Introduction
In December 2019, a new infectious respiratory disease emerged in Wuhan, Hubei province, China (Huang et al., 2020; Wang et al., 2020; Zhu et al., 2020). It diffused rapidly worldwide and became a pandemic. The World Health Organization (WHO) has officially named it coronavirus disease 2019 (COVID-19), and the virus has been classified as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 causes a severe clinical picture, ranging from mild malaise to death by sepsis/acute respiratory distress syndrome (Gabutti et al., 2020). The epidemiology of COVID-19 highlights differences either in the susceptibility to the infection or in death rates among populations (https://covid19.who.int/). As for other multifactorial conditions, this variability may be related to environmental differences among countries, such as access to medical care and the age structure of the population (Pareek et al., 2020); nevertheless, it may be related also to human genetic variability. From this perspective, angiotensin-converting enzyme 2 (ACE2) and serine protease 2 (TMPRSS2) genes are good candidates due to their role in the viral infection. Indeed, ACE2 was reported to be the main entry receptor for SARS-CoV-2 (Wang et al., 2020). Entry depends on the binding of the surface unit, S1, of the spike (S) protein of the virus to the receptor. SARS-CoV-2 engages ACE2 as the entry receptor and employs the host cellular TMPRSS2 for S-protein priming (Matsuyama et al., 2010; Hoffmann et al., 2020). TMPRSS2 is important for the spread of several viruses, including influenza A viruses and other coronaviruses (Glowacka et al., 2011; Gierer et al., 2013; Zhou et al., 2015; Shirato et al., 2016, 2018; Iwata-Yoshikawa et al., 2019).
Different studies have already investigated the potential associations between genetic variants of the ACE2 gene and COVID-19 (Asselta et al., 2020; Benetti et al., 2020; Cao et al., 2020; Darbani, 2020; Devaux et al., 2020). Here, we have analyzed the genetic markers of the TMPRSS2 gene and the differences in their alternative allele frequencies (AFs) among populations to identify possible susceptibility loci to COVID-19 and to correlate them with disease epidemiology.
Tissue Expression of TMPRSS2 Gene
TMPRSS2 is highly expressed in ileal absorptive enterocytes, nasal goblet secretory cells, epithelial cells of bronchi, as well as type I and type II alveolar cells (Bertram et al., 2012; Collin et al., 2020; Ziegler et al., 2020). TMPRSS2 expression in type I alveolar cells increases with aging in mice and humans (Schuler et al., 2020). Additionally, TMPRSS2 is highly expressed in corneal epithelium and conjunctival specimens, suggesting that ocular surface cells could be the gateway of SARS-CoV-2 as well as a reservoir for person-to-person transmission (Collin et al., 2020).
Accordingly, in-silico expression analysis of the TMPRSS2 supported the high TMPRSS2 expression in tissues of the respiratory tract, such as the bronchus, pharyngeal mucosa, and lung. However, no difference in gene expression between males and females was observed for non-gender-specific tissues (Figures S1A,B).
Analysis of Genetic Variants of TMPRSS2 Locus
We analyzed 1,025 variants in the TMPRSS2 gene region (chr21:42836478-42903043, 66.566 Kb) from the gnomAD v2.1.1 database. Annotation of TMPRSS2 variants was performed with ANNOVAR by using 34 pathogenic variant scores (Table S1) and the AFs of 17 populations (Table S5; Russo et al., 2020). Genomic coordinates were based on the GRCh37/hg19 build. The classification of non-synonymous variants was performed using the following predictor tools: M-CAP (score >0.025), MutationTester (A-D, disease-causing), CADD v1.3 (Phred score >15) for the pathogenic variants. VEST3 (score <0.5), REVEL (score <0.5), and RadialSVM (T, tolerated) for the benign variants (Rajarshi et al., 2017). Variants with conflicting interpretations were excluded from further analyses.
The locus region comprises 496 non-coding and 520 coding variants (Table S2). Forty-three variants were classified as loss-of-function (LoF), while 88/334 (26%) non-synonymous variants were predicted as pathogenic. All of them exhibit very low AFs (Figure 1A). Accordingly, a recent study identified a few functional ultra-rare variants in TMPRSS2, all of them with AFs < 0.001 (Gupta et al., 2020). These findings agree with the recommended benign frequency cut-off of 0.0001 for the TMPRSS2 gene, as from the Varsome database.
Figure 1.
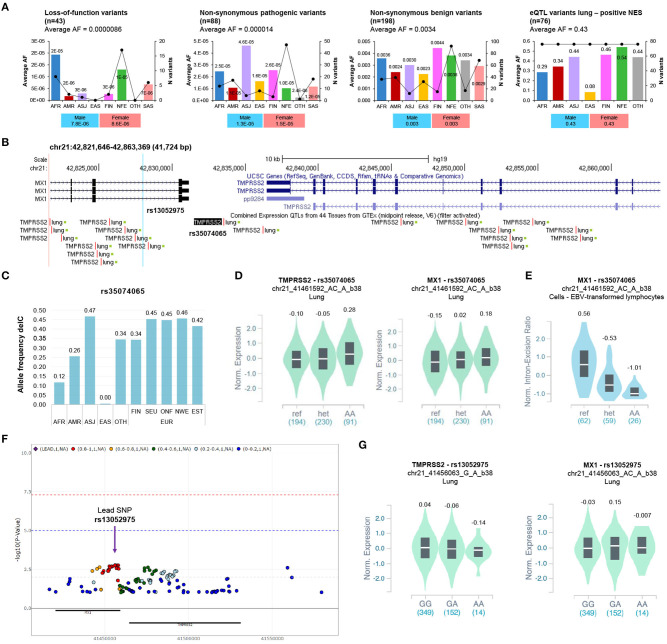
Analysis of the coding variants and the eQTLs of TMPRSS2 locus. (A) The alternative allele frequency (AF) distribution of non-synonymous pathogenic, benign, loss-of-function, and eQTL-lung variants (positive NES) of TMPRSS2 in different populations. The Y axis of each bar plot shows average allele frequency (AF) in each population. The second right axis displays the number of variants within each population. The bar graph below shows the AF distribution in the overall gnomAD population stratified according to gender. AFR, African/African American; AMR, Latino/Admixed American; ASJ, Ashkenazi Jewish; EAS, East Asian; FIN, Finnish; NFE, Non-Finnish European; SAS, South Asian; OTH, Other (population not assigned). (B) Schematics of the genomic region encompassing eQTL lung variants of TMPRSS2 locus (NES positive ≥ 0.1) by Genome Browser (GRCh37/hg19, https://genome.ucsc.edu/). The most significant eQTL-lung (rs35074065) and the eQTL rs13052975, associated with severe COVID-19, are highlighted. (C) The allele frequencies of delC variant rs35074065 were annotated by the gnomAD database (WGS data). AFR, African/African American; AMR, Latino/Admixed American; ASJ, Ashkenazi Jewish; EAS, East Asian; OTH, Other (population not assigned); FIN, Finnish; SEU, Southern European; ONF, Other non-Finnish European; NEW, North-Western European; EST, Estonian. (D) Violin plot showing the effect of the eQTL rs35074065 on TMPRSS2 and MX1 expression in lung (TMPRSS2: p = 3.9e-11; NES = 0.13; MX1: p = 0.000010; NES = 0.20). (E) Violin plot showing the effect of the sQTL rs35074065 on MX1 splicing isoform expression (p = 1.6e-13; NES = −0.83). (F) Regional association plot (P < 0.1) of variants at TMPRSS2 locus (proximity: ± 50Kb; analysis II, adjusted for top 10 PCs, age, sex) and respiratory failure in COVID-19 (data from COVID-19 GWAS results browser, https://ikmb.shinyapps.io/COVID-19_GWAS_Browser/). (G) Violin plot showing the effect of the eQTL rs13052975 on TMPRSS2 and MX1 expression in lung (TMPRSS2: P = 0.0063; NES = −0.072; MX1: P = 0.15; NES = −0.090).
Africans (AFR) showed the highest AFs for the LoF variants across populations. Similarly, the Swedish population exhibited the highest AF for LoF variants among the Europeans (Figure S2A). Regarding the non-synonymous pathogenic variants, we observed the highest AF among the Ashkenazi Jewish (ASJ) population (Figure 1A), while the Finnish (FIN) showed the highest AF among European subpopulations (Figure S2A). The AFs of non-synonymous variants classified as benign (198/334, 59.3%) were similarly distributed among the different populations (Figure 1A).
Analysis of the Genetic Regulatory Variants Driving TMPRSS2 Expression
To analyze the distribution of expression quantitative trait loci (eQTL) for TMPRSS2, we used the data from the Genotype Tissue Expression (GTEx) database (Table S3). The reference transcript for TMPRSS2 annotation was NM_001135099 (ENST00000398585) (Figure S1C).
We found 203 unique and significant (FDR<0.05) eQTLs for TMPRSS2 in five different tissues: 136 (66.9%) in lung, 56 (27.6%) in testis, 9 (4.4%) in prostate, 1 (0.5%) in ovarian, and 1 in thyroid (0.5%) tissue. The AFs of the 136 eQTLs with the strongest association in the lung tissue (eQTLs-lung) showed no substantial differences among different populations (Figure S2B). Nevertheless, the average AF of 76 eQTLs-lung with a positive normalized effect size (NES) was higher in European populations (FIN, 0.463; NFE, 0.541) compared to the average AF observed in the East Asian population (EAS) (0.085) (Figure 1A, Table S4). Of note, Europe displayed the highest median prevalence of COVID-19 cases among the WHO regions, while South East Asia showed a low prevalence of the disease (Figure S3).
Interestingly, the top 25 variants (NES > 0.1) localize in a genomic region that includes both TMPRSS2 and MX1 genes. The most significant eQTL, rs35074065, is in the intergenic region between the two genes (distance = 2379 from MX1; distance = 2958 from TMPRSS2) (Figure 1B) and shows the lowest AF in EAS (delC, 0.0049) (Figure 1C). Notably, the alternative allele delC seems to be associated with high expression of both TMPRSS2 and MX1 in lung tissue (Figure 1D). Moreover, the same variant is also a splicing QTL (sQTL) associated with low expression of MX1 splicing isoform in different tissues (Figure 1E).
A recent analysis of nasal gene expression and genome-wide genetic variation data yielded three independent TMPRSS2 eQTLs located in the downstream region of the gene: the SNP rs1475908, whose alternative allele (A) is associated with low TMPRSS2 expression, and the two variants rs74659079 (allele T) and rs2838057 (allele A), both associated with high TMPRSS2 expression. Interestingly, the eQTL rs1475908 shows the highest AF among EAS (A:0.38) and EUR (A:0.35) and the lowest frequency among Latinos (0.17) (Sajuthi et al., 2020).
These findings agree with a previous study that demonstrated the association of two high TMPRSS2 expression-variants, rs2070788 (allele G) and rs383510 (allele T), with increased susceptibility to the influenza virus infection A (H7N9) (Cheng et al., 2015). Of note, the SNP rs2070788 was recently included in a haplotype associated with high TMPRSS2 expression, whose AF is significantly increased in Europeans (Asselta et al., 2020).
We also verified the association between variants at TMPRSS2 locus and respiratory failure in patients with COVID-19 using the summary statistics of a recently published GWAS (Ellinghaus et al., 2020). We found 13 SNPs showing a level of significance less than or equal to 0.002 in high-linkage disequilibrium (LD) (r2:0.8-1) and a total 89 SNPs with P < 0.05 (Figure 1F, Table S6).
The most significant eQTL-lung for TMPRSS2 (rs35074065) was found to be not significant (P = 0.30, OR = 1.07) and was located outside the LD block of the most significant rs13052975 variant (Figure S4, Table S6), whose alternative allele (A) correlates with low TMPRSS2 in the lung (p = 0.0063), albeit with a non-significant p-value after multiple testing correction (Figure 1G). However, while current GWAS results do not support a role for the eQTL in respiratory failure associated with COVID-19, we cannot exclude its role in general susceptibility or other definitions of severity, such type of symptoms shown and duration of illness.
Discussion
TMPRSS2 plays an important role in initiating SARS-CoV-2 and other respiratory viral infections (Glowacka et al., 2011). It has been suggested recently that the SARS-CoV-2 sequence has evolved by generating a unique four amino acid insertion between S1 and S2 domains of the spike protein, which created potential furin or TMPRSS2 cleavage site (Wu et al., 2020).
TMPRSS2 is highly expressed in tissues of the aerodigestive tract (Bertram et al., 2012; Collin et al., 2020; Ziegler et al., 2020). Epidemiological data showed that the incidence and severity of diagnosed COVID-19 may be higher in men than women. Nevertheless, TMPRSS2 gene expression data from the GTEx database do not highlight any difference between males and females. Importantly, developmental regulation of the expression of the TMPRSS2 gene has been suggested that may underlie the relative protection of infants and children from severe respiratory illness (Schuler et al., 2020).
To investigate the genetic features of TMPRSS2 locus among different populations, we analyzed both the coding-region variants of TMPRSS2 and the eQTLs, which may regulate its gene expression. Our findings do not support the existence of common coding pathogenic variants for TMPRSS2 among different populations. Accordingly, recent studies identified only a few pathogenic, ultra-rare variants in this gene (Gupta et al., 2020; Paniri et al., 2020; Sajuthi et al., 2020). Therefore, given their rarity, we do not believe that coding variants, except for some rare cases, can determine the diverse susceptibility to viral infection and diverse clinical manifestations.
Conversely, it seems that the genetic regulatory variants driving TMPRSS2 expression may have a role in the different susceptibility to SARS-CoV-2 and other respiratory viral infections among populations. Our in-silico analysis shows that common eQTLs-lung for TMPRSS2 are less frequent in EAS but more frequent in EUR. Interestingly, the allele delC of the top significant eQTL (rs35074065) associates with higher expression of TMPRSS2 compared to the reference allele and is less frequent among East Asians, suggesting a protective role against the infection in this population. The potential protective role of genetic variants associated with reduced TMPRSS2 expression is supported by a GWAS. This demonstrated that rs35074065 is near to another eQTL, rs13052975, nominally significantly associated with severe COVID-19 (Ellinghaus et al., 2020). The alternative allele (A) of rs13052975 confers protection against severe clinical phenotype of the disease, correlates with low expression of TMPRSS2, and is common among EAS (A:0.45), while its frequency in other populations is below 0.26 (AFR, A:0.26; AMR, A:0.21; NFE, A:0.16).
Interestingly, the top eQTL variants derived from the GTEx database were in the intergenic region between TMPRSS2 and MX1 genes. This locus has already been shown to harbor common genetic variants with pleiotropic effects on age-related diseases like heart failure, stroke, coronary heart disease, and atrial fibrillation (He et al., 2016). MX1 is an interferon (IFN)-α/β-inducible gene that codifies a guanosine triphosphate-metabolizing protein involved in the cellular antiviral response. It is widely recognized as an influenza susceptibility gene (Ciancanelli et al., 2016). Of note, the downregulation of MX1 has been documented in non-responder patients to interferon-based antiviral therapy of chronic hepatitis C virus infection (Persico et al., 2008). GTEx data suggested that the eQTL rs35074065 (delC) is associated with high expression of TMPRSS2 but also with a low expression of the MX1-splicing isoform (ENSG00000157601.13) in the esophagus, LCL, adipose tissue, whole blood, breast, small intestine, and lung (P ranging from 2 × 10–15 to 6 × 10–7). However, the strongest MX1-splicing QTLs at these tissues (P ranging from 2 × 10–45 to 1.9 × 10–16) were located within the coding region of MX1, and independent analyses thus need to be performed to verify if the effect of intergenic variant rs35074065 on MX1 isoform still remains significant after accounting for the stronger variants.
We hypothesize that common variants driving TMPRSS2 expression might have a mild-to-moderate effect in the susceptibility to SARS-CoV-2 infection. Particularly, genetic variants associated with reduced TMPRSS2 expression might confer less individual susceptibility to SARS-CoV-2 infection and favor a better outcome. This hypothesis agrees with epidemiological data that show higher COVID-19 prevalence and mortality rates in Europe and the Americas (with a greater frequency of high-TMPRSS2 expression-associated alleles). Conversely, a lower prevalence of the disease and mortality rates are found in South East Asia characterized by a high frequency of low-TMPRSS2 expression-associated alleles. However, genetic studies in large cohorts of COVID-19 cases and appropriate controls are needed to confirm this hypothesis.
Unraveling the role of regulatory variants of this locus could represent an interesting starting point for the treatment of COVID-19. Indeed, targeting TMPRSS2 expression and/or activity could be a promising candidate for potential interventions against COVID-19 (Kawase et al., 2012; Stopsack et al., 2020). Of note, the camostat mesylate (CM), a serine protease inhibitor that blocks TMPRSS2 activity, has been already introduced in Japan for the treatment of unrelated disorders (Gierer et al., 2013; Zhou et al., 2015). CM showed a protecting role against death in mice following a lethal SARS-CoV infection. Moreover, the advantage is its low cost (Uno, 2020). Unfortunately, to date, there are no clinical data on CM in COVID-19, and so human clinical trials are desirable.
Author Contributions
IA, RR, and MC designed and conducted the study, and prepared the manuscript. MC, VL, and RR analyzed the data. AI provided a critical review of the manuscript. All the authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank their colleagues Francesco Manna, Barbara Eleni Rosato, Annalaura Montella, and Roberta Marra for continuing their lab work with the same dedication as always during this troubled period of COVID-19 pandemic. This manuscript has been released as a pre-print at https://www.biorxiv.org/ (Russo et al., 2020).
Footnotes
Funding. This study was supported by the project CEINGE TASK-FORCE COVID19, code D64I200003800 by Regione Campania for the fight against Covid-19 (DGR n. 140 del 17 marzo 2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00872/full#supplementary-material
References
- Asselta R., Paraboschi E. M., Mantovani A., Duga S. (2020). ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging 12, 10087–10098. 10.18632/aging.103415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., et al. (2020). ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur J Hum Genet. 1–13. 10.1038/s41431-020-0691-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Heurich A., Lavender H., Gierer S., Danisch S., Perin P., et al. (2012). Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS ONE. 7:e35876. 10.1371/journal.pone.0035876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., et al. (2020). Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 6:11. 10.1038/s41421-020-0147-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Zhou J., To K. K., Chu H., Li C., Wang D., et al. (2015). Identification of TMPRSS2 as a susceptibility gene for severe 2009 pandemic A(H1N1) influenza and A(H7N9) influenza. J. Infect. Dis. 212, 1214–1221. 10.1093/infdis/jiv246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli M. J., Abel L., Zhang S. Y., Casanova J. L. (2016). Host genetics of severe influenza: from mouse Mx1 to human IRF7. Curr. Opin. Immunol. 38, 109–120. 10.1016/j.coi.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin J., Queen R., Zerti D., Dorgau B., Georgiou M., Djidrovski I., et al. (2020). Co-expression of SARS-CoV-2 entry genes in the superficial adult human conjunctival, limbal and corneal epithelium suggests an additional route of entry via the ocular surface. Ocul Surf. 10.1016/j.jtos.2020.05.013. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbani B. (2020). The expression and polymorphism of entry machinery for COVID-19 in human: juxtaposing population groups, gender, and different tissues. Int. J. Environ. Res. Public Health. 17:3433. 10.3390/ijerph17103433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C. A., Rolain J. M., Raoult D. (2020). ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 53, 425–435. 10.1016/j.jmii.2020.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., et al. (2020). Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 10.1056/NEJMoa2020283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabutti G., d'Anchera E., Sandri F., Savio M., Stefanati A. (2020). Coronavirus: update related to the current outbreak of COVID-19. Infect Dis Ther. 8, 1–13. 10.1007/s40121-020-00295-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Krämer-Kühl A., et al. (2013). The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 87, 5502–5511. 10.1128/JVI.00128-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Müller M. A., Allen P., Soilleux S., Pfefferle S., et al. (2011). Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85, 4122–4134. 10.1128/JVI.02232-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Charron J., Stenger C. L., Painter J., Steward H., Cook T. W., et al. (2020). SARS-CoV2 (COVID-19) structural/evolution dynamicome: insights into functional evolution and human genomics. bioRxiv [preprint]. 10.1101/2020.05.15.098616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Kernogitski Y., Kulminskaya I., Loika Y., Arbeev K. J., Loiko E., et al. (2016). Pleiotropic meta-analyses of longitudinal studies discover novel genetic variants associated with age-related diseases. Front. Genet. 7:179. 10.3389/fgene.2016.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181, 271–280. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan. Lancet. 395, 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. (2019). TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 93, e01815–e01818. 10.1128/JVI.01815-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. (2012). Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 86, 6537–6545. 10.1128/JVI.00094-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. (2010). Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 84, 12658–12664. 10.1128/JVI.01542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniri A., Hosseini M. M., Akhavan-Niaki H. (2020). First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. J. Biomol. Struct. Dyn. 1–18. 10.1080/07391102.2020.1767690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek M., Bangash M. N., Pareek N., Pan D., Sze S., Minhas J. S., et al. (2020). Ethnicity and COVID-19: an urgent public health research priority. Lancet. 395, 1421–1422. 10.1016/S0140-6736(20)30922-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persico M., Capasso M., Russo R., Persico E., Croc,è L., Tiribelli C., et al. (2008). Elevated expression and polymorphisms of SOCS3 influence patient response to antiviral therapy in chronic hepatitis C. Gut. 57, 507–515. 10.1136/gut.2007.129478 [DOI] [PubMed] [Google Scholar]
- Rajarshi G., Ninad O., Sharon E. P. (2017). Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biol. 18:225. 10.1186/s13059-017-1353-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo R., Andolfo I., Lasorsa V. A., Iolascon A., Capasso M. (2020). Genetic analysis of the novel SARS-CoV-2 host receptor TMPRSS2 in different populations. bioRxiv [preprint]. 10.1101/2020.04.23.057190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajuthi S. P., DeFord P., Jackson N. D., Montgomery M. T., Everman J. l., et al. (2020). Type 2 and interferon inflammation strongly regulate SARS-CoV-2 related gene expression in the airway epithelium. bioRxiv [preprint]. 10.1101/2020.04.09.034454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler B. A., Habermann A. C., Plosa E. J., Taylor C. J., Jetter C., Kapp M. E., et al. (2020). Age-related expression of SARS-CoV-2 priming protease TMPRSS2 in the developing lung. bioRxiv [preprint]. 10.1101/2020.05.22.111187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kanou K., Kawase M., Matsuyama S. (2016). Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J. Virol. 91, e01387–e01316. 10.1128/JVI.01387-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. (2018). Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology 517, 9–15. 10.1016/j.virol.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopsack K. H., Mucci L. A., Antonarakis E. S., Nelson P. S., Kantoff P. W. (2020). TMPRSS2 and COVID-19: serendipity or opportunity for intervention? Cancer Discov. 10, CD-20–0451. 10.1158/2159-8290.CD-20-0451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y. (2020). Camostat mesilate therapy for COVID-19. Intern. Emerg. Med. 29, 1–2. 10.1007/s11739-020-02345-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P. W., Hayden F. G., Gao G. F. (2020). A novel coronavirus outbreak of global health concern. Lancet 395, 470–473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Niu P., Wang L., Zhou H., Zhao X., Wang W., et al. (2020). Mutations, Recombination and insertion in the evolution of 2019-nCoV. bioRxiv [preprint]. 10.1101/2020.02.29.971101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Jr, Nunneleyet J. W., et al. (2015). Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 116, 76–84. 10.1016/j.antiviral.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Jingdong S., et al. (2020). A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C. G. K., Allon S. J., Nyquist S. K., Mbano I. M., Miao V. N., Tzouanas C. N., et al. (2020). SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 181, 1016–1035. 10.1016/j.cell.2020.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.