The symbiotic bacterium Sinorhizobium meliloti contributes greatly to growth of the agriculturally valuable host plant alfalfa by fixing atmospheric nitrogen. Chemotaxis of S. meliloti cells toward alfalfa roots mediates this symbiosis. The present study establishes programmed proteolysis as a factor in the maintenance of the S. meliloti chemotaxis system. Knowledge about cell cycle-dependent, targeted, and selective proteolysis in S. meliloti is important to understand the molecular mechanisms of maintaining a suitable chemotaxis response. While the role of regulated protein turnover in the cell cycle progression of Caulobacter crescentus is well understood, these pathways are just beginning to be characterized in S. meliloti. In addition, our study should alert about the cautionary use of epitope tags for protein quantification.
KEYWORDS: ClpXP protease, alphaproteobacteria, cell cycle, chemotaxis, epitope tags
ABSTRACT
Chemotaxis and motility are important traits that support bacterial survival in various ecological niches and in pathogenic and symbiotic host interaction. Chemotactic stimuli are sensed by chemoreceptors or methyl-accepting chemotaxis proteins (MCPs), which direct the swimming behavior of the bacterial cell. In this study, we present evidence that the cellular abundance of chemoreceptors in the plant symbiont Sinorhizobium meliloti can be altered by the addition of several to as few as one amino acid residues and by including common epitope tags such as 3×FLAG and 6×His at their C termini. To further dissect this phenomenon and its underlying molecular mechanism, we focused on a detailed analysis of the amino acid sensor McpU. Controlled proteolysis is important for the maintenance of an appropriate stoichiometry of chemoreceptors and between chemoreceptors and chemotactic signaling proteins, which is essential for an optimal chemotactic response. We hypothesized that enhanced stability is due to interference with protease binding, thus affecting proteolytic efficacy. Location of the protease recognition site was defined through McpU stability measurements in a series of deletion and amino acid substitution mutants. Deletions in the putative protease recognition site had similar effects on McpU abundance, as did extensions at the C terminus. Our results provide evidence that the programmed proteolysis of chemotaxis proteins in S. meliloti is cell cycle regulated. This posttranslational control, together with regulatory pathways on the transcriptional level, limits the chemotaxis machinery to the early exponential growth phase. Our study identified parallels to cell cycle-dependent processes during asymmetric cell division in Caulobacter crescentus.
IMPORTANCE The symbiotic bacterium Sinorhizobium meliloti contributes greatly to growth of the agriculturally valuable host plant alfalfa by fixing atmospheric nitrogen. Chemotaxis of S. meliloti cells toward alfalfa roots mediates this symbiosis. The present study establishes programmed proteolysis as a factor in the maintenance of the S. meliloti chemotaxis system. Knowledge about cell cycle-dependent, targeted, and selective proteolysis in S. meliloti is important to understand the molecular mechanisms of maintaining a suitable chemotaxis response. While the role of regulated protein turnover in the cell cycle progression of Caulobacter crescentus is well understood, these pathways are just beginning to be characterized in S. meliloti. In addition, our study should alert about the cautionary use of epitope tags for protein quantification.
INTRODUCTION
A number of bacterial species can transition between motile and sessile lifestyles. Factors that influence this behavior include nutrient availability, cell density, the presence of solid surfaces, and their eukaryotic hosts (1–5). Many bacteria use flagella for motility to travel within their environment. By controlling flagellar rotational mode, bacteria can move toward beneficial attractants and away from detrimental repellents in a process called chemotaxis (6).
Sinorhizobium meliloti is a soil-dwelling alphaproteobacterium that has the ability to live freely in the soil, in a biofilm on the surface of plant roots, or as a differentiated bacteroid within specialized root organs called nodules (7, 8). The symbiosis of S. meliloti with select legumes involves multiple developmental changes, leading to the final development of nitrogen-fixing bacteroids (8–10). Biological nitrogen fixation provides the host plant with ammonia, which greatly promotes plant growth. Before the symbiotic relationship can be initiated, the bacterial symbiont uses chemotaxis to sense chemical compounds exuded by the host, moves toward host roots, and colonize them (11). The chemotaxis process is mediated by a two-component signaling system using methyl-accepting chemotaxis proteins (MCPs) as chemoreceptors (for recent reviews, see Wadhams et al. [6], Porter et al. [12], Hazelbauer et al. [13] and Salah Ud-Din et al. [14]). Typically, MCPs are integral membrane proteins with a periplasmic ligand binding domain (LBD), two transmembrane helices, and a cytoplasmic signaling domain. The LBDs are highly variable between receptors; this variation is reflected in a diverse set of ligands (14, 15). In contrast, the cytoplasmic domains are highly conserved, allowing them to interact with the same set of proteins in the cytoplasm. MCPs assemble into a highly organized array architecture at the cell poles that allows for cooperativity between receptors within the chemotaxis system (16, 17). Binding of a ligand to the LBD changes the conformation of the MCP, thereby altering the activity of the histidine autokinase CheA. Phosphorylated CheA transfers its phosphate group to the response regulator CheY, which interacts with components of the flagellar motor. This alters its rotational mode, resulting in a change in swimming behavior. CheR and CheB are two enzymes utilized to maintain baseline activity of CheA. CheR is a constitutively active methyltransferase, which adds methyl groups to conserved glutamate residues on the cytoplasmic signaling domain of MCPs. CheB serves as an antagonistic methylesterase when activated through phosphorylation by CheA-P.
Since the motility and chemotaxis machinery is not required during sessile life or in planta, protein production and degradation in S. meliloti are strictly regulated (18–20). One of the most important regulators in this transcriptional hierarchy is Rem. This OmpR-like protein is a class IB regulator, controlled by the LuxR-type master regulator VisNR (class IA), and is only synthesized during the exponential growth phase (18, 19). Rem controls expression of class II motility genes, which dictate if class III genes are expressed. Class III genes are responsible for assembly of the flagellum and the chemotaxis machinery (11, 19). It is vital for bacterial survival that cellular differentiation processes and cell cycle regulation are closely linked. Thus, the cell cycle master regulator CtrA indirectly regulates the majority of genes involved in chemotaxis and motility and also directly controls four MCPs and three flagellin-encoding genes (21).
Alphaproteobacteria employ a regulatory network in cell cycle-dependent differentiation and expression of motility genes (22–25). In Caulobacter crescentus, a freshwater alphaproteobacterium with a motile and a sessile lifestyle, CtrA is regulated through phosphorylation and targeted proteolysis upon the onset of DNA replication by ClpXP (26). In addition, CtrA-dependent proteins in C. crescentus, such as the chemoreceptors McpA and McpB, are also targets of ClpXP and are degraded during the course of the cell cycle (27, 28). Notably, C. crescentus expresses two chemotaxis clusters (referred to as major and alternate), both of which play a role in biofilm formation. However, only the major chemotaxis cluster is cell cycle regulated and involved in the chemotactic response (29). Furthermore, the degradation of proteins in C. crescentus is spatially organized through the regulatory protein CpdR, which controls polar localization of the protease (30, 31). In S. meliloti, CtrA is a key protein in cell cycle progression; its cellular abundance is regulated not only on the transcriptional level but also posttranslationally through proteolysis. While ctrA is an essential gene in S. meliloti, mutations prohibiting CtrA proteolysis are also lethal (32, 33). CtrA is unstable during exponential growth, and its degradation is regulated by the DifJ/PleC-like histidine kinase CbrA (31, 34). Proper timing of the S. meliloti cell cycle is critical to cell fate during asymmetric cell division and in symbiosis (31, 35, 36).
The protease complex ClpXP is strictly required for growth and normal cell division in C. crescentus, and the ClpX subunit has been shown to also be essential in S. meliloti (20, 31, 37). This ubiquitous AAA+ protease uses ATP binding and hydrolysis to degrade proteins within the cell. During regulatory proteolysis, ClpX targets specific degradation motifs, termed degrons, unfolds the target protein into a useable substrate, and delivers it to ClpP, which hydrolyzes the unfolded chain. ClpXP has been studied as model for other AAA+ proteases like ClpAP, ClpCP, and HslUV (38). Degrons for ClpXP are accessible and are preferentially positioned at the N or C terminus of a target protein (39). One well-characterized degron is the C-terminal SsrA tag, which is 11 amino acid residues (sequence: AANDENYALAA) in length. It is fused to nascent polypeptides to rescue a stalled ribosome that is in the process of translating a truncated mRNA (40). While this system was discovered in Escherichia coli, it is used in almost all bacteria and in some organelles to specifically direct the marked substrates to ClpXP and ClpAP. Binding of the SsrA-tagged substrates to the principal protease ClpXP is specifically mediated through interactions between the last three residues (LAA) of the SsrA tag and the ClpX subunit (38, 40, 41). This short motif at the C terminus appears to be common to an entire class of degrons. Several bacterial species use ClpXP to degrade chemotaxis-related proteins, such as CtrA, McpA, and McpB in C. crescentus and CheW in E. coli and Bacillus subtilis (42–44). C. crescentus McpA and McpB both possess hydrophobic regions characterized by alanine residues near the C terminus, which are commonly found in degrons (27, 28, 45). However, little is known about the degradation of chemotaxis and flagellar proteins by ClpXP in S. meliloti, with the exception of one study that has identified flagellar proteins associated with ClpXP through coimmunoprecipitation experiments. The same study reported a link between CtrA and the protease HslUV (20).
In this work, we provide evidence that chemotaxis proteins in S. meliloti undergo proteolysis through protease recruitment by specific recognition sites near their C terminus and that this targeted proteolysis is cell cycle dependent. Using a combinatory approach of quantitative immunoblot assays and targeted mutagenesis, we showed that the C-terminal fusion of epitope tags frequently used for protein detection, as well as the addition of single amino acid residues, enhance the stability of chemotaxis proteins. Focusing on the amino acid receptor McpU, we further dissected and defined the protease recognition site near the C terminus. Knowledge accrued from this study will aid in a better understanding of the regulation of the chemotaxis machinery during cell cycle progression and inform future research using epitope tags for cellular protein quantification in alphaproteobacteria.
RESULTS
C-terminal fusion of commonly used epitope tags increases the abundance of McpU in S. meliloti.
During our analysis of chemoreceptor stoichiometry in S. meliloti (46), we attempted to utilize epitope tags for the assisted quantification of individual MCPs. However, we observed that the fusion of a 3×FLAG tag to the C terminus of McpU, which adds 22 amino acid residues, substantially increased its abundance. To determine whether this phenomenon was specific to the 3×FLAG tag, we constructed two more epitope tag strains, one expressing McpU with a 1×FLAG tag (8 amino acids; BS222) and one with another commonly used tag, the 6-histidine (6×His) tag (6 amino acids; BS220) (Fig. 1). In each case, abundance of McpU bearing a C-terminal epitope tag was greater than that of native McpU in the wild-type strain (RU11/001). Next, we used comparative immunoblotting with purified anti-McpU polyclonal antibodies to quantify the amount of McpU in each mutant strain. The McpU band intensity was compared directly either to the intensity of native McpU or to that of mutant strain lysates with added lysates of an mcpU deletion strain (RU11/828) in known ratios (Fig. 2 and 3). This experimental approach kept the amount of cellular material in each cell lysate constant, which prevented the overestimation of McpU in mutant strains (47). The increase in McpU abundance was expressed as a fold change compared to the amount of McpU in the wild type (Fig. 4). All three tags significantly increased the amount of McpU; the 3×FLAG and 6×His tags increased abundance by 6.2-fold (±0.8) and 6.0-fold (±1.9), respectively, and the 1×FLAG tag caused an 8.2-fold (±1.8) increase over that of the wild type.
FIG 1.
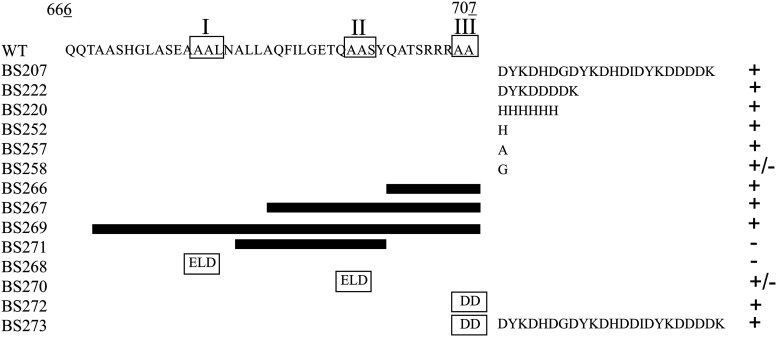
Mutational analysis of the C-terminal region of McpU and its effect on stability. Depicted is a diagram of the C-terminal region of McpU (amino acids [aa] 666 to 707) and the extensions, deletions, and substitutions made in its native chromosomal locus (Table 1). Boxes in the wild-type sequence and the respective mutants indicate specific motifs and their substitutions, respectively. Black bars represent extent of deletions. The column on the right symbolizes McpU abundance relative to that in the wild type; +, increase, −, decrease; +/−, no change.
FIG 2.
Representative immunoblots for direct comparison of McpU abundance in cell lysates of wild type compared to that in mutant strains. Each lane contains cell lysates from 1 ml of culture at an optical density at 600 nm (OD600) of 0.25. Each panel is the result of different film exposures; therefore, comparisons can be made only within a panel.
FIG 3.
Representative immunoblots used to quantify the relative abundance of McpU in cell lysates of the wild type compared to mutant strains. Lanes 1 to 3 contain RU11/001 lysates (WT) from 1 ml of culture at an OD600 of 0.25. Lanes denoted by a wedge shape contain a mix of each mutant strain and RU11/828 (ΔmcpU) at ratios of 1:1, 3:1, 5.7:1, 7:1, and 9:1, yielding a total volume of 1 ml of culture at an OD600 of 0.25. The last lane in each panel contains lysates from 1 ml of culture of the respective mutant strain at an OD600 of 0.25.
FIG 4.
Relative abundance of McpU in mutant strains compared to that in the wild type. Asterisks indicate quantifications obtained through direct comparison (Fig. 2); all other values were obtained through generation of a standard curve (Fig. 3). The dashed line represents wild-type abundance of McpU. Values and error bars are the means and standard deviations from three biological replicates. Statistical significance was determined by a two-tailed Student t test (P < 0.05). All values but those marked by an octothorpe denote statistically significant differences from the wild type.
Addition of the single amino acid residue histidine or alanine but not glycine increases McpU abundance.
To test if less extensive modifications such as single amino acid additions would elicit an increase in McpU abundance, three mutant strains were constructed that added a single histidine (BS252), alanine (BS257), or glycine residue (BS258) to the C terminus of McpU (Fig. 1). BS252 and BS257 both exhibited a significant increase in McpU abundance, by 4.7-fold (±1.0) and 3.2-fold (±0.9), respectively (Fig. 3). In contrast, BS258 displayed no significant difference in McpU abundance (Fig. 2). In conclusion, changes of the C terminus as minor as the addition of an individual amino acid residue can increase McpU stability. However, it appears that the size and/or nature of the modification is important, as the addition of an amino acid without side chain did not alter McpU abundance.
Deciphering the putative protease recognition site in the C-terminal domain of McpU.
Evidence from McpU epitope tag fusions and single amino acid additions suggested that the C terminus of McpU is critical in controlling its abundance, which determined that the protein is specifically targeted by a protease. To decipher the mechanism of target recognition, we identified three potential protease recognition sites based on evidence from McpA and McpB in C. crescentus and known E. coli degrons (48). In C. crescentus McpA and McpB, a short, alanine-rich motif is required for its degradation by ClpXP (27, 28). We found two such putative protease recognition motifs in the C-terminal domain of McpU, namely, motif I (AAL) and motif II (AAS) (Fig. 1). In analogy to Tsai and Alley (2001) (28), both motifs were mutated to ELD, resulting in mutant strains BS268 and BS270. A third motif, named motif III (AA) and encompassing the last two residues, was mutated to DD (BS272) (Fig. 1). While motif I and II variants were less (0.2-fold [±0.2]) or equally (1.5-fold [±0.3]) stable than the wild-type McpU, the motif III variant exhibited a small but significant increase in protein abundance by about 2.2-fold (±0.2) (Fig. 2). However, the increase was about three times smaller than that of the maximally stabilizing tag fusion variants (Fig. 3). Finally, we tested whether a combination of the motif III variant with the 3×FLAG epitope fusion had an additive effect on McpU stability (BS273), but this was not the case. The dialanine mutation in combination with the 3×FLAG tag increased McpU stability approximately to the same level as that of the 3×FLAG epitope fusion alone, namely, 4.8-fold (±3.1) (Fig. 3). In conclusion, the C-terminal dialanine (di-Ala peptide) is involved in McpU degradation, while the two other putative protease recognition sites are not.
Identifying regions in the C-terminal domain affecting McpU stability.
Since specific mutations in the three protease recognition sites had no or only a moderate effect on McpU stability, we hypothesized that additional residues within these regions contribute to protease recognition. To test this theory, we deleted regions within the C-terminal domain of McpU. In analogy to the C. crescentus MS-ring protein FliF, regions were chosen to encompass the three dialanine-containing motifs putatively associated with proteolysis (27, 28, 49). Initially, three mutant strains were created that expressed variants of McpU that were C-terminally truncated by 9 (Δ amino acids [aa] 699 to 707; BS266), 25 (Δ aa 683 to 707; BS267), or 39 (Δ aa 668 to 707; BS269) amino acid residues (Fig. 1). The abundance of McpU truncated by 9 amino acids (BS266) was 6.6-fold (±0.4) increased, which was comparable to that of epitope-tagged mutants. However, truncating McpU by 25 amino acids increased its abundance by only 3.2-fold (±0.4), while a 39-amino-acid truncation yielded a protein that was just 1.8-fold (±0.3) more abundant than wild-type McpU (Fig. 2 and 4). These results indicate that the last 9 residues play a central role in McpU stability. To further decipher the putative protease recognition site, we deleted 16 amino acids preceding the 9 most C-terminal amino acids (Δ aa 683 to 698; BS271). Interestingly, the modification resulted in a 0.14-fold (±0.1) decrease in McpU abundance (Fig. 2 and 3). It is conceivable that this 16-amino-acid region contains features supporting McpU stability, which would explain the gradually decreased stability of McpU lacking the most C-terminal 25 or 39 amino acids, compared to a 9-amino-acid truncation. However, the possibility cannot be excluded that the internal 16-amino-acid deletion disrupted secondary structure elements and thus proper folding of the mutated protein, leading to its instability. To summarize, deleting the 9 most C-terminal residues, including motif III (di-Ala peptide), induced the most dramatic increase in McpU abundance, suggesting that important determinants of the protease recognition site are contained in this region.
Effect of C-terminal 3×FLAG tag fusions on the abundance of four additional chemoreceptors and the coupling protein CheW1.
We next asked whether the positive effect of epitope tags on protein stability was unique to the S. meliloti chemoreceptor McpU. Therefore, we chose two cytosolic chemoreceptors, McpY and IcpA, the high-abundance receptor McpV, and a receptor that was below the limit of detection, McpT (46), as well as the major receptor-CheA coupling protein CheW1, for stability assays. We constructed strains through allelic exchange that encode proteins with C-terminal 3×FLAG tag fusions in their native chromosomal locus and quantified protein abundance in cell extracts as described above (Fig. 5A to D). Table 1 lists the number of molecules per cell and the ratio between native and tagged proteins. Four of the five proteins were more abundant with the C-terminal epitope tag, with ratios ranging from 2-fold (±0.5) to 45-fold (±19). The change in abundance of McpT could only be estimated to be at least 210-fold (±118) because the amount of untagged protein was below the limit of detection (Table 1) (46). In contrast, a direct comparison of band intensities in cell extracts from strains expressing McpV or McpV-3×FLAG indicated the presence of very similar protein amounts (Fig. 5E). In conclusion, the fusion of a C-terminal 3×FLAG tag increases the in vivo abundance of at least four S. meliloti chemoreceptors and one chemotaxis protein.
FIG 5.
Representative immunoblots used to quantify chemotaxis proteins with C-terminal 3×FLAG fusions. (A) Lanes 1 to 3 contain BS193 (McpY-3×FLAG expressing strain) cell lysate from 1 ml of culture at an OD600 of 0.25, and wild type (WT) + McpV-3×FLAG lanes contain RU11/001 (WT) cell lysate from 1 ml of culture at an OD600 of 0.25 with McpV-LBD-3×FLAG (0.4, 0.3, 0.2, 0.01, 0.05, and 0 ng). (B) Lanes 1 to 3 contain BS194 (McpT-3×FLAG-expressing strain) cell lysate from 1 ml of culture at an OD600 of 0.25, and WT + McpV-3×FLAG lanes contain RU11/001 (WT) cell lysate from 1 ml of culture at an OD600 of 0.25 with McpV-LBD-3×FLAG (1.8, 1.5, 1, 0.5, 0.3, and 0 ng). (C) Lanes 1 to 3 contain BS201 (CheW1-3×FLAG-expressing strain) cell lysate from 1 ml of culture at an OD600 of 0.25, and WT + McpV-3×FLAG lanes contain RU11/001 (WT) cell lysate from 1 ml of culture at an OD600 of 0.25 with McpV-LBD-3×FLAG (8, 6.5, 5.2, 4, 2.6, and 0 ng). (D) Lanes 1 to 3 contain BS203 (IcpA-3×FLAG-expressing strain) cell lysate from 1 ml of culture at an OD600 of 0.25, and WT + McpV-3×FLAG lanes contain RU11/001 (WT) cell lysate from 1 ml of culture at an OD600 of 0.25 with McpV-LBD-3×FLAG (10, 8, 6, 4, 2, and 0 ng). (E) Direct comparison of McpV abundance in cell lysates of the wild type compared to those of BS221. Each lane contains cell lysates from 1 ml of culture at an OD600 of 0.25.
TABLE 1.
Absolute protein abundance of chemotaxis proteins with C-terminal 3×FLAG epitope
| Native protein | No. of molecules per cell | Epitope-tagged protein | No. of molecules per cell | Fold increase |
|---|---|---|---|---|
| CheW1 | 189a | CheW1-3×FLAG | 367 ± 97 | 1.9 ± 0.5 |
| IcpA | 17b | IcpA-3×FLAG | 468 ± 155 | 27 ± 9 |
| McpT | <0.36 (BDLb ,c ) | McpT-3×FLAG | 76 ± 42 | 210 ± 118 |
| McpU | 47b | McpU-3×FLAG | 291 ± 36 | 6.2 ± 0.8 |
| McpV | 299b | McpV-3×FLAG | 314d ±3 | 1.05 ± 0.1d |
| McpY | 0.33b | McpY-3×FLAG | 15 ± 6 | 45 ± 19 |
Value from Arapov et al. (82).
Value from Zatakia et al. (46). BDL, below detection limit.
Value represents the maximum amount of McpT (Zatakia et al. [46]). This value was deduced from the minimal amount of enhanced green fluorescent protein (EGFP) required for detection, namely, 5 pg or the equivalent of 0.36 molecule per cell, detected in immunoblot assays performed under standardized conditions compared to the lack of an McpT-EGFP band in cell extracts.
Value is derived from direct comparison of band intensities between native McpV and McpV-3×FLAG (Fig. 5E) and does not a represent a statistically significant change. Number of molecules per cell is based on this value.
Chemotaxis in strains with increased McpU abundance is unaltered.
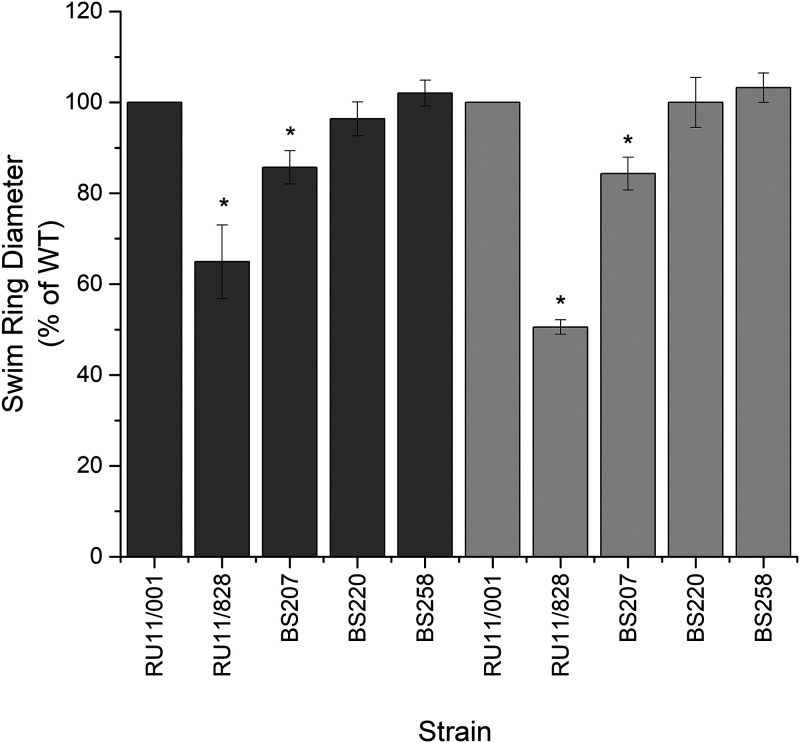
To investigate whether increased McpU stability and abundance due to the addition of epitope tags enhances the chemotaxis response to attractants senses by McpU, we tested the behavior of S. meliloti wild-type and stability mutant strains in low-percentage-agar swim plate assays with tryptone-yeast extract (Bromfield) and with defined Rhizobium basal (RB) medium containing 10−4 M lysine. Lysine was chosen as the carbon source as it is known to be a sole ligand for McpU (50, 51). As previously described, the deletion of mcpU (RU11/828) resulted in a reduced swim ring diameter (50, 52). Although McpU abundance increased 6-fold with C-terminal 3×FLAG and 6×His-epitope tags (Fig. 3), swim ring diameters were either slightly reduced (BS207) or similar to those of the wild type (BS220) (Fig. 6). The slight decrease in chemotaxis of BS207 with 3×FLAG-tagged McpU versus BS220 with 6×His-tagged McpU might be due to the larger size of the 3×FLAG tag (22 amino acids). We observed previously that a C-terminal fusion of enhanced green fluorescent protein (EGFP) to McpU caused a small reduction in swim ring size (53). In conclusion, abundance of McpU does not correlate with chemotactic performance.
FIG 6.
Relative swim ring diameter of S. meliloti mutant strains in Bromfield and in Rhizobium basal medium containing 10−4 M lysine. Plates containing 0.3% agar were inoculated with 3 μl stationary-phase TYC culture and incubated for 3 and 5 days, respectively, at 30°C. Dark and light gray bars represent results obtained in Bromfield and Rhizobium basal medium with 10−4 M lysine, respectively. Values are the means, and error bars reflect the standard deviation from five replicates. Statistical significance was determined by a two-tailed Student t test (P < 0.05). Asterisks denote statistically significant differences from the wild type.
DISCUSSION
Proteolysis is an important process in all living cells that allows for a fast but irreversible response to a changing environment. General and regulatory proteolysis in bacteria is essential for many physiological processes. As a result, it is tightly controlled. Contributing proteases are known to bind and degrade only a subset of protein substrates (54). During regulatory proteolysis, specific removal of native proteins is controlled through degrons, which are freely accessible motifs at either N or C termini (39). We documented that introduction of epitope tags to the C terminus of several S. meliloti chemotaxis proteins effectively disrupted normal in vivo proteolysis, likely by shielding of the degron at the C terminus.
The role of the C-terminal domain in McpU degradation.
Addition of charged epitope tags and single-amino-acid residues to the C terminus of McpU increased abundance of the protein up to 8-fold. Larger and charged tags had a greater effect on abundance than single amino acids, and larger and charged amino acids had a greater effect than smaller ones (+ His > Ala > Gly). An analogous effect has been described for C. crescentus FliF (55). We therefore hypothesize that charged amino acids abolish degradation by blocking access of the protease to the hydrophobic degradation signal. This is supported by the fact that the addition of glycine, which lacks a side chain, did not stabilize McpU (Fig. 2). The presence of a hydrophobic degradation signal containing an AAL or AVA motif has been reported for C. crescentus McpA and McpB (27, 28). Of the three putative motifs in McpU, only mutations of the C-terminal di-Ala peptide affected McpU stability (Fig. 2 and 3). The importance of this motif for protein stability in conjunction with residues in the preceding region allowed the conclusion that a protease recognition site exists in the C-terminal domain of McpU. Yet it is unclear which exact residues are critical for recognition. Deletion of the di-Ala peptide together with the preceding 7-amino-acid region is sufficient to promote protein stability, a result that is in agreement with the degradation scheme of C. crescentus FliF (55). However, we cannot dismiss the possibility that additional residues in the larger aliphatic C-terminal region of McpU are contributing to an extended protease recognition site. Analyses of targeted protein degradation in E. coli and C. crescentus suggest that the terminal di-Ala peptide in McpU could be part of a larger SsrA-degron like motif (41, 56). ClpXP, as well as other proteases such as HslUV, is known to bind to the 11-amino-acid SsrA degron (20, 57). A di-Ala peptide motif is present in the C-terminal domain of S. meliloti and C. crescentus CtrA and is important for its ClpXP-dependent proteolysis (35, 49). The nature of the degradation signal does not seem to be based on a distinct primary sequence. Rather, ClpXP appears to recognize short clusters of aliphatic residues in the hydrophobic, disordered C-terminal domain of McpU, similar to C. crescentus FliF (55).
Effect of epitope tags on other S. meliloti chemotaxis proteins.
The presence of C-terminal epitope tags impacted not only the stability of McpU but that of four of the five S. meliloti chemotaxis proteins assayed. The abundances of McpT, McpY, IcpA, and CheW1 were increased to various degrees between 2-fold (CheW1) and an estimated 210-fold (McpT) when a 3×FLAG tag was fused to their respective C termini (Fig. 5, Table 2). An alignment of the C-terminal domains revealed that only CheW1 possesses a C-terminal di-Ala peptide that was determined to mediate degradation of McpU (Fig. 7). However, the C-terminal domain of all analyzed proteins are rich in aliphatic residues (Fig. 7), which could explain their tendency to targeted proteolytic degradation. Additionally, we identified putative protease recognition sites near the C terminus of McpY and McpT, analogous to C. crescentus McpA and McpB (27, 28) (Fig. 7). In general, the prediction of specific protease recognition sites in S. meliloti chemotaxis proteins is still challenging.
TABLE 2.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| ER2566 | ion ompT lacZ::T7 | New England Biolabs |
| S17-1 | Tpr Smr recA endA thi hsdR RP4-2 Tc::Mu::Tn7 | Simon et al. (83) |
| S. meliloti | ||
| RU11/001 | Smr; spontaneous streptomycin-resistant wild-type strain | Pleier et al. (76) |
| RU11/828 | Smr ΔmcpU | Meier et al. (50) |
| BS193 | Smr McpT-3×FLAG | This work |
| BS194 | Smr McpY-3×FLAG | This work |
| BS201 | Smr CheW1-3×FLAG | This work |
| BS203 | Smr IcpA-3×FLAG | This work |
| BS207 | Smr McpU-3×FLAG | This work |
| BS220 | Smr McpU-6×His | This work |
| BS221 | Smr McpV-3×FLAG | This work |
| BS222 | Smr McpU-1×FLAG | This work |
| BS252 | Smr McpU-1×His | This work |
| BS256 | Smr McpU-2×His | This work |
| BS257 | Smr McpU-1×Ala | This work |
| BS258 | Smr McpU-1×Gly | This work |
| BS266 | Smr; McpU Δ aa 699–707 | This work |
| BS267 | Smr; McpU Δ aa 683–707 | This work |
| BS268 | Smr; McpU aa 679–681 AAL mutated to ELD | This work |
| BS269 | Smr; McpU Δ aa 668–707 | This work |
| BS270 | Smr; McpU aa 695–697 AAS mutated to ELD | This work |
| BS271 | Smr; McpU Δ aa 683–698 | This work |
| BS272 | Smr; McpU aa 706–707 AA mutated to DD | This work |
| BS273 | Smr; McpU-3×FLAG Δ aa 706–707 AA mutated to DD | This work |
| Plasmids | ||
| pK18mobsacB | Kmr mob sacB, vector used for homologous allelic exchange | Schäfer et al. (84) |
| pBS459 | pTYB11-McpV-LBD-3×FLAG | This work |
aTpr, trimethoprim resistant; Smr, streptomycin resistant; Kmr, kanamycin resistant; aa, amino acid(s).
FIG 7.
Comparison of the region comprising 47 amino acid residues proximal to the carboxy terminus of C. crescentus McpA and McpB, five S. meliloti MCPs, and CheW1. Residues critical for correct proteolysis are indicated by shaded boxes according to Tsai et al. (28), Potocka et al. (27), and this work. Putative residues important for proteolysis are marked by unshaded boxes. Numbers at the end of each sequence reflect overall protein length.
Chemoreceptor degradation and cell cycle control.
The majority of the chemoreceptors in S. meliloti are unstable during exponential cell culture growth. While this posttranslational regulation appears to be inefficient, recent evidence from E. coli suggests that the investment in chemotaxis and motility is proportionally increased with the reproductive fitness advantage derived from the ability to follow a nutrient gradient (58). Therefore, the cost of protein synthesis and degradation should not be higher than the potential benefit that a specific chemoreceptor can provide. In fact, C. crescentus not only degrades the chemoreceptor array during the course of the cell cycle but releases an entire macromolecular machinery by ejecting its flagellum (59, 60). While these studies were performed with synchronized C. crescentus cells, a comprehensive investigation of the cell cycle-dependent degradation of chemotaxis proteins in S. meliloti remains an aim for future investigations.
The pathways for regulating cell division in S. meliloti continue to be explored in detail, but similarities to C. crescentus exist, such as asymmetric cell division and regulation of CtrA stability via proteolysis (33, 34, 49, 61). Timing of the S. meliloti cell cycle is critical to cell fate during division and in symbiosis (31, 35, 36). In particular, S. meliloti downregulates motility and chemotaxis genes when attaching to root hairs and, at later stages, in nodules (62, 63). However, negative regulation of these genes occurs much sooner during planktonic growth, specifically during early exponential growth. We have shown that transcription, and presumably posttranslational regulation, of Rem, the main activator of motility, is growth phase dependent (18). Remarkably, the two chemoreceptors whose stability were most greatly affected by the presence of the 3×FLAG tag, McpT and McpY, exhibit a certain degree of uncoupling from Rem regulation (46). It would be interesting to assess the molecular mechanisms for the differential degradation of these chemotaxis proteins in the future. Detailed knowledge about the molecular mechanisms of cell cycle-dependent, targeted, and selective proteolysis in S. meliloti is important to understand the mechanisms of maintaining receptor stoichiometry, which is pertinent for receptor cooperativity and signal propagation.
McpV stability and cell cycle control.
The only chemotaxis protein in our assay that did not significantly increase in abundance as a C-terminal 3×FLAG tag fusion was McpV (Fig. 5, Table 1), although it terminates in a di-Ala peptide, and its C-terminal domain contains several other putative degradation motifs (Fig. 7). Interestingly, McpV is the most abundant MCP within the S. meliloti cell, accounting for approximately 70% of all MCPs (46). A study by Pini et al. (2015) (21) indicated that transcription of mcpV is not regulated by CtrA, while other chemoreceptor genes (icpA, mcpT, mcpU, mcpX, and mcpZ) are under direct or indirect control of CtrA (21, 32). Commonly, proteins that are encoded by genes in a regulon under the control of the same transcriptional regulator are also degraded by a shared protease (48). For instance, E. coli ftsZ, dps, katE, and glpD are all transcribed under the control of σS, and their gene products, including σS, are degraded by ClpXP (48). It is conceivable to propose that the lack of cell cycle-related control of mcpV transcription by CtrA exempts it from the proteolytic mechanism governing the other receptors, which would explain its high abundance. In C. crescentus and related alphaproteobacteria, a complex proteolytic hierarchy exists, which allows for targeted degradation of proteins (64). Since S. meliloti motility and chemotaxis are limited to early exponential growth, transcriptional and posttranslational regulatory controls are in place to negatively regulate the chemotaxis machinery at later stages of growth (18, 53). We envision a mechanism in the S. meliloti chemotaxis system to selectively target a subset of receptors for proteolysis. At this point, however, it is challenging to make predictions based on primary sequences alone.
Implications of regulated proteolysis for cell cycle control.
Alphaproteobacteria, including S. meliloti, divide asymmetrically, producing two genetically identical daughter cells of distinct morphology (22, 65–67). C. crescentus displays the best-known example of asymmetric division, with dramatic morphological differences between swarmer and stalked cell types. The larger stalked cell can immediately enter the prokaryotic S phase, while the smaller swarmer cell stays in the G1 phase before differentiation into a stalked cell. (25). S. meliloti does not undergo such an obvious differentiation, but asymmetric cell division produces two progeny cells of different sizes (22, 67). The underlying molecular mechanisms associated with asymmetric division appear to be conserved and tied to the cell cycle within the class of alphaproteobacteria (33, 61, 68). This type of differentiation potentially allows bacteria to increase their fitness when exposed to stress by enriching the diversity of their phenotypes. Asymmetric cell division is an important individual-level bet-hedging strategy for planktonic S. meliloti cells (69). The same cell cycle control mechanisms that regulate asymmetric division in S. meliloti are influential in expression of motility and chemotaxis genes (21, 22, 32, 34). Thus, it is plausible that chemotaxis and motility could also be important for bet-hedging (21, 68). The previous observation of a mixed population of motile and nonmotile cells during exponential growth could be an evidence for bet hedging (46). Programmed proteolysis of the chemoreceptor array is an important step in C. crescentus cell type differentiation (27, 28, 44). Differentiation of asymmetrically dividing S. meliloti cells has received relatively little attention outside the symbiosis process (62, 63). The current study presents evidence that regulated proteolysis of the S. meliloti chemotaxis machinery is tied to the cell cycle and therefore linked to asymmetric cell division in alphaproteobacteria.
General conclusions on the use of epitope tag fusions.
Engineered peptide epitope tags are sensitively and specifically recognized by commercially available antibodies to facilitate the detection, localization, and purification of proteins (70, 71). Mostly, epitope tags have either no effect or a detrimental effect on protein stability (72). However, it has been reported that the presence of a tag can also increase the thermal stability of a particular protein, suggesting that tag fusions have a variety of outcomes (73). Although epitope tags typically do not disrupt normal protein functions, we showed that they can alter protein stability in the chemotaxis system of S. meliloti. Our present study asserts that the presence of C-terminal epitope tags disrupts cell cycle-regulated proteolysis in the chemotaxis system of S. meliloti and induces an accumulation of the tagged proteins. These results are supported by similar findings in another alphaproteobacterium, C. crescentus, specifically the flagellar protein FliF (55). The C. crescentus receptor McpA has been widely used as a model substrate for protein degradation during the course of the C. crescentus cell cycle (30, 74, 75). Researchers interested in pursuing similar cell cycle experiments in S. meliloti or other alphaproteobacteria should be aware of the possible effect of epitope tags on their experimental outcomes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Derivatives of E. coli K-12 and S. meliloti MV II-1 and the plasmids used are listed in Table 1. RU11/001 is a spontaneous streptomycin-resistant derivative of MVII-1 (76).
Media and growth conditions.
E. coli strains were grown in lysogeny broth (LB) (77) at indicated temperatures. S. meliloti strains were grown in TYC medium (0.5% tryptone, 0.3% yeast extract, and 0.13% CaCl2·6H2O [pH 7.0]) (78) or Sinorhizobium motility medium (SMM) {RB [6.1 mM K2HPO4, 3.9 mM KH2PO4, 1 mM MgSO4, 1 mM (NH4)2SO4, 0.1 mM CaCl2, 0.1 mM NaCl, 0.01 mM Na2MoO4, 0.001 mM FeSO4, 2 μg/liter biotin, and 10 μg/liter thiamine] (79), 0.2% mannitol, and 2% tryptone-yeast extract (TY)} or Bromfield medium (0.04% tryptone, 0.01% yeast extract, and 0.01% CaCl2·2H2O) (18). Motile cells for immunoblots were grown in SMM for 2 days, diluted to an optical density at 600 nm (OD600) of 0.02 and incubated at 30°C to an OD600 of 0.25. The following antibiotics were used in their final concentrations: ampicillin at 100 μg/ml and kanamycin at 50 μg/ml (for E. coli) and neomycin at 120 μg/ml and streptomycin at 600 μg/ml (for S. meliloti). S. meliloti cell culture used within the motility assays was grown in 3 ml TYC for 2 days at 30°C.
Motility assays.
Swim plates (0.3% Bacto agar) containing Bromfield medium or RB minimal medium supplemented with lysine (10−4 M) were inoculated with 3 μl stationary-phase bacterial cell culture grown in TYC to an OD600 of approximately 2.0. Bromfield plates were incubated for 3 days and RB-lysine plates for 5 days, both at 30°C.
Genetic and DNA manipulations.
S. meliloti mutants listed in Table 2 were created by allelic replacement essentially as laid out previously (46, 80, 81).
Purification of recombinant McpV-LBD-3×FLAG.
McpV-LBD-3×FLAG was purified using the Impact (NEB) system. McpV-LBD-3×FLAG was expressed from pBS0459 in E. coli ER2566 as described in Zatakia et al. (46). Expression was induced at an OD600 of 0.6 to 0.8 by addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and growth was continued at 16°C for 16 h. Cells were harvested, suspended in Impact buffer (500 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 20 mM Tris-HCl [pH 8.0]), and lysed by three passages through a French pressure cell at 20,000 lb/in2 (SLM Aminco, Silver Spring, MD). The cleared lysate was loaded on a chitin-agarose (New England Biolabs) column (6 cm by 5 cm), and intein-mediated cleavage was induced by addition of Impact buffer with 50 mM dithiothreitol and incubation overnight at 4°C. The protein was eluted with Impact buffer. Pooled protein fractions were then subject to size exclusion chromatography using a HiPrep 26/60 Sephacryl S-200 high-resolution (HR) column (GE Healthcare). The column was developed in phosphate-buffered saline (PBS) (100 mM NaCl, 80 mM Na2HPO4, and 20 mM NaH2PO4 [pH 7.5]) with 5% (vol/vol) glycerol. Protein concentrations were obtained by quantitative amino acid analyses after total acid hydrolysis performed at the Protein Chemistry Lab, Texas A&M University.
Immunoblotting.
Polyclonal antibodies were raised against the ligand binding domain of McpU or McpV (McpU-LBD or McpV-LBD) and then subsequently purified as described in Scharf et al. (46, 78). Cell extracts were prepared as follows. Wild-type and deletion strains were grown in SMM to an OD600 of 0.25 ± 0.05. Aliquots (1 ml) were then pelleted by centrifugation at 21,000 × g for 10 min at room temperature, suspended in approximately 15 μl of the supernatant and 15 μl of Laemmli buffer (4.5% SDS, 18.75 mM Tris-HCl [pH 6.5], 43.5% glycerol, 0.0125% bromophenol blue, and 5% β-mercaptoethanol). Samples were boiled for 10 min and stored at −20°C. For the relative quantification of McpU abundance, mutant and deletion strains were grown to an OD600 of 0.25 ± 0.05, mixed in specified ratios prior to centrifugation, and then treated as described above. Relative differences in abundance were derived from three biological replicates for each mutant. For the quantification of IcpA-3×FLAG, McpT-3×FLAG, McpY-3×FLAG, and CheW1-3×FLAG in cell extracts, defined amounts of purified McpV-LBD-3×FLAG were added to wild-type extracts to create a standard curve. Means were derived from six biological replicates.
Blotting was performed in the same manner as described by Zatakia et al. (46). Cell extracts were separated by SDS-polyacrylamide gel electrophoresis and transferred onto 0.45-μm nitrocellulose membranes. Membranes were blocked for 2 h or overnight with 5% nonfat dry milk in PBS-0.1% Tween 20 and probed with a 1:200 dilution of affinity-purified antibodies or a 1:20,000 dilution of monoclonal anti-FLAG M2 antibody (Sigma-Aldrich). Blots were washed for 30 min with PBS-0.1% Tween 20 with four buffer changes and then probed with a 1:1,500 dilution of donkey anti-rabbit polyclonal antibodies linked to horseradish peroxidase or a 1:4,000 dilution of sheep anti-mouse antibodies linked to horseradish peroxidase. The blots were then washed for 30 min with PBS-0.1% Tween 20 with four buffer changes. Detection was performed by chemiluminescence (Amersham ECL Western blotting detection kit; GE Healthcare) using Hyperfilm ECL (GE Healthcare). Images were scanned with an Epson Perfection 160SU scanner, and pixel intensities were then quantified with ImageJ. The number of molecules per cell was calculated using the quantified number of cells per volume for S. meliloti previously determined in Zatakia et al. (46).
ACKNOWLEDGMENTS
This study was supported by NSF grants MCB-1253234 and MCB-1817652 to Birgit E. Scharf, the VT Fralin Summer Undergraduate Research Fellowship to Jiwoo Kim, and the VT Multicultural Academics Opportunity Program Fellowship to Maya Pahima.
We are indebted to Hardik Zatakia for purification of McpV-LBD-3×FLAG, to Rafael Castañeda Saldaña for quantification of CheW1-3×FLAG, and to Hiba Baaziz, K. Karl Compton, Floricel Gonzalez, and Richard Sobe for critical reading and comments.
REFERENCES
- 1.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 2.Locke J. 2013. How bacteria choose a lifestyle. Nature 503:476–477. doi: 10.1038/nature12837. [DOI] [PubMed] [Google Scholar]
- 3.Davey ME, O'Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raina J-B, Fernandez V, Lambert B, Stocker R, Seymour JR. 2019. The role of microbial motility and chemotaxis in symbiosis. Nat Rev Microbiol 17:284–294. doi: 10.1038/s41579-019-0182-9. [DOI] [PubMed] [Google Scholar]
- 5.Oliveira NM, Foster KR, Durham WM. 2016. Single-cell twitching chemotaxis in developing biofilms. Proc Natl Acad Sci U S A 113:6532–6537. doi: 10.1073/pnas.1600760113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 7.Amaya-Gómez CV, Hirsch AM, Soto MJ. 2015. Biofilm formation assessment in Sinorhizobium meliloti reveals interlinked control with surface motility. BMC Microbiol 15:58–58. doi: 10.1186/s12866-015-0390-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Rhijn P, Vanderleyden J. 1995. The Rhizobium-plant symbiosis. Microbiol Rev 59:124–142. doi: 10.1128/MMBR.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van de Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, Kevei Z, Farkas A, Mikulass K, Nagy A, Tiricz H, Satiat-Jeunemaitre B, Alunni B, Bourge M, Kucho K, Abe M, Kereszt A, Maroti G, Uchiumi T, Kondorosi E, Mergaert P. 2010. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327:1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- 10.Brewin NJ. 1991. Development of the legume root nodule. Annu Rev Cell Biol 7:191–226. doi: 10.1146/annurev.cb.07.110191.001203. [DOI] [PubMed] [Google Scholar]
- 11.Scharf BE, Hynes MF, Alexandre GM. 2016. Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol Biol 90:549–559. doi: 10.1007/s11103-016-0432-4. [DOI] [PubMed] [Google Scholar]
- 12.Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol 9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 13.Hazelbauer GL, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci 33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salah Ud-Din AIM, Roujeinikova A. 2017. Methyl-accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea. Cell Mol Life Sci 74:3293–3303. doi: 10.1007/s00018-017-2514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falke JJ, Hazelbauer GL. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci 26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briegel A, Ladinsky MS, Oikonomou C, Jones CW, Harris MJ, Fowler DJ, Chang YW, Thompson LK, Armitage JP, Jensen GJ. 2014. Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling. Elife 3:e02151. doi: 10.7554/eLife.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang W, Briegel A. 2019. Diversity of bacterial chemosensory arrays. Trends Microbiol 28:68–80. doi: 10.1016/j.tim.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Rotter C, Mühlbacher S, Salamon D, Schmitt R, Scharf B. 2006. Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti. J Bacteriol 188:6932–6942. doi: 10.1128/JB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sourjik V, Muschler P, Scharf B, Schmitt R. 2000. VisN and VisR are global regulators of chemotaxis, flagellar, and motility genes in Sinorhizobium (Rhizobium) meliloti. J Bacteriol 182:782–788. doi: 10.1128/jb.182.3.782-788.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogden AJ, McAleer JM, Kahn ML. 2019. Characterization of the Sinorhizobium meliloti HslUV and ClpXP protease systems in free-living and symbiotic states. J Bacteriol 201:e00498-18. doi: 10.1128/JB.00498-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pini F, De Nisco NJ, Ferri L, Penterman J, Fioravanti A, Brilli M, Mengoni A, Bazzicalupo M, Viollier PH, Walker GC, Biondi EG. 2015. Cell cycle control by the master regulator CtrA in Sinorhizobium meliloti. PLoS Genet 11:e1005232. doi: 10.1371/journal.pgen.1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. 2004. Morphological and functional asymmetry in α-proteobacteria. Trends Microbiol 12:361–365. doi: 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Bird TH, MacKrell A. 2011. A CtrA homolog affects swarming motility and encystment in Rhodospirillum centenum. Arch Microbiol 193:451–459. doi: 10.1007/s00203-011-0676-y. [DOI] [PubMed] [Google Scholar]
- 24.Francis N, Poncin K, Fioravanti A, Vassen V, Willemart K, Ong TA, Rappez L, Letesson JJ, Biondi EG, De Bolle X. 2017. CtrA controls cell division and outer membrane composition of the pathogen Brucella abortus. Mol Microbiol 103:780–797. doi: 10.1111/mmi.13589. [DOI] [PubMed] [Google Scholar]
- 25.Curtis PD, Brun YV. 2010. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev 74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith SC, Joshi KK, Zik JJ, Trinh K, Kamajaya A, Chien P, Ryan KR. 2014. Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proc Natl Acad Sci U S A 111:14229–14234. doi: 10.1073/pnas.1407862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potocka I, Thein M, Østerås M, Jenal U, Alley M. 2002. Degradation of a Caulobacter soluble cytoplasmic chemoreceptor is ClpX dependent. J Bacteriol 184:6635–6641. doi: 10.1128/jb.184.23.6635-6642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai JW, Alley MR. 2001. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol 183:5001–5007. doi: 10.1128/jb.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berne C, Brun YV. 2019. The two chemotaxis clusters in Caulobacter crescentus play different roles in chemotaxis and biofilm regulation. J Bacteriol 201:e00071-19. doi: 10.1128/JB.00071-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. 2006. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A 103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi H, De Nisco NJ, Chien P, Simmons LA, Walker GC. 2009. Sinorhizobium meliloti CpdR1 is critical for co-ordinating cell cycle progression and the symbiotic chronic infection. Mol Microbiol 73:586–600. doi: 10.1111/j.1365-2958.2009.06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Nisco NJ, Abo RP, Wu CM, Penterman J, Walker GC. 2014. Global analysis of cell cycle gene expression of the legume symbiont Sinorhizobium meliloti. Proc Natl Acad Sci U S A 111:3217–3224. doi: 10.1073/pnas.1400421111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnett MJ, Hung DY, Reisenauer A, Shapiro L, Long SR. 2001. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J Bacteriol 183:3204–3210. doi: 10.1128/JB.183.10.3204-3210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schallies KB, Sadowski C, Meng J, Chien P, Gibson KE. 2015. Sinorhizobium meliloti CtrA stability is regulated in a CbrA-dependent manner and influenced by CpdR1. J Bacteriol 197:2139–2149. doi: 10.1128/JB.02593-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue S, Biondi EG. 2019. Coordination of symbiosis and cell cycle functions in Sinorhizobium meliloti. Biochim Biophys Acta Gene Regul Mech 1862:691–696. doi: 10.1016/j.bbagrm.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Hallez R, Delaby M, Sanselicio S, Viollier PH. 2017. Hit the right spots: cell cycle control by phosphorylated guanosines in alphaproteobacteria. Nat Rev Microbiol 15:137–148. doi: 10.1038/nrmicro.2016.183. [DOI] [PubMed] [Google Scholar]
- 37.Vass RH, Nascembeni J, Chien P. 2017. The essential role of ClpXP in Caulobacter crescentus requires species constrained substrate specificity. Front Mol Biosci 4:28–28. doi: 10.3389/fmolb.2017.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker TA, Sauer RT. 2012. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta 1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt R, Bukau B, Mogk A. 2009. Principles of general and regulatory proteolysis by AAA+ proteases in Escherichia coli. Res Microbiol 160:629–636. doi: 10.1016/j.resmic.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 40.Fritze J, Zhang M, Luo Q, Lu X. 2020. An overview of the bacterial SsrA system modulating intracellular protein levels and activities. Appl Microbiol Biotechnol 104:5229–5213. doi: 10.1007/s00253-020-10623-x. [DOI] [PubMed] [Google Scholar]
- 41.Gottesman S, Roche E, Zhou Y, Sauer RT. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev 12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexander RP, Zhulin IB. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci U S A 104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerth U, Kock H, Kusters I, Michalik S, Switzer RL, Hecker M. 2008. Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J Bacteriol 190:321–331. doi: 10.1128/JB.01233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joshi KK, Chien P. 2016. Regulated proteolysis in bacteria: Caulobacter. Annu Rev Genet 50:423–445. doi: 10.1146/annurev-genet-120215-035235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. 2001. Overlapping recognition determinants within the SsrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci U S A 98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zatakia HM, Arapov TD, Meier VM, Scharf BE. 2017. Cellular stoichiometry of methyl-accepting chemotaxis proteins in Sinorhizobium meliloti. J Bacteriol 200:e00614-17. doi: 10.1128/JB.00614-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scharf BE, Fahrner KA, Turner L, Berg HC. 1998. Control of direction of flagellar rotation in bacterial chemotaxis. Proc Natl Acad Sci U S A 95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flynn JM, Neher SB, Kim Y-I, Sauer RT, Baker TA. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell 11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 49.Ryan KR, Judd EM, Shapiro L. 2002. The CtrA response regulator essential for Caulobacter crescentus cell-cycle progression requires a bipartite degradation signal for temporally controlled proteolysis. J Mol Biol 324:443–455. doi: 10.1016/s0022-2836(02)01042-2. [DOI] [PubMed] [Google Scholar]
- 50.Meier VM, Muschler P, Scharf BE. 2007. Functional analysis of nine putative chemoreceptor proteins in Sinorhizobium meliloti. J Bacteriol 189:1816–1826. doi: 10.1128/JB.00883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webb BA, Compton KK, del Campo JSM, Taylor D, Sobrado P, Scharf BE. 2017. Sinorhizobium meliloti chemotaxis to multiple amino acids is mediated by the chemoreceptor McpU. Mol Plant Microbe Interact 30:770–777. doi: 10.1094/MPMI-04-17-0096-R. [DOI] [PubMed] [Google Scholar]
- 52.Webb BA, Helm RF, Scharf BE. 2016. Contribution of individual chemoreceptors to Sinorhizobium meliloti chemotaxis towards amino acids of host and nonhost seed exudates. Mol Plant Microbe Interact 29:231–239. doi: 10.1094/MPMI-12-15-0264-R. [DOI] [PubMed] [Google Scholar]
- 53.Meier VM, Scharf BE. 2009. Cellular localization of predicted transmembrane and soluble chemoreceptors in Sinorhizobium meliloti. J Bacteriol 191:5724–5733. doi: 10.1128/JB.01286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gur E, Biran D, Ron EZ. 2011. Regulated proteolysis in Gram-negative bacteria—how and when? Nat Rev Microbiol 9:839–848. doi: 10.1038/nrmicro2669. [DOI] [PubMed] [Google Scholar]
- 55.Grünenfelder B, Tawfilis S, Gehrig S, Østerås M, Eglin D, Jenal U. 2004. Identification of the protease and the turnover signal responsible for cell cycle-dependent degradation of the Caulobacter FliF Motor Protein. J Bacteriol 186:4960–4971. doi: 10.1128/JB.186.15.4960-4971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herman C, Thévenet D, Bouloc P, Walker GC, D'Ari R. 1998. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev 12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sundar S, McGinness KE, Baker TA, Sauer RT. 2010. Multiple sequence signals direct recognition and degradation of protein substrates by the AAA+ protease HslUV. J Mol Biol 403:420–429. doi: 10.1016/j.jmb.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ni B, Colin R, Link H, Endres RG, Sourjik V. 2020. Growth-rate dependent resource investment in bacterial motile behavior quantitatively follows potential benefit of chemotaxis. Proc Natl Acad Sci U S A 117:595–601. doi: 10.1073/pnas.1910849117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stallmeyer MB, Hahnenberger KM, Sosinsky GE, Shapiro L, DeRosier DJ. 1989. Image reconstruction of the flagellar basal body of Caulobacter crescentus. J Mol Biol 205:511–518. doi: 10.1016/0022-2836(89)90222-2. [DOI] [PubMed] [Google Scholar]
- 60.Shapiro L, Maizel J. 1973. Synthesis and structure of Caulobacter crescentus flagella. J Bacteriol 113:478–485. doi: 10.1128/JB.113.1.478-485.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. 2010. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol 4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oke V, Long SR. 1999. Bacteroid formation in the Rhizobium–legume symbiosis. Curr Opin Microbiol 2:641–646. doi: 10.1016/s1369-5274(99)00035-1. [DOI] [PubMed] [Google Scholar]
- 63.Mergaert P, Uchiumi T, Alunni B, Evanno G, Cheron A, Catrice O, Mausset A-E, Barloy-Hubler F, Galibert F, Kondorosi A, Kondorosi E. 2006. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium–legume symbiosis. Proc Natl Acad Sci U S A 103:5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahmoud SA, Chien P. 2018. Regulated proteolysis in bacteria. Annu Rev Biochem 87:677–696. doi: 10.1146/annurev-biochem-062917-012848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lam H, Matroule J-Y, Jacobs-Wagner C. 2003. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell 5:149–159. doi: 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- 66.Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. 2012. Polar growth in the alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci U S A 109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greif D, Pobigaylo N, Frage B, Becker A, Regtmeier J, Anselmetti D. 2010. Space-and time-resolved protein dynamics in single bacterial cells observed on a chip. J Biotechnol 149:280–288. doi: 10.1016/j.jbiotec.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 68.Fields AT, Navarrete CS, Zare AZ, Huang Z, Mostafavi M, Lewis JC, Rezaeihaghighi Y, Brezler BJ, Ray S, Rizzacasa AL, Barnett MJ, Long SR, Chen EJ, Chen JC. 2012. The conserved polarity factor PodJ1 impacts multiple cell envelope-associated functions in Sinorhizobium meliloti. Mol Microbiol 84:892–920. doi: 10.1111/j.1365-2958.2012.08064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ratcliff WC, Denison RF. 2010. Individual-level bet hedging in the bacterium Sinorhizobium meliloti. Curr Biol 20:1740–1744. doi: 10.1016/j.cub.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 70.Jarvik JW, Telmer CA. 1998. Epitope tagging. Annu Rev Genet 32:601–618. doi: 10.1146/annurev.genet.32.1.601. [DOI] [PubMed] [Google Scholar]
- 71.Shevtsova Z, Malik JM, Michel U, Scholl U, Bahr M, Kugler S. 2006. Evaluation of epitope tags for protein detection after in vivo CNS gene transfer. Eur J Neurosci 23:1961–1969. doi: 10.1111/j.1460-9568.2006.04725.x. [DOI] [PubMed] [Google Scholar]
- 72.Booth WT, Schlachter CR, Pote S, Ussin N, Mank NJ, Klapper V, Offermann LR, Tang C, Hurlburt BK, Chruszcz M. 2018. Impact of an N-terminal polyhistidine tag on protein thermal stability. ACS Omega 3:760–768. doi: 10.1021/acsomega.7b01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aslantas Y, Surmeli NB. 2019. Effects of N-terminal and C-terminal polyhistidine tag on the stability and function of the thermophilic P450 CYP119. Bioinorg Chem Appl 2019:8080697. doi: 10.1155/2019/8080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lau J, Hernandez-Alicea L, Vass RH, Chissen P. 2015. A phosphosignaling adaptor primes the AAA+ protease ClpXP to drive cell cycle-regulated proteolysis. Mol Cell 59:104–116. doi: 10.1016/j.molcel.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pleier E, Schmitt R. 1991. Expression of two Rhizobium meliloti flagellin genes and their contribution to the complex filament structure. J Bacteriol 173:2077–2085. doi: 10.1128/jb.173.6.2077-2085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. doi: 10.1128/JB.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scharf B, Schuster-Wolff-Bühring H, Rachel R, Schmitt R. 2001. Mutational analysis of the Rhizobium lupini H13-3 and Sinorhizobium meliloti flagellin genes: importance of flagellin A for flagellar filament structure and transcriptional regulation. J Bacteriol 183:5334–5342. doi: 10.1128/jb.183.18.5334-5342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Götz R, Limmer N, Ober K, Schmitt R. 1982. Motility and chemotaxis in two strains of Rhizobium with complex flagella. J Gen Microbiol 128:789–798. doi: 10.1099/00221287-128-4-789. [DOI] [Google Scholar]
- 80.Sourjik V, Schmitt R. 1996. Different roles of CheY1 and CheY2 in the chemotaxis of Rhizobium meliloti. Mol Microbiol 22:427–436. doi: 10.1046/j.1365-2958.1996.1291489.x. [DOI] [PubMed] [Google Scholar]
- 81.Compton KK, Hildreth SB, Helm RF, Scharf BE. 2018. Sinorhizobium meliloti chemoreceptor McpV senses short-chain carboxylates via direct binding. J Bacteriol 200:e00519-18. doi: 10.1128/JB.00519-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arapov TD, Castaneda Saldana R, Sebastian AL, Keith Ray W, Helm RF, Scharf BE. 2020. Cellular stoichiometry of chemotaxis proteins in Sinorhizobium meliloti. J Bacteriol 200:JB.00141-20. doi: 10.1128/jb.00141-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simon R, O’Connell M, Labes M, Pühler A. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other Gram-negative bacteria. Methods Enzymol 118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 84.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]