Abstract
The emergence of SARS-CoV-2 has prompted a worldwide health emergency. There is an urgent need for therapeutics, both through the repurposing of approved drugs and the development of new treatments. In addition to the viral drug targets, a number of human drug targets have been suggested. In theory, targeting human proteins should provide an advantage over targeting viral proteins in terms of drug resistance, which is commonly a problem in treating RNA viruses. This paper focuses on the human protein TMPRSS2, which supports coronavirus life cycles by cleaving viral spike proteins. The three-dimensional structure of TMPRSS2 is not known and so we have generated models of the TMPRSS2 in the apo state as well as in complex with a peptide substrate and putative inhibitors to aid future work. Importantly, many related human proteases have 80% or higher identity with TMPRSS2 in the S1–S1’ subsites, with plasminogen and urokinase-type plasminogen activator (uPA) having 95% identity. We highlight 376 approved, investigational or experimental drugs targeting S1A serine proteases that may also inhibit TMPRSS2. Whilst the presence of a relatively uncommon lysine residue in the S2/S3 subsites means that some serine protease inhibitors will not inhibit TMPRSS2, this residue is likely to provide a handle for selective targeting in a focused drug discovery project. We discuss how experimental drugs targeting related serine proteases might be repurposed as TMPRSS2 inhibitors to treat coronaviruses.
Keywords: Molecular modelling, Serine protease, TMPRSS2, Homology modelling, SARS-CoV-2, Drug design
Graphical abstract
Highlights
-
•
TMPRSS2 is an S1A serine protease that represents a good drug target to treat SARS-CoV-2.
-
•
Virus cannot develop resistance mutations against Non-viral drug targets such as TMPRSS2.
-
•
Drugbank has 376 approved, investigational or experimental drugs targeting S1A serine proteases that may inhibit TMPRSS2.
-
•
A homology model of TMPRSS2 based on the crystal structure of TMPRSS15 suggests that many of these may also inhibit TMPRSS2.
-
•
S1A serine protease inhibitors should be tested in the existing assays for in vitro inhibition of TMPRSS2.
The emergence of SARS-CoV-2 has prompted a worldwide health emergency. There is an urgent need for therapeutics, both through the repurposing of approved drugs and the development of new treatments. In this paper we highlight existing experimental drugs might be repurposed to treat coronaviruses.
1. Introduction
There is an urgent need to identify drugs and drug targets which are effective in treating COVID-19. Numerous drug targets have been suggested [1] and a plethora of drug repurposing efforts are underway [2,3]. Transmembrane Serine Protease 2 (TMPRSS2) is a membrane bound serine protease also known as Epitheliasin. TMPRSS2 belongs to the S1A class of serine proteases alongside proteins such as factor Xa and trypsin. Whilst there is evidence that TMPRSS2 autoclaves to generate a secreted protease [4], its physiological function has not been clearly identified, However, it is known to play a crucial role in facilitating entry of coronavirus particles into cells by cleaving the spike protein [5,6]. Coronavirus spike proteins are thought to be cleaved at two sites, termed S1/S2 and S2’ [[7], [8], [9]]. The proteases furin, trypsin, cathepsin, TMPRSS2, TMPRSS4, TMPRSS11, and human airway trypsin-like protease have all been implicated in these cleavages [[9], [10], [11], [12], [13]]. For SARS-CoV, the S2′ cleavage site has a sequence motif (PTKR|S) that appears suitable for cleavage by trypsin-like proteases such as TMPRSS2 whereas the S1/S2 cleavage site has a sequence motif (SLLR|S) that appears suitable for cleavage by cathepsin or trypsin-like proteases. Whilst the SARS-CoV-2 S2’ cleavage site has a similar sequence motif to SARS-CoV (PSKR|S) and would thus be suitable for cleavage by trypsin-like proteases, insertions of additional arginine residues at the SARS-CoV-2 S1/S2 cleavage site (RRAR|S) clearly generate a furin cleavage site [14,15]. Interestingly, this difference has been implicated in viral transmissibility of SARS-CoV-2 [16]. In theory, preventing cleavage at either site should be deleterious to viral invasion.
There is good evidence that TMPRSS2 represents a good drug target for coronaviruses. TMPRSS2-expressing cells are more susceptible to SARS-CoV-2 infection and knockout mouse models show that lack of TMPRSS2 in the airways reduces the severity of lung pathology after SARS-CoV and MERS-CoV infection [17]. For this reason, it has been suggested as a potential drug target for coronaviruses [18,19] such as SARS-CoV-2 [20]. TMPRSS2 is highly expressed in lung tissue [21] and it has been suggested that differential expression in males may lead to higher risk in male patients [20,22,23] Peptidic inhibitors of TMPRSS2 have been described [12] and the covalent TMPRSS2 inhibitor Camostat is being tested in a clinical trial against COVID-19. However, in this study we identify a number of experimental drugs with the potential to target TMPRSS2. MEROPS, the peptide database, lists 219 members of the S1A family in humans [24]. Many of the members in this family have been studied in detail, yielding numerous high resolution crystal structures, known inhibitors, and licensed drugs [25]. Importantly, the S1A family has a conserved fold with an arginine binding site that is targeted by the majority of small molecule inhibitors and drugs with its three catalytic residues (aspartate, histidine, and serine) in close proximity to the arginine binding site. In this study we generate homology models of human TMPRSS2 in the apo state as well as in complex with substrate peptide and a number of small molecule experimental drugs.
2. Materials and methods
2.1. Development of a TMPRSS2 homology model
TMPRSS2 is in the S1A serine protease family. Sequences for all the human members of the family are available on Github: https://github.com/djhuggins/TMPRSS2/tree/master/Sequences.
We aligned the complete sequences with Clustal Omega [26]. The overall sequence identity between TMPRSS2 and the other S1A family proteases is available on Github: https://github.com/djhuggins/TMPRSS2/tree/master/Alignments.
We selected a subset of sequences for alignment based on availability of structural data and similarity between sequences in the active site. We looked in particular at the identity in the active site of the enzymes to inform model and compound selection. We identified 20 residues close to the active site (S1–S1′) as well as a larger set of 34 residues spanning the whole binding site (S4–S3–S2–S1–S1′-S2′-S3′-S4′). These residues are identified in Fig. 1 . The percentage identities between TMPRSS2 and this subset are given in Table 1 :
Fig. 1.
Sequence alignment between the serine protease domains of TMPRSS2 and TMPRSS15. The loop regions that were remodelled are highlighted by green bars. TMPRSS2 residues in the S4–S3–S2–S1–S1′-S2′-S3′-S4′ subsites are colored red. TMPRSS2 residues in the S1–S1′ subsites are further highlighted in bold. The lysine residue between the S2/S3 subsites is highlighted with a blue arrow. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
Table 1.
A subset of human S1A serine proteases and the percentage identity with TMPRSS2 of their protease domains, S1–S1′ subsites, and S4–S3–S2–S1–S1′-S2′-S3′-S4′ subsites.
| Protein Name | Uniprot Name | TMPRSS2 Protease Domain % Identity | TMPRSS2 S4–S4’ % Identity | TMPRSS2 S1–S1’ % Identity |
|---|---|---|---|---|
| Plasminogen | PLMN_HUMAN | 41.07 | 64.71 | 95.00 |
| uPA | UROK_HUMAN | 33.77 | 58.82 | 95.00 |
| Trypsin-1 | TRY1_HUMAN | 38.60 | 58.82 | 90.00 |
| Plasma kallikrein | KLKB1_HUMAN | 41.99 | 61.76 | 85.00 |
| Coagulation factor VII | FA7_HUMAN | 38.16 | 61.76 | 85.00 |
| Hepsin | HEPS_HUMAN | 41.88 | 58.82 | 85.00 |
| TMPRSS15 | TMPRSS15_HUMAN | 41.30 | 58.82 | 85.00 |
| Coagulation factor XI | FA11_HUMAN | 42.17 | 55.88 | 85.00 |
| TMPRSS11E | TM11E_HUMAN | 40.27 | 55.88 | 80.00 |
| Tryptase gamma | TRYG1_HUMAN | 39.82 | 52.94 | 80.00 |
| Coagulation factor IX | FA9_HUMAN | 39.19 | 52.94 | 80.00 |
| Coagulation factor XII | FA12_HUMAN | 36.89 | 50.00 | 80.00 |
| Coagulation factor X | FA10_HUMAN | 36.77 | 47.06 | 80.00 |
| Chymotrypsin B | CTRB2_HUMAN | 40.91 | 44.12 | 65.00 |
Importantly, many of the proteases have 80% or higher identity with TMPRSS2 in the S1–S1′ subsites, with plasminogen and urokinase-type plasminogen activator (uPA) having 95% identity. We selected the protein TMPRSS15 to generate a homology model (also known as enteropeptidase). TMPRSS15 provides a good template for building a homology model of TMPRSS2, with a deletion of two residues and an insertion of one residue. It also features a lysine residue in the S2/S3 subsites, which is unique to TMPRSS2 and TMPRSS15. The alignment of the TMPRSS2 and TMPRSS15 protease domains is shown in Fig. 1.
To build a homology model, the structure of TMPRSS15 was taken from PDB ID 4DGJ [27]. Selenomethionines were changed to methionines and missing side-chains were added using Schrödinger’s Preparation Wizard, which was also used to evaluate the orientations of the asparagine, glutamine, and histidine residues, as well as the protonation state of all ionizable residues. All heteroatomic species such as buffer solvents and ions were removed. Water molecules in the arginine binding site were retained. The TMPRSS2 loop sequences EKPLNNPWH, QSFMSY, and VYDNLITPA (see Fig. 1) were remodelled using Schrodinger’s Prime and the whole protein was then energy minimized with the heavy atoms converged to an RMSD of 0.3 Å. The homology model of TMPRS22 is available on Github overlaid with the benzamidine molecule from PDBID 2OQ5 [28]: https://github.com/djhuggins/TMPRSS2/blob/master/HomologyModels/TMPRSS2_HomologyModel_Enteropeptidase4DGJ_BenzamidineOverlay.pdb.
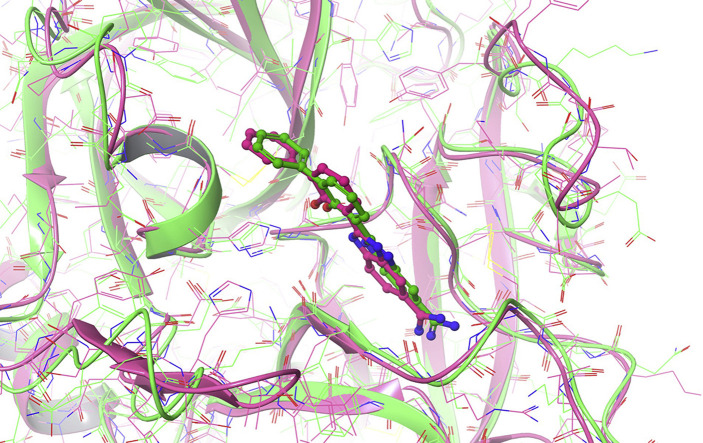
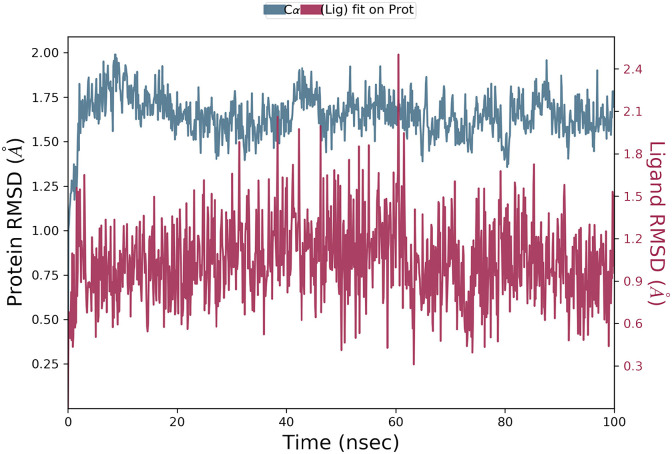
To assess the quality of the homology model, we performed 100 ns of molecular dynamics using the Desmond package. The system was set up as an orthorhombic box with a minimum distance of 10 Å from the protein to the box edge. Water was modelled with the SPC water model and the OPLS3e forcefield was used for the protein and ligand. 100 ns of simulation was then performed in the NPT ensemble at 300 K and 1.01325 bar. Fig. 2 shows the RMSD of the protein and the ligand during the simulation. Both stabilize within the first 10 ns? The protein remains stable, with an alpha carbon RMSD around 1.7 Å relative to the original structure and always below 2.0 Å. The ligand also remains stable, with an RMSD around 0.9 Å relative to the original structure and always below 2.5 Å.
Fig. 2.
The RMSD of the protein alpha carbons relative to the original structure (blue) and the RMSD of the ligand relative to the original structure (red) across the 100 ns simulation. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
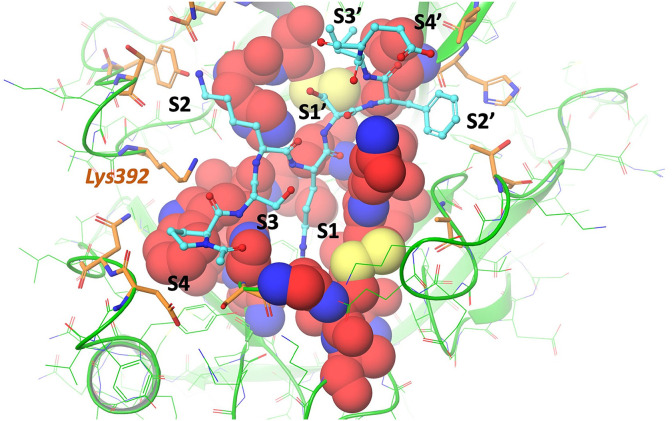
We also generated a model of TMPRS22 bound to an eight-residue sequence from the SARS-CoV-2 S2′ cleavage site (PSKR|SFIE). The structure of the peptide was based on the structure of bovine pancreatic trypsin inhibitor (BPTI) bound to anionic trypsin from PDBID 3FP6 [29]. We used the backbone residues of BPTI and generated a homology model of the SARS-CoV-2 S2’ sequence. The structure of the peptide was then energy minimized. Fig. 3 shows the model, highlighting the eight subsites.
Fig. 3.
Predicted structure of the SARS-CoV-2 S2′ cleavage site sequence PSKRSFIE bound to the homology model of TMPRS22. The TMPRSS2 protein is displayed as green ribbons with atoms in green wire. The peptide is displayed as cyan balls and sticks. Residues within 4 Ångström of the S1–S1′ sites are displayed as red space filling. Residues within 4 Ångström of the S4–S3–S2–S1–S1′-S2′-S3′-S4′ sites are displayed as orange sticks. The eight subsites and residue Lys392 are denoted. The homology model of TMPRS22 overlaid with the SARS-CoV-2 S2′ cleavage site PSKRSFIE 8-mer is available on Github: https://github.com/djhuggins/TMPRSS2/blob/master/HomologyModels/TMPRSS2_HomologyModelFrom4DGJ_SubstrateOverlay.pdb. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
2.2. Identification of small molecule drugs targeting S1A serine proteases
In order to identify potential drugs that might inhibit TMPRSS2, we exploited structural modelling data from related serine proteases. We used the MEROPS database to identify all proteins in the S1A family [24]. We then used Drugbank [30] to identify 36 drug targets in the S1A family for which approved, investigational or experimental drugs are available. A list of the target names, UniProt identifiers, and Drugbank ID of these proteins is available on Github: https://github.com/djhuggins/TMPRSS2/blob/master/DrugBankData/S1ASerineProtease_DrugBank_TargetIDs.csv.
A list of all the 376 Drugbank drugs targeting S1A serine proteases is also available on Github: https://github.com/djhuggins/TMPRSS2/blob/master/DrugBankData/S1ASerineProtease_DrugBank_DrugIDs.csv.
Included with this is the list of DrugBank molecules for each target and a list of the Protein Databank PDB identifiers [31] of these drugs bound to the targets. Of these 36 targets, the 32 targets for which PDB structural data is available are reported in Table 2 .
Table 2.
Human protein targets in the S1A serine protease family with structural data for approved, investigational or experimental drugs.
| Protein Name | UniProt ID | UniProt Gene Name | DrugBank Target ID |
|---|---|---|---|
| Apolipoprotein(a) | P08519 | APOA_HUMAN | 1283 |
| Cathepsin G | P08311 | CATG_HUMAN | 1010 |
| Chymase | P23946 | CMA1_HUMAN | 1038 |
| Coagulation factor IX | P00740 | FA9_HUMAN | 364 |
| Coagulation factor VII | P08709 | FA7_HUMAN | 333 |
| Coagulation factor X | P00742 | FA10_HUMAN | 216 |
| Coagulation factor XI | P03951 | FA11_HUMAN | 1021 |
| Coagulation factor XII | P00748 | FA12_HUMAN | 4672 |
| Complement C1r subcomponent | P00736 | C1R_HUMAN | 2093 |
| Complement C1s subcomponent | P09871 | C1S_HUMAN | 1529 |
| Complement factor B | P00751 | CFAB_HUMAN | 1701 |
| Complement factor D | P00746 | CFAD_HUMAN | 1840 |
| Complement factor I | P05156 | CFAI_HUMAN | 8979 |
| Haptoglobin | P00738 | HPT_HUMAN | 10,260 |
| Hepatocyte growth factor | P14210 | HGF_HUMAN | 1121 |
| Kallikrein-6 | Q92876 | KLK6_HUMAN | 1586 |
| Kallikrein-7 | P49862 | KLK7_HUMAN | 4150 |
| Myeloblastin | P24158 | PRTN3_HUMAN | 954 |
| Neutrophil elastase | P08246 | ELNE_HUMAN | 394 |
| Plasma kallikrein | P03952 | KLKB1_HUMAN | 2440 |
| Plasminogen | P00747 | PLMN_HUMAN | 211 |
| Prostasin | Q16651 | PRSS8_HUMAN | 3746 |
| Prostate-specific antigen | P07288 | KLK3_HUMAN | 8908 |
| Prothrombin | P00734 | THRB_HUMAN | 48 |
| Serine protease hepsin | P05981 | HEPS_HUMAN | 2128 |
| Tissue-type plasminogen activator | P00750 | TPA_HUMAN | 1088 |
| Trypsin-1 | P07477 | TRY1_HUMAN | 1739 |
| Trypsin-3 | P35030 | TRY3_HUMAN | 1583 |
| uPA | P00749 | UROK_HUMAN | 895 |
| Vitamin K-dependent protein C | P04070 | PROC_HUMAN | 380 |
| Vitamin K-dependent protein Z | P22891 | PROZ_HUMAN | 547 |
We identified 250 approved, investigational or experimental drugs targeting S1A proteases for which structural data is available. These drugs correspond to 479 PDB structures. We downloaded the structural data for these drugs in complex with their targets and aligned them in the same reference frame. The aligned structures are available on Github: https://github.com/djhuggins/TMPRSS2/tree/master/AlignedPDBs.
We overlaid the homology model of TMPRSS2 with the PDB structures containing the approved, investigational or experimental small-molecule drugs for S1A serine proteases. We selected a subset of these molecules based on fit within the homology model of TMPRSS2. We then used Embrace minimization with GBSA solvation to test whether all molecules fit with the homology model of TMPRSS2 [32]. The use of minimization ensures that the resulting complexes to do contain any clashes or bad contacts.
3. Results
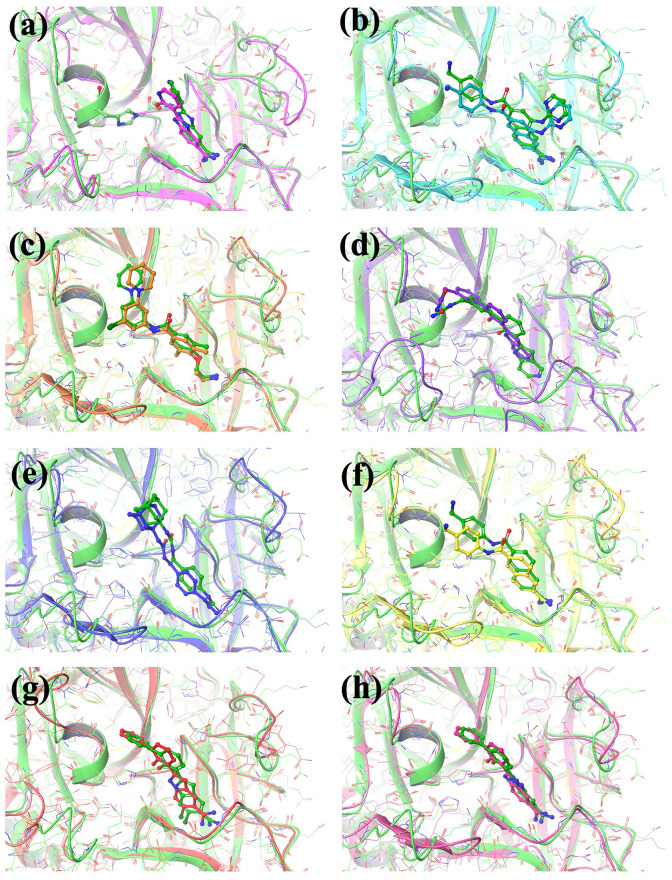
Based on the homology model described above, the S2–S3 subsites of TMPRSS2 appear to be different than that of related proteases due to the presence of a charge residue Lys392 (see Fig. 3). Whilst this suggests that many existing serine protease inhibitors which fill these subsites will not bind to TMPRSS2, it presents an opportunity to develop selective inhibitors in the future to exploit TMPRSS2 selectively in therapeutic settings. Fig. 4 presents the predicted TMPRSS2 binding modes for a set of non-covalent S1A serine protease experimental drugs overlaid with the experimental crystal structures of the drugs bound to their known target. These drugs target the S1–S1′ subsites where identity between the S1A serine proteases is very high (see Table 2).
Fig. 4.
Putative model of TMPRSS2 bound to the experimental drugs from (a) Trypsin in PDBID 1GHV [33], (b) uPA in PDBID 1SQA [34], (c) uPA in PDBID 2VIV [35], (d) factor VII in PDBID 2FLR [36], (e) uPA in PDBID 1EJN [37], (f) uPA in PDBID 1OWH [38], (g) hepsin in PDBID 1O5E [39], and (h) uPA in PDBID 1GJC [40]. The TMPRSS2 protein is displayed as green ribbons with atoms in green wire and the predicted binding modes of the experimental drugs in TMPRSS2 are shown in green balls and sticks. The experimental protein structures are displayed as different colored ribbons with wire atoms and the binding modes of the experimental drugs are shown as balls and sticks. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The predicted complexes and a file with their Embrace MMGBSA scores are available on Github: https://github.com/djhuggins/TMPRSS2/tree/master/Embrace.
The S1–S1′ subsites of TMPRSS2 has a high similarity to the known targets of all these drugs (see Table 1). Table 3 reports further details for the drugs in Fig. 4.
Table 3.
A set of experimental drugs targeting S1A serine proteases that may inhibit TMPRSS2 and are modelled in Fig. 3.
| Drugbank ID | Crystallized Protein | PDB ID | Notes |
|---|---|---|---|
| DB04442 | Trypsin | 1GHZ | |
| DB03082 | uPA | 1SQA | Abbott compound |
| DB08697 | uPA | 2VIV | Astex compound |
| DB07247 | Factor VII | 2FLR | |
| DB03782 | uPA | 1EJN | |
| DB02398 | uPA | 1OWH | |
| DB03865 | Hepsin | 1O5E | Trypsin/thrombin/uPA/hepsin inhibitor |
| DB01725 | uPA | 1GJC | Trypsin/thrombin/uPA inhibitor |
All the small molecule drugs with an experimental crystal structure are reported on Github: https://github.com/djhuggins/TMPRSS2/blob/master/DrugBankData/S1ASerineProtease_DrugBank_DrugIDs_WithPDBData.csv.
We predict that a number of these may also inhibit TMPRSS2 and could prove effective as treatments for COVID-19. If hitting numerous host proteases is important then simple molecules such as DB04442 may prove more effective.
4. Discussion
A number of drug targets have been suggested for coronaviruses [41] such as SARS-CoV-2. A recent study highlights that three covalent inhibitors of the drug target TMPRSS2 blocked SARS-CoV-2 infection of human lung cells, with nafamostat [42] showing better than camostat or gabexate [43]. Non-viral drug targets such as TMPRSS2 have the advantage that the virus cannot develop resistance mutations that reduce the affinity of the drug for the target. Mutations that allow the virus to utilize alternative host proteases are possible, but the corresponding change in pathogenesis leads to a higher likelihood of being deleterious to viral fitness. As a drug target TMPRSS2 has the additional advantage that the drug discovery community has significant experience in developing drugs targeting serine proteases. For instance, many diverse and high affinity inhibitors have been synthesized for widely studied S1A serine protease targets such as Thrombin and Factor Xa [44].
A number of host proteases have been implicated in cleavage of the coronavirus including furin, trypsin, cathepsins, TMPRSS2, TMPRSS4, and human airway trypsin-like protease [45]. Given the furin cleavage in SARS-CoV-2 it seems likely that furin-targeted drugs would prove useful in treating the virus [14]. Whilst there are no drugs that specifically target furin, it is possible that Camostat targets furin in addition to S1A proteases given a mechanism of action where it covalently binds to the arginine binding site. However, at this stage there is significantly more evidence that targeting TMPRSS2 will effectively treat SARS-CoV-2. The known TMPRSS2 inhibitor Camostat is being assessed in a clinical trial against COVID-19 and other inhibitors, such as Namafostat, look to be effective in cell-based studies. However, it seems highly likely that these simple covalent binders may inhibit other S1A serine proteases and this may lead to unwanted side effects for these drugs. TMPRSS2 knockout mice develop normally with no observable phenotype suggesting that it may be safely targeted [46]. To maximize safety, it would be very useful to identify exactly which proteases are key to cleaving the SARS-CoV-2 spike protein. Recent work suggests that TMPRSS2 and TMPRSS4 may both be important [47]. This work identifies a number of experimental drugs for serine proteases that may effectively inhibit TMPRSS2 based on homology and Embrace binding free energy calculations. Three are shown in Table 4 .
Table 4.
Three of the TMPRSS2 Embrace top scoring serine protease inhibitors.
| Drugbank ID | Structure | Protein Target |
|---|---|---|
| DB03782 | 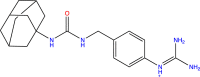 |
Urokinase-type plasminogen activator |
| DB03213 | 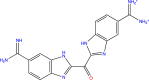 |
Trypsin-1 |
| DB04107 | 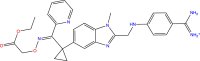 |
Trypsin-1 |
All three molecules in Table 4 contain a positively charged warhead similarly to nafamostat, camostat, and gabexate (guanidine in the case of to DB03782, nafamostat, camostat, and gabexate versus amidine in the case of DB03213 and DB04107). The vast majority of the 376 serine protease inhibitors contain such a warhead.
In this study we have highlighted a large number of S1A serine protease inhibitors with experimental drug status that may inhibit TMPRSS2. Assays have previously been reported that would allow these molecules to be tested against TMPRSS2(12) [48] and one or more of these may have the appropriate PK properties to attain sufficiently high concentrations in the lung and effectively treat COVID-19.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Acknowledgements go to Neil Rawlings for help with the MEROPS database and to Fraser Glickman, Patti Aha, Nigel Liverton, and Peter Meinke for careful reading of the manuscript. The author gratefully acknowledges the support to the project generously provided by the Tri-Institutional Therapeutics Discovery Institute (TDI), a 501(c) (3) organization. TDI receives financial support from Takeda Pharmaceutical Company, TDI’s parent institutes (Memorial Sloan Kettering Cancer Center, The Rockefeller University and Weill Cornell Medicine) and from a generous contribution from Mr. Lewis Sanders and other philanthropic sources.
References
- 1.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020 Mar;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 2.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020 May 4 doi: 10.1021/acs.jcim.0c00179. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. vol. 6. Cell Discovery; 2020. Network-based Drug Repurposing for Novel Coronavirus 2019-nCoV/SARS-CoV-2. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afar D.E., Vivanco I., Hubert R.S., Kuo J., Chen E., Saffran D.C. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Canc. Res. 2001 Feb 15;61(4):1686–1692. [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 Apr 16;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bestle D., Heindl M.R., Limburg H., Pilgram O., Moulton H. TMPRSS2 and furin are both essential for proteolytic activation and spread of SARS-CoV-2 in human airway epithelial cells and provide promising drug targets. bioRxiv. 2020 doi: 10.26508/lsa.202000786. https://www.biorxiv.org/content/10.1101/2020.04.15.042085v1.abstract Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleine-Weber H., Elzayat M.T., Hoffmann M., Pöhlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018 Nov 9;8(1):16597. doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015 Apr 16;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009 Apr 7;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kam Y.-W., Okumura Y., Kido H., Ng L.F.P., Bruzzone R., Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS One. 2009 Nov 17;4(11) doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zang R., Castro M.F.G., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M. vol. 5. Science Immunology; 2020. (TMPRSS2 and TMPRSS4 Promote SARS-CoV-2 Infection of Human Small Intestinal Enterocytes). eabc3582. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer D., Sielaff F., Hammami M., Böttcher-Friebertshäuser E., Garten W., Steinmetzer T. Identification of the first synthetic inhibitors of the type II transmembrane serine protease TMPRSS2 suitable for inhibition of influenza virus activation. Biochem. J. 2013 Jun 1;452(2):331–343. doi: 10.1042/BJ20130101. [DOI] [PubMed] [Google Scholar]
- 13.Bosch B.J., Bartelink W., Rottier P.J.M. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008 Sep;82(17):8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020 Apr;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q., Qiu Y., Li J.-Y., Zhou Z.-J., Liao C.-H., Ge X.-Y. A unique protease cleavage site predicted in the spike protein of the novel pneumonia coronavirus (2019-nCoV) potentially related to viral transmissibility. Virol. Sin. 2020 Mar 20 doi: 10.1007/s12250-020-00212-7. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019 Mar 15;93(6) doi: 10.1128/JVI.01815-18. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen L.W., Mao H.J., Wu Y.L., Tanaka Y., Zhang W. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017 Nov;142:1–10. doi: 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012 Jun;86(12):6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stopsack K.H., Mucci L.A., Antonarakis E.S., Nelson P.S., Kantoff P.W. TMPRSS2 and COVID-19: serendipity or opportunity for intervention? Canc. Discov. 2020 Apr 10 doi: 10.1158/2159-8290.CD-20-0451. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020 doi: 10.15252/embj.20105114. https://www.embopress.org/doi/abs/10.15252/embj.20105114 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asselta R., Paraboschi E.M., Mantovani A., Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. 2020. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3559608 Available from: [DOI] [PMC free article] [PubMed]
- 23.Qi J., Zhou Y., Hua J., Zhang L., Bian J., Liu B. The scRNA-seq expression profiling of the receptor ACE2 and the cellular protease TMPRSS2 reveals human organs susceptible to COVID-19 infection. 2020. Available from: [DOI] [PMC free article] [PubMed]
- 24.Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018 Jan 4;46(D1):D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maryanoff B.E. Inhibitors of serine proteases as potential therapeutic agents: the road from thrombin to tryptase to cathepsin G. J. Med. Chem. 2004 Feb 12;47(4):769–787. doi: 10.1021/jm030493t. [DOI] [PubMed] [Google Scholar]
- 26.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7(1) doi: 10.1038/msb.2011.75. https://www.embopress.org/doi/abs/10.1038/msb.2011.75 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simeonov P., Zahn M., Sträter N., Zuchner T. Crystal structure of a supercharged variant of the human enteropeptidase light chain. Proteins. 2012 Jul;80(7):1907–1910. doi: 10.1002/prot.24084. [DOI] [PubMed] [Google Scholar]
- 28.Kyrieleis O.J.P., Huber R., Ong E., Oehler R., Hunter M., Madison E.L. Crystal structure of the catalytic domain of DESC1, a new member of the type II transmembrane serine proteinase family. FEBS J. 2007 Apr;274(8):2148–2160. doi: 10.1111/j.1742-4658.2007.05756.x. [DOI] [PubMed] [Google Scholar]
- 29.Zakharova E., Horvath M.P., Goldenberg D.P. Structure of a serine protease poised to resynthesize a peptide bond. Proc. Natl. Acad. Sci. U. S. A. 2009 Jul 7;106(27):11034–11039. doi: 10.1073/pnas.0902463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018 Jan 4;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guvench O., Weiser J., Shenkin P., Kolossváry I., Still W.C. Application of the frozen atom approximation to the GB/SA continuum model for solvation free energy. J. Comput. Chem. 2002 Jan 30;23(2):214–221. doi: 10.1002/jcc.1167. [DOI] [PubMed] [Google Scholar]
- 33.Katz B.A., Elrod K., Luong C., Rice M.J., Mackman R.L., Sprengeler P.A. A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site. J. Mol. Biol. 2001 Apr 13;307(5):1451–1486. doi: 10.1006/jmbi.2001.4516. [DOI] [PubMed] [Google Scholar]
- 34.Wendt M.D., Geyer A., McClellan W.J., Rockway T.W., Weitzberg M., Zhao X. Interaction with the S1β-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors. Bioorg. Med. Chem. Lett. 2004;14 doi: 10.1016/j.bmcl.2004.04.030. 3063–8. Available from: [DOI] [PubMed] [Google Scholar]
- 35.Frederickson M., Callaghan O., Chessari G., Congreve M., Cowan S.R., Matthews J.E. Fragment-based discovery of mexiletine derivatives as orally bioavailable inhibitors of urokinase-type plasminogen activator. J. Med. Chem. 2008 Jan 24;51(2):183–186. doi: 10.1021/jm701359z. [DOI] [PubMed] [Google Scholar]
- 36.Riggs J.R., Hu H., Kolesnikov A., Leahy E.M., Wesson K.E., Shrader W.D. Novel 5-azaindole factor VIIa inhibitors. Bioorg. Med. Chem. Lett. 2006 Jun 15;16(12):3197–3200. doi: 10.1016/j.bmcl.2006.03.049. [DOI] [PubMed] [Google Scholar]
- 37.Sperl S., Jacob U., de Prada N.A., Sturzebecher J., Wilhelm O.G., Bode W. vol. 97. 2000. (4-Aminomethyl)phenylguanidine derivatives as nonpeptidic highly selective inhibitors of human urokinase; pp. 5113–5118. (Proceedings of the National Academy of Sciences). Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wendt M.D., Rockway T.W., Geyer A., McClellan W., Weitzberg M., Zhao X. Identification of novel binding interactions in the development of potent, selective 2-naphthamidine inhibitors of urokinase. Synthesis, structural analysis, and SAR of N-phenyl amide 6-substitution. J. Med. Chem. 2004 Jan 15;47(2):303–324. doi: 10.1021/jm0300072. [DOI] [PubMed] [Google Scholar]
- 39.Katz B.A., Luong C., Ho J.D., Somoza J.R., Gjerstad E., Tang J. Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA) J. Mol. Biol. 2004 Nov 19;344(2):527–547. doi: 10.1016/j.jmb.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 40.Katz B.A., Sprengeler P.A., Luong C., Verner E., Elrod K., Kirtley M. Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets. Chem. Biol. 2001 Nov;8(11):1107–1121. doi: 10.1016/s1074-5521(01)00084-9. [DOI] [PubMed] [Google Scholar]
- 41.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses — drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujii S., Hitomi Y. New synthetic inhibitors of C1r, C1 esterase, thrombin, plasmin, kallikrein and trypsin. Biochim. Biophys. Acta. 1981 Oct 13;661(2):342–345. doi: 10.1016/0005-2744(81)90023-1. [DOI] [PubMed] [Google Scholar]
- 43.Hoffmann M., Schroeder S., Kleine-Weber H., Müller M.A., Drosten C., Pöhlmann S. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob. Agents Chemother. 2020 Apr 20 doi: 10.1128/AAC.00754-20. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vukovic S., Huggins D.J. Quantitative metrics for drug–target ligandability. Drug Discov. Today. 2018 doi: 10.1016/j.drudis.2018.02.015. https://www.sciencedirect.com/science/article/pii/S1359644617305895 Available from: [DOI] [PubMed] [Google Scholar]
- 45.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-15562-9. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim T.S., Heinlein C., Hackman R.C., Nelson P.S. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol. Cell Biol. 2006 Feb;26(3):965–975. doi: 10.1128/MCB.26.3.965-975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zang R., Castro M.F.G., McCune B.T., Zeng Q. TMPRSS2 and TMPRSS4 mediate SARS-CoV-2 infection of human small intestinal enterocytes. bioRxiv. 2020 doi: 10.1126/sciimmunol.abc3582. https://www.biorxiv.org/content/10.1101/2020.04.21.054015v1.abstract Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shrimp An enzymatic TMPRSS2 assay for assessment of clinical candidates and discovery of Inhibitors as potential treatment of COVID-19. Biorxiv. 2020 doi: 10.1021/acsptsci.0c00106. [DOI] [PMC free article] [PubMed] [Google Scholar]