Abstract
Background
Immunological life-threatening complications frequently occur in post-hematopoietic stem cell transplantation (HSCT), despite matching recipient and donor (R/D) pairs for classical human leukocyte antigens (HLA). Studies have shown that R/D non-HLA disparities within the major histocompatibility complex (MHC) are associated with adverse effects post-HSCT.
Methods
We investigated the impact of mismatches of single-nucleotide polymorphisms (SNPs) in C4A/C4B genes, for showing the highest diversity in the MHC gamma block, on 238 patients who underwent HLA 10/10 unrelated donor (URD) HSCT. The endpoints were acute graft-versus-host disease (aGVHD), chronic graft-versus-host disease (cGVHD) and mortality. One hundred and twenty-nine R/D pairs had 23 C4-SNPs typed by PCR-SSP (Gamma-Type™v.1.0), and 109 R/D pairs had these 23 SNPs identified by next-generation sequencing (NGS) using the Illumina platform.
Results
The percentage of patients who received HSC from HLA 10/10 donors with 1–7 mismatches was 42.9%. The R/D pairs were considered C4 mismatched when bearing at least one disparity. These mismatches were not found to be risk factors for aGVHD, cGVHD or mortality after unrelated HSCT when SNPs were analyzed together (matched or mm ≥ 1), independently or according to the percentage of incompatibilities (full match for 23 SNPs; 1–3 mm and >3 mm). An exception was the association between 1–3 mismatches at the composite of SNPs C13193/T14952/T19588 with the development of aGVHD (P = 0.012) and with grades III-IV of this disease (P = 0.004).
Conclusion
Our data are not consistent with the hypothesis that disparities in C4A/C4B SNPs increase the risks of post-HSCT adverse effects for the endpoints investigated in this study.
Keywords: SNPs, C4A/C4B genes, MHC gamma block, Unrelated hematopoietic stem cell transplantation, GVHD
Introduction
Hematopoietic stem cell transplantation (HSCT) from an unrelated donor (URD) is the treatment of choice for several hematological diseases. Matching recipient and donor (R/D) pairs for classical transplantation antigens (HLA-A, -B, -C, -DR, -DQ and -DP) has been a major concern but is insufficient to avoid post-HSCT immunological complications such as graft-versus-host disease, which occurs in 35–69% of patients.1, 2
The central region of the major histocompatibility complex (MHC) is populated with many genes related to immune and inflammatory responses, leaving much undetected genetic variability when pairing recipients and URDs.3 Therefore, HLA compatibility indicates neither non-HLA loci identity nor MHC haplotypes sharing by R/URDs. The development of methods for MHC haplotype assignment enabled the investigation of the impact of haplotype mismatching in HSCT. These studies revealed a higher risk of acute graft-versus-host-disease (aGVHD) in transplants with HLA-identical R/D pairs, but not haplotype-matched.4, 5, 6
However, it is not feasible to utilize these laborious methods in clinical routine. Many studies have searched for genetic markers within the MHC that can predict post-unrelated HSCT adverse outcomes. Differences between R/D pairs at a single-nucleotide polymorphism (SNP) in coding, intronic or 3′ untranslated regions, in microsatellites and in MHC haplotypes, have been associated with post-transplant complications.6, 7, 8, 9, 10
MHC genes are commonly inherited as a haplotype, which is a set of different alleles that are physically linked and inherited from each parent. Simple typing of HLA genes does not provide information on which HLA alleles are paternal or maternal. Analyses of the inheritance of HLA genes show the existence of recombination events within the MHC and suggest the existence of a structure divided into 4 genetic blocks.11 HLA and complement genes are housed within these blocks: the alpha block contains HLA-A, the beta block harbors the HLA-B and -C genes, the gamma block houses the complement system genes C4A and C4B and the delta block comprises the HLA-DRB1 and -DQB1 genes. Compatibility for gamma block genes is not evaluated in donor selection. Although identical HLA siblings have the same genetic variants in this region, in transplants with URD, the gamma block gene content may be different from that in the recipient. As already demonstrated by previous studies,12 this different genetic load may influence the outcomes of HSCT. Some previous studies focusing on the effect of C4A/C4B mismatches have suggested positive associations with aGVHD,13, 14, 15 while others have not corroborated these data.16, 17
For the above-mentioned reasons, this retrospective study was aimed to further investigation of the impact of C4A/C4B genetic variability on the development of aGVHD (as the primary endpoint), cGVHD and mortality in the unrelated HSCT setting.
Methods
Patients
This retrospective study included 238 patients (104 adults/134 pediatric patients) who received hematopoietic stem cells (HSCs) from unrelated HLA-A, -B, -C, -DRB1 and -DQB1-matched donors. Transplants were performed at four Brazilian centers between September 1996 and August 2015: Hospital de Clínicas/Universidade Federal do Paraná (N = 162), Hospital Amaral Carvalho (N = 48), Hospital Israelita Albert Einstein (N = 20) and Hospital Nossa Senhora das Graças (N = 8). Patients, donors and transplant characteristics are summarized in Table 1. This study was performed in accordance with the Declaration of Helsinki and was approved by the Medical Ethical Committees.
Table 1.
Patient, donor, and transplant characteristics.
| Characteristics | N | % |
|---|---|---|
| Malignant disease | 124 | 52.1 |
| Acute lymphoblastic leukemia | 45 | 18.91 |
| Acute myeloid leukemia | 34 | 14.29 |
| Chronic myelogenous leukemia | 17 | 7.14 |
| Myelodysplastic syndrome | 12 | 5.04 |
| Others malignant diseases | 16 | 6.72 |
| Non-malignant disease | 114 | 47.9 |
| Aplastic anemia | 39 | 16.39 |
| Fanconi anemia | 40 | 16.81 |
| Others non-malignant diseases | 35 | 14.71 |
| Hematopoietic stem cell source | ||
| Bone marrow | 199 | 83.6 |
| Peripheral blood | 39 | 16.4 |
| Conditioning | ||
| Myeloablative | 145 | 61 |
| Reduced intensity | 81 | 34 |
| Non-myeloablative | 12 | 5 |
| Anti-thymocyte globulin | 198 | 83.2 |
| GVHD prophylaxis | ||
| CsA + MTX | 214 | 90 |
| Others | 28 | 12 |
| Gender | ||
| Patient Male/Female | 141/97 | 59/41 |
| Donor Male/Female | 147/91 | 62/38 |
| Age | ||
| Recipient (M) | 4 m–74 y (M = 15) | – |
| Donor (M) | 18–54 y (M = 30) | – |
M: median; m: month; y: year; GVHD: graft-versus-host disease; CsA: cyclosporin A; MTX: methotrexate.
HLA-DPB1 typing
All R/D pairs were DPB1-typed by SSOP (LABType®SSO; OneLambda, USA) and sequence based typed (SeCore®SBT, OneLambda, USA). The DPB1 T-Cell Epitope Algorithm v.2.0 was utilized to determine the permissiveness of mismatches.
C4A and C4B single-nucleotide polymorphism (SNP) typing
Twenty-three C4A/C4B SNPs were typed by PCR-SSP in 129 R/D pairs using Gamma-Type™ v.1.0 (Conexio Genomics, Australia), following the manufacturer’s instructions.
The remaining 109 R/D pairs had the same 23 Gamma-Type™ SNPs, identified by NGS using reagents by Conexio Genomics in the Illumina platform. The DNA fragment purification steps in the library preparation were performed using Beckman Ampure XP (Beckman Coulter, USA). The amplicon concentrations were measured using Qubit dsHS reagents, according to the manufacturer’s recommendations (Thermo Fisher Scientific, USA). The Nextera-XT DNA Sample Preparation (Illumina, USA) was utilized for double-stranded DNA libraries, and sequencing was performed in the MiSeq-platform using Kits-v3 (Illumina, USA). NGS data were analyzed using Assign™MPS software (Conexio Genomics, Australia) and genomic reference NG_011638.1 to determine variants in the C4A/C4B genes. R/D pairs with a difference ≥1 among the 23 SNPs were considered mismatched. The characteristics of SNPs are presented in Table 2.
Table 2.
Characteristics of the C4A/C4B SNPs.
| SNP identification (SSP Kit) | Exon/intron | Base change | Amino acid substitution | SNP Positiona | Identification in DBSNPb |
|---|---|---|---|---|---|
| T9763 | E12 | C > T | Arginine > Tryptophan | 6:g.31959596 | – |
| C9796 | E12 | A > C | Threonine > Proline | 6:g.31959629 | – |
| T9881 | I12 | C > T | Intronic | 6:g.31959714 | – |
| T10289 | I13 | G > T | Intronic | 6:g.31960122 | – |
| T10309 | I13 | C > T | Intronic | 6:g.31960142 | – |
| C10676 | I14 | T > C | Intronic | 6:g.31960509 | – |
| A11437 | E17 | C > A | Arginine > Serine | 6:g.31961270 | – |
| A11483 | E17 | G > A | Arginine > Glutamine | 6:g.31961316 | – |
| G12071 | I19 | A > G | Intronic | 6:g.31961904 | rs144749273 |
| A12152 | I19 | G > A | Intronic | 6:g.31961985 | rs12524856 |
| G12749 | I21 | A > G | Intronic | 6:g.31962582 | rs428963 |
| A12568 | E21 | G > A | Alanine > Threonine | 6:g.31962401 | rs429329 |
| A13189 | I23 | G > A | Intronic | 6:g.31963022 | – |
| C13193 | I23 | T > C | Intronic | 6:g.31963026 | rs149464899 |
| T14563 | I28 | C > DEL | Intronic | 6:g.31964395 | rs564578360 |
| T14757 | I28 | C > T | Intronic | 6:g.31964590 | – |
| A14831 | E29 | G > A | Proline > Proline | 6:g.31964664 | rs368403366 |
| T14952 | E29 | T > G | Alanine > Serine | 6:g.31964785 | rs201016130 |
| G15108 | E30 | T > G | Aspartic Acid > Glutamic Acid | 6:g.31964941 | – |
| C16954 | E33 | G > C | Alanine > Proline | 6:g.31966787 | – |
| T17316 | E34 | C > T | Histidine > Histidine | 6:g.31967149 | – |
| T19588 | I38 | C > T | Intronic | 6:g.31969421 | rs149763320 |
| A20170 | E40 | G > A | Leucine > Leucine | 6:g.31970003 | – |
Genomic Sequence: NC_000006.11 Chromosome 6 Reference GRCh37.p13 Primary Assembly.
SNPs DataBase (DBSNP – Short Genetic Variation www.ncbi.nlm.nih.gov/projects/SNP).
Statistics
The results of quantitative variables were described by means, standard deviations, medians, minimum and maximum values. Categorical variables were presented by frequencies and percentages. The associations of clinical variables and SNP-related variables with mortality were evaluated by adjusted Cox regression models, and hazard ratio values were estimated with 95% confidence intervals. In this model, aGVHD and cGVHD were considered time-dependent variables. The associations of clinical variables and SNP-related variables with aGVHD and cGVHD were analyzed by adjusted Fine and Gray models, and the estimated association measurement was the subdistribution hazard ratio.
Univariate analysis was performed for factors associated with the endpoints of interest, considering each of the clinical variables: patient age, gender, cytomegalovirus (CMV), diagnosis, HSC source, conditioning, GVHD prophylaxis, number of infused cells and each of the SNP-related variables (match: compatible for 23 SNPs; mismatch: incompatible for ≥1 SNP). Individual impact was investigated for SNPs with a mismatch status in ≥10 R/D pairs.
Multivariate models were adjusted for each variable related to SNPs, including as covariables the clinical variables that showed significance in univariate analysis. The significance level was P < 0.05, except for the individual effect of C4A/C4B SNPs, where Bonferroni correction was applied (P < 0.002 = statistical significance). Stata v.14.1 (Stata Corp LP, USA) and free statistical software EZR18 were utilized for data analysis.
Results
Effect of HLA-DPB1
DPB1 typing showed that 35/238 (14.7%) HLA-A-, B-, C-, DRB1- and DQB1-identical R/URD pairs were also matched for this gene (12/12), 99 (41.6%) having one or two permissive DPB1 mismatches, and 104 (43.7%) having one or two non-permissive mismatches. The impact of these mismatches on HSCT was analyzed in two groups of patients, the first comprising 134 (56.3%) DPB1-matched and DPB1-permissive mismatched pairs, the second comprising 104 pairs with at least one non-permissive mismatch (43.7%). None of the groups showed significant associations with mortality (HR = 1.25; 95% CI = 0.81–1.93; P = 0.321), aGVHD (SHR = 0.86; 95% CI = 0.56–1.32; P = 0.494) and cGVHD (SHR = 0.73; 95% CI = 0.43–1.25; P = 0.250).
C4A and C4B SNPs
One hundred and thirty-six (57.1%) R/D pairs were matched for 23 SNPs, while 102 (42.9%) had at least one difference and were mismatched (Table 3). The isolated impact of C4A/C4B SNPs on the clinical outcomes after HSCT was analyzed in 12 of 23 SNPs. They were selected for having at least 10 R/D mismatched pairs to allow statistical testing (Table 4).
Table 3.
C4A/C4B matching status in recipient/donor pairs.
| Number of MM SNPs | N (238) | % |
|---|---|---|
| 0 | 136 | 57.1 |
| 1 | 34 | 14.3 |
| 2 | 22 | 9.2 |
| 3 | 16 | 6.7 |
| 4 | 14 | 5.9 |
| 5 | 10 | 4.2 |
| 6 | 3 | 1.3 |
| 7 | 3 | 1.3 |
MM: mismatched; SNPs: single-nucleotide polymorphisms.
Table 4.
Matching status of the 23 C4A/C4B SNPs in recipient/donor pairs (N = 238).
| SNP Identificationa | Matched |
Mismatched |
||
|---|---|---|---|---|
| N | % | N | % | |
| A20170 | 199 | 83.6 | 39 | 16.4 |
| A14831 | 208 | 87.4 | 30 | 12.6 |
| A12152 | 211 | 88.7 | 27 | 11.3 |
| T9881 | 215 | 90.3 | 23 | 9.7 |
| T14952 | 217 | 91.2 | 21 | 8.8 |
| T14757 | 219 | 92.0 | 19 | 8.0 |
| G12071 | 220 | 92.4 | 18 | 7.6 |
| T14563 | 220 | 92.4 | 18 | 7.6 |
| T19588 | 220 | 92.4 | 18 | 7.6 |
| G12749 | 224 | 94.1 | 14 | 5.9 |
| G15108 | 227 | 95.4 | 11 | 4.6 |
| C13193 | 228 | 95.8 | 10 | 4.2 |
| T10289 | 233 | 97.9 | 5 | 2.1 |
| T17316 | 233 | 97.9 | 5 | 2.1 |
| A11437 | 234 | 98.3 | 4 | 1.7 |
| A12568 | 235 | 98.7 | 3 | 1.3 |
| T9763 | 236 | 99.2 | 2 | 0.8 |
| C10676 | 236 | 99.2 | 2 | 0.8 |
| T10309 | 237 | 99.6 | 1 | 0.4 |
| A11483 | 237 | 99.6 | 1 | 0.4 |
| C9796 | 238 | 100.0 | 0 | 0.0 |
| A13189 | 238 | 100.0 | 0 | 0.0 |
| C16954 | 238 | 100.0 | 0 | 0.0 |
SNP identification is based on the reference sequence NG_011638.1.
Acute graft-versus-host disease
Acute GVHD was associated with malignant diseases (SHR = 1.55; 95% CI = 1.01–2.38; P = 0.046) and with peripheral blood as the stem cell source (SHR = 1.85; 95% CI = 1.14–3.00; P = 0.013). Patients under myeloablative conditioning developed more GVHD than those under other regimens (SHR = 1.82; 95% CI = 1.13–2.93; P = 0.013). Patients who received cyclosporin A and methotrexate for GVHD prophylaxis had less aGVHD than those receiving other treatments (SHR = 2.19; 95% CI = 1.34–3.56; P = 0.002). The median age of recipients who developed aGVHD (18.9 yrs.) was higher than that of those who did not manifest this disease (13.5 yrs.) (HR = 1.02; 95% CI = 1.01–1.03; P = 0.004).
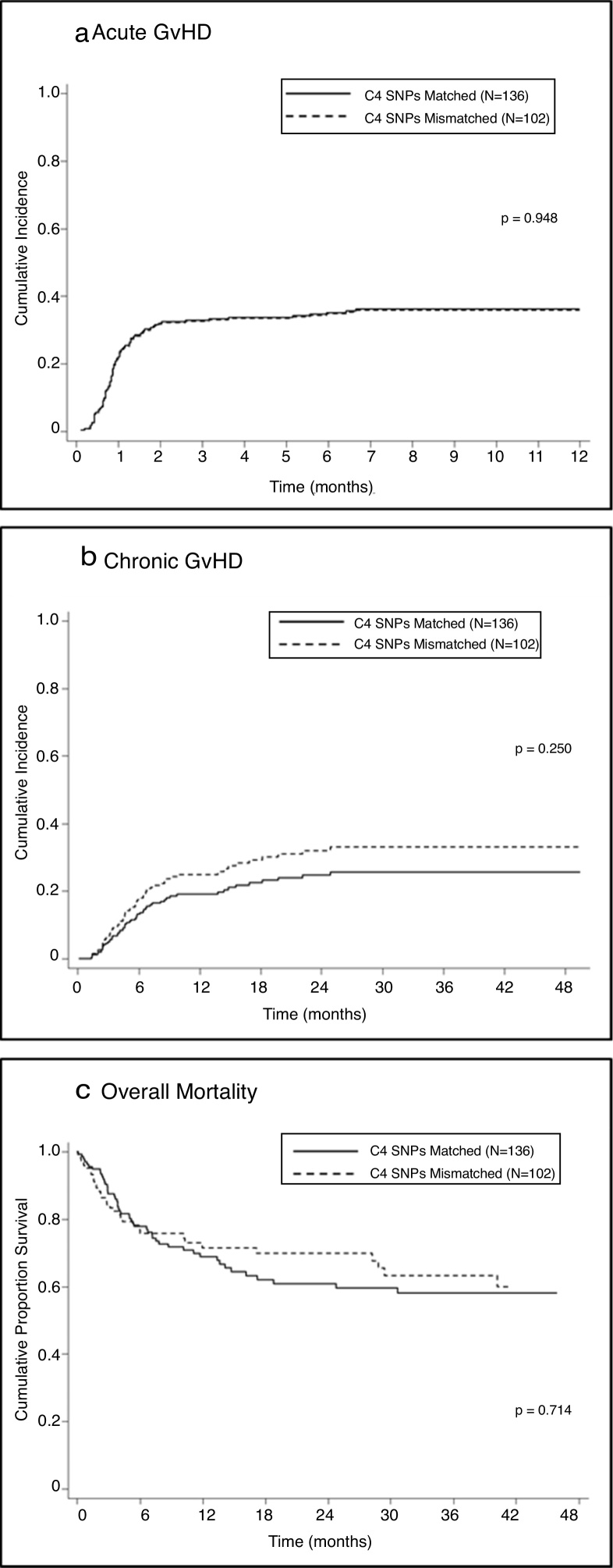
Univariate analysis showed no association between mismatches in C4-SNPs and the development of aGVHD in either the total sample (SHR = 0.99; 95% CI = 0.64–1.52; P = 0.948 — Figure 1A) or stratified according to disease status, malignant diseases (SHR = 0.98; 95% CI = 0.56–1.72; P = 0.96) or non-malignant diseases (SHR = 0.98, 95%CI = 0.50–1.93; P = 0.95).
Figure 1.
Associations between SNPs mismatched and clinical outcomes.
The independent effect of R/D mismatches for each of the 12 SNPs on the development of aGVHD was performed using a Fine and Gray model, but no association was found. SNPs C13193, T14952 and T19588 showing P < 0.2 and SHR > 1.0 were combined into a composite variable, and mismatches were associated with an increased risk for aGVHD in both univariate (Table 5) and multivariate analyses (Table 6); 45.9% of mismatched patients developed forms II–IV versus 26.4% of matched patients (p = 0.029). Similar results were observed when evaluating only aGVHD grades III-IV (32.4% mismatched vs. 11.4% matched; p = 0.004).
Table 5.
| Cohort | Composite variablea | Patients | aGVHDb (%) | SHRc | 95% CIc | P valuec |
|---|---|---|---|---|---|---|
| Total (N = 238) | Matched | 201 | 71 (35.3%) | |||
| Mismatched | 37 | 20 (54.0%) | 1.906 | 1.152–3.155 | 0.012 | |
| Malignant diseases (N = 124) | Matched | 102 | 43 (42.1%) | |||
| Mismatch | 22 | 13 (59.1%) | 1.74 | 0.9392–3.223 | 0.078 | |
| Non-malignant diseases (N = 114) | Match | 99 | 28 (28.3%) | |||
| Mismatch | 15 | 7 (46.7%) | 1.984 | 0.851–4.625 | 0.110 |
Table 6.
Multivariate analysis to evaluate the impact of C4A/C4B composite variable mismatches on acute GVHD.
| Cohort | SNP composite variablea match versus mismatch |
Recipient age at transplant |
Diagnosis (malignant vs. non-malignant diseases) |
|||
|---|---|---|---|---|---|---|
| SHR (95% CI)b | P value b | SHR (95% CI)b | P value b | SHR (95% CI)b | P value b | |
| Total | 1.723 (1.023–2.901) | 0.041 | 1.011 (0.999–1.022) | 0.059 | 1.401 (0.914–2.149) | 0.120 |
| Malignant diseases | 1.700 (0.999–2.891) | 0.050 | 1.014 (1.003–1.025) | 0.013 | – | – |
| Non-malignant diseases | 1.795 (0.635–5.076) | 0.27 | 1.007 (0.983–1.032) | 0.58 | – | – |
The analysis stratified according to disease status showed that a higher proportion of patients with malignant diseases who developed aGVHD had mismatches for the composite of SNPs, but the difference did not reach statistical significance in univariate or multivariate analysis (Tables 5,6). No association was shown between the development of aGVHD grades II–IV and incompatibilities at the composite SNPs (P = 0.23). However, a higher proportion of patients with malignant diseases who manifested aGVHD III–IV were mismatched for the composite variable compared with the matched group (P = 0.04). Univariate and multivariate analyses of recipients with non-malignant diseases showed no association between aGVHD grades II–IV and mismatches at the composite variable. When analysis was restricted to aGVHD grades III–IV, a higher incidence of this disease was seen in the mismatched group (26.7% mismatched vs. 7.1% matched pairs; P = 0.04).
Chronic graft-versus-host disease
Univariate analysis showed no association between one or more mismatches at C4 SNPs and the development of cGVHD (SHR = 1.36; 95% CI = 0.81–2.30; p = 0.25 — Figure 1B); a lack of association was also observed when the analysis was stratified according to disease status: malignant (SHR = 1.34; 95% CI = 0.62–2.90; P = 0.45) and non-malignant (SHR = 1.33; 95% CI = 0.65–2.71; P = 0.44).
None of the 12 SNPs analyzed independently showed an effect on the development of cGVHD. The 5 SNPs (T14757, T14831, T14952, G15108, A20170) with P < 0.2 and SHR > 1.0 were pooled into a composite variable, but no association was found between these mismatches and an increased risk of developing cGVHD in the total sample (SHR = 1.51; 95% CI = 0.95–2.40; P = 0.081); no association was also seen in the analysis stratified according to disease status: malignant (SHR = 1.84; 95% CI = 0.91–3.72; P = 0.088) and non-malignant (SHR = 1.83; 95% CI = 0.85–3.94; P = 0.120).
Mortality
Univariate analysis of the demographics data showed an association between mortality and malignant disease (HR = 1.85; 95% CI = 1.17–2.92; P = 0.009), peripheral blood as the stem cell source (HR = 2.08; 95% CI = 1.25–3.45; P = 0.005) and myeloablative conditioning (HR = 1.70; 95% CI = 1.01–2.86; P = 0.047). The median age of the deceased recipients (23 yrs.) was higher than that of the survivors (12.4 yrs.) (HR = 1.03; 95% CI = 1.02–1.04; p < 0.001).
One or more mismatches at C4-SNPs were not associated with post-HSCT mortality (HR = 0.92; 95% CI = 0.59–1.44; P = 0.714 – Figure 1C) in the total sample or in the sample stratified by disease status: malignant (HR = 1.04; 95% CI = 0.60–1.81; P = 0.879) and non-malignant (HR=0.73; 95% CI=0.34-1.58; P = 0.431).
Independent analysis of the 12 SNPs showed no association with mortality post-HSCT. The variable comprising SNPs G12071 and T14563 (P < 0.2; HR < 1.0) suggested an association with lower mortality in mismatched patients (HR = 0.43; 95% CI = 0.19–1.00; P = 0.05), but this trend was not seen in multivariate analysis after adjusting for clinical variables (HR = 0.46; 95% CI = 0.19–1.06; P = 0.07). Univariate analysis of the sample stratified according to the diagnosis showed no association with mortality (malignant disease: HR = 0.66; 95%CI = 0.26–1.66; P = 0.38; non-malignant: HR = 0.17; 95% CI = 0.23–1.26; P = 0.08), and multivariate analysis also revealed no effect of the composite variable mismatches on this endpoint.
Discussion
This retrospective study included patients who received HSC from HLA-A-, B-, C-, DRB1- and DQB1-matched URDs, and 32.8% developed post-transplant GVHD (14.7% grade II; 14.7% grades III–IV). The fact that the HLA match is not enough to avoid post-transplant complications, such as life-threatening GVDH, has been well documented by robust studies. Other R/D disparities inside and/or outside MHC also contributed to the allogeneic response leading to post-transplant adverse effects,6, 10, 19 and an HLA match does not assure haplotype sharing in unrelated HSCT.6 The present study investigated whether the matching status of 23 SNPs of C4A/C4B genes, located in the MHC gamma block, would impact the outcomes of post-HSCT. If so, they would be useful markers of identity at other MHC genetic loci between patients and HLA 10/10 matched URDs, thus lowering the risk of post-transplant immunological complications.
The endpoints of this study were aGVHD, cGVHD and mortality. Considering that DPB1 mismatches have been associated with these outcomes20, 21, 22 and could confound C4-SNP analysis, all recipients/donors were high-resolution typed for this gene. Fifty-six percent of R/D pairs had DP-compatible or -permissive mismatches, while 44% had non-permissive mismatches; however, in this cohort, non-permissive DPB1 mismatches were not associated with an increased risk of GVHD or mortality. Different factors could have led to this result, such as the small size of the studied population, inclusion of adult and pediatric patients, malignant and non-malignant diseases, different permissiveness21 and expression levels of DP,22 conditioning and GVHD prophylaxis. Pre-transplant anti-thymocyte globulin was given to 83.2% of our patients, and this strategy for GVHD control has been associated with a decrease in the risk of GVHD post-HSCT, possibly for overcoming the R/D genetic diversity.23
Characterization of 23 C4A/C4B SNPs showed that, despite the HLA 10/10 compatibility, 42.9% of our patients received HSC from donors with 1–7 mismatches, among whom 37.2% were HLA 10/12, 49% were 11/12 and 13.8% were 12/12. This result is within the range of other studies that found 20–73% of R/D disparities in these same complement cascade genes, and differences may be due to sample characteristics and frequencies of SNPs in different populations.15, 16, 17, 24, 25 The allele and genotypic frequencies of these C4A/C4B SNPs were determined based on the data of individuals typed by NGS (N = 109), but there was no statistical power to identify associations with the transplant outcomes in this cohort (data not shown).
All but one of the eight SNPs with mutations that resulted in an amino acid substitution were considered tolerable by in silico analysis utilizing SIFT and POLYPHEN-2 tools (C4B pre-protein was the reference for amino acid changes). The investigation of the possible impact of the variant A11437 on the GVHD development, assigned as probably harmful by POLYPHEN-2, was not feasible due to the low allele frequency and absence of a homozygous genotype of this variant (AA) in this small cohort. The independent effect of mismatches at SNP 11437 on clinical outcomes was not investigated because the number of patients with mismatched donors for this polymorphism (4/238) was insufficient for statistical testing.
Regarding acute GVHD, no association was found with one or more mismatches of the C4A/C4B SNPs in the total sample, either by univariate or multivariate analysis (Fine and Gray model; P = 0.948). This absence of an association was confirmed when the analysis was stratified by type of disease (malignant vs. non-malignant) and percentage of mismatches in the 23 SNPs (full match; 1–3 mm and >3 mm). This result does not confirm the findings of a previous study, in 2014, showing a significant association between gamma-type mismatches and post-HSCT aGVHD grades II–IV with HLA10/10 URDs.13 A possible explanation for the conflicting results is that HLA matching was based on medium-resolution typing for some of the loci and therefore, undetected HLA mismatching could have contributed to GVHD. Another group, in 2016, showed a weak association between mismatches at C4 SNPs and a higher incidence of aGVHD grades II–IV, not observed in multivariate analysis.15 Their patients received HSCs from peripheral blood that may have contributed to this trend of association, while 83.6% of our patients received bone marrow grafts.
The isolated effect of SNPs on the development of aGVHD grades I–IV showed that patients mismatched for SNP 14952 had a higher incidence of the acute form of this disease (P = 0.04), although this difference lost significance after correction for multiple comparisons. This independent effect was evaluated only in 12/23 SNPs that were mismatched in at least 10 R/D pairs to allow statistical testing. Univariate analysis of patients transplanted with one or more mismatches in the composite variable (SNPs C13193, T14952 and T19588) showed a higher incidence of GVHD grades II–IV in the total sample (P = 0.004), as well as in samples stratified according to malignant (P = 0.04) and non-malignant (P = 0.04) status; multivariate analysis revealed similar results. Another study also found an association between incompatibilities in the composite variable (SNPs 12749, 16986 and C4A isotype) and an increased risk of aGVHD grades III–IV in both univariate (RR 2.43; P = 0.004) and multivariate analyses (RR 2.54; P = 0.002).14 Although the selection criteria for the composite variable were the same—i.e., values of P < 0.20 and SHR > 1.0 in the isolated SNP analysis—our results cannot be compared because the SNPs included in the composites were different. In our study, SNP 12749 did not meet this requirement (SHR = 0.59, 95% CI = 0.19–1.85; P = 0.363); moreover, SNPs 16986 and C4A isotype were not included in our panel.
Concerning chronic GVHD, no significant difference was observed between matched vs. mismatched for C4-SNPs in the total sample or when stratified by disease status (malignant and non-malignant). An absence of associations was also seen when data analysis was performed according to the percentage of mismatches in the C4-SNPs (full match; 1–3 mm and >3 mm). The individual effect of these SNPs on the development of cGVHD showed a positive association with a mismatch at SNP 14757 (P = 0.015), but significance was lost after the Bonferroni correction. Jimenez et al. (2016) found a borderline association with cGVHD (p = 0.048), while Askar et al. (2015) did not evaluate this outcome. The players of cGVHD are alloreactive cells, dysregulated T and B lymphocytes and innate immune cells that lead to autoimmune disease-related traits.26 The copy number variation (CNV) of C4A/C4B has been associated with the development of autoimmune diseases,27, 28 but analysis of the copy number diversity in the C4 genes was not within the scope of this study; however, it is worthwhile to investigate the possible impact of CNV on the chronic manifestation of GVHD.
In the present study, disparities determined by C4A/C4B SNPs did not impact mortality when performed in the total sample, stratified according to disease status or based on the percentage of C4 mismatches. One borderline association with a variable comprising SNPs G12071/T14563 was found, but it was probably spurious or limited by sample size.
A recent single-center retrospective study analyzed 25 SNPs in the C4A/C4B genes utilizing PCR-SSP (Conexio Genomics); 23/25 of these SNPs were also investigated in our study, but the reagents for SNPs 16986 and C4A isotype were not included in the panel when we performed C4 typing. They found that disparities between recipients and their HLA 10/10 URD had no impact on survival, relapse and acute or chronic GVHD.17 A larger CIBMTR retrospective study investigated the MHC gamma-block diversity using the Illumina NGS platform and identified 338 SNPs in recipients of HSC from HLA 10/10 URDs. They found no significant association between mismatches in this region of MHC, including C4A/C4B genes, and post-HSCT outcomes, such as aGVHD, cGVHD, overall survival, disease-free survival, transplant-related mortality and engraftment.16
The primary endpoint in these latter studies, as well as in ours, is aGVHD. Despite the differences in the cohort (patient/donor characteristics and sample size), single vs. multicenter and methods for SNP characterization, the results of these 23 C4-SNPS investigated in all studies are consistent with the absence of significant associations between mismatches and the development of post-HSCT acute and/or chronic GVHD.
Conclusion
Despite the high proportion of patients and URDs with differences in the C4A/C4B SNPs (42.9%), we found that these mismatches are not risk factors for the acute or chronic forms of GVHD or mortality after unrelated HSCT. This conclusion is applied to SNPs evaluated together (matched or ≥1 mm), independently or additively considering the number of incompatibilities (full match for 23 SNPs; 1–3 mm and >3 mm). An exception was the association between 1–3 mismatches at the composite of SNPs C13193/T14952/T19588 with the development of aGVHD (P = 0.012) and more severe manifestations of this disease (P = 0.004). This result needs confirmation in a much larger cohort because the finding could be either spurious due to the limited sample size or each of these three alleles could be in linkage disequilibrium with the same allele at a transplant determinant locus.
The MHC is a targeted genomic region that is used to investigate clinically relevant genetic diversity due to the high density of genes with immune and/or inflammatory functions. Our hypothesis regarding post-HSCT outcomes being impacted by R/URD incompatibilities in the C4A/C4B SNPs was not confirmed, but this does not mean that transplant determinant loci are not present in the MHC central region. Further studies in larger cohorts, allowing for the investigation of SNP-associated risks, not only by R/D mismatching but also by patient or donor genotype, may reveal genetic markers in linkage disequilibrium with transplant determinants. Reliable markers of non-HLA genetic loci identity are needed to optimize unrelated donor selection and improve HSCT outcomes.
Funding
This work was supported by Programa Nacional de Apoio à Atenção Oncológica (PRONON) [grant number 25000.159310/2014-74].
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We are grateful for the support provided by Associação Alírio Pfiffer (Instituto TMO) and Associação dos Amigos do Hospital de Clínicas (AAHC) for this project.
References
- 1.Moyer A.M., Hashmi S.K., Kroning C.M., Kremers W.K., De Goey S.R., Patnaik M. Clinical outcomes of HLA-DPB1 mismatches in 10/10 HLA-matched unrelated donor-recipient pairs undergoing allogeneic stem cell transplant. Eur J Haematol. 2017;99(3):275–282. doi: 10.1111/ejh.12916. [DOI] [PubMed] [Google Scholar]
- 2.Devillier R., Dalle J.H., Kulasekararaj A., D’aveni M., Clément L., Chybicka A. Unrelated alternative donor transplantation for severe acquired aplastic anemia: a study from the french society of bone marrow transplantation and cell therapies and the EBMT severe aplastic anemia working party. Haematologica. 2016;101(7):884–890. doi: 10.3324/haematol.2015.138727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart C.A. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14(6):1176–1187. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tay G.K., Witt C.S., Christiansen F.T., Charron D., Baker D., Herrmann R. Matching for MHC haplotypes results in improved survival following unrelated bone marrow transplantation. Bone Marrow Transplant. 1995;15(3):381–385. [PubMed] [Google Scholar]
- 5.Witt C., Sayer D., Trimboli F., Saw M., Herrmann R., Cannell P. Unrelated donors selected prospectively by block-matching have superior bone marrow transplant outcome. Hum Immunol. 2000;61(2):85–91. doi: 10.1016/s0198-8859(99)00147-0. [DOI] [PubMed] [Google Scholar]
- 6.Petersdorf E.W., Malkki M., Gooley T.A., Martin P.J., Guo Z. MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med. 2007;4(1):e8. doi: 10.1371/journal.pmed.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogunia-kubik K., Lange A. HSP70-hom gene polymorphism in allogeneic hematopoietic stem-cell transplant recipients correlates with the development of acute graft-versus-host disease. Transplantation. 2005;79(7):815–820. doi: 10.1097/01.tp.0000153157.97736.2c. https://www.ncbi.nlm.nih.gov/pubmed/15818324 PMID: 15818324; Pubmed website: [DOI] [PubMed] [Google Scholar]
- 8.Harkensee C., Oka A., Onizuka M., Middleton P.G., Inoko H., Nakaoka H. Microsatellite scanning of the immunogenome associates MAPK14 and ELTD1 with graft-versus-host disease in hematopoietic stem cell transplantation. Immunogenetics. 2013;65(6):417–427. doi: 10.1007/s00251-013-0691-z. [DOI] [PubMed] [Google Scholar]
- 9.Petersdorf E.W., Malkki M., Gooley T.A., Spellman S.R., Haagenson M.D., Horowitz M.M. MHC-resident variation affects risks after unrelated donor hematopoietic cell transplantation. Sci Transl Med. 2012;4(144) doi: 10.1126/scitranslmed.3003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersdorf E.W., Malkki M., Horowitz M.M., Spellman S.R., Haagenson M.D., Wang T. Mapping MHC haplotype effects in unrelated donor hematopoietic cell transplantation. Blood. 2013;121(10):1896–1905. doi: 10.1182/blood-2012-11-465161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaudieri S., Leelayuwat C., Tay G.K., Townend D.C., Dawkins R.L. The major histocompatability complex (MHC) contains conserved polymorphic genomic sequences that are shuffled by recombination to form ethnic-specific haplotypes. J Mol Evol. 1997;45(1):17–23. doi: 10.1007/pl00006194. [DOI] [PubMed] [Google Scholar]
- 12.Dorak M.T., Chalmers E.A., Sproul A.M., Mills K.I., Wilson D.W., Galbraith I. MHC class III polymorphisms in selection of donors for BMT. Bone Marrow Transplant. 1993;11(1):37–41. [PubMed] [Google Scholar]
- 13.Getz J., Sayer D.C., Pasquini R., Funke V., Bonfim C.S., Colturato V.R. Impact of MHC Gamma Block (GAMMA-TYPE) mismatching in the outcome of unrelated hematopoietic stem cell transplantation. Tissue Antigens. 2014;84(1):40–41. [Google Scholar]
- 14.Askar M., Majhail N.S., Rybicki L., Zhange A., Thomas D., Chen D. Single Nucleotide gene polymorphisms (SNP) in the gamma block of the major histocompatibility complex (MHC) are independent risk factors for severe acute graft versus host disease (GVHD) in unrelated donor hematopoietic cell transplantation (HCT) Biol Blood Marrow Transplant. 2015;21(2):S326–7. [Google Scholar]
- 15.Jimenez A., Mitchell R.L., Kongtim P. Major histocompatibility complex (MHC) gamma-block (GB) mismatch is associated with graft-versus-host disease (GVHD) in allogeneic hematopoietic stem cell transplantation (HSCT) Bone Marrow Transplant. 2016;51(September):S379–S380. doi: 10.1038/bmt.2016.49. [DOI] [Google Scholar]
- 16.Askar M., Sayer D., Wang T., Haagenson M., Spellman S.R., Lee S.J. Analysis of single nucleotide polymorphisms in the gamma block of the major histocompatibility complex in association with clinical outcomes of hematopoietic cell transplantation: a center for international blood and marrow transplant research Study. Biol Blood Marrow Transplant. 2019;25(4):664–672. doi: 10.1016/j.bbmt.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moyer A.M., Hashmi S.K., Kroning C., De Goey S.R., Patnaik M., Litzow M. Does matching for SNPs in the MHC gamma block in 10/10 HLA-matched unrelated donor-recipient pairs undergoing allogeneic stem cell transplant improve outcomes? Hum Immunol. 2018;79(7):532–536. doi: 10.1016/j.humimm.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spierings E., Kim Y.H., Hendriks M., Borst E., Sergeant R., Canossi A. Multicenter analyses demonstrate significant clinical effects of minor Histocompatibility Antigens on GvHD and GvL after HLA-matched related and unrelated Hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2013;19(8):1244–1253. doi: 10.1016/j.bbmt.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Pidala J., Lee S.J., Ahn K.W., Spellman S., Wang H.L., Aljurf M. Non-permissive -DPB1 mismatch among otherwise HLA-matched donor-recipient pairs results in increased overall mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation for hematologic malignancies. Blood. 2014;124(16):2596–2606. doi: 10.1182/blood-2014-05-576041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleischhauer K., Shaw B.E., Gooley T., Malkki M., Bardy P., Bignon J.D. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13(4):366–374. doi: 10.1016/S1470-2045(12)70004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersdorf E.W., Malkki M., O’hUigin C., Carrington M., Gooley T., Haagenson M.D. High HLA-DP expression and graft-versus-host disease. N Engl J Med. 2015;373(7):599–609. doi: 10.1056/NEJMoa1500140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baron F., Mohty M., Blaise D., Socié G., Labopin M., Esteve J. Anti-thymocyte globulin as graft-versus-host disease prevention in the setting of allogeneic peripheral blood stem cell transplantation: a review from the acute leukemia working party of the european society for blood and marrow transplantation. Haematologica. 2017;102(2):224–234. doi: 10.3324/haematol.2016.148510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogan H.M., Candy L., DeSantis D., Sayer D.C. Gamma-type identifies MHC haplotype mimsmatched donors that are HLA matched at HLA-A -B -C -DRB1 and DQB1. Tissue Antigens. 2013;81(5):291–292. [Google Scholar]
- 25.Gendzekhadze K., Hogan H., Wirtz C., Garcia-Gomez J., Sayer D., Senitzer D. P067 How different is gamma block in HLA matched (10/10) unrelated pairs detected by next generation sequencing (NGS)? Hum Immunol. 2016;77(2016):86. [Google Scholar]
- 26.Cooke K.R., Luznik L., Sarantopoulos S., Hakim F.T., Jagasia M., Fowler D.H. The biology of chronic graft-versus-host disease: a task force report from the national institutes of health consensus development project on criteria for clinical trials in chronic graft-versus-host disease. Biol Blood Marrow Transplant. 2017;23(February (2)):211–234. doi: 10.1016/j.bbmt.2016.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lintner K.E., Wu Y.L., Yang Y., Spencer C.H., Hauptmann G., Hebert L.A. Early components of the complement classical activation pathway in human systemic autoimmune diseases. Front Immunol. 2016;7(February):1–22. doi: 10.3389/fimmu.2016.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li N., Zhang J., Liao D., Yang L., Wang Y., Hou S. Association between C4, C4A, and C4B copy number variations and susceptibility to autoimmune diseases: a meta-analysis. Sci Rep. 2017 doi: 10.1038/srep42628. [DOI] [PMC free article] [PubMed] [Google Scholar]