 h darkness at 26 °C. Four-week old plants were harvested and immediately frozen in liquid nitrogen.
Gene identification, sequence analysis, and dendrogram analysis
Using poplar TPS9 (Irmisch et al. 2014) as a query, we conducted a TBLASTN analysis against 6 Poaceae gene sets (Zea mays, Sorghum bicolor, Setaria italica, Panicum virgatum, Oryza sativa, Brachypodium distachyon) and 27 angiosperm gene sets (Aquilegia coerulea, Arabidopsis lyrata, A. thaliana, Brassica rapa, Capsella rubella, Carica papaya, Citrus clementina, Citrus x sinensis, Cucumis sativus, Eucalyptus grandis, Eutrema salsugineum, Fragaria vesca, Glycine max, Gossypium raimondii, Linum usitatissimum, Malus x domestica, Manihot esculenta, Medicago truncatula, Mimulus guttatus, Phaseolus vulgaris, Populus trichocarpa, Prunus persica, Ricinus communis, Solanum lycopersicum, S. tuberosum, Theobroma cacao, Vitis vinifera) available in the Phytozome 9.1 database (phytozome.jgi.doe.gov). In addition, the NCBI database (www.ncbi.nlm.nih.gov) was similarly screened for TPS genes from monocotyledonous species outside the Poaceae (species of Arecales, Asparagales, Liliales, and Zingiberales). Sequences longer than 1499 bp were considered as full-length genes and subjected to phylogenetic analysis. For the estimation of a phylogenetic tree, we used the MUSCLE (codon) algorithm (gap open, − 2.9; gap extend, 0; hydrophobicity multiplier, 1.2; clustering method, UPGMB) implemented in MEGA6 (Tamura et al. 2011) to compute a nucleotide codon alignment with the 932 obtained full-length TPS sequences. Based on the MUSCLE alignment, the tree was reconstructed with MEGA6 using a maximum likelihood algorithm (model/method, General Time Reversible model; substitutions type, nucleotide; rates among sites, gamma distributed with invariant sites (G + I); gamma parameters, 5; gaps/missing data treatment, partial deletion; site coverage cutoff, 50%). Guided by the resulting TPS tree, 163 out of the 932 sequences could be identified as monocot-specific TPS-a2 genes and were subjected to deeper phylogenetic analysis using the same methods and parameters as described above, except with a site coverage cutoff of 80%. A bootstrap resampling analysis with 1000 replicates was performed to evaluate the topology of the tree. FASTA files are available as Supplemental data set 1 (angiosperm TPS) and Supplemental data set 2 (Poaceae TPS).
Ancestor reconstruction
Codon alignments of characterized TPS-a genes from the clades I, II, and III were created using the PRANK algorithm provided by the Guidance web-based interface (https://guidance.tau.ac.il/) (MSA scores are given in Supplemental Table S1; FASTA files are available as Supplemental data sets 3 (clade I), 4 (clade II), and 5 (clade III)). The PRANK alignments were loaded into MEGA6 and model tests were performed to search for the optimal substitution models and parameters. Maximum likelihood (ML) trees (Supplemental Figures S5-S7) were generated with MEGA6 using the models and parameters given in Supplemental Table S1. All sites of the sequences were considered for the generation of the trees. The most likely protein ancestor sequences were extracted from the ML trees using the program ExtAncSeqMEGA (Hall 2011) (for accuracy scores see Supplemental Table S1).
Signal peptide prediction
Signal peptides were predicted with the programs ChloroP (https://www.cbs.dtu.dk/services/ChloroP/), TargetP (https://www.cbs.dtu.dk/services/TargetP/), and Predotar v1.04 (https://urgi.versailles.inra.fr/predotar/).
Cloning and gene synthesis
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Single-stranded cDNA was synthesized from total RNA using SuperScript™ III Reverse Transcriptase (Invitrogen, Carlsbad, CA) and oligo(dT) primers. The complete open reading frames (ORF) of ZmTPS20-Del, ZmTPS22-Del, and ZmTPS15-B73 from maize were amplified from cDNA and cloned into the sequencing vector pCR4-TOPO (Invitrogen). After complete sequencing, ZmTPS20-Del and ZmTPS22-Del were subcloned as BsaI fragments into the expression vector pASK-IBA7 (IBA-GmbH, Göttingen, Germany). Before subcloning, an internal BsaI restriction site in ZmTPS22-Del was mutated by site-directed mutagenesis as described in Köllner et al. 2006. The ORF of ZmTPS15-B73 lacking the first 68 codons, which encode the N-terminal signal peptide, was subcloned into the expression vector pET100/D-TOPO (Invitrogen). The complete ORFs of Bradi3g14710 and Bradi3g15956 from Brachypodium distachyon were amplified from cDNA and cloned into the expression vector pEXP5-CT/TOPO (Invitrogen). The reconstructed ancestor sequences of clades I–III were synthesized as codon-optimized genes and subsequently cloned into the expression vector pET100/D-TOPO. The N-terminal signal peptide (33 amino acids) of the clade III ancestor was truncated before heterologous expression. All primers used for amplification, subcloning, and mutagenesis are listed in Supplemental Table S2. Sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov) with the accession numbers MT294265 (ZmTPS20-Del), MT294266 (ZmTPS22-Del), MT294267 (ZmTPS15-B73), MT294268 (Bradi3g14710), MT294269 (Bradi3g15956), MT294270 (clade I ancestor), MT294271 (clade II ancestor), and MT294272 (clade III ancestor). The codon-optimized sequences of the TPS ancestors are given in Supplemental Figure S8.
Heterologous expression of TPS genes in Escherichia coli
For heterologous expression in E. coli, pASK-IBA7-constructs were introduced into the strain TOP10 (Invitrogen), while pET100/D-TOPO-constructs were expressed in the strain BL21 (DE3). Liquid cultures of the bacteria harboring the expression constructs were grown at 37 °C to an OD600 of 0.6. Expression of the recombinant proteins from pET100/D-TOPO constructs in BL21 (DE3) was induced by addition of isopropyl-b-thiogalactopyranoside to a final concentration of 1 mM, and the expression of pASK-IBA7 constructs in TOP10 cells was induced with 200 mg/l of anhydrotetracycline (IBA, Göttingen, Germany). Cultures were incubated for 20 h at 18 °C and cells were collected by centrifugation and disrupted by a 4 × 30 s treatment with a sonicator (Bandelin UW2070, Berlin, Germany) in chilled extraction buffer (50 mM Tris–HCl, pH 7.5, with 5 mM dithiothreitol and 10% (v/v) glycerol). The cell fragments were removed by centrifugation at 14,000 g and the supernatant was desalted into assay buffer (10 mM Tris–HCl, pH 7.5, 1 mM dithiothreitol, 10% (v/v) glycerol) by passage through a Econopac 10DG column (BioRad, Hercules, CA, USA).
TPS enzyme assays
To determine the catalytic activity of the different terpene synthases and the reconstructed TPS ancestors, enzyme assays containing 40 μl of the bacterial extract and 60 µl assay buffer with 10 μM (E,E)-FPP or GPP and 10 mM MgCl2, in a Teflon-sealed, screw-capped 1 ml GC glass vial were performed. A SPME (solid phase microextraction) fiber consisting of 100 µm polydimethylsiloxane (Supelco, Bellefonte, PA, USA) was placed into the headspace of the vial for 0.5 h incubation at 30 °C. For analysis of the adsorbed reaction products, the SPME fiber was directly inserted into the injector of the gas chromatograph.
Terpene synthase product analysis
Qualitative analysis of TPS enzyme products was conducted using an Agilent 6890 Series gas chromatograph coupled to an Agilent 5973 quadrupole mass selective detector (interface temp.: 270 °C; quadrupole temp.: 150 °C, source temp.: 230 °C, electron energy: 70 eV). The constituents of the product blends were separated with a DB-5MS column (Agilent, Santa Clara, CA, USA, 30 m × 0.25 mm × 0.25 µm) and helium (1 ml min−1) as carrier gas. The SPME fiber was injected without split at an initial oven temperature of 60 °C. The temperature was held for 2 min and then increased to 220 °C with a gradient of 7 °C min−1, followed by a further increase to 300 °C with 60 °C min−1 and a hold for 3 min. Compounds were identified by comparison of retention times and mass spectra to those of authentic standards obtained from Fluka (Seelze, Germany), Roth (Karlsruhe, Germany), Sigma (St, Louis, MO, USA), or Bedoukian (Danbury, CT, USA), or by reference spectra in the Wiley and National Institute of Standards and Technology libraries.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Open access funding provided by Projekt DEAL. This research was funded by the Max Planck Society.
Author contributions
KL, XC, AMN, TGK, and FC carried out the experimental work. FC, JG, and TGK participated in the design of the study and improved the manuscript. TGK conceived of the study and drafted the manuscript. All authors read and approved the final manuscript.
Data availability
All data generated or analyzed during this study are included in the main text or supplement of this article. Sequences of functionally characterized terpene synthases and terpene synthase ancestors are available on GenBank: MT294265-MT294272.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Katrin Luck, Email: kluck@ice.mpg.de.
Xinlu Chen, Email: xchen24@utk.edu.
Ayla M. Norris, Email: ayla.norris@usda.gov
Feng Chen, Email: fengc@utk.edu.
Jonathan Gershenzon, Email: gershenzon@ice.mpg.de.
Tobias G. Köllner, Email: koellner@ice.mpg.de
References
- Bathe U, Tissier A. Cytochrome P450 enzymes: a driving force of plant diterpene diversity. Phytochemistry. 2019;161:149–162. doi: 10.1016/j.phytochem.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Block AK, Vaughan MM, Schmelz EA, et al. Biosynthesis and function of terpenoid defense compounds in maize (Zea mays) Planta. 2019;249:21–30. doi: 10.1007/s00425-018-2999-2. [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Meyergauen G, Croteau R. Plant terpenoid synthases—molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, et al. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66:212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Li GL, Kollner TG, et al. Positive Darwinian selection is a driving force for the diversification of terpenoid biosynthesis in the genus Oryza. BMC Plant Biol. 2014;14:1–12. doi: 10.1186/s12870-014-0239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Chen H, Yuan JS, et al. The rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol J. 2018;16:1778–1787. doi: 10.1111/pbi.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kollner TG, Li GL, et al. Combinatorial evolution of a terpene synthase gene cluster explains terpene variations in Oryza. Plant Physiol. 2020;182:480–492. doi: 10.1104/pp.19.00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AX, Xiang CY, Li JX, et al. The rice (E)-beta-caryophyllene synthase (OsTPS3) accounts for the major inducible volatile sesquiterpenes. Phytochemistry. 2007;68:1632–1641. doi: 10.1016/j.phytochem.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Degenhardt J, Köllner TG, Gershenzon J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry. 2009;70:1621–1637. doi: 10.1016/j.phytochem.2009.07.030. [DOI] [PubMed] [Google Scholar]
- Ding YZ, Huffaker A, Köllner TG, et al. Selinene volatiles are essential precursors for maize defense promoting fungal pathogen resistance. Plant Physiol. 2017;175:1455–1468. doi: 10.1104/pp.17.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong LM, Jongedijk E, Bouwmeester H, et al. Monoterpene biosynthesis potential of plant subcellular compartments. New Phytol. 2016;209:679–690. doi: 10.1111/nph.13629. [DOI] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E, Gershenzon J. Biochemistry of plant volatiles. Plant Physiol. 2004;135:1893–1902. doi: 10.1104/pp.104.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana A, Held M, Fantaye CA, et al. Attractiveness of constitutive and herbivore-induced sesquiterpene blends of maize to the parasitic wasp Cotesia marginiventris (Cresson) J Chem Ecol. 2011;37:582–591. doi: 10.1007/s10886-011-9967-7. [DOI] [PubMed] [Google Scholar]
- Garms S, Köllner TG, Boland W. A multiproduct terpene synthase from Medicago truncatula generates cadalane sesquiterpenes via two different mechanisms. J Org Chem. 2010;75:5590–5600. doi: 10.1021/jo100917c. [DOI] [PubMed] [Google Scholar]
- Gutensohn M, Orlova I, Nguyen TTH, et al. Cytosolic monoterpene biosynthesis is supported by plastid-generated geranyl diphosphate substrate in transgenic tomato fruits. Plant J. 2013;75:351–363. doi: 10.1111/tpj.12212. [DOI] [PubMed] [Google Scholar]
- Hall BG. Phylogenetic trees made easy. Sunderland: Sinauer Associates; 2011. [Google Scholar]
- Hartwig S, Frister T, Alemdar S, et al. SUMO-fusion, purification, and characterization of a (+)-zizaene synthase from Chrysopogon zizanioides. Biochem Biophys Res Commun. 2015;458:883–889. doi: 10.1016/j.bbrc.2015.02.053. [DOI] [PubMed] [Google Scholar]
- Hong YJ, Irmisch S, Wang SC, et al. Theoretical and experimental analysis of the reaction mechanism of MrTPS2, a triquinane-forming sesquiterpene synthase from chamomile. Chem Eur J. 2013;19:13590–13600. doi: 10.1002/chem.201301018. [DOI] [PubMed] [Google Scholar]
- Irmisch S, Krause ST, Kunert G, et al. The organ-specific expression of terpene synthase genes contributes to the terpene hydrocarbon composition of chamomile essential oils. BMC Plant Biol. 2012;12:84. doi: 10.1186/1471-2229-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch S, Jiang YF, Chen F, et al. Terpene synthases and their contribution to herbivore-induced volatile emission in western balsam poplar (Populus trichocarpa) BMC Plant Biol. 2014;14:1–16. doi: 10.1186/s12870-014-0270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanithi PS, Zerbe P. Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front Plant Sci. 2019;10:1166. doi: 10.3389/fpls.2019.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, Schnee C, Gershenzon J, et al. The variability of sesquiterpenes emitted from two Zea mays cultivars is controlled by allelic variation of two terpene synthase genes encoding stereoselective multiple product enzymes. Plant Cell. 2004;16:1115–1131. doi: 10.1105/tpc.019877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, O’maille PE, Gatto N, et al. Two pockets in the active site of maize sesquiterpene synthase TPS4 carry out sequential parts of the reaction scheme resulting in multiple products. Arch Biochem Biophys. 2006;448:83–92. doi: 10.1016/j.abb.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Köllner TG, Schnee C, Li S, et al. Protonation of a neutral (S)-beta-bisabolene intermediate is involved in (S)-beta-macrocarpene formation by the maize sesquiterpene synthases TPS6 and TPS11. J Biol Chem. 2008;283:20779–20788. doi: 10.1074/jbc.M802682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, Held M, Lenk C, et al. A maize (E)-beta-caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most American maize varieties. Plant Cell. 2008;20:482–494. doi: 10.1105/tpc.107.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, Gershenzon J, Degenhardt J. Molecular and biochemical evolution of maize terpene synthase 10, an enzyme of indirect defense. Phytochemistry. 2009;70:1139–1145. doi: 10.1016/j.phytochem.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Liang J, Liu J, Brown R, et al. Direct production of dihydroxylated sesquiterpenoids by a maize terpene synthase. Plant J. 2018;94:847–856. doi: 10.1111/tpj.13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CF, Shen BZ, Xu ZN, et al. Characterization of the monoterpene synthase gene tps26, the ortholog of a gene induced by insect herbivory in maize. Plant Physiol. 2008;146:940–951. doi: 10.1104/pp.107.109553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchlinski A, Chen XL, Lovell JT, et al. Biosynthesis and emission of stress-induced volatile terpenes in roots and leaves of switchgrass (Panicum virgatum L.) Front Plant Sci. 2019;10:1144. doi: 10.3389/fpls.2019.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazouki L, Niinemets U. Multi-substrate terpene synthases: their occurrence and physiological significance. Front Plant Sci. 2016;7:1019. doi: 10.3389/fpls.2016.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu QY, Liang J, Shen QQ, et al. A wheat beta-patchoulene synthase confers resistance against herbivory in transgenic arabidopsis. Genes. 2019;10:441. doi: 10.3390/genes10060441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Mao HJ, Liang J, et al. Functional characterization of ZmTPS7 reveals a maize tau-cadinol synthase involved in stress response. Planta. 2016;244:1065–1074. doi: 10.1007/s00425-016-2570-y. [DOI] [PubMed] [Google Scholar]
- Sallaud C, Rontein D, Onillon S, et al. A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell. 2009;21:301–317. doi: 10.1105/tpc.107.057885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Schauvinhold I, Larson M, et al. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA. 2009;106:10865–10870. doi: 10.1073/pnas.0904113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee C, Köllner TG, Held M, et al. The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc Natl Acad Sci USA. 2006;103:1129–1134. doi: 10.1073/pnas.0508027103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele CL, Crock J, Bohlmann J, et al. Sesquiterpene synthases from grand fir (Abies grandis)—comparison of constitutive and wound-induced activities, and cDNA isolation, characterization and bacterial expression of delta-selinene synthase and gamma-humulene synthase. J Biol Chem. 1998;273:2078–2089. doi: 10.1074/jbc.273.4.2078. [DOI] [PubMed] [Google Scholar]
- Stromberg CAE (2011) Evolution of grasses and grassland ecosystems. In: Jeanloz R, Freeman KH (eds) Annual review of earth and planetary sciences, Vol 39. pp 517–544
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D. Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol. 2015;148:63–106. doi: 10.1007/10_2014_295. [DOI] [PubMed] [Google Scholar]
- Unsicker SB, Kunert G, Gershenzon J. Protective perfumes: the role of vegetative volatiles in plant defense against herbivores. Curr Opin Plant Biol. 2009;12:479–485. doi: 10.1016/j.pbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Yuan JS, Köllner TG, Wiggins G, et al. Molecular and genomic basis of volatile-mediated indirect defense against insects in rice. Plant J. 2008;55:491–503. doi: 10.1111/j.1365-313X.2008.03524.x. [DOI] [PubMed] [Google Scholar]
- Zhuang XF, Köllner TG, Zhao N, et al. Dynamic evolution of herbivore-induced sesquiterpene biosynthesis in sorghum and related grass crops. Plant J. 2012;69:70–80. doi: 10.1111/j.1365-313X.2011.04771.x. [DOI] [PubMed] [Google Scholar]
- Zi JC, Matsuba Y, Hong YJ, et al. Biosynthesis of lycosantalonol, a cis-Prenyl derived diterpenoid. J Am Chem Soc. 2014;136:16951–16953. doi: 10.1021/ja508477e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in the main text or supplement of this article. Sequences of functionally characterized terpene synthases and terpene synthase ancestors are available on GenBank: MT294265-MT294272.