Two-component signal transduction systems are widely used by bacteria to adequately respond to environmental changes by adjusting cellular parameters, including the cell cycle. In Caulobacter crescentus, PleC acts as a phosphatase that indirectly protects the response regulator CtrA from premature inactivation during the G1 phase of the cell cycle. Here, we provide genetic and biochemical evidence that PleC is seconded by another phosphatase, CckN. The activity of PleC and CckN phosphatases is restricted to the G1 phase since both proteins are degraded by ClpXP protease before the G1-S transition. Degradation is independent of any known proteolytic adaptors and relies, in the case of CckN, on an unsuspected N-terminal degron. Our work illustrates a typical example of redundant functions between two-component proteins.
KEYWORDS: two-component system, phosphorelay, phosphatase, cell cycle
ABSTRACT
In the model organism Caulobacter crescentus, a network of two-component systems involving the response regulators CtrA, DivK, and PleD coordinates cell cycle progression with differentiation. Active phosphorylated CtrA prevents chromosome replication in G1 cells while simultaneously regulating expression of genes required for morphogenesis and development. At the G1-S transition, phosphorylated DivK (DivK∼P) and PleD (PleD∼P) accumulate to indirectly inactivate CtrA, which triggers DNA replication initiation and concomitant cellular differentiation. The phosphatase PleC plays a pivotal role in this developmental program by keeping DivK and PleD phosphorylation levels low during G1, thereby preventing premature CtrA inactivation. Here, we describe CckN as a second phosphatase akin to PleC that dephosphorylates DivK∼P and PleD∼P in G1 cells. However, in contrast to PleC, no kinase activity was detected with CckN. The effects of CckN inactivation are largely masked by PleC but become evident when PleC and DivJ, the major kinase for DivK and PleD, are absent. Accordingly, mild overexpression of cckN restores most phenotypic defects of a pleC null mutant. We also show that CckN and PleC are proteolytically degraded in a ClpXP-dependent way before the onset of the S phase. Surprisingly, known ClpX adaptors are dispensable for PleC and CckN proteolysis, raising the possibility that as yet unidentified proteolytic adaptors are required for the degradation of both phosphatases. Since cckN expression is induced in stationary phase, depending on the stress alarmone (p)ppGpp, we propose that CckN acts as an auxiliary factor responding to environmental stimuli to modulate CtrA activity under suboptimal conditions.
IMPORTANCE Two-component signal transduction systems are widely used by bacteria to adequately respond to environmental changes by adjusting cellular parameters, including the cell cycle. In Caulobacter crescentus, PleC acts as a phosphatase that indirectly protects the response regulator CtrA from premature inactivation during the G1 phase of the cell cycle. Here, we provide genetic and biochemical evidence that PleC is seconded by another phosphatase, CckN. The activity of PleC and CckN phosphatases is restricted to the G1 phase since both proteins are degraded by ClpXP protease before the G1-S transition. Degradation is independent of any known proteolytic adaptors and relies, in the case of CckN, on an unsuspected N-terminal degron. Our work illustrates a typical example of redundant functions between two-component proteins.
INTRODUCTION
The alphaproteobacterium Caulobacter crescentus divides asymmetrically to generate two daughter cells with different cell fates, a sessile stalked cell and a motile swarmer cell. While the newborn stalked cell can immediately reenter the S phase and initiate chromosome replication, the smaller swarmer cell engages in an obligatory motile and chemotactic but nonreplicative G1 phase. Concomitantly with its entry into the S phase (G1-S transition), the swarmer cell differentiates into a stalked cell (swarmer-to-stalked cell transition). A complex regulatory network controlling the activity of the central and essential response regulator CtrA coordinates different cell cycle stages with accompanying morphological changes and development. CtrA activity is carefully regulated throughout the cell cycle at the transcriptional and posttranslational levels. CtrA protein levels and its phosphorylation status are mostly determined by the action of a phosphorelay involving the hybrid kinase CckA and its cognate histidine phosphotransferase (HPt) ChpT (1–4). In the swarmer cell, the kinase activity of CckA is stimulated at the flagellated pole by the physical contact with the nonconventional histidine kinase (HK) DivL (5–8). DivL is free to activate CckA since its inhibitor—the response regulator DivK—is dephosphorylated (i.e., inactivated) by the phosphatase PleC (PleCP). Hence, CckA promotes the ChpT-dependent phosphorylation of CtrA, thereby stimulating its activity. At the same time, the CckA/ChpT phosphorelay also protects CtrA from its proteolytic degradation by phosphorylating CpdR, a response regulator whose unphosphorylated form primes the ClpXP protease for CtrA degradation (4, 9). Active CtrA (CtrA∼P) binds the single chromosomal origin of replication (Cori) to prevent DNA replication initiation (Fig. 1a). As a transcription factor, CtrA∼P also directly activates or represses the expression of more than 200 genes involved in multiple biological processes, including cell cycle, cell differentiation, and cell division (10).
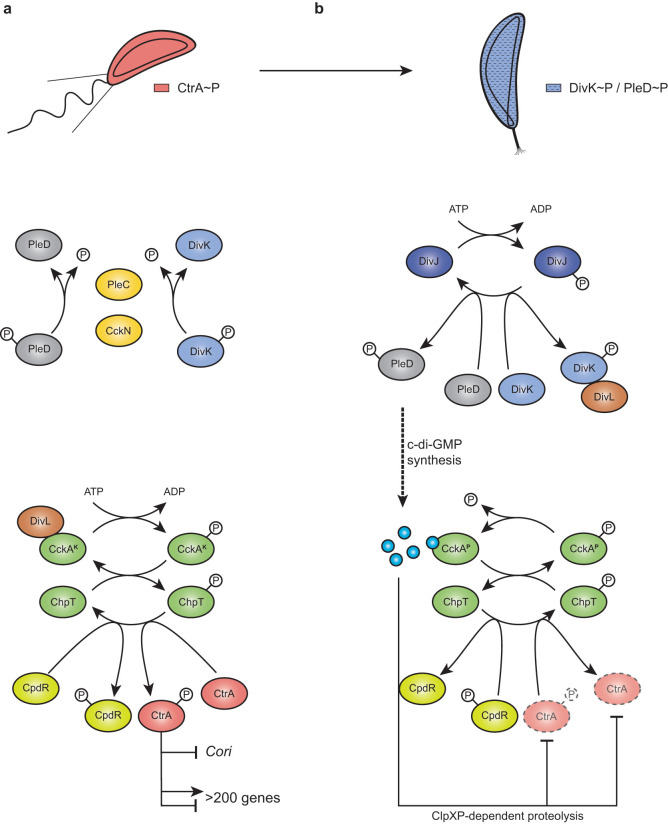
FIG 1.
(a and b) CtrA regulation pathway in Caulobacter crescentus in swarmer (a) and stalked (b) cells. In swarmer cells (a), actively dephosphorylated by PleC and CckN, DivK is therefore not able to interact with DivL. Free DivL activates the phosphorelay, culminating in CtrA and CpdR phosphorylation. Active CtrA (CtrA∼P) regulates the expression of more than 200 genes and inhibits DNA replication initiation by binding the single chromosomal origin of replication (Cori). At the G1-S transition (b), CckN and PleC are cleared from the cells, while DivK and PleD are phosphorylated by their cognate histidine kinase DivJ. Phosphorylated DivK (DivK∼P) interacts with DivL and reduces its affinity for CckA, leading to an inhibition of its kinase activity on CtrA and CpdR. Phosphorylation of PleD promotes its diguanylate cyclase activity, resulting in an increased synthesis of c-di-GMP. High levels of c-di-GMP not only stimulate CckA phosphatase activity on both CpdR∼P and CtrA∼P but also drive, concomitantly with unphosphorylated CpdR, ClpXP-dependent degradation of CtrA. Together, these effects result in the rapid inactivation of CtrA, allowing DNA replication initiation to proceed.
At the G1-S transition, DivK becomes highly phosphorylated (11, 12). This results from (i) a switch from PleC phosphatase to kinase activity before proteolytic removal of PleC and (ii) posttranslational stimulation of DivJ, the major HK responsible for DivK and PleD phosphorylation (15). Once phosphorylated, DivK∼P physically interacts with DivL and strongly reduces its affinity for CckA (7, 8, 13). Hence, the kinase activity of CckA is no longer stimulated. Simultaneously, CckA phosphatase activity is directly stimulated by c-di-GMP, the levels of which strongly and rapidly rise due to activation of the diguanylate cyclase PleD and inactivation of the phosphodiesterase PdeA. PleD becomes highly phosphorylated (i.e., activated) by DivJ at the differentiating pole (14, 15), whereas PdeA is degraded by ClpXP (16). High levels of c-di-GMP also drive ClpXP-dependent degradation of CtrA directly by binding to the proteolytic adaptor PopA (17, 18). Together, these events result in the rapid inactivation of CtrA during the G1-S transition and trigger an irreversible program leading to chromosome replication and cell differentiation (Fig. 1b). Inactivation of PleC phosphatase activity and the resulting increase in DivK∼P (and PleD∼P) are, to our knowledge, the earliest known events in this G1-S transition signaling pathway. We thus wondered whether other factors besides PleC and DivJ could influence DivK and PleD (de)phosphorylation.
Almost 20 years ago, interaction partners of DivK were identified in a yeast two-hybrid screen (13). Apart from PleC, DivJ, and DivL, which were unsurprisingly found as prominent hits, another HK, called CckN, was discovered in this study as a physical partner of DivK (13), but the role played by this actor in the CtrA regulatory network has not been characterized in detail so far. Here, we show that similar to PleC, CckN displays phosphatase activity toward DivK and PleD during the G1/swarmer phase of the cell cycle. However, in contrast to PleC, the kinase activity of CckN cannot be activated by DivK∼P at the G1-S transition. Both phosphatases are required to sustain optimal CtrA activity in the nonreplicative swarmer cells before being inactivated by proteolysis at the G1-S transition. Interestingly, we also show that both CckN and PleC are the earliest CtrA regulatory proteins to concomitantly disappear, likely by ClpXP-dependent proteolysis. Surprisingly, these degradations do not rely on any known proteolytic adaptors for ClpXP. In addition, we show that cckN expression is stimulated in the stationary phase, depending on (p)ppGpp. We propose a model in which CckN influences CtrA activity under nonoptimal growth conditions.
RESULTS
CckN is a second phosphatase for DivK and PleD.
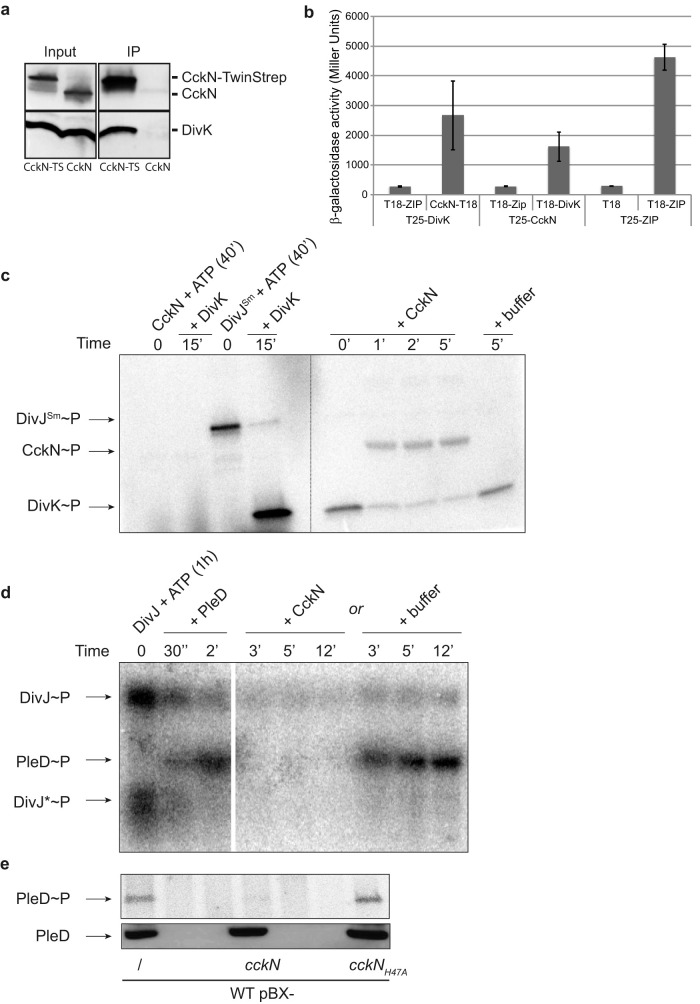
CckN was previously identified as an interaction partner of DivK in a yeast two-hybrid screen (13). The interaction of CckN with DivK was confirmed by coimmunoprecipitation (Fig. 2a) and bacterial two-hybrid assays (Fig. 2b). We next tested whether CckN displayed kinase activity, i.e., could autophosphorylate in vitro in the presence of ATP. Purified CckN with either an N-terminal His6 or a His6-MBP tag did not show autokinase activity in vitro in our experiments, despite the presence of a predicted histidine kinase-like ATPase (HATPase) domain (pfam02518) and all the catalytic residues in the DHp and CA domains conserved in prototypical HisKA-type histidine kinases (19). In contrast, we detected robust autophosphorylation of the soluble cytoplasmic catalytic histidine kinase (HK) core region of His6-MBP-tagged purified C. crescentus DivJ and PleC proteins or His6-tagged purified Sinorhizobium meliloti DivJ (DivJSm) (Fig. 2c; see Fig. S1a and b in the supplemental material). A nonphosphorylatable variant of DivK (DivKD53N) stimulated C. crescentus DivJ and PleC autokinase activity, as reported before (15), but did not show any stimulatory effect on CckN autophosphorylation (Fig. S1a and b). Accordingly, DivK could be phosphorylated with the noncognate DivJSm but not with CckN. In contrast, CckN was able to efficiently dephosphorylate DivK∼P, with CckN becoming simultaneously phosphorylated (Fig. 2c). Since PleC and DivJ are known to also (de)phosphorylate another response regulator akin to DivK, PleD, we next tested whether CckN could dephosphorylate PleD. As shown in Fig. 2d, CckN was able to rapidly dephosphorylate PleD. In the presence of DivKD53N in reaction mixtures containing DivJ and CckN, PleD dephosphorylation was still observed (Fig. S1c), suggesting that in contrast to PleC (15), the kinase activity of CckN is not subject to stimulation by DivKD53N. Finally, we measured PleD∼P levels in vivo in strains overexpressing cckN from a multicopy plasmid under the control of the xylose-inducible PxylX promoter (pBX-cckN). In line with in vitro data, we found that the phosphorylated form of PleD (PleD∼P) was strongly reduced upon overexpression of wild-type cckN compared to a control strain harboring an empty vector, whereas overexpression of a mutant variant of cckN (cckNH47A) that has the predicted phosphorylatable His47 residue mutated to Ala did not influence PleD∼P levels (Fig. 2e). Together, these results suggest that CckN is a phosphatase but not a kinase for both DivK and PleD.
FIG 2.
CckN is a phosphatase for DivK and PleD. (a) Coimmunoprecipitation (Co-IP) experiments showing that CckN and DivK are part of the same protein complex. Co-IP was performed on protein extracts of wild-type cells harboring the pBX-cckN-TwinStrep (RH2235) or the pBX-cckN (RH2070) plasmid. CckN and DivK were detected by Western blotting using, respectively, anti-CckN and anti-DivK antibodies before (input) and after (IP) immunoprecipitation with Strep-Tactin coated magnetic beads. CckN-TS, CckN-TwinStrep. (b) Bacterial two-hybrid assay showing that CckN and DivK interact with each other. β-Galactosidase assays were performed on MG1655 cyaA::frt (RH785) strains coexpressing T18 fused to ZIP, cckN, or divK with T25 fused to ZIP, cckN, or divK. T18 and T25 alone were used as negative controls, while coexpression of T18-ZIP and T25-ZIP was used as a positive control. Error bars indicate standard deviations (SD); n ≥ 3. (c) In vitro phosphorylation assay showing that CckN cannot phosphorylate DivK but can dephosphorylate DivK∼P. CckN or DivJSm was incubated alone for 40 min with [γ-32P]ATP before the addition of DivK for 15 min. Then, DivK phosphorylated by DivJSm was washed to remove excess [γ-32P]ATP (dotted line) and incubated with or without CckN for the indicated time. (d) In vitro phosphorylation assay showing that CckN can dephosphorylate PleD∼P. DivJ was incubated alone for 1 h with [γ-32P]ATP before the addition of PleD for 2 min followed by the addition of CckN or buffer and incubation for the indicated time. DivJ* likely corresponds to a degradation product of DivJ. (e) In vivo phosphorylation assay showing that overexpression of functional cckN decreases PleD phosphorylation. Wild-type (RH50) cells harboring the pBX, pBX-cckN, or cckNH47A plasmid were grown for 3 h in M5G with 0.05 mM phosphate supplemented with 0.1% xylose. The phosphorylation (up) and protein (down) levels of PleD were determined in vivo as described in Materials and Methods.
CckN impacts CtrA activity through DivK.
As a regulator of DivK phosphorylation, CckN is expected to affect CtrA activity. To test this hypothesis, we monitored the activity of CtrA-dependent promoters in different genetic backgrounds using lacZ transcriptional reporters and β-galactosidase assays. We found that inactivating cckN in an otherwise wild-type background did not substantially change CtrA activity (Fig. S2a). Likewise, ΔcckN did not interfere with the hyper-activity of CtrA measured in a ΔdivJ background (Fig. S2a). Whereas CtrA-dependent promoters displayed almost no activity in ΔpleC cells (Fig. S2a), the concomitant inactivation of divJ in ΔdivJ ΔpleC cells restored a detectable activity (Fig. 3a; Fig. S2b). Interestingly, cckN inactivation in this ΔdivJ ΔpleC background diminished activity of CtrA-dependent promoters, with a decrease ranging from 20% to 80% depending on the promoter, but did not influence the activity of promoters that are not regulated by CtrA (Fig. 3a; Fig. S2b). These results suggest that PleC constitutes the main phosphatase of DivK and thus needs to be inactivated—together with DivJ—to unmask the effects of CckN. We also found that ΔcckN modulated activity of CtrA in a pleCF778L background (Fig. S3a). This PleC variant was described to lack kinase but not phosphatase activity (PleCK–P+) in vitro and was shown to complement motility, phage sensitivity, and stalk biogenesis defects of a ΔpleC mutant when expressed on a multicopy plasmid (20). However, in cells in which pleCF778L was expressed as the only copy at the endogenous pleC locus, the activity of CtrA-dependent promoters was less active than in wild-type cells but more active than in ΔpleC cells (Fig. S3a). These data suggest that when expressed from its native genomic context, the phosphatase activity displayed by PleCF778L is reduced compared to wild-type PleC. Accordingly, the activity of CtrA-dependent promoters was further decreased in pleCF778L ΔcckN cells to levels observed with fully inactive alleles of pleC, such as ΔpleC or pleCH610A (Fig. S3a). In line with these effects, cckN inactivation in a pleCF778L background further decreased motility and attachment compared to the parental pleCF778L strain (Fig. S3b). Together, these data support the idea that PleC masks the phosphatase activity of CckN on DivK under standard laboratory conditions.
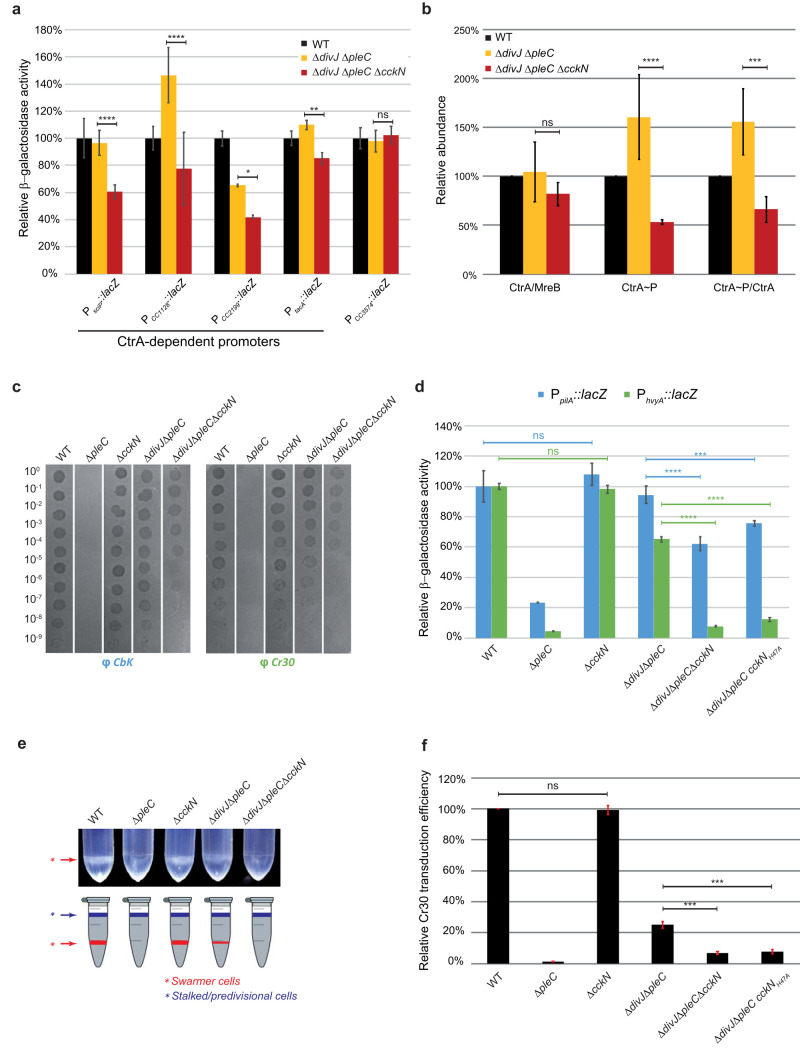
FIG 3.
CckN controls development by regulating CtrA activity. (a) β-Galactosidase assays were performed on wild-type (RH50), ΔdivJ ΔpleC (RH1103), and ΔdivJ ΔpleC ΔcckN (RH1111) strains harboring lacZ fusions to CtrA-dependent (PsciP, PCC1128, PCC2199, and PtacA) and CtrA-independent (PCC3574) promoters grown in complex medium (PYE) and normalized to the wild type (WT) (100%). Error bars indicate SD; n = 4. (b) The protein and the phosphorylation levels of CtrA were measured in wild-type (RH50), ΔdivJ ΔpleC (RH1103), and ΔdivJ ΔpleC ΔcckN (RH1111) strains and normalized to the WT (100%). The CtrA protein levels normalized to the MreB levels were determined by Western blotting. The CtrA phosphorylation levels were determined in vivo as described in Materials and Methods. The CtrA∼P/CtrA ratios were obtained by dividing black values by yellow values. Error bars indicate SD; n = 3. (c) Bacteriophage sensitivity assays were performed with CbK and Cr30 on wild-type (RH50), ΔpleC (RH217), ΔcckN (RH1106), ΔdivJ ΔpleC (RH1103), and ΔdivJ ΔpleC ΔcckN (RH1111) strains on PYE agar plates. (d) β-Galactosidase assays were performed on wild-type (RH50), ΔpleC (RH217), ΔcckN (RH1106), ΔdivJ ΔpleC (RH1103), ΔdivJ ΔpleC ΔcckN (RH1111), and ΔdivJ ΔpleC cckNH47A (RH1800) strains harboring PpilA::lacZ or PhvyA::lacZ fusions grown in complex medium (PYE) and normalized to the WT (100%). Error bars indicate SD; n = 3. (e) Buoyancy was evaluated by mixing 550 μl of Ludox LS colloidal silica (30%) with 1 ml of wild-type (RH50), ΔpleC (RH217), ΔcckN (RH1106), ΔdivJ ΔpleC (RH1103), and ΔdivJ ΔpleC ΔcckN (RH1111) strains grown in PYE. Then, the mix was centrifuged for 15 min at 9,000 rpm. (f) Cr30-dependent transduction efficiency was measured by transducing Cr30 lysates (LHR73 or LHR75) in wild-type (RH50), ΔpleC (RH217), ΔcckN (RH1106), ΔdivJ ΔpleC (RH1103), ΔdivJ ΔpleC ΔcckN (RH1111), and ΔdivJ ΔpleC cckNH47A (RH1800) strains grown in complex medium (PYE) and normalized to the WT (100%). Error bars indicate SD; n = 2. Means were statistically compared using a two-way ANOVA (a, b, and d) or a one-way ANOVA (f), followed by Tukey’s multiple-comparison test; ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Given that DivK∼P negatively affects CtrA phosphorylation and protein levels (4, 21), we monitored phosphorylation and protein abundance of CtrA in ΔdivJ ΔpleC versus ΔdivJ ΔpleC ΔcckN cells. Whereas CtrA abundance did not vary substantially, the phosphorylation state of CtrA was strongly diminished in the triple mutant (ΔdivJ ΔpleC ΔcckN) in comparison to the parental ΔdivJ ΔpleC strain (Fig. 3b). Thus, CckN impacts CtrA activity mainly by promoting its phosphorylation, presumably via dephosphorylation of DivK. Comparative chromatin immunoprecipitation experiments coupled to deep sequencing (ChIP-seq) in ΔdivJ ΔpleC ΔcckN versus ΔdivJ ΔpleC cells showed that cckN inactivation impacts the entire CtrA regulon (Fig. S3c, Table S1).
Finally, we checked whether the effect of CckN on CtrA phosphorylation and activity was fully dependent on DivK by monitoring the activity of CtrA in the absence of divK. As divK is essential but was shown to be dispensable in a cpdRD51A background (22), we measured the activity of CtrA-regulated promoters in cpdRD51A ΔdivK with or without cckN. In contrast to divK+ cells (Fig. 3a and d), inactivating cckN did not influence CtrA activity in the absence of DivK any further (Fig. S3d), suggesting that CckN acts on CtrA mostly—if not entirely—through DivK. Altogether, these results suggest that CckN is a phosphatase of DivK akin to PleC, ultimately keeping CtrA active in G1 cells.
CckN controls development in a CtrA-dependent way.
Next, we tested whether cckN inactivation displayed developmental defects. A ΔpleC strain is known to be fully resistant to both CbK and Cr30 bacteriophages (Fig. 3c) (23, 24). This is due to pilA and hvyA transcription being directly activated by CtrA∼P and not being sufficiently expressed in the absence of PleC (Fig. 3d) (24, 25). pilA codes for the major pilin subunit of polar pili used by phage CbK as a receptor (26). hvyA encodes a transglutaminase homolog that specifically protects swarmer cells from capsulation, thereby allowing the Cr30 bacteriophage to reach its receptor, the S-layer (27). Thus, neither CbK nor Cr30 can infect ΔpleC cells since the polar pili are absent and the S-layer is inaccessible. Because the differential capsulation of Caulobacter daughter cells is also responsible for their specific buoyancy properties, with the noncapsulated swarmer cells being heavy and the other capsulated cell types light (24), ΔpleC swarmer cells lack their typical low buoyancy feature by becoming capsulated (Fig. 3e). As expected and as reported before, full resistance to both bacteriophages, high buoyancy, and low promoter activity for pilA (PpilA) and hvyA (PhvyA) genes displayed by the single ΔpleC strain were restored by inactivating divJ (Fig. 3c to e) (11, 24, 28). Interestingly, we found that a nonfunctional allele of cckN (ΔcckN or cckNH47A) reduced sensitivity to bacteriophages, significantly decreased the activity of PpilA and PhvyA, and lost low buoyancy in a ΔdivJ ΔpleC or pleCF778L background but not in an otherwise wild-type background (Fig. 3c to e; Fig. S3e). In line with the lower sensitivity to Cr30 infection, the relative Cr30-mediated transduction efficiency was also significantly reduced in ΔdivJ ΔpleC ΔcckN and ΔdivJ ΔpleC cckNH47A mutants compared to the parental ΔdivJ ΔpleC strain (Fig. 3f). In addition, expressing cckN from the chromosomal xylose-inducible promoter PxylX could complement loss of cckN in the ΔdivJ ΔpleC ΔcckN mutant, as sensitivity to both CbK and Cr30 infections increased upon xylose induction (Fig. S3f). Together, these data suggest that cckN controls development by sustaining optimal CtrA activity.
Overexpression of cckN suppresses ΔpleC defects by enhancing CtrA activity.
Our results suggested that cckN overexpression should lead to an increase of CtrA phosphorylation. To test this hypothesis, cckN was first mildly overexpressed in a ΔpleC mutant from the endogenous xylX locus. According to the data presented above, we found that the xylose-induced expression of cckN in a ΔpleC background (ΔpleC PxylX::cckN) restored sensitivity to CbK and Cr30 phages (Fig. S4a) and significantly increased hvyA and pilA expression (Fig. S4b). Thus, CckN is capable of replacing PleC function under conditions where PleC is absent. However, the same mild overexpression of cckN did not restore attachment of ΔpleC cells (Fig. S4c), likely because the holdfast production required for irreversible attachment essentially relies on PleC kinase rather than phosphatase activity on PleD (15). In support of this, mild overexpression of cckN in wild-type cells significantly decreased binding to abiotic surfaces, likely by dephosphorylating PleD∼P (Fig. S4c). Furthermore, cckN inactivation in an otherwise wild-type or a ΔdivJ ΔpleC background did not significantly decrease attachment (Fig. S4d). In contrast, cckN inactivation in a pleCF778L background—which displayed a partial attachment defect compared to the complete loss of attachment of a pleC null mutant—led to a significant decrease of attachment (Fig. S3b). These effects could be due to the decrease of CtrA∼P, which results in reduced expression of pilA (Fig. S3a) involved in primary attachment (29) and hfa genes involved in holdfast attachment (10).
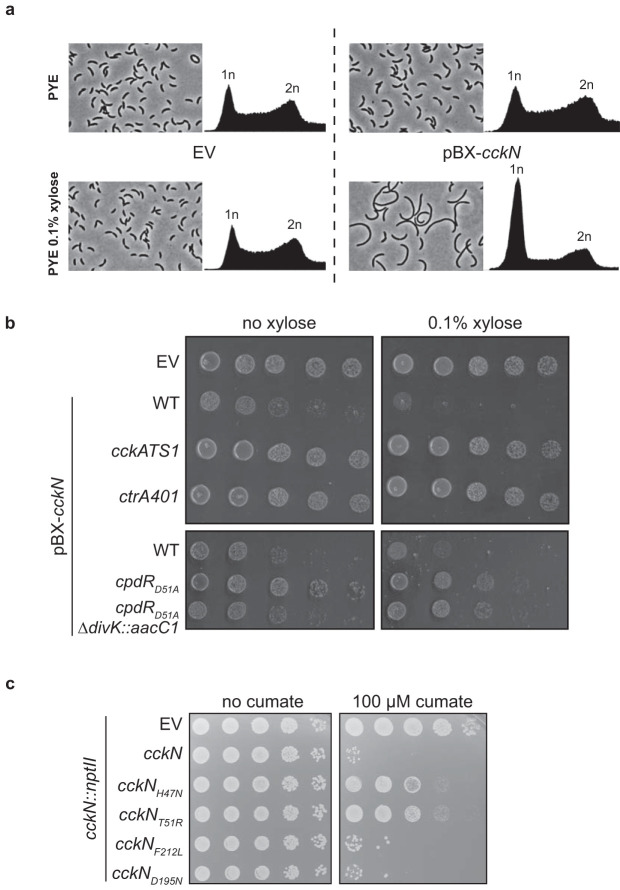
To further characterize the role played by cckN, we generated a multicopy plasmid on which cckN was placed under the control of the xylose-inducible PxylX promoter (pBX-cckN). In comparison to the control wild-type strain harboring an empty plasmid (EV), wild-type cells harboring pBX-cckN were filamentous and arrested in G1 when xylose was added to the medium (Fig. 4a). This result suggests that a strong overexpression of cckN led to hyperactivation of CtrA∼P, which interfered with DNA replication initiation, and consequently to a reduced viability as estimated by CFU count (Fig. 4b). The toxicity associated with cckN overexpression was not observed in cells with reduced CtrA activity (cckATS1 and ctrA401) (1, 3) or abundance (cpdRD51A and cpdRD51A ΔdivK) (22) (Fig. 4b). Indeed, the thermosensitive cckATS1 allele harbors two mutations (I484N and P485A) that compromise the kinase activity of CckA, thereby decreasing CtrA phosphorylation (3, 30), while the ctrA401 allele harbors a T170I substitution in CtrA that decreases its activity (1, 31). On the other hand, CtrA activity is reduced in a cpdRD51A background since this phosphoablative variant of CpdR leads to constant degradation of CtrA along the cell cycle (22). Similarly, CckN variants that are predicted to lack kinase and phosphatase activities (CckNK–P–)—cckNH47N and cckNT51R, the latter of which was designed based on the PleCK–P– variant T561R (20)—were less toxic upon overexpression than wild-type CckN (Fig. 4c). In contrast, overexpression of CckN variants predicted to have lost only kinase activity (CckNK–P+)—cckNF212L, designed based on PleCF778L (20), and cckND195N, harboring a mutation in the G1 box required for ATP binding—were still as toxic as the wild type (Fig. 4c). Thus, our data suggest that the major role of CckN in vivo is not to phosphorylate, but to dephosphorylate DivK and PleD and thereby to stimulate CtrA activity.
FIG 4.
Overexpression of cckN leads to a CtrA-dependent toxic G1 block. (a) Phase-contrast imaging and FACS profiles of wild-type (RH50) cells harboring either the empty pBX vector (EV) or a pBX-cckN plasmid grown for 3 h in PYE supplemented with 0.1% xylose. (b) Serial dilutions of the wild-type (RH50) strain harboring the empty pBX vector (EV) and the wild-type (RH50), cckATS1 (RH340), ctrA401 (RH212), cpdRD51A (RH347), and cpdRD51A ΔdivK::aacC1 (RH1411) strains harboring pBXMCS-2-cckN grown in PYE were spotted on PYE (left) or PYE supplemented with 0.1% xylose (right) and incubated for 2 days at 30°C. (c) Serial dilutions of the cckN::nptII strain harboring the empty pQF vector (EV) or pQF vector expressing cckN variants grown in PYE were spotted on PYE (left) or PYE supplemented with 100 μM cumate (right) and incubated for 2 days at 30°C.
Transcription of cckN is stimulated by CtrA∼P and (p)ppGpp.
Since our ChIP-Seq data suggested that cckN might be a direct target of CtrA (Table S1), we monitored the promoter activity of cckN (PcckN) using a transcriptional fusion to lacZ on a replicative plasmid (pRKlac290-PcckN) in mutant strains harboring higher or lower CtrA activity. We found that the activity of PcckN was decreased or increased in strains harboring lower (ΔpleC, ctrA401, and cckATS1) or higher (ΔdivJ and divK341) CtrA activity, respectively (Fig. S5a and b). These data suggest that cckN is subjected to a positive feedback loop by CtrA∼P, but this regulation might be indirect, since we could not identify a predicted consensus CtrA binding site in the promoter region of cckN. We also found that PcckN activity was induced upon entry into stationary phase (Fig. S5c). In agreement with recent data showing that (p)ppGpp is required to sustain optimal activity of CtrA-dependent promoters during stationary phase (31), induction of PcckN was not observed in (p)ppGpp0 (ΔspoT) cells (Fig. S5c). In addition, ectopic production of (p)ppGpp—by a functional truncated version of the Escherichia coli (p)ppGpp synthetase RelA expressed from the xylose-inducible promoter (PxylX::relA′) at the xylX locus—increased PcckN activity in exponentially growing cells only when xylose was supplemented to the medium (Fig. S5d). In contrast, inducing the expression of the corresponding catalytically inactive variant of RelA (PxylX::relA′E335Q) with xylose did not change PcckN activity (Fig. S5d). Together, these results suggest that induction of cckN expression in stationary phase by CtrA∼P is (p)ppGpp dependent.
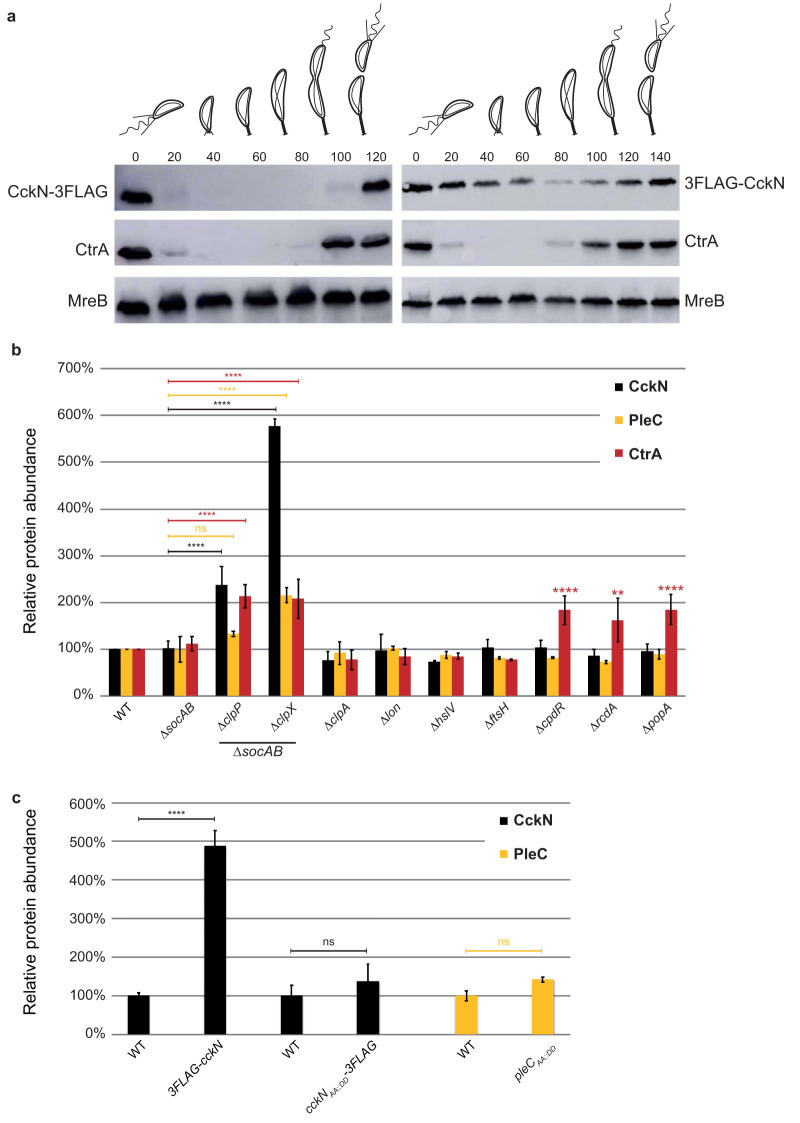
PleC and CckN are cleared from G1 cells in a ClpXP-dependent way.
Based on the results presented above, PleC and CckN prevent premature inactivation of CtrA during the G1 phase of the cell cycle by maintaining the low phosphorylation level of DivK. At the G1-S transition, DivK∼P levels rise to indirectly trigger CtrA dephosphorylation and ClpXP-dependent proteolysis. This suggests that both CckN and PleC should be inactivated first. Thus, we tested whether CckN abundance fluctuates along the cell cycle by monitoring levels of CckN fused to a triple FLAG tag at either its N or C terminus expressed as the only copy from the native chromosomal locus. Interestingly, CckN-3FLAG was present only in G1/swarmer cells, whereas 3FLAG-CckN remained roughly constant throughout the cell cycle (Fig. 5a). The strains expressing a 3FLAG tag at the 5′ or 3′ extremity of cckN did not display any particular morphological or growth defects in comparison to the wild-type strain. The rapid disappearance of CckN-3FLAG during the G1-S transition (Fig. S6a and b) suggests the involvement of ATP-dependent proteolysis. To identify the protease responsible for CckN proteolysis, CckN-3FLAG abundance was quantified in a set of known C. crescentus protease and proteolytic adaptor mutant strains. CtrA was used as a control since it is degraded during the G1-S transition by the ClpXP protease with the help of the adaptor proteins CpdR, RcdA, and PopA bound to c-di-GMP (32). As expected, CtrA levels increased in ΔclpX and ΔclpP strains, as well as in strains lacking its known proteolytic adaptors CpdR, RcdA, and PopA (9, 17, 33) (Fig. 5b). Note that deletion of clpX and clpP genes was done in a ΔsocAB background that suppresses their essentiality (34). CckN-3FLAG levels increased in the ΔclpX and ΔclpP mutants, and this effect was independent of ClpXP proteolytic adaptors required for degradation of cell cycle regulators (Fig. 5b) (2, 35–38). In agreement with this result, we found that CckN-3FLAG levels properly fluctuated throughout the cell cycle in ΔcpdR cells, in contrast to CtrA, whose degradation strictly depends on CpdR (Fig. S6d). As PleC abundance was also suggested to vary along the cell cycle (39) by disappearing together with CtrA at the G1-S transition (Fig. S6a and b), we also determined PleC abundance in the same protease and proteolytic adaptor mutants. Similarly to CckN, PleC abundance increased in ΔclpX and in ΔclpP mutants but not in strains lacking CpdR, RcdA, and PopA adaptors (Fig. 5b).
FIG 5.
The ClpXP protease is responsible for the cell cycle oscillation of CckN and PleC. (a) Immunoblotting of protein samples extracted from synchronized cckN-3FLAG (RH1881) or 3FLAG-cckN (RH1929) cells to follow CckN, CtrA, and MreB abundance throughout the cell cycle. The times at which the samples were withdrawn after synchrony are indicated in minutes. (b) The relative abundance of CckN-3FLAG, PleC, and CtrA was measured in wild-type (RH50 or RH1881), ΔsocAB (RH1671 or RH737), ΔsocAB ΔclpP (RH2279 or RH1063), ΔsocAB ΔclpX (RH1674 or RH995), ΔclpA (RH864 or RH1093), Δlon (RH2472 or RH1228), ΔhslV (RH864 or RH1247), ΔftsH (RH865 or RH1229), ΔcpdR (RH339 or RH1133), ΔrcdA (RH323 or RH1149), and ΔpopA (RH315 or RH1151) strains. Means were statistically compared using a two-way ANOVA, followed by Dunnett’s multiple-comparison test; ns, not significant; **, P < 0.01; ****, P < 0.0001. (c) The relative abundance of CckN-3FLAG and PleC and CtrA was measured in wild-type (RH1881), 3FLAG-cckN (RH1929), cckNAA::DD-3FLAG (RH1576), and pleCAA::DD (RH2548) strains grown in complex medium (PYE) and normalized to the corresponding WT (100%). Error bars indicate SD; n = 3. Means were statistically compared using a one-way ANOVA, followed by Sidak’s multiple-comparison test; ns, not significant; ****, P < 0.0001. Only significant differences are indicated on the graph.
Previous studies demonstrated that substituting the two C-terminal hydrophobic residues (AA, AG, or VA) of cell cycle regulators proteolyzed by ClpXP with two aspartate residues (DD)—CtrAAA::DD, KidOVA::DD, TacAAG::DD, GdhZAA::DD, and ShkAAG::DD—led to their stabilization (2, 35–38). Since both CckN and PleC displayed two alanine residues at their C-terminal extremity (Fig. S6c), we tested their potential involvement in the recognition of PleC and CckN by ClpX by creating pleCAA::DD and cckNAA::DD-3FLAG mutants and monitoring protein abundance in asynchronous and synchronized populations. CckNAA::DD-3FLAG was not protected from degradation along the cell cycle (Fig. 5b; Fig. S6e), suggesting that in contrast to most ClpXP substrates, CckN degradation does not rely on a C-terminal degron. In contrast, PleCAA::DD levels increased and did not oscillate anymore over the cell cycle in comparison to wild-type PleC (Fig. 5b; Fig. S6f and g), suggesting that the C-terminal motif Ala-Ala is critical for PleC degradation. Given that a 3FLAG fusion at the N-terminal extremity of CckN led to protein stabilization, whereas the same tag at the C-terminal end did not interfere with proteolysis (Fig. 5a), an N-terminal motif instead of the two C-terminal hydrophobic residues is likely involved in the recognition of CckN by ClpX. Altogether, our data show that CckN and PleC are degraded in a ClpXP-dependent manner early in G1 phase, before the inactivation of the master regulator CtrA at the G1-S transition by dephosphorylation and proteolysis. However, in contrast to the cell cycle regulators described so far to be proteolyzed by ClpXP, including CtrA itself, degradation of CckN and PleC does not rely on known proteolytic adaptors and involves for CckN an N-terminal motif. Possibly, as yet unidentified ClpXP adaptor proteins are required for timely degradation of PleC and CckN at the G1-S transition. Alternatively, PleC and CckN could be directly recognized by ClpXP without the help of any auxiliary factors.
DISCUSSION
CckN was identified almost 2 decades ago in a yeast two-hybrid screen as an interaction partner of the response regulator DivK (13). Here, we confirmed this interaction and also uncovered CckN as a second functional phosphatase for DivK and PleD (Fig. 1). In contrast to the primary phosphatase PleC, whose inactivation leads to pleiotropic effects, cckN loss-of-function mutants (ΔcckN or cckNH47A) did not display any detectable phenotype in an otherwise wild-type background. In contrast, deletion of cckN in a ΔdivJ ΔpleC background led to a decrease of CtrA activity (Fig. 3; see Fig. S1 and S3 and Table S1 in the supplemental material) as well as developmental defects. In addition, a mild overexpression of cckN suppresses ΔpleC defects associated with its phosphatase, but not kinase, activity (Fig. S4), while strong cckN overexpression leads to a CtrA-dependent G1 block and toxicity (Fig. 4). Thus, our data suggest that PleC phosphatase might be seconded by CckN upon specific conditions that still need to be uncovered.
Despite the redundancy of their phosphatase activity, CckN and PleC display different subcellular localizations since PleC operates at the swarmer pole while CckN is diffused into the cytosol (Fig. S7). It is noteworthy that a CckN-green fluorescence protein (GFP) fusion expressed from the native cckN locus was almost undetectable, suggesting that the expression level of cckN is low. It is likely that the polar localization of PleC (11) has been selected for regulating its kinase rather its phosphatase activity. Indeed, PleCK is allosterically activated at the differentiating pole by polar DivK∼P (15), suggesting that the kinase activity of PleC is restricted to that pole. In contrast, DivK∼P can still be efficiently dephosphorylated by PleC without being localized at the pole in ΔspmX cells (28). In ΔspmX, the activity of the σ54-dependent transcriptional activator TacA is exacerbated (40). A comparative experiment involving transposon (Tn) insertions coupled to deep sequencing (Tn-Seq) done on ΔspmX versus wild-type cells supported that conclusion, since insertions into genes coding for positive regulators of TacA activity or expression were overrepresented (40). This was the case for shkA coding for the hybrid kinase ShkA activating TacA by phosphorylation or rpoN encoding σ54 (40). Interestingly, Tn insertions also strongly accumulated in pleC and cckN in this experiment, likely because inactivation of their phosphatase activity downregulates the CtrA-dependent PtacA promoter, thereby limiting tacA expression. Alternatively, or in addition, a reduction in CtrA activity might also result in decreased expression of other genes affecting the ShkA-TacA pathway, such as pleD or other factors that could influence the phosphorylation state of the PleD/ShkA/TacA axis. Since TacA expression largely relies on the phosphatase activity of PleC (38), these Tn-Seq data support the redundancy between CckN and PleC. Notwithstanding, it has been shown that TacA phosphorylation by ShkA during the G1 phase requires c-di-GMP synthesized by PleD (38). Since TacA activity—monitored with a PspmX::lacZ translational fusion—was not decreased in ΔdivJ ΔpleC cells but was strongly diminished in ΔpleD cells, it has been proposed that a third histidine kinase (HK) phosphorylates PleD in a ΔdivJ ΔpleC background (38). In agreement with the fact that CckN does not display kinase activity (Fig. S1), we found that PspmX activity was as high in ΔdivJ ΔpleC ΔcckN cells as in wild-type cells (Fig. S8), thereby excluding CckN as the missing kinase for PleD. Thus, other phosphodonors for PleD, as well as for DivK, as already suggested (11), remain to be uncovered. Indeed, the fact that cckN inactivation causes DivK-dependent phenotypes in a ΔdivJ ΔpleC background demonstrates that, in the absence of DivJ and PleC, DivK is largely functional. The fact that CckN does seem to play a modulatory role on DivK/CtrA activity, rather than an essential role, suggests the presence of yet other regulators affecting DivK.
Akin to DivK, PleC and DivJ also modulate PleD phosphorylation (14, 15, 41). We show here that PleD is completely dephosphorylated in vivo upon cckN overexpression (Fig. 2e), very likely as a result of two complementary effects, first, by decreasing phosphorylation levels of DivK, which was shown to allosterically affect DivJ and PleC kinase activity and thereby PleD phosphorylation (15), and second, by directly dephosphorylating PleD∼P (Fig. 2d). Concomitantly maintaining low PleD∼P and DivK∼P levels is important for coordinating cell cycle and developmental control. Indeed, strong activation of PleD in G1 cells would trigger premature abandonment of motility without starting DNA replication. Interestingly, no autokinase activity was detected with CckN even in the presence DivKD53N, yet DivKD53N was able to strongly induce kinase activity of DivJ and PleC (15) (Fig. S1).
Surprisingly, we observed phosphorylated CckN in the presence of DivJ and ATP (Fig. S1c). Thus, CckN and DivJ might interact with each other to form heterodimers in which CckN is transphosphorylated by DivJ. If true, this would even further protect DivK and PleD from premature activation during G1 or the G1-S transition by draining phosphoryl groups away from DivJ to CckN. CckN could act as a prototypical phosphatase, i.e., catalyzing hydrolysis of phospho-aspartate on the receiver domain (REC-Asp∼P) of DivK and PleD via coordination of a water molecule without being phosphorylated itself. Alternatively, CckN could serve as a sort of histidine phosphotransferase (HPt) that accepts phosphoryl groups from REC-Asp∼P in a back-transfer reaction, thereby leading to REC dephosphorylation while being phosphorylated itself. In fact, the phosphorylatable His residue required for autophosphorylation is dispensable for phosphatase activity in the prototypical bifunctional histidine kinase/phosphatase EnvZ, where His243 can be replaced by several amino acids still allowing significant dephosphorylation of its cognate substrate, OmpR (42). In contrast, His47 of CckN seems essential for its proposed function in dephosphorylating DivK and PleD, since replacement of His47 in CckN by Ala or Asn essentially phenocopies a cckN null allele in all assays tested. Of note, the idea that bifunctional histidine kinases/phosphatases with an intact CA domain can mainly function as HPts to distribute phosphoryl groups between up- and downstream components is not unprecedented. It was recently proposed that LovK and PhyK involved in the general stress response in C. crescentus exert their main functions by acting as HpPts (43). PhyK and LovK share similarity in their DHp domains but belong to different HK subfamilies, HisKA2 and HWE, respectively, the former of which essentially has a CA domain identical to HisKA kinases with all known residues important for autophosphorylation conserved (43–45). PhyP from the alphaproteobacterium Sphingomonas melonis Fr1 harbors a DHp domain similar to PhyK/LovK but fails to be classified as either HisKA2 or HWE kinase due to its degenerate CA domain (46). PhyP was initially described as a phosphatase for PhyR, the response regulator universally controlling the general stress response in alphaproteobacteria (46, 47). However, PhyP was recently shown to act as an HPt rather than a true phosphatase, shuttling the phosphoryl group toward or away from PhyR (48). Thus, histidine kinases/phosphatases exist that employ back-transfer of phosphoryl groups as a means to dephosphorylate the response regulator. Whether CckN (and PleC) belongs to this group of enzymes employing such a mechanism remains to be elucidated in the future.
Our data strongly suggest that both CckN and PleC are proteolyzed in a ClpXP-dependent manner in early G1 cells (Fig. 5; Fig. S6). Unexpectedly and in contrast to any ClpXP substrates described to date, it seems that none of the known ClpX-dependent adaptors is required for CckN or PleC degradation. This is surprising knowing that CckN was found as a potential partner of RcdA in a pulldown assay (49). RcdA is a proteolytic adaptor that delivers some ClpXP substrates (e.g., TacA) to the CpdR-primed ClpXP protease (49). Thus, it is possible that CckN interacts with RcdA to limit its availability as a protease adaptor during early G1 phase, thereby avoiding premature RcdA functions in the hierarchical proteolytic events. Although we cannot exclude the possibility that PleC and CckN are directly recognized and bound by ClpX to be degraded, their rapid turnover in the G1 phase suggests the existence of a novel adaptor protein primed to ClpXP.
Since CckN is restricted to G1 cells in C. crescentus, its functions could be important in conditions requiring an extended G1 phase. In its natural oligotrophic environment, C. crescentus is expected to encounter extended periods of nutrient starvation during which the swarmer cells might not initiate DNA replication. Indeed, we know that limiting nitrogen or carbon leads to a (p)ppGpp-dependent G1 block in C. crescentus (50, 51) and that cckN expression is positively regulated by (p)ppGpp (Fig. S5c and d). In these stressful conditions, CckN might therefore be required to second PleC in maintaining DivK low phosphorylation low and avoiding premature inactivation of CtrA∼P. We tested this hypothesis by comparing the viability of wild-type and ΔcckN cells maintained in the stationary phase of growth. However, despite a (p)ppGpp-dependent induction of cckN expression under these conditions, ΔcckN cells remained as viable as the wild type when maintained in the stationary phase for days (Fig. S5e). Likewise, we did not find any particular stressful conditions (nutrient starvation, oxidative stress, heavy metal exposure, temperatures, etc.) that decreased viability of ΔcckN cells, whether pleC was present and whether the strains were incubated individually or mixed equally in the same culture (data not shown). However, the laboratory conditions in which we measured the fitness cost of cckN inactivation remain far from the natural habitats.
In alphaproteobacteria, DivK phosphorylation is known to be regulated by multiple paralogous HKs, which based on the similarity of their DHp domains, belong to the PleC/DivJ-like family, with numbers ranging from one in the obligate intracellular pathogen Rickettsia prowazekii to four in the facultative host-associated bacteria Sinorhizobium meliloti and Agrobacterium tumefaciens (52–58). Interestingly, the role played by these proteins on DivK phosphorylation varies between species. For instance, B. abortus PdhS is a bona fide bifunctional HK regulating positively and negatively the phosphorylation of DivK (59). In contrast, neither DivJ nor PleC was able to modulate DivK phosphorylation (N. Francis, personal communication) or localization (53). However, as both HKs can physically interact with DivK, it has been hypothesized that DivJ and PleC could display kinase or phosphatase activity in specific conditions, such as in Brucella-containing vacuoles during cellular infection. In S. meliloti, DivK can be phosphorylated by DivJ and CbrA and dephosphorylated by PleC (56), whereas the function of PdhSA remains unknown. Thus, the multiple PleC/DivJ-like proteins that interact with DivK likely refer to the various environments encountered by these alphaproteobacteria, depending on their lifestyles. The challenge now will be to identify the specific stimuli regulating the activity of these DivK-associated HKs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli TOP10 was used for cloning purposes and grown aerobically in Luria-Bertani (LB) broth (Sigma). All Caulobacter crescentus strains used in this study are derived from the synchronizable wild-type strain NA1000 and were grown at 30°C in peptone-yeast extract (PYE) or synthetic medium supplemented with glucose (M2G or M5G) as described in reference 51. Genes expressed from the inducible xylX promoter (PxylX) were induced with 0.1% to 0.2% xylose in an xylX+ background. Motility was assayed on PYE plates containing 0.3% agar. Generalized transduction was performed with phage Cr30 according to the procedure described in reference 60.
For E. coli, antibiotics were used at the following concentrations (μg/ml; in liquid/solid medium): 100/100 kanamycin, 30/50 ampicillin, 12.5/12.5 oxytetracycline, 100/100 spectinomycin, 50/100 streptomycin, and 20/30 chloramphenicol. For C. crescentus, media were supplemented with kanamycin (5/20), oxytetracycline (2.5/2.5), spectinomycin (25/50), streptomycin (5/5), hygromycin (100/100), nalidixic acid (20), chloramphenicol (1/2), or gentamicin (0.5/5) when appropriate. Cumate (4-isopropylbenzoic acid) was dissolved in 100% ethanol to result in a 1,000× 100 mM stock solution and used at a final concentration of 100 μM in the plates. For “no cumate” controls, an equal volume of ethanol was added to the plates.
Motility assay.
Five microliters of saturated overnight cultures was inoculated into PYE 0.3% swarm agar plates, and the plates were incubated at 30°C for 3 to 5 days. ImageJ software was then used to quantify areas of the swarm colonies as described in reference 51.
Bacteriophage sensitivity assays.
The sensitivity to CbK (LHR1) and CR30 (LHR2) bacteriophages was determined as follows. First, 200 μl overnight culture of Caulobacter cells grown in PYE was mixed to 4 to 5 ml of prewarmed PYE top agar (0.45% agar) medium and poured on a PYE plain agar plate. Then, CbK and Cr30 bacteriophage lysates were serially diluted and spotted (5-μl drops) on the top agar once solidified and incubated overnight at 30°C.
Construction of plasmids and strains.
Detailed descriptions of bacterial strains are included in the supplemental material. The strains, plasmids, and oligonucleotides used for plasmids and strain construction are listed in Tables S2 to S4 in the supplemental material. E. coli S17-1 and E. coli MT607 helper strains were used for transferring plasmids to C. crescentus by biparental and triparental matings, respectively. In-frame deletions were created by using the pNPTS138-derivative plasmids as follows. The plasmids in the C. crescentus genome after single homologous recombination were selected on PYE plates supplemented with kanamycin. Three independent recombinant clones were inoculated in PYE medium without kanamycin and incubated overnight at 30°C. Then, dilutions were spread on PYE plates supplemented with 3% sucrose and incubated at 30°C. Single colonies were picked and transferred onto PYE plates with and without kanamycin. Finally, to discriminate between mutated and wild-type loci, kanamycin-sensitive clones were tested by PCR on colonies using locus-specific oligonucleotides.
Attachment assays.
Overnight cultures were diluted in a 96-well microplate and incubated for 18 h to 24 h at 30°C before measuring absorbance at the optical density of 660 nm (OD660). Unattached cells were discarded, and the microplate was washed 3 times with distilled H2O (dH2O). Then, 0.1% crystal violet (CV) was added for 15 min under agitation before the wells were washed with dH2O. Finally, CV was dissolved in a 20% acetic acid solution for 15 min under agitation, and absorbance at 595 nm (OD595) was measured. To normalize attachment to growth, the ratio between OD595 and OD660 was used.
Spotting assays.
For the experiments shown in Fig. 4b and c, 10-fold serial dilutions (in PYE) were prepared in 96-well plates from 5-ml cultures in standard glass tubes grown overnight at 30°C in PYE Kan (Fig. 4b) or Tet (Fig. 4c), and dilution series were spotted on PYE Kan and PYE Kan 0.1% xylose (Fig. 4b) or replica spotted on PYE Tet and PYE Tet Q100 plates using an in-house-made 8-by-6 (48-well) metal pin replicator (Fig. 4c). Plates were incubated at 30°C for 2 days, and pictures were taken.
Synchronization of cells.
Synchronization of cells was performed as described in reference 37. Briefly, cells were grown in 200 ml of PYE to an OD660 of 0.6, harvested by centrifugation for 20 min at 5,000 × g and 4°C, resuspended in 60 ml of ice-cold phosphate (PO43–) buffer, and combined with 30 ml of Ludox LS colloidal silica (30%) (Sigma-Aldrich). Cells resuspended in Ludox were centrifuged for 40 min at 9,000 × g and 4°C. Swarmer cells, corresponding to the bottom band, were isolated, washed twice in ice-cold PO43– buffer, and finally, resuspended in prewarmed PYE medium for growth at 30°C. Samples were collected every 15 min for Western blot, microscopy, and fluorescence-activated cell sorter (FACS) analyses.
Protein purification.
In order to immunize rabbits for production of polyclonal antibodies and/or to perform in vitro phosphorylation assays, DivJSm, CckN, DivK, and His6-PleC505-842 were purified as follows. A BL21(DE3) strain harboring plasmid pML31-TRX-His6-divJHKSm (56), pET-28a-cckN, pET-28a-divK, or pET-28a-pleC505-842 was grown in LB medium supplemented with kanamycin until an OD600 of 0.7 was reached. IPTG (isopropyl-β-d-thiogalactopyranoside) was added at a final concentration of 1 mM, and the culture was incubated at 37°C for 4 h. Then, cells were harvested by centrifugation for 20 min at 5,000 × g and 4°C. The pellet was resuspended in 20 ml BD buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 10% glycerol, 10 mM MgCl2, 12.5 mM imidazole) supplemented with complete EDTA-free protease cocktail inhibitor (Roche), 400 mg lysozyme (Sigma), and 10 mg DNase I (Roche) and incubated for 30 min on ice. Cells were then lysed by sonication and the lysate by centrifugation (12,000 rpm for 30 min at 4°C) was loaded on a Ni-nitrilotriacetic acid (Ni-NTA) column and incubated for 1 h at 4°C with end-over-end shaking. The column was then washed with 5 ml BD buffer, 3 ml Wash1 buffer (BD buffer with 25 mM imidazole), 3 ml Wash2 buffer (BD buffer with 50 mM imidazole), and 3 ml Wash3 buffer (BD buffer with 75 mM imidazole). Proteins bound to the column were eluted with 3 ml elution buffer (BD buffer with 100 mM imidazole) and aliquoted in 300-μl fractions. All the fractions containing the protein of interest (checked by Coomassie blue staining) were pooled and dialyzed in dialysis buffer (50 mM Tris [pH 7.4], 12.5 mM MgCl2).
For the experiments shown in Fig. 2d and Fig. S1, proteins were expressed in E. coli BL21(DE3) harboring plasmid pETHisMBP-cckN, pETHisMBP-divJ, or pETHisMBP-pleC or in E. coli BL21(DE3) pLys harboring plasmids pCC2 or pRP112. Strains were grown overnight in 5 ml LB-Miller at 37°C with appropriate antibiotics, diluted 100-fold in 500 ml LB-Miller with appropriate antibiotics, and grown at 37°C until the cultures reached an OD600 of 0.5 to 0.8. Cultures were then shifted to 23°C and incubated for another hour, after which protein expression was induced by addition of 0.2 mM IPTG. After incubation overnight, cells were harvested by centrifugation (5,000 × g, 20 min, 4°C), washed once with 20 ml of 1× phosphate-buffered saline (PBS), flash-frozen in liquid N2, and stored at −80°C until purification. For purification, the pellet was resuspended in 8 ml of buffer A (2× PBS containing 500 mM NaCl, 10 mM imidazole, and 2 mM β-mercaptoethanol) supplemented with DNase I (AppliChem) and complete protease inhibitor (Roche). After one passage through a French press cell, the suspension was centrifuged (10,000 × g, 30 min, 4°C), and the supernatant was mixed with 800 μl of Ni-NTA slurry, prewashed with buffer A, and incubated for 1 to 2 h on a rotary wheel at 4°C. Ni-NTA–agarose was loaded on a polypropylene column and washed with at least 50 ml of buffer A, after which the protein was eluted with 2.5 ml of buffer A containing 500 mM imidazole. The eluate was immediately loaded on a PD-10 column preequilibrated with kinase buffer (10 mM HEPES-KOH [pH 8.0], 50 mM KCl, 10% glycerol, 0.1 mM EDTA, 5 mM MgCl2, and 5 mM β-mercaptoethanol). The protein was then eluted with 3.5 ml of kinase buffer and stored at 4°C until further use. All experiments were performed within 1 week of purification.
Immunoblot analysis.
Protein crude extracts were prepared by harvesting cells from the exponential growth phase (OD660, ∼0.3). The pellets were then resuspended in SDS-PAGE loading buffer by normalizing to the OD660 before lysing cells by incubating them for 10 min at 95°C. The equivalent of 0.5 ml of culture (OD660, 0.3) was loaded, and proteins were subjected to electrophoresis in a 12% SDS-polyacrylamide gel, transferred onto a nitrocellulose membrane, and then blocked overnight in 5% (wt/vol) nonfat dry milk in phosphate-buffered saline (PBS) with 0.05% Tween 20. The membrane was immunoblotted for ≥3 h with the following primary antibodies: anti-M2 (1:5,000) (Sigma), anti-CtrA (1:5,000), anti-MreB (1:5,000), anti-PleC (1:5,000), anti-CckN (1:2,000), and anti-DivK (1:2,000). This was followed by immunoblotting for ≤1 h with the following secondary antibodies: 1:5,000 anti-mouse (for anti-FLAG) or 1:5,000 anti-rabbit (for all the others) antibodies linked to peroxidase (Dako Agilent) and visualized thanks to Clarity Western ECL substrate chemiluminescence reagent (Bio-Rad) and an Amersham Imager 600 (GE Healthcare).
FACS analysis.
Bacterial cells were incubated at 30°C until they reached the mid-log phase, and 100 μl cells was added to 900 μl ice-cold 70% (vol/vol) ethanol solution and stored at –20°C for 4 h or until further use. For rifampin treatment, the mid-log-phase cells were grown in the presence of 20 μg/ml rifampin at 30°C for 3 h. Then, 1 ml of these cells was fixed as mentioned above. For staining and analysis, 2 ml fixed cells was pelletized and washed once with 1 ml staining buffer (10 mM Tris · HCl [pH 7.2], 1 mM EDTA, 50 mM sodium citrate plus 0.01% Triton X-100). Then, the cells were resuspended in 1 ml staining buffer containing 0.1 mg/ml RNaseA (Roche Life Sciences) and incubated at room temperature (RT) for 30 min. The cells were then harvested by centrifugation at 6,000 × g for 2 min, and the pellets were resuspended in 1 ml staining buffer supplemented with 0.5 μM SYTOX Green nucleic acid stain (Molecular Probes, Life Technologies), followed by incubation in the dark for 5 min. These cells were then analyzed immediately in a flow cytometer (FACSCalibur; BD Biosciences) at laser excitation of 488 nm. At least 1 × 104 cells were recorded in triplicate for each experiment.
Microscopy.
All strains were imaged during the exponential phase of growth (OD660 between 0.1 and 0.4) after immobilization on 1.5% PYE–agarose pads. Microscopy was performed using an Axioskop microscope (Zeiss), an Orca-Flash 4.0 camera (Hamamatsu), and Zen 2.3 software (Zeiss). Images were processed using ImageJ.
In vivo 32P labeling.
A single colony of cells picked from a PYE agar plate was washed with M5G medium lacking phosphate and was grown overnight in M5G with 0.05 mM phosphate to an OD660 of 0.3. Then, 1 ml of culture was harvested and resuspended in the same volume of SDS-PAGE loading buffer to determine the relative protein content by immunoblot analysis, and 1 ml of culture was labeled for 4 min at 30°C using 30 μCi [γ-32P]ATP. Following lysis, proteins were immunoprecipitated with 3 μl of polyclonal antisera (anti-CtrA or anti-PleD). The precipitate proteins were resolved by SDS-PAGE, and 32P-labeled proteins were visualized using a super-resolution screen (Perkin Elmer) and quantified using a Cyclone Plus storage phosphor system (Perkin Elmer). The signal was normalized to the relative protein content determined by immunoblotting of whole-cell lysates probed with antibodies. Note that we checked on cold samples that immunoprecipitation of CtrA or PleD was comparable from one strain to the other.
In vitro phosphorylation assays.
For the experiments shown in Fig. 2c, autophosphorylation was performed on 5 μM DivJSm kinase in 50 mM Tris-HCl (pH 7.4) supplemented with 2 mM DTT, 5 mM MgCl2, 500 μM ATP, and 2 μCi [γ-32P]ATP at 30°C for 40 min. Then, 5 μM DivK was added to the mix supplemented with 5 mM MgCl2, and phosphotransfer was performed at RT for 15 min. To remove the excess ATP, the sample was washed twice in an Amicon column with a cutoff of 10 kDa by adding 450 μl of 50-mM Tris-HCl (pH 7.4) and centrifuging this for 10 min at 12,000 rpm. CckN was then added at a final concentration of 5 μM, and the mix was incubated for 1, 2, and 5 min at RT. The reaction was stopped by adding SDS-PAGE loading buffer, and samples were resolved by SDS-PAGE; the dried gel was visualized using a Super Resolution screen (Perkin Elmer) and quantified using a Cyclone Plus storage phosphor system (Perkin Elmer).
For the experiments shown in Fig. 2d and Fig. S1, all reactions were performed in kinase buffer supplemented with 433 μM ATP and 5 to 20 μCi γ-[32P]ATP (3,000 Ci/mmol; Hartmann Analytic) at room temperature. For the experiments shown in Fig. S1a, reaction mixtures containing His6-MBP-PleC (5 μM), His6-MBP-DivJ (5 μM), or His6-MBP-CckN (5 μM) without or with DivKD53N (10 μM) were prepared and incubated for 20 min at RT before autophosphorylation was started by addition of ATP. Aliquots were withdrawn from the reactions as indicated in the figure, stopped by addition of 5× SDS sample buffer, and stored on ice. For the experiments shown in Fig. S2b, reaction mixtures containing His6-MBP-DivJ (9 μM), His6-MBP-CckN (11 μM), PleD (28 μM), and/or DivKD53N (28 μM) were prepared and incubated for 30 min at RT before autophosphorylation was started. Reactions were stopped after 60 min by addition of 5× SDS sample buffer and stored on ice. For the reactions shown in Fig. 2d, His-MBP-DivJ (10 μM) was autophosphorylated for 60 min in a reaction volume of 150 μl, and then 20 μl of PleD (8 μM final) was added; the reaction was split in 40-μl aliquots, and after allowing PleD phosphorylation for 2 min, 10 μl of His6-MBP-CckN (4 μM final) or kinase buffer (control) was added. Aliquots were withdrawn from the reactions as indicated in the figure, stopped by addition of 5× SDS sample buffer, and stored on ice. Reactions were run on precast Mini-Protean TGX (Bio-Rad) gels; wet gels were exposed to a phosphor screen, which was subsequently scanned using a Typhoon FLA 7000 imaging system (GE Healthcare).
Quantification of DivJ and PleC phosphorylation.
For quantification of DivJ and PleC phosphorylation in vitro, the upper bands in the gels shown in Fig. S1a, corresponding to full-length MBP-DivJ and MBP-PleC, were subjected to measurements (“integrated density”) with a “rectangle” of fixed size using FIJI. For each gel, a rectangle of the same size to the left of lane 1 (a part of the gel that had no proteins/sample loaded) was used to measure the background, the value of which was subtracted from all other measured band intensities to obtain “background-corrected” absolute values. Values were normalized to the signal obtained after 60 min of autophosphorylation of DivJ or PleC without DivKD53N (expressed in percentage in Fig. S1b). Note that reactions with and without DivKD53N for DivJ or PleC were run on the same gel to ensure proper comparison of results with and without DivKD53N.
β-Galactosidase assays.
Overnight saturated cultures of Caulobacter cells harboring lacZ reporter plasmids were diluted ≥50× in fresh medium and incubated at 30°C until an OD660 of 0.3 to 0.5 was reached. For β-galactosidase assays done in stationary phase (Fig. S5b), cells were incubated at 30°C for 24 h (final OD660, ∼1.3). Then, 50-μl aliquots of the cells were treated with a few drops of chloroform. To this, 750 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgSO4 [pH 7.0]) was added, followed by 200 μl of 4 mg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG). Then, the reaction mixture was incubated at 30°C until a yellow color developed and was stopped by addition of 500 μl of 1 M Na2CO3, and the OD420 of the supernatant was measured. The activity of the β-galactosidase expressed in Miller units (MU) was calculated using the following equation: MU = (OD420 × 1,000)/(OD660 × t × v), where t is the time of the reaction (minutes) and v is the volume of cultures used in the assays (milliliters). Experimental values were the average of three independent experiments.
Bacterial two-hybrid (BTH) assays.
BTH assays were performed as described in reference 51. Briefly, 2 μl of MG1655 cyaA::frt (RH785) strains expressing T18 and T25 fusions were spotted on MacConkey agar base plates supplemented with ampicillin, kanamycin, maltose (1%), and IPTG (1 mM) and incubated for 3 to 4 days at 30°C. All proteins were fused to the T25 at their N-terminal extremity (pKT25) or to the T18 at their N-terminal (pUT18C) or C-terminal (pUT18) extremity. The β-galactosidase assays were performed on 50 μl E. coli BTH strains cultivated overnight at 30°C in LB medium supplemented with kanamycin, ampicillin, and IPTG (1 mM) as described in reference 61.
Coimmunoprecipitation assays.
C. crescentus cells were grown in 200 ml of PYE (supplemented with 0.1% xylose if required) to an OD660 of 0.7 to 0.9 and harvested by centrifugation for 20 min at 5,000 × g and 4°C. The pellets were washed once with PBS and resuspended in 10 ml PBS containing 2 mM DTSP [dithiobis(succinimidyl propionate)] for cross-linking. After 30 min at 30°C, cross-linking was quenched by the addition of Tris-HCl to a final concentration of 0.150 M for 30 min. Cells were then washed twice with PBS and once with 20 ml coimmunoprecipitation (Co-IP) buffer (20 mM HEPES, 150 mM NaCl, 20% glycerol, 80 mM EDTA). The pellets were resuspended in lysis buffer (1× CelLytic B [Sigma], 10 mM MgCl2, 67.5U Ready-Lyse lysozyme [Epicentre], 10 U/ml DNase I, 2% NP-40 Surfact-Amps detergent [Thermo Scientific], one-half tablet Complete EDTA-free antiproteases [Roche]) and incubated under soft agitation at RT for 30 min. Cells were then lysed by sonication, and lysates were cleared by centrifugation and incubated for 2 h at 4°C with MagStrep type 3 XT beads (Iba). The beads were washed 6 times with W buffer (Iba), and precipitated proteins were released by 15 min incubation in BX buffer (Iba). SDS loading buffer was added to samples and boiled for 10 min. Equal volumes of coimmunoprecipitates and cell lysates from equal numbers of cells were analyzed by SDS-PAGE and Western blotting. Membranes were probed with anti-DivK and anti-CckN primary antibodies.
Chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq) assays.
ChIP-Seq assays were performed as described in reference 62. Briefly, 80 ml of mid-log-phase cells (OD660 of 0.6) were cross-linked in 1% formaldehyde and 10 mM sodium phosphate (pH 7.6) at room temperature for 10 min and then for 30 min on ice. Cross-linking was stopped by addition of 125 mM glycine and incubated for 5 min on ice. Cells were washed thrice in PBS, resuspended in 450 μl TES buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, and 100 mM NaCl), and lysed with 2 μl of Ready-lyse lysozyme solution for 5 min at RT. Protease inhibitors (Roche) were added, and the mixture was incubated for 10 min. Then, 550 μl of ChIP buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], and 167 mM NaCl, plus protease inhibitors) was added to the lysate and incubated at 37°C for 10 min before sonication (2 × 8 bursts of 30 sec on ice using a Diagenode Bioruptor) to shear DNA fragments to a length of 300 to 500 bp. Lysate was cleared by centrifugation for 10 min at 12,500 rpm at 4°C, and protein content was evaluated by measuring the OD280. Then, 7.5 mg of proteins was diluted in ChIP buffer supplemented with 0.01% SDS and precleared for 1 h at 4°C with 50 μl of protein A-agarose beads (Bio-Rad) and 100 μg bovine serum albumin (BSA). Two microliters of polyclonal anti-CtrA antibodies was added to the supernatant before overnight incubation at 4°C under gentle agitation. Next, 80 μl of BSA presaturated protein A-agarose beads was added to the solution and incubated for 2 h at 4°C with rotation, washed once with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl), once with high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), once with LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]), and once with TE buffer (10 mM Tris-HCl [pH 8.1] 1 mM EDTA) at 4°C, followed by and a second wash with TE buffer at RT. The DNA-protein complexes were eluted twice in 250 μl freshly prepared elution buffer (0.1 M NaHCO3, 1% SDS). NaCl was added at a concentration of 300 mM to the combined eluates (500 μl) before overnight incubation at 65°C to reverse the cross-link. The samples were treated with 20 μg of proteinase K in 40 mM EDTA and 40 mM Tris-HCl (pH 6.5) for 2 h at 45°C. DNA was extracted using a Nucleospin PCR cleanup kit (Macherey-Nagel) and resuspended in 50 μl elution buffer (5 mM Tris-HCl [pH 8.5]). DNA sequencing was performed using an Illumina HiSeq 4000 instrument (Genomicscore KULeuven, Belgium).
Next-generation sequencing data analysis.
Around 2 × 107 single-end sequence reads (1 × 50) were first mapped on the genome of C. crescentus NA1000 (GenBank accession number NC_011916.1) and converted to Sequence Alignment/Map (SAM) format using the Burrows-Wheeler Aligner (BWA) (63) and SAMtools (64), respectively, from the Sourceforge server (https://sourceforge.net/). The MACS2 (65) algorithm was used to model the length of DNA fragments and the shift size. Next, the number of reads overlapping each genomic position was computed using custom Python scripts and the previously modeled DNA fragment and shift sizes. A peak was defined as the genomic region where each position has more reads than the 97th percentile. The candidate peaks were annotated using custom Python scripts. For comparing strains, the total number of reads was normalized by the ratio of the number of reads between the two strains.
Statistical analyses.
The significance of differences between mean values was determined by one-way or two-way analysis of variance (ANOVA) with a Tukey’s, Dunnett’s, or Sidak’s multiple-comparison posttest. All the analyses were performed using GraphPad Prism 8 software. A P value of <0.05 was considered significant.
Data availability.
ChIP-Seq data have been deposited in the Gene Expression Omnibus (GEO) database with the accession number GSE152025.
ACKNOWLEDGMENTS
We are grateful to Patrick Viollier and Peter Chien for providing strains and plasmids, Emanuele Biondi for anti-CtrA antibodies, and Fanny Zola and Aurélie Mayard for help with proteins purification. We thank the members of the BCcD team for critical reading of the manuscript and helpful discussions.
The work conducted in the laboratory of U.J. was supported by the Swiss National Science Foundation (grants 31003A_166503 and 310030B_185372). This work was supported by a Research Credit (CDR J.0169.16) and Incentive Grant for Scientific Research (MIS F.4516.19) from the Fonds de la Recherche Scientifique-FNRS (F.R.S.-FNRS) to R.H., and R.H. is a research associate of F.R.S.-FNRS.
J.C., A.K., U.J., and R.H. conceived and designed the experiments. J.C. performed all the experiments except as otherwise stated. A.K. performed the in vitro phosphorylation assays shown in Fig. 2d and Fig. S1 as well as the growth assays upon overexpression of cckN variants shown in Fig. 4c. K.P. designed the bioinformatic tool to analyze the ChIP-Seq data. T.B. characterized the ClpXP-dependent degradation of PleC (Fig. 5b and c and Fig. S6f and g). J.C., A.K., U.J., and R.H. analyzed the data. J.C., A.K., and R.H. wrote the paper.
We declare no competing financial interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 2.Domian IJ, Quon KC, Shapiro L. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415–424. doi: 10.1016/S0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs C, Domian IJ, Maddock JR, Shapiro L. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 4.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 5.Wu J, Ohta N, Zhao JL, Newton A. 1999. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc Natl Acad Sci U S A 96:13068–13073. doi: 10.1073/pnas.96.23.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reisinger SJ, Huntwork S, Viollier PH, Ryan KR. 2007. DivL performs critical cell cycle functions in Caulobacter crescentus independent of kinase activity. J Bacteriol 189:8308–8320. doi: 10.1128/JB.00868-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iniesta AA, Hillson NJ, Shapiro L. 2010. Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc Natl Acad Sci U S A 107:7012–7017. doi: 10.1073/pnas.1001767107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Childers WS, Xu Q, Mann TH, Mathews II, Blair JA, Deacon AM, Shapiro L. 2014. Cell fate regulation governed by a repurposed bacterial histidine kinase. PLoS Biol 12:e1001979. doi: 10.1371/journal.pbio.1001979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. 2006. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A 103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laub MT, Chen SL, Shapiro L, McAdams HH. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci U S A 99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheeler RT, Shapiro L. 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell 4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs C, Hung D, Shapiro L. 2001. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc Natl Acad Sci U S A 98:4095–4100. doi: 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta N, Newton A. 2003. The core dimerization domains of histidine kinases contain recognition specificity for the cognate response regulator. J Bacteriol 185:4424–4431. doi: 10.1128/jb.185.15.4424-4431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol Microbiol 47:1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- 15.Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, Biondi EG, Laub MT, Jenal U. 2008. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell 133:452–461. doi: 10.1016/j.cell.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. 2011. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell 43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith SC, Joshi KK, Zik JJ, Trinh K, Kamajaya A, Chien P, Ryan KR. 2014. Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proc Natl Acad Sci U S A 111:14229–14234. doi: 10.1073/pnas.1407862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 20.Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C. 2004. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell 118:579–590. doi: 10.1016/j.cell.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Hung DY, Shapiro L. 2002. A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression. Proc Natl Acad Sci U S A 99:13160–13165. doi: 10.1073/pnas.202495099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iniesta AA, Shapiro L. 2008. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc Natl Acad Sci U S A 105:16602–16607. doi: 10.1073/pnas.0808807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang SP, Sharma PL, Schoenlein PV, Ely B. 1993. A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc Natl Acad Sci U S A 90:630–634. doi: 10.1073/pnas.90.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ardissone S, Fumeaux C, Berge M, Beaussart A, Theraulaz L, Radhakrishnan SK, Dufrene YF, Viollier PH. 2014. Cell cycle constraints on capsulation and bacteriophage susceptibility. Elife 3:e03587. doi: 10.7554/eLife.03587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fumeaux C, Radhakrishnan SK, Ardissone S, Theraulaz L, Frandi A, Martins D, Nesper J, Abel S, Jenal U, Viollier PH. 2014. Cell cycle transition from S-phase to G1 in Caulobacter is mediated by ancestral virulence regulators. Nat Commun 5:4081. doi: 10.1038/ncomms5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagenaur C, Farmer S, Agabian N. 1977. Adsorption properties of stage-specific Caulobacter phage phiCbK. Virology 77:401–407. doi: 10.1016/0042-6822(77)90436-6. [DOI] [PubMed] [Google Scholar]
- 27.Edwards P, Smit J. 1991. A transducing bacteriophage for Caulobacter crescentus uses the paracrystalline surface layer protein as a receptor. J Bacteriol 173:5568–5572. doi: 10.1128/jb.173.17.5568-5572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radhakrishnan SK, Thanbichler M, Viollier PH. 2008. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev 22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodenmiller D, Toh E, Brun YV. 2004. Development of surface adhesion in Caulobacter crescentus. J Bacteriol 186:1438–1447. doi: 10.1128/jb.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs C, Ausmees N, Cordwell SJ, Shapiro L, Laub MT. 2003. Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol Microbiol 47:1279–1290. doi: 10.1046/j.1365-2958.2003.03379.x. [DOI] [PubMed] [Google Scholar]
- 31.Delaby M, Panis G, Viollier PH. 2019. Bacterial cell cycle and growth phase switch by the essential transcriptional regulator CtrA. Nucleic Acids Res 47:10628–10644. doi: 10.1093/nar/gkz846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vass RH, Zeinert RD, Chien P. 2016. Protease regulation and capacity during Caulobacter growth. Curr Opin Microbiol 34:75–81. doi: 10.1016/j.mib.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. 2006. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell 124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 34.Aakre CD, Phung TN, Huang D, Laub MT. 2013. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol Cell 52:617–628. doi: 10.1016/j.molcel.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radhakrishnan SK, Pritchard S, Viollier PH. 2010. Coupling prokaryotic cell fate and division control with a bifunctional and oscillating oxidoreductase homolog. Dev Cell 18:90–101. doi: 10.1016/j.devcel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 36.Bhat NH, Vass RH, Stoddard PR, Shin DK, Chien P. 2013. Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Mol Microbiol 88:1083–1092. doi: 10.1111/mmi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beaufay F, Coppine J, Mayard A, Laloux G, De Bolle X, Hallez R. 2015. A NAD-dependent glutamate dehydrogenase coordinates metabolism with cell division in Caulobacter crescentus. EMBO J 34:1786–1800. doi: 10.15252/embj.201490730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaczmarczyk A, Hempel AM, von Arx C, Bohm R, Dubey BN, Nesper J, Schirmer T, Hiller S, Jenal U. 2020. Precise timing of transcription by c-di-GMP coordinates cell cycle and morphogenesis in Caulobacter. Nat Commun 11:816. doi: 10.1038/s41467-020-14585-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen JC, Viollier PH, Shapiro L. 2005. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol 55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- 40.Janakiraman B, Mignolet J, Narayanan S, Viollier PH, Radhakrishnan SK. 2016. In-phase oscillation of global regulons is orchestrated by a pole-specific organizer. Proc Natl Acad Sci U S A 113:12550–12555. doi: 10.1073/pnas.1610723113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsing W, Silhavy TJ. 1997. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol 179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lori C, Kaczmarczyk A, de Jong I, Jenal U. 2018. A single-domain response regulator functions as an integrating hub to coordinate general stress response and development in Alphaproteobacteria. mBio 9:e00809-18. doi: 10.1128/mBio.00809-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lourenco RF, Kohler C, Gomes SL. 2011. A two-component system, an anti-sigma factor and two paralogous ECF sigma factors are involved in the control of general stress response in Caulobacter crescentus. Mol Microbiol 80:1598–1612. doi: 10.1111/j.1365-2958.2011.07668.x. [DOI] [PubMed] [Google Scholar]
- 45.Foreman R, Fiebig A, Crosson S. 2012. The LovK-LovR two-component system is a regulator of the general stress pathway in Caulobacter crescentus. J Bacteriol 194:3038–3049. doi: 10.1128/JB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaczmarczyk A, Campagne S, Danza F, Metzger LC, Vorholt JA, Francez-Charlot A. 2011. Role of Sphingomonas sp. strain Fr1 PhyR-NepR-sigmaEcfG cascade in general stress response and identification of a negative regulator of PhyR. J Bacteriol 193:6629–6638. doi: 10.1128/JB.06006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaczmarczyk A, Vorholt JA, Francez-Charlot A. 2013. Cumate-inducible gene expression system for sphingomonads and other Alphaproteobacteria. Appl Environ Microbiol 79:6795–6802. doi: 10.1128/AEM.02296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gottschlich L, Bortfeld-Miller M, Gabelein C, Dintner S, Vorholt JA. 2018. Phosphorelay through the bifunctional phosphotransferase PhyT controls the general stress response in an alphaproteobacterium. PLoS Genet 14:e1007294. doi: 10.1371/journal.pgen.1007294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joshi KK, Berge M, Radhakrishnan SK, Viollier PH, Chien P. 2015. An adaptor hierarchy regulates proteolysis during a bacterial cell cycle. Cell 163:419–431. doi: 10.1016/j.cell.2015.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorbatyuk B, Marczynski GT. 2005. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol 55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- 51.Ronneau S, Petit K, De Bolle X, Hallez R. 2016. Phosphotransferase-dependent accumulation of (p)ppGpp in response to glutamine deprivation in Caulobacter crescentus. Nat Commun 7:11423. doi: 10.1038/ncomms11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. 2004. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol 12:361–365. doi: 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Hallez R, Mignolet J, Van Mullem V, Wery M, Vandenhaute J, Letesson JJ, Jacobs-Wagner C, De Bolle X. 2007. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. EMBO J 26:1444–1455. doi: 10.1038/sj.emboj.7601577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. 2010. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol 4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J, Heindl JE, Fuqua C. 2013. Coordination of division and development influences complex multicellular behavior in Agrobacterium tumefaciens. PLoS One 8:e56682. doi: 10.1371/journal.pone.0056682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pini F, Frage B, Ferri L, De Nisco NJ, Mohapatra SS, Taddei L, Fioravanti A, Dewitte F, Galardini M, Brilli M, Villeret V, Bazzicalupo M, Mengoni A, Walker GC, Becker A, Biondi EG. 2013. The DivJ, CbrA and PleC system controls DivK phosphorylation and symbiosis in Sinorhizobium meliloti. Mol Microbiol 90:54–71. doi: 10.1111/mmi.12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sadowski CS, Wilson D, Schallies KB, Walker G, Gibson KE. 2013. The Sinorhizobium meliloti sensor histidine kinase CbrA contributes to free-living cell cycle regulation. Microbiology 159:1552–1563. doi: 10.1099/mic.0.067504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heindl JE, Crosby D, Brar S, Pinto JF, Singletary T, Merenich D, Eagan JL, Buechlein AM, Bruger EL, Waters CM, Fuqua C. 2019. Reciprocal control of motility and biofilm formation by the PdhS2 two-component sensor kinase of Agrobacterium tumefaciens. Microbiology 165:146–162. doi: 10.1099/mic.0.000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francis N, Poncin K, Fioravanti A, Vassen V, Willemart K, Ong TA, Rappez L, Letesson JJ, Biondi EG, De Bolle X. 2017. CtrA controls cell division and outer membrane composition of the pathogen Brucella abortus. Mol Microbiol 103:780–797. doi: 10.1111/mmi.13589. [DOI] [PubMed] [Google Scholar]
- 60.Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol 204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 61.Ronneau S, Caballero-Montes J, Coppine J, Mayard A, Garcia-Pino A, Hallez R. 2019. Regulation of (p)ppGpp hydrolysis by a conserved archetypal regulatory domain. Nucleic Acids Res 47:843–854. doi: 10.1093/nar/gky1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pini F, De Nisco NJ, Ferri L, Penterman J, Fioravanti A, Brilli M, Mengoni A, Bazzicalupo M, Viollier PH, Walker GC, Biondi EG. 2015. Cell cycle control by the master regulator CtrA in Sinorhizobium meliloti. PLoS Genet 11:e1005232. doi: 10.1371/journal.pgen.1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
ChIP-Seq data have been deposited in the Gene Expression Omnibus (GEO) database with the accession number GSE152025.