Abstract
The parasitic plant genus Cuscuta is notoriously difficult to transform and to propagate or regenerate in vitro. With it being a substantial threat to many agroecosystems, techniques allowing functional analysis of gene products involved in host interaction and infection mechanisms are, however, in high demand. We set out to explore whether Agrobacterium‐mediated transformation of different plant parts can provide efficient alternatives to the currently scarce and inefficient protocols for transgene expression in Cuscuta. We used fluorescent protein genes on the T‐DNA as markers for transformation efficiency and transformation stability. As a result, we present a novel highly efficient transformation protocol for Cuscuta reflexa cells that exploits the propensity of the infection organ to take up and express transgenes with the T‐DNA. Both, Agrobacterium rhizogenes and Agrobacterium tumefaciens carrying binary transformation vectors with reporter fluorochromes yielded high numbers of transformation events. An overwhelming majority of transformed cells were observed in the cell layer below the adhesive disk’s epidermis, suggesting that these cells are particularly susceptible to infection. Cotransformation of these cells happens frequently when Agrobacterium strains carrying different constructs are applied together. Explants containing transformed tissue expressed the fluorescent markers in in vitro culture for several weeks, offering a future possibility for development of transformed cells into callus. These results are discussed with respect to the future potential of this technique and with respect to the special characteristics of the infection organ that may explain its competence to take up the foreign DNA.
Keywords: adhesive disk, Agrobacterium transformation, Cuscuta, haustorium, parasitic plants
1. INTRODUCTION
Parasitic plants account for considerable agricultural losses of almost all food and fodder crops around the world. Many plant lineages contain one or more genera that live a parasitic live. Some are highly specialized and are only found in certain biotopes while others are widely distributed generalists. The obligate parasitic plant genus Cuscuta, commonly known as “dodder” is found almost worldwide and infects a broad range of host plants and, thus, belongs to the latter group. The parasitic attack by all Cuscuta species starts by winding around the host stem. A multicellular feeding structure termed the haustorium, which can reach 2–3 mm in size in some species and originates from a secondary meristem in the cortex close to the vascular bundles, develops within a few days and breaches the host tissue integrity using mechanical pressure and enzymatic digestion of cell walls (Nagar et al., 1984; Johnsen et al., 2015; Kaiser et al., 2015). To make the penetration possible, a sucker‐shaped attachment organ provides the necessary counterforce. The development of this organ, termed “adhesive disk” or “upper haustorium” (Vaughn, 2002; Lee, 2007; Kaiser et al., 2015), is easily visible as a swelling of the host‐facing side of the parasite’s stem. A bio‐adhesive substance, secreted by the epidermal cells of the adhesive disk, anchors the parasite to the host (Vaughn, 2002; Galloway et al., 2020). Morphologically, the development of the adhesive disk is marked by major local growth processes and shape changes of the involved cells: cells in the parasite’s cortex around the developing haustorium elongate significantly and epidermal cells that are normally rectangular and stretched in a longitudinal direction reshape into digitate cells that appear slightly rounded on the surface (Shimizu and Aoki, 2019).
In light of the detrimental effect to important crops exerted by Cuscuta worldwide, it is imperative that mechanisms guiding the parasitic attack are better understood. One prerequisite for achieving a better understanding of the molecular processes and mechanisms of a Cuscuta attack is the knowledge of the genomic disposition. Recently, complete genome sequences have been published for two Cuscuta species, C. campestris and C. australis (Sun et al., 2018; Vogel et al., 2018), covering this need. However, two significant bottlenecks to perform in situ protein studies or even empower targeted genome manipulations do remain. These are low transformation efficiencies on one hand and recalcitrance of callus propagation and shoot regeneration in vitro on the other hand. Few approaches to transform C. trifolii (Borsics et al., 2002) and C. europaea (Svubova and Blehova, 2013) have been reported. Explants from seedlings were transformed using Agrobacterium tumefaciens, and expression of the transgenes was demonstrated in the calli. However, a lack in efficiency of these approaches have so far precluded the use of transient or stable transformation for functional studies of the proteins and cells involved in establishing the parasitic interaction.
Besides A. tumefaciens, Agrobacterium rhizogenes has become a popular agent for transforming plant genomes (Ozyigit et al., 2013). A. rhizogenes is a soil‐borne bacterium infecting many angiosperms and causing them to produce a copious number of roots which became known as the “hairy root syndrome” (Bahramnejad et al., 2019). Like A. tumefaciens, A. rhizogenes transfers a segment of DNA known as T‐DNA into its hosts. The transfer process is controlled by virulence (vir) genes that are induced by phenolic signal molecules (Gelvin, 2003). The A. rhizogenes T‐DNA is stably integrated into the plant nuclear genome where it expresses the rol (rooting locus) genes required for excessive adventitious root growth (Ozyigit et al., 2013). What has made these hairy roots popular for plant biotechnology is that they can be propagated in the absence of exogenous plant hormones. Very recently, it was shown that an A. rhizogenes gene coding for the mikimopine synthase was horizontally transferred into several Cuscuta species (Zhang et al., 2020), including Cuscuta campestris (Vogel et al., 2018) and Cuscuta australis (Sun et al., 2018), suggesting that Cuscuta species may be susceptible to infection by this Agrobacterium species despite their lack of roots.
With this study, we set out to test the applicability of the hairy root transformation protocol in Cuscuta. Although hairy roots as such were not obtained, we were able to obtain high numbers of transformed cells in the species Cuscuta reflexa, particularly in its adhesive disks. We describe here the simple and highly efficient protocol that can yield hundreds of transformed cells within a week based on the use of A. rhizogenes. We further show that the protocol is applicable to use with A. tumefaciens with equally high success, which widens possibilities for single or cotransformation of different constructs, thus allowing functional studies of gene products like protein localization and interaction studies or expression of heterologous or synthetic transgenes within these cells that play a decisive role for Cuscuta’s pathogenicity.
2. MATERIALS AND METHODS
2.1. Plant material and Agrobacterium strains
Cuscuta reflexa, Cuscuta campestris, and Cuscuta platyloba were grown in a greenhouse on the host Pelargonium zonale under continuous illumination and a constant temperature of 21°C (Förste et al., 2020). The bacteria and binary T‐DNA‐containing vectors pRedRoot and XM82, respectively, were kindly contributed by Prof. Harro Bouwmeester (University of Amsterdam, Netherlands) (A. rhizogenes) and Prof. Tessa Burch‐Smith (University of Tennessee, USA) (A. tumefaciens) and are described in more detail in other studies (Limpens et al., 2004; Libiakova et al., 2018; Bobik et al., 2019). Cultures of A. rhizogenes MSU440 and A. tumefaciens GV3101 without binary plasmids were grown on LB medium (tryptone 10 g/L, NaCl 10 g/L, yeast extract 5 g/L, agar 7,5 g/L) supplemented with 100 mg/L Spectinomycin or 50 mg/L Rifampicin plus 50 mg/L Gentamycin, respectively. For bacteria containing the respective binary vectors, Kanamycin at 50 mg/L was added to the growth medium.
2.2. Induction of infection structure formation by far‐red light
For induction of infection structures, Cuscuta apical shoots of approximately 12 cm were harvested from the stock plant and exposed to far‐red light (740 nm) in an otherwise dark room for 90–120 min as described before (Olsen et al., 2016) with modifications. The steps were conducted in far‐red light to not reverse the induction (Tada et al., 1996). To provide a tactile stimulus, four shoots of roughly equal diameter were placed next to each other between two layers of bench paper with one‐sided plastic coating (Whatman® Benchkote® surface protector; Cat. # 2300731) that was moistened with tap water (filter‐paper side facing the shoots). This set‐up was carefully placed between two back‐to‐back facing petri dish halves (with the filter paper and the cut ends both protruding into a container with tap water [see Figure 1]). Moderate pressure was applied by taping the two petri dish plates together. When kept at room temperature in darkness, infection structures started to develop after about 3 days (Olsen et al., 2016).
FIGURE 1.
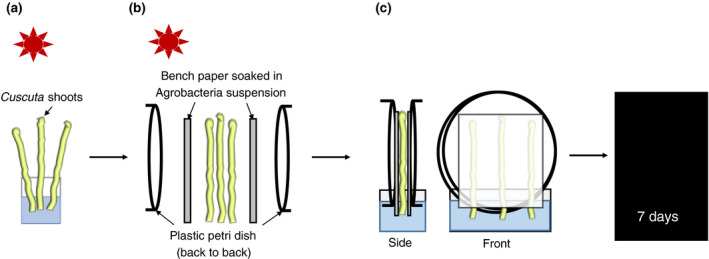
Overview of the experimental setup. (a) Exposure of Cuscuta shoots to far‐red light (740 nm) for 90–120 min. (b) Placement of Cuscuta shoots between Agrobacterium‐soaked bench paper sheets and two petri dish halves (back to back). (c) Incubation in darkness with shoots and bench paper placed in a water reservoir
2.3. Transformation of Cuscuta cells
An Agrobacterium culture was grown overnight in selective media (see above) and adjusted to an OD (600 nm) of 1–1.6, before using 2 ml of this suspension to soak the paper side of the bench paper (approximately 8 × 8 cm area). For mock controls, agrobacteria lacking a T‐DNA‐encoding binary vector were used. The assembly with far‐red light‐treated Cuscuta stems was done as described above and the whole setup was then incubated in a dark incubator set to room temperature for 7 days. After disassembling the setup, shoots were briefly rinsed under tap water, remnants of filter paper sticking to the adhesive disks were carefully removed without damaging the plant tissue, and stems were kept for up to 2 days in water or wrapped in wet paper towels before being subjected to microscopical analysis.
For exchange of water to Agrobacterium culture and vice versa (see Tables S1 and S2), the setup was disassembled on day 3 under far‐red light (740 nm) and the bench paper layer exchanged before the setup was re‐assembled and subjected to further incubation in the dark for another 5 days.
2.4. Microscopical imaging
Fluorescence localization in the Cuscuta stems exposed to agrobacteria and in the corresponding controls was documented using a StereoLumar V12 stereo microscope or an Axiovert M200 inverted microscope (both from Zeiss) using Zeiss filter sets 43 (for dsRed) and 38 (for GFP). Images were taken using the Axiovision Software (Version 4.8.2). The same exposure times were used for the different fluorescence filter sets for a given sample or magnification, unless specified otherwise. FIJI/ImageJ (V 2.0.0) was used to analyze the pictures, add scale bars, and produce overlays. When adjusting brightness, contrast, minimum and maximum intensities, all fluorescence images of one set were treated alike.
Fluorescence intensity scanning was performed on the marked area containing the region of interest (see Figure S5) using the histogram function of FIJI/ImageJ. Intensity counts were exported to Microsoft Excel for visualization in one joint colored graph.
2.5. Vibratome sectioning
Transformed infection sites were cross‐sectioned using a Vibratome (Leica VT1000 E vibrating blade microtome). Section thickness was 100 µm. Sections were viewed and documented using a StereoLumar V12 stereo microscope or an Axiovert M200 inverted microscope (Zeiss) using Zeiss filter sets 43 (for dsRed) and 38 (for GFP).
2.6. Life cell staining with 5‐carboxyfluorescein di‐acetate (CFDA)
To evaluate the vitality of the transformed cells, the vital stain CFDA (50 mM in DMSO) was diluted immediately prior to use to a final working concentration of 1 mM in water. Stems were covered with a thin layer of CFDA by spreading small drops of a few µl each evenly over the Cuscuta stem and infection sites (adjusted from “Drop‐And‐See assay” [Cui et al., 2015]). After incubation for 10 minutes in the dark, the CFDA was removed by briefly rinsing with tap water, gently dried with paper, and viewed under a StereoLumar V12 stereo microscope using Zeiss filter set 38.
2.7. Cultivation of explants
Cuscuta reflexa stems with infection sites and confirmed transformation events were sterilized for 2–5 min in 70% Ethanol and during this time gently cleaned using a brush. After a subsequent 15 min incubation step in 1.2% Sodium hypochloride, the stems were incubated on a shaker in sterile tap water containing 400 mg/L Cefotaxime overnight, then cut into pieces that contained one or two infection structures each and transferred to MS (Murashige and Skoog) Basal Medium supplemented with 0.8% Agar, 3% Sucrose, MS Vitamin solution, and 400 mg/L Cefotaxime. Plates were covered with aluminum foil to avoid photobleaching and were kept at 23°C. Explants were transferred to fresh medium approximately every 4 weeks.
3. RESULTS
3.1. Agrobacterium rhizogenes does not induce hairy roots in Cuscuta species
A. rhizogenes is typically applied to the roots (Ho‐Plagaro et al., 2018), hypocotyl (Alagarsamy et al., 2018) or cotyledons (Ron et al., 2014) of dicotyledonous angiosperms by cutting, puncturing, or otherwise wounding these tissues. In the absence of proper roots, cotyledons, or other leaves, we first tested hairy root induction on young germinating seedlings of C. campestris and on different parts of the shoots from different Cuscuta species using A. rhizogenes with and without the pRedRoot T‐DNA. Occasionally, transformed cells exhibiting an intense orange fluorescence from expression of the reporter protein dsRed have indeed been observed, particularly in shoot tips (Figure S1). However, no root development could be observed in any of our trials with the strain MSU440, independent of whether it carried the pRedRoot T‐DNA or not and transformation success reliability was poor.
3.2. A. rhizogenes transforms adhesive disk cells of C. reflexa with high efficiency
We next decided to expose developing infection structures to a pRedRoot‐containing A. rhizogenes culture. For this we used the method for induction of haustoriogenesis in C. reflexa described by Olsen et al. (2016) that uses a combination of far‐red light illumination and tactile stimuli to synchronize haustorial development (Tada et al., 1996). The stems on which the infection sites developed where exposed to A. rhizogenes for as long as it took for haustoria to emerge (7–8 days) (Figure 1). Upon microscopical analysis, a large number of intensely orange‐fluorescing cells expressing dsRed were revealed that were almost exclusively located in the adhesive disks around the protruding haustorium (Figure 2a–f). The dsRed fluorescence was visible in distinct spots often consisting of 5–15 clustered cells, but single transformed cells and bigger clusters were also observed (Figure 2). Cross sections through sites where transformation had occurred revealed that the cells expressing the dsRed were mostly not located at the very surface but rather in a cell layer directly below the elongated epidermal cells (Figure 2g–l). Fifty‐two percent of the adhesive disks exhibited one or more spots with dsRed fluorescence (based on 52 infected shoots with 426 infection structures) (Table 1), but there was considerable variation between individual shoots. While some green and blue autofluorescence was observed in the central haustorial tissue (Figure S2), the adhesive disk of C. reflexa exhibited little to no autofluorescence, as demonstrated by mock transformations with A. rhizogenes cells that lack the pRedRoot T‐DNA (Figure 2m,n). Experiments where A. rhizogenes was removed or added after 2 days, showed that the uptake of the T‐DNA in the first 2 days is minimal to absent, and seems to happen only once the development of the infection sites has commenced (Tables S1 and S2).
FIGURE 2.
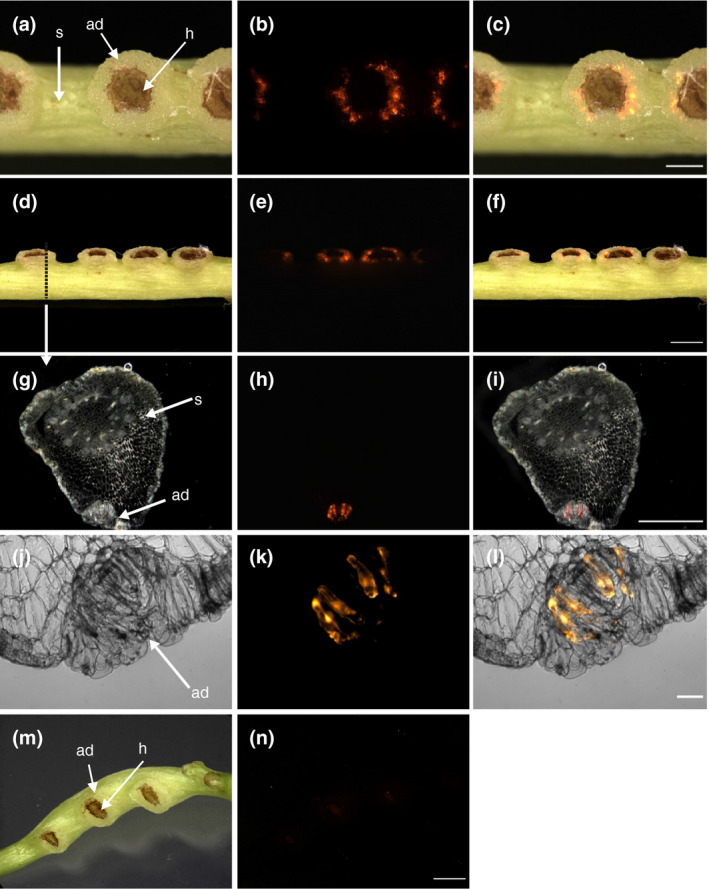
Transformation of C. reflexa adhesive disk cells by A. rhizogenes containing the binary vector pRedRoot. (a‐f) Intact infection sites after transformation. Topview (a‐c) and sideview (d‐f) of transformed adhesive disks are shown. (g‐l) Semi‐thin vibratome sections of transformed adhesive disk tissue in the region of the stippled line (in d) show subepidermal localization of transformed cells in the adhesive disk (ad). (m, n) Mock transformation with A. rhizogenes lacking the binary pRedRoot plasmid. Darkfield or brightfield pictures (first column) are shown alongside the fluorescence images taken with a Cy3 filter (middle column). Overlays of both are shown in the right column. Adhesive disks (ad), haustoria (h), and stems (s) are indicated by arrows. Scale bars are 1,000 µm (c and i), 2,000 µm (f and n), and 100 µm (l)
TABLE 1.
Transformation efficiency overview
| Number of shoots | # infection sites total | # adhesive disks transformed | Agrobacterium strain | Reporter fluorochrome |
|---|---|---|---|---|
| 6 | 61 | 46 | A. rhizogenes MSU440 | dsRed |
| 4 | 16 | 13 | A. rhizogenes MSU440 | dsRed |
| 6 | 37 | 33 | A. rhizogenes MSU440 | dsRed |
| 4 | 77 | 37 | A. rhizogenes MSU440 | dsRed |
| 8 | 41 | 11 | A. rhizogenes MSU440 | dsRed |
| 12 | 57 | 38 | A. rhizogenes MSU440 | dsRed |
| 12 | 137 | 43 | A. rhizogenes MSU440 | dsRed |
| 3 | 21 | 11 | A. tumefaciens GV3101 | GFP |
| 3 | 25 | 8 | A. tumefaciens GV3101 | GFP |
| 8 | 63 | 15 | A. tumefaciens GV3101 | GFP |
| 8 | 52 | 15 | A. tumefaciens GV3101 | GFP |
| 8 | 113 | 74 | A. tumefaciens GV3101 | GFP |
| 8 | 93 | 49 | A. tumefaciens GV3101 | GFP |
| 5 | 38 | 0 | A. rhizogenes MSU440 | None |
| 4 | 28 | 0 | A. rhizogenes MSU440 | None |
| 4 | 50 | 0 | A. rhizogenes MSU440 | None |
3.3. Adhesive disk cells maintain their integrity and viability during the transformation process
Life cell stains like CFDA are membrane permeable and are hydrolyzed in the cytoplasm to the green fluorescent carboxyfluorescein. In the present case, CFDA fluorescence in the adhesive disks would be evidence that the transformed cells maintain their integrity and viability during the treatment and transformation. When shoots that showed dsRed fluorescence in the adhesive disks where exposed to a CFDA solution, the adhesive disks and often also the haustoria exhibited green fluorescence (Figure 3). CFDA fluorescence was also observed regularly in young side‐shoot buds but only very rarely in the intact stems (Figure S1). The fluorescence was observed within 1 minute in the adhesive disks (Figure S3). This indicates that the cuticle that protects Cuscuta stems and that hinders the uptake of the stain into stems is most likely “leaky” or absent in the adhesive disks and haustoria, thus permitting the stain to enter the respective tissue with such efficiency.
FIGURE 3.
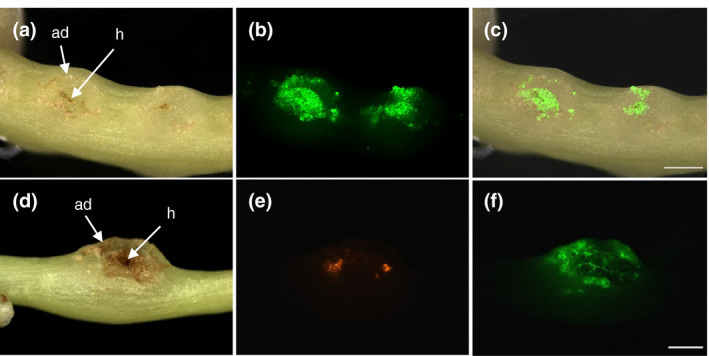
Uptake of 5‐Carboxyfluorescein‐diacetate (CFDA) into adhesive disks of C. reflexa. (a–c) Green CFDA fluorescence is shown in a shoot segment containing two developing infection sites. A superimposition of the images from (a) and (b) is shown in (c). (d–f) CFDA uptake into an adhesive disk containing dsRed‐expressing cells. Darkfield images (left column) and fluorescence images (middle column and lower right image) are shown. Adhesive disks (ad) and haustoria (h) are indicated. Scale bars are 1,000 µm
3.4. Application of the transformation protocol is not limited to A. rhizogenes
In order to reveal whether the high transformation rates were a result of a specific susceptibility of C. reflexa to A. rhizogenes, we exposed far‐red light induced stems to Agrobacterium tumefaciens carrying a GFP gene in the T‐DNA of a binary plasmid (Bobik et al., 2019) using the same setup. Only very weak background fluorescence was seen in this case in the orange channel and in the blue channel, while the high intensity of green fluorescence in a ring corresponding to the adhesive disk indicated that the GFP was expressed in this tissue as a result of the transformation (Figure 4A–I). As with the dsRed, GFP was expressed in elongated cells beneath the layer of epidermal cells in the adhesive disk (Figure S4). The infection frequency was on average 43% for A. tumefaciens (based on 38 infected shoots with 367 infection structures) (Table 1).
FIGURE 4.
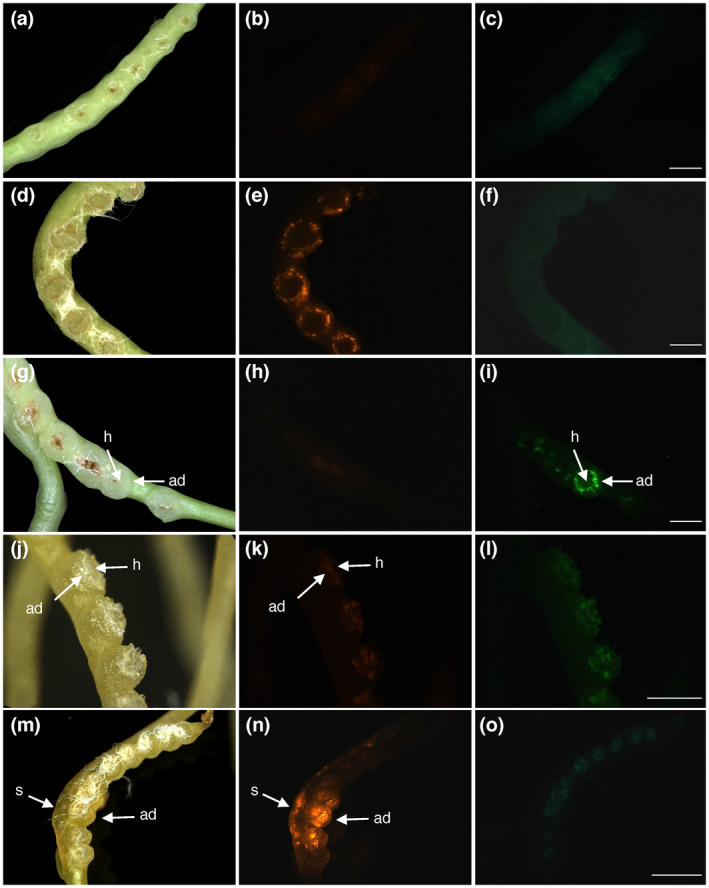
Extension of the protocol to A. tumefaciens and C. campestris. (a–f) Negative (a–c) and positive (d–f) controls using the combination of C. reflexa and A. rhizogenes (see also Figure 2). (g–i) Transformation after combining C. reflexa with A. tumefaciens containing a binary GFP‐expressing vector. (j–o) Negative control (j–l) and pRedRoot transformation (m–o) using the combination of C. campestris and A. rhizogenes. Scale bars represent 2,000 µm (c, f, i and o) and 1,000 µm (l), respectively. White fibers of the bench paper from the experimental setup can be seen adhering strongly to the adhesive disks in some darkfield images (d, g, j and m)
3.5. Transformation of other Cuscuta species
Within the genus Cuscuta, three subgenera are distinguished: subgenus Monogyna, which includes C. reflexa, subgenus Grammica, and subgenus Cuscuta (Yuncker, 1932). To test whether our protocol is applicable to C. campestris whose sequenced genome (Vogel et al., 2018) would make it a very interesting target for genome modifications, we repeated the same transformation setup with this species (Figure 4) and a third Cuscuta species, C. platyloba (Figure S1), both belonging to the subgenus Grammica. With both Agrobacterium species, a higher degree of necrotic tissue was observed in these two species as a result of this treatment, which, in turn, created a higher amount of unspecific autofluorescence. While adhesive disk transformation could be observed in C. campestris (Figure 4K–L), it was by far not as frequent as in C. reflexa and was often weaker than in stem tissue adjacent haustoria‐forming sites.
3.6. Simultaneous exposure to both Agrobacterium strains yields a high number of cotransformation events
A desired feature of transformation protocols is the possibility to express multiple transgenes in the same cell. This can be achieved by time‐consuming sequential transformation or the cloning of suites of genes into the T‐DNA of one vector, often yielding large unwieldy constructs. However, the high susceptibility of the same C. reflexa tissue to both, A. rhizogenes and A. tumefaciens, opens for the possibility of introducing multiple constructs into the same cell by co‐infection. To achieve this, both species of Agrobacterium carrying each their respective fluorescent reporter construct (dsRed in A. rhizogenes and GFP in A. tumefaciens) were mixed in a 1:1 ratio (based on their ODs at 595 nm) prior to exposing the C. reflexa stems to them in our transformation setup. Fluorescence microscopy revealed that both, dsRed and GFP, were visible with similar yields in the adhesive disks as in single transformation experiments. A considerable amount of overlapping fluorescence indicated that cotransformation did in fact occur at a high rate (Figure 5A–D). In order to see whether the same cells (and not just cells in the same area) indeed expressed both fluorescent proteins, we prepared semi‐thin cross sections through transformed regions and documented the fluorescence location with microphotography (Figure 5E–H) and by densitometry scanning of fluorescence intensities over an area containing several transformed clusters (Figure 5I, Figure S5). Both revealed an exact coincidence of the two fluorophores in several cells, suggesting that there are hot spots of susceptible tissue that is frequently co‐transformed. At the same time, the occurrence of cells transformed with only one fluorochrome shows that each fluorescence signal is specific.
FIGURE 5.
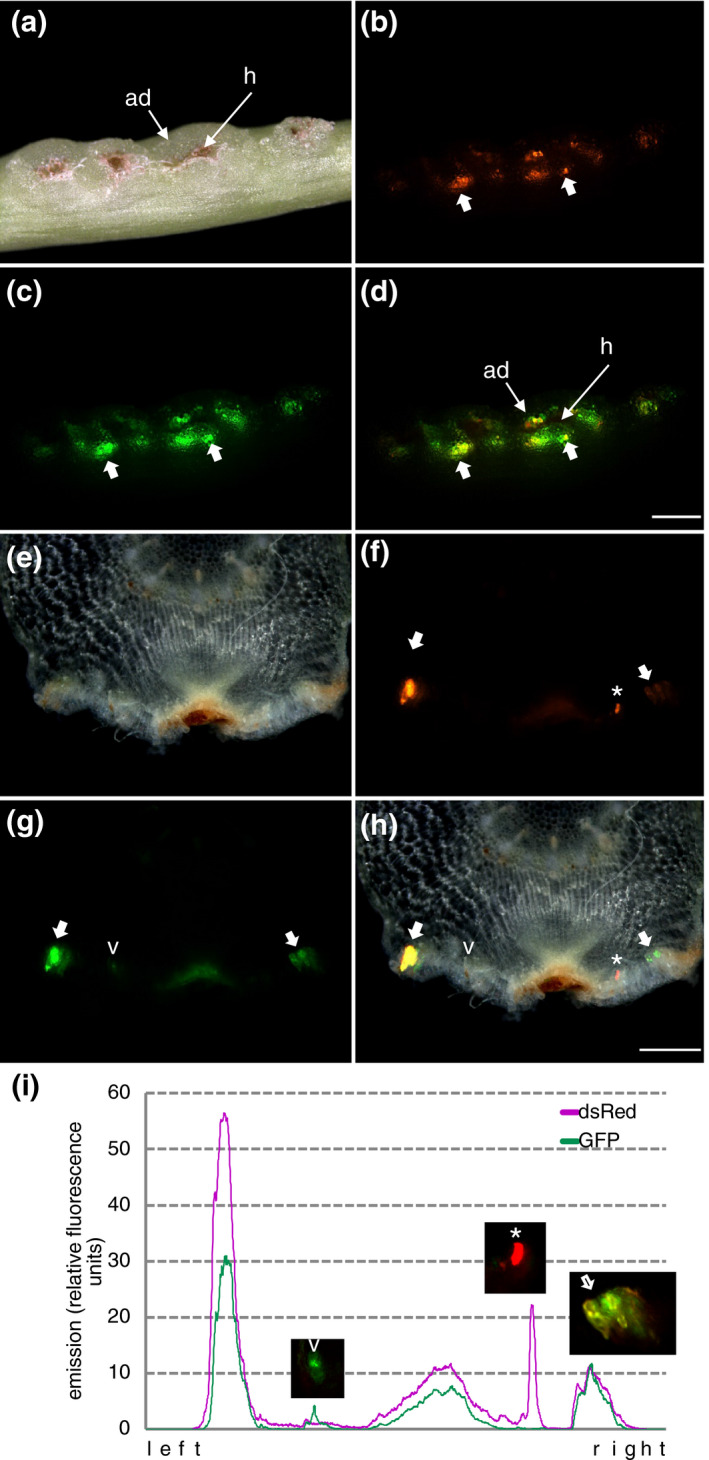
Cotransformation of different reporter proteins in C. reflexa adhesive disks. DsRed was introduced with A. rhizogenes while GFP was introduced with A. tumefaciens. (a–d) Side view of intact stem with darkfield (a), red fluorescence (b), and green fluorescence (c) images. Figure (d) shows a superimposed image of the two fluorescence images with thick arrows pointing at spots where both reporter proteins coincide (yellow color due to overlay). The scale bar measures 1,000 µm. (e–h) Cross section through a transformed infection site with darkfield (e), red fluorescence (f), and green fluorescence (g) images. Figure (h) shows a superimposed image of all three images. The scale bar measures 500 µm. The asterisk and the arrowhead indicate cells that are transformed with only one reporter protein. The thick arrow indicates cells were both reporter proteins coincide. (i) Intensity scan performed on the two single fluorescent images with the purple line representing the dsRed fluorescence and the green line representing the GFP fluorescence
3.7. Transgene expression after a transformation event is preserved over several weeks
The frequent occurrence of transformed cell clusters raised the question whether these arose through cell division and propagation of single transformed cells, indicating not only a stable insertion of the transgenes but, moreover, also the possibility to regenerate vegetative or reproductive tissue by in vitro propagation from the transformation events. To test this, we sterilized explants containing transformed tissue and maintained them in in vitro cultures. The explants showed slight growth of cells at the edges of the adhesive disks, including the transformed regions, but significant propagation was not observed over a period of up to 8 weeks. The fluorescence was consistently high for at least 4 weeks (Figure 6) but started to decrease during longer cultivation times.
FIGURE 6.
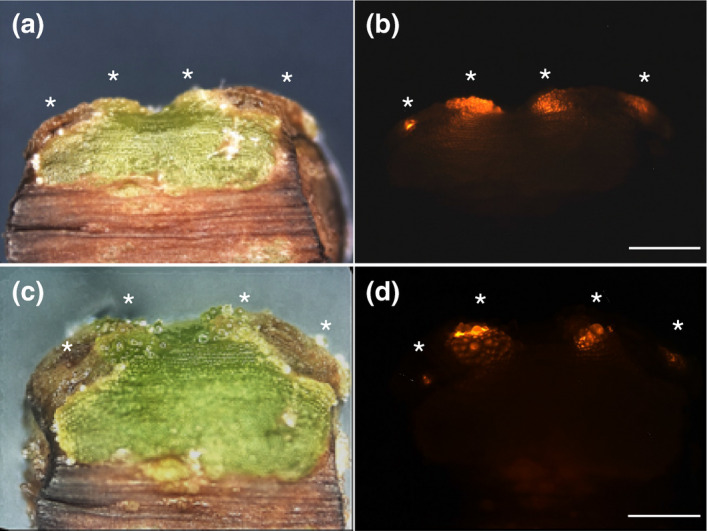
Cultivation of pRedRoot‐expressing explants from C. reflexa. The shown explant contains two infection structures with transformed adhesive disks. Asterisks indicate clusters of transformed cells. Images were taken after 5 days (a and b) and 4 weeks (c–d) in sterile culture. Brightfield images are shown next to the red fluorescence. The scale bars represent 1,000 µm
4. DISCUSSION
In the last few years, research on Cuscuta has seen a rapid increase and with the publication of the first two genome sequences of C. campestris (Vogel et al., 2018) and C. australis (Sun et al., 2018), our knowledge on these parasites has experienced a significant leap forward. Nevertheless, there are still several obstacles that need to be overcome before the possibilities offered by these genomes can be fully exploited. The main bottleneck is the lack of an efficient and reproducible protocol with which transgene expression or even genetic manipulations can be performed. Previous attempts to transform Cuscuta have shown that A. tumefaciens is able to transform callus cells (Borsics et al., 2002; Svubova and Blehova, 2013). However, in our hands these transformation events were very scarce, explaining maybe why this approach has not yielded greater success.
With the work presented here, we show that transformation of Cuscuta reflexa works very efficiently when developing infection sites are targeted. As our experiments show, the protocol works with both A. rhizogenes and A. tumefaciens, and allows the expression of reporter proteins from different binary vectors and under the control of different promoters. The T‐DNA of pRedRoot, a binary vector developed for A. rhizogenes, contains the dsRed protein under control of the Ubi10 promoter (Libiakova et al., 2018), while the A. tumefaciens line used by Bobik et al. (2019) contains a GFP gene controlled by the 35S promoter. Both promoters allow strong constitutive expression of transgenes. It remains to be shown if other – in particular inducible – promoters work in Cuscuta as well as the standard constitutive promoters do, or if adaptations are required for use with the parasite. The simplicity of the method (low‐tech, cheap, and easy‐to‐implement) (Figure 1) and the high reproducibility make it suitable for larger‐scale screening approaches or to multiplex functional studies. The high numbers of transformed cells and the longevity of transgene expression, furthermore, permit investigations of transgene product behavior, protein localization or secretion over time, and in situ interaction assays between molecules or cells.
The high transformation rates specifically of adhesive disk cells are certainly noteworthy. These cells are naturally in close contact with the host and may thus exhibit a higher propensity to take up different types of substances, but the same is true for the haustorium cells which did not show the same aptness for transformation. It is not yet clear what structural or metabolic characteristics are responsible for this behavior, but some speculative scenarios are more likely than others and shall be discussed here.
Agrobacteria are known to be attracted to polyphenolic substances exuded by plant roots (Ozyigit et al., 2013). Haustorial sites are also rich in hydroxycinnamic acid derivatives, caffeic acid depsides and other polyphenols (Löffler et al., 1995), which is evident by the rapid browning of infection sites upon their exposure to air (Johnsen et al., 2015). Comparably high concentrations of phenolic substances, albeit in a slightly different composition, are interestingly also found in the meristematic apex of Cuscuta shoots (Löffler et al., 1995). After adhesive disks, shoot tips were the second tissue that showed a heightened susceptibility toward agrobacterial infection tentatively suggesting that infection success can in part be explained by a metabolically driven attraction of the agrobacteria to these sites. However, attraction by polyphenols does not explain why only the adhesive disks and not the haustoria are transformed as the latter exhibit high phenolic substance production as well (Löffler et al., 1995). Both, haustoria as well as the surrounding adhesive disks, are characterized in general by a high metabolic activity, so the differential transgene expression is likely not caused by differences in transcription and translation activities. The fluorescent cell integrity indicator CFDA stains adhesive disk cells when applied externally to the surface (Figure 3 and Figure S3), indicating their viability throughout the transformation process. The uptake of the CFDA happens with such high speed (Figure S3) that it is reasonable to conclude that the protective cuticular layer that ostensibly slows down its uptake in stems is altered or missing. Although CFDA follows a different uptake route than the T‐DNA, which is delivered by agrobacterial cells through a type IV secretion system, the similarities in staining and transformation patterns suggest that the nature of the surface and its permeability may influence the ability of agrobacteria to infect the cells beneath. This as well as the presence of potentially other mechanisms in Cuscuta that can regulate agrobacterial infection success should be investigated further in the future.
None of the species that belong to the genus Cuscuta possesses roots and there are, therefore, no obvious natural targets of the plant pathogenic bacterium Agrobacterium rhizogenes. Nevertheless, some Cuscuta species were recently shown to have acquired a gene coding for the Mikimopine synthase (mis gene) (Zhang et al., 2020) that is typically transferred to plants during A. rhizogenes infection in order to supply the bacterium with opines. Plant homologues of the mis gene are found only in a handful of plant species belonging to the genera Nicotiana and Linnaria where they are believed to have originated by three independent horizontal gene transfer (HGT) events (Kovacova et al., 2014). These species do not display the hairy root syndrome, which hypothetically could be attributed to the HGT‐derived mis gene. By way of small interfering RNAs from the HGT‐derived mis that may degrade T‐DNA‐borne mis transcripts during an infection, these acquired genes could potentially prevent A. rhizogenes growth. Their evolution under selective pressure and the coverage of mis‐derived siRNAs, at least, seem to corroborate this possibility for Nicotiana (Kovacova et al., 2014). It can be debated whether this could also be the case in Cuscuta. However, the mis gene was so far only found in species belonging to the subgenus Grammica (Zhang et al., 2020), but was not detected in a transcriptome database (Olsen et al., 2016) of C. reflexa. Therefore, it is more likely that the loss of key genes involved in root development is responsible for the failure to produce hairy roots in A. rhizogenes‐infected Cuscuta tissue.
5. CONCLUSIONS
Using fluorescent reporter proteins and far‐red light mediated infection structure induction, we have shown that the adhesive disk of C. reflexa is highly susceptible to Agrobacterium‐mediated transformation. With the high number of transformation events that were observed using our protocol and with the stability of transgene expression, it will be possible to perform transformations with a high number of constructs. Our trials with other Cuscuta species showed some transformation success but species‐specific adaptations may be required to apply the technique to other dodder species. Our protocol is very useful to provide further functional insight on Cuscuta and its enigmatic infection structure because it is cheap, low‐tech, easily scalable, and suitable for transformations of genes at low or high throughput. Moreover, if the transformed cells can be induced to produce callus and ultimately whole regenerated plants, it will enable plant scientists to harness the genome sequence information and create Cuscuta mutants.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
AUTHOR CONTRIBUTIONS
L.G. and K.K. designed the transformation setup; L.L. and L.G. performed the transformation experiments; L.L. performed the life cell staining experiments, evaluated all data, and performed image analyses; K.K. conceived the project and wrote the article with contributions of all the authors.
Supporting information
Fig S1‐S5
Table S1‐S2
ACKNOWLEDGMENTS
This work is part of the doctoral thesis of LL. Financial support from the Tromsø Research Foundation (Mohn Foundation) (grant 16‐TF‐KK to KK) and the Department of Arctic and Marine Biology (AMB, UiT The Arctic University of Norway) is gratefully acknowledged. We thank Prof. Harro Bouwmeester (University of Amsterdam, Netherlands) for stimulating discussions that provided the initial impulse to this study. We further thank the “Norway‐Armenia cooperation in plant molecular biology and biotechnology for agricultural development” project (CPEA‐LT‐2016/10092), funded by the Eurasia program, Norwegian Agency for International Cooperation and Quality Enhancement in Higher Education (DIKU) for supporting the research visit of LG to Tromsø. Staff at the Climate Laboratory Holt (especially Leidulf Lund) is thanked for Cuscuta maintenance. Agrobacterium strains and binary reporter plasmids were kindly provided by Prof. Tessa Burch‐Smith (University of Tennessee, USA) and Prof. Harro Bouwmeester (University of Amsterdam, Netherlands). Alessandra Guerrieri (University of Amsterdam, Netherlands) is thanked for sharing tips on agrobacterial growth and nonparasitic plant transformation. Prof. Karsten Fischer and Dr. Stian Olsen (both from UiT The Arctic University of Tromsø, Norway) have contributed with numerous suggestions and discussions to the work and the manuscript.
Anna-Maria Lachner L, Galstyan L, Krause K. A highly efficient protocol for transforming Cuscuta reflexa based on artificially induced infection sites. Plant Direct. 2020;4:1–11. 10.1002/pld3.254
Link to the manuscript on the biorxiv preprint server: https://doi.org/10.1101/2020.04.06.028191
REFERENCES
- Alagarsamy, K. , Shamala, L. F. , & Wei, S. (2018). Protocol: High‐efficiency in‐planta Agrobacterium‐mediated transgenic hairy root induction of Camellia sinensis var. sinensis . Plant Methods, 14, 17 10.1186/s13007-018-0285-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahramnejad, B. , Naji, M. , Bose, R. , & Jha, S. (2019). A critical review on use of Agrobacterium rhizogenes and their associated binary vectors for plant transformation. Biotechnology Advances, 37, 107405 10.1016/j.biotechadv.2019.06.004 [DOI] [PubMed] [Google Scholar]
- Bobik, K. , Fernandez, J. C. , Hardin, S. R. , Ernest, B. , Ganusova, E. E. , Staton, M. E. , & Burch‐Smith, T. M. (2019). The essential chloroplast ribosomal protein uL15c interacts with the chloroplast RNA helicase ISE2 and affects intercellular trafficking through plasmodesmata. New Phytologist, 221, 850–865. 10.1111/nph.15427 [DOI] [PubMed] [Google Scholar]
- Borsics, T. , Mihalka, V. , Oreifig, A. S. , Barany, I. , Lados, M. , Nagy, I. , … Toldi, O. (2002). Methods for genetic transformation of the parasitic weed dodder (Cuscuta trifolii Bab. et Gibs) and for PCR‐based detection of early transformation events. Plant Science, 162, 193–199. 10.1016/S0168-9452(01)00536-2 [DOI] [Google Scholar]
- Cui, W. , Wang, X. , & Lee, J. Y. (2015). Drop‐ANd‐See: A simple, real‐time, and noninvasive technique for assaying plasmodesmal permeability. Methods in Molecular Biology, 1217, 149–156. 10.1007/978-1-4939-1523-1_10 [DOI] [PubMed] [Google Scholar]
- Förste, F. , Mantouvalou, I. , Kanngiesser, B. , Stosnach, H. , Lachner, L. A. , Fischer, K. , & Krause, K. (2020). Selective mineral transport barriers at Cuscuta‐host infection sites. Physiologia Plantarum, 168 (4), 934–947. 10.1111/ppl.13035 [DOI] [PubMed] [Google Scholar]
- Galloway, A. F. , Knox, P. , & Krause, K. (2020). Sticky mucilages and exudates of plants: Putative microenvironmental design elements with biotechnological value. New Phytologist, 225, 1461–1469. 10.1111/nph.16144 [DOI] [PubMed] [Google Scholar]
- Gelvin, S. B. (2003). Agrobacterium‐mediated plant transformation: The biology behind the "gene‐jockeying" tool. Microbiology and Molecular Biology Reviews, 67, 16–37. 10.1128/MMBR.67.1.16-37.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho‐Plagaro, T. , Huertas, R. , Tamayo‐Navarrete, M. I. , Ocampo, J. A. , & Garcia‐Garrido, J. M. (2018). An improved method for Agrobacterium rhizogenes‐mediated transformation of tomato suitable for the study of arbuscular mycorrhizal symbiosis. Plant Methods, 14, 34 10.1186/s13007-018-0304-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen, H. R. , Striberny, B. , Olsen, S. , Vidal‐Melgosa, S. , Fangel, J. U. , Willats, W. G. , … Krause, K. (2015). Cell wall composition profiling of parasitic giant dodder (Cuscuta reflexa) and its hosts: A priori differences and induced changes. New Phytologist, 207, 805–816. 10.1111/nph.13378 [DOI] [PubMed] [Google Scholar]
- Kaiser, B. , Vogg, G. , Furst, U. B. , & Albert, M. (2015). Parasitic plants of the genus Cuscuta and their interaction with susceptible and resistant host plants. Frontiers in Plant Science, 6, 45 10.3389/fpls.2015.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacova, V. , Zluvova, J. , Janousek, B. , Talianova, M. , & Vyskot, B. (2014). The evolutionary fate of the horizontally transferred agrobacterial mikimopine synthase gene in the genera Nicotiana and Linaria . PLoS One, 9, e113872 10.1371/journal.pone.0113872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. B. (2007). Structure and development of the upper haustorium in the parasitic flowering plant Cuscuta japonica (Convolvulaceae). American Journal of Botany, 94, 737–745. 10.3732/ajb.94.5.737 [DOI] [PubMed] [Google Scholar]
- Libiakova, D. , Ruyter‐Spira, C. , Bouwmeester, H. J. , & Matusova, R. (2018). Agrobacterium rhizogenes transformed calli of the holoparasitic plant Phelipanche ramosa maintain parasitic competence. Plant Cell, Tissue and Organ Culture (PCTOC), 135, 321–329. 10.1007/s11240-018-1466-x [DOI] [Google Scholar]
- Limpens, E. , Ramos, J. , Franken, C. , Raz, V. , Compaan, B. , Franssen, H. , … Geurts, R. (2004). RNA interference in Agrobacterium rhizogenes‐transformed roots of Arabidopsis and Medicago truncatula . Journal of Experimental Botany, 55, 983–992. 10.1093/jxb/erh122 [DOI] [PubMed] [Google Scholar]
- Löffler, C. , Sahm, A. , Wray, V. , Czygan, F.‐C. , & Proksch, P. (1995). Soluble phenolic constituents from Cuscuta reflexa and Cuscuta platyloba . Biochemical Systematics and Ecology, 23, 121–128. 10.1016/0305-1978(95)93846-U [DOI] [Google Scholar]
- Nagar, R. , Singh, M. , & Sanwal, G. G. (1984). Cell wall degrading enzymes in Cuscuta reflexa and its hosts. Journal of Experimental Botany, 35, 8 10.1093/jxb/35.8.1104 [DOI] [Google Scholar]
- Olsen, S. , Striberny, B. , Hollmann, J. , Schwacke, R. , Popper, Z. , & Krause, K. (2016). Getting ready for host invasion: Elevated expression and action of xyloglucan endotransglucosylases/hydrolases in developing haustoria of the holoparasitic angiosperm Cuscuta . Journal of Experimental Botany, 67, 695–708. 10.1093/jxb/erv482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozyigit, I. I. , Dogan, I. , & Tarhan, E. A. (2013). Agrobacterium rhizogenes‐mediated transformation and its biotechnological applications in crops In Hakeem K. R., Ahmad P., & Ozturk M. (Eds.), Crop improvement (pp. 1–48). New York, Dordrecht, Heidelberg, London: Springer Science + Business Media. [Google Scholar]
- Ron, M. , Kajala, K. , Pauluzzi, G. , Wang, D. , Reynoso, M. A. , Zumstein, K. , … Brady, S. M. (2014). Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type‐specific gene expression and function using tomato as a model. Plant Physiology, 166, 455–469. 10.1104/pp.114.239392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K. , & Aoki, K. (2019). Development of parasitic organs of a stem holoparasitic plant in genus Cuscuta . Frontiers in Plant Science, 10, 1435 10.3389/fpls.2019.01435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, G. , Xu, Y. , Liu, H. , Sun, T. , Zhang, J. , Hettenhausen, C. , … Wu, J. (2018). Large‐scale gene losses underlie the genome evolution of parasitic plant Cuscuta australis . Nature Communications, 9, 2683 10.1038/s41467-018-04721-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svubova, R. , & Blehova, A. (2013). Stable transformation and actin visualization in callus cultures of dodder (Cuscuta europaea). Biologia, 68, 633–640. 10.2478/s11756-013-0188-0 [DOI] [Google Scholar]
- Tada, Y. , Sugai, M. , & Furuhashi, K. (1996). Haustoria of Cuscuta japonica, a holoparasitic flowering plant, are induced by the cooperative effects of raf‐red light and tactile stimuli. Plant and Cell Physiology, 37, 1049–1053. 10.1093/oxfordjournals.pcp.a029052 [DOI] [Google Scholar]
- Vaughn, K. C. (2002). Attachment of the parasitic weed dodder to the host. Protoplasma, 219, 227–237. 10.1007/s007090200024 [DOI] [PubMed] [Google Scholar]
- Vogel, A. , Schwacke, R. , Denton, A. K. , Usadel, B. , Hollmann, J. , Fischer, K. , … Krause, K. (2018). Footprints of parasitism in the genome of the parasitic flowering plant Cuscuta campestris . Nature Communications, 9, 2515 10.1038/s41467-018-04344-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuncker, T. G. (1932). The genus Cuscuta . Memoirs of the Torrey Botanical Club, 18, 113–331. [Google Scholar]
- Zhang, Y. , Wang, D. , Wang, Y. , Dong, H. , Yuan, Y. , Yang, W. , … Li, Z. (2020). Parasitic plant dodder (Cuscuta spp.): A new natural Agrobacterium‐to‐plant horizontal gene transfer species. Science China Life Sciences, 63, 312–316. 10.1007/s11427-019-1588-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S5
Table S1‐S2


