Abstract
Background
Serological assays for the determination of the immune status of patients that have tested positive for infection with SARS-CoV-2 by RT-PCR are required for, e.g., contact tracing and epidemiological studies. However, data concerning the performance parameters of commercially available high-throughput ELISA tests are still not available on a large scale.
Study design
In our study, we have evaluated an in-house developed ELISA for the detection of the immunoglobulin classes A, G and M directed against the full-length spike glycoprotein from SARS-CoV-2. For this analysis, we have included 110 sera from patients presenting with COVID-19 symptoms or blood donors without symptoms collected at the Austrian Red Cross, Blood Transfusion Service for Upper Austria, Linz. In addition, we have selected four commercially available IgG-based ELISAs as well as one IgA/IgG-based ELISA for the detection of SARS-CoV-2 antigens as well as a multiplexed IgG-based micro-ELISA assay developed for rapid Point of Care testing applications.
Conclusions
All assays evaluated in the course of this study demonstrated suitable sensitivity and specificity values for the identification of patients that have experienced a past infection with SARS-CoV-2. However, testing for the presence of additional immunoglobulins (IgA and IgM) as well as using combinations of different viral antigens is highly advised to improve the predictive values of serological assays.
Keywords: SARS-CoV-2, COVID-19, ELISA, Point of care, Immunoglobulin, Evaluation
1. Background
SARS-CoV-2 belongs to the group of beta-coronaviruses and represents the causative agent for coronavirus disease 2019 (COVID-19), an acute respiratory disease that emerged in the Chinese province of Wuhan in December 2019 and has since rapidly spread leading to an ongoing pandemic [1,2]. The gold standard to diagnose an infection with SARS-CoV-2 is reverse transcriptase polymerase chain reaction (RT-PCR) allowing for the identification of asymptomatic carriers as well as patients with an acute infection [3]. While this represents an extremely important step that facilitates curbing the actual spread of the infection, the identification of patients with past episodes of COVID-19 is another cornerstone contributing to the successful handling of the ongoing pandemic and is an area of intensive research [[4], [5], [6]]. This is especially important in light of the fact that patients that have recovered from a previous SARS-CoV-2 infection might harbor immunoglobulins that could protect them from future infections with this virus, opening also the possibility of using convalescent plasma as a treatment option for COVID-19 patients [7,8]. Additional aspects during the management of pandemic situations also depend on the information of reliably identifying individuals recovered from COVID-19. Examples include the assessment of herd immunity, strategies to protect vulnerable groups like elderly or people with medical preconditions, or even for pharmacological questions like characterizing antibodies specific for viral antigens exhibiting neutralizing effects that could be used for designing and developing vaccines [9,10].
This study characterizes the basic performance parameters for an in-house developed ELISA and compares it with five commercially available assays as well as a recently launched microfluidic-chip based, multiplexed micro-ELISA assay designed for rapid Point of Care (POC) testing applications. For this analysis, we have included 110 sera from patients and regular blood donors that presented with or without COVID-19 symptoms collected at the Austrian Red Cross, Blood Transfusion Service for Upper Austria, Linz.
2. Materials and methods
2.1. Serum samples
110 serum samples from patients presenting with COVID-19 symptoms or blood donors without symptoms have been collected at the Austrian Red Cross, Blood Transfusion Service Upper Austria, Linz and written consent was obtained at the time of donation to use sample material also for research purposes (54 male participants with a median age of 44,11 years and 56 female participants with a median age of 43,04 years). Sera of patients with a positive SARS-CoV-2 RT-PCR test result were collected at least 3 week post recovery to ensure enough time to develop a proper immune response. In addition, samples from regular blood donors without the classical COVID-19 symptoms have been obtained to assess possible cross-reactive events that could eventually lead to false positive results. Since there have also been reports on the presence of SARS-CoV-2 RNA in blood and serum samples obtained from infected patients [11], heat inactivation of the samples in this study has been performed at 56 °C for 30 min prior to analysis to minimize any residual risk for the laboratory personnel. No heat denaturation was done for the samples used for the Epitope diagnostics and the Abbott Architect ELISA analyses.
2.2. Immunoassay platforms
We have evaluated an in-house developed ELISA (Division of Pathophysiology, Linz, Austria) that allows for the detection of immunoglobulin classes A, G and M directed against the full-length spike glycoprotein of SARS-CoV-2 (NAC-REC31828; The Native Antigen Company, Kidlington, Oxford, United Kingdom). Additional assay components have been purchased from Merck KGaA, Darmstadt, Germany (e.g., HRP-labeled anti-immunoglobulin antibodies A0295, A0170, A6907 and TMB detection reagent ES001). For the in-house ELISA, the Limit-of Detection (LoD) for the individual immunoglobulin classes was determined as OD450 arbitrary units (A.U.). We have calculated the ratio of the mean of the absorption of the specific signals versus blank controls measured at 450 nanometers for 20 negative samples and added 3 times the standard deviation of the mean to set the LoD for the individual immunoglobulin classes.
Samples were also analyzed with the following commercially available immunoassay platforms according to manufacturer’s instructions: anti-SARS-CoV-2 ELISA IgG/IgA (www.euroimmun.de, Euroimmun AG, Germany), the EDI new Coronavirus COVID-19 IgG ELISA (www.epitopediagnostics.com, Epitope diagnostics Inc., USA), the Vircell COVID-19 ELISA IgG (en.vircell.com, Vircell Spain S.L.U., Spain), recomWell SARS-CoV-2 IgG (www.mikrogen.de, Mikrogen GmbH, Germany) and the SARS-CoV-2 IgG ELISA (www.abbott.com, Abbott GmbH, Germany). In addition, we have also included a microfluidic chip based multiplexed micro-ELISA platform for POC testing to detect IgG antibodies against SARS-CoV-2 antigens in this study (www.genspeed-biotech.com, Genspeed Biotech GmbH, Austria). This assay is currently in the process of final certification and will eventually allow for single sample analysis including discrimination of different viral antigens within approximately 15 min. In Table 1 the specifications of the respective ELISA kits in terms of immunoglobulin classes used for detection of virus-specific antibodies and the respective SARS-CoV-2 antigens are summarized.
Table 1.
| Assay Manufacturer | Immunoglobulin classes detected | Antigens used | ||||
|---|---|---|---|---|---|---|
| Epitope diagnostics Inc. | IgG | Recombinant full length nucleocapsid protein | ||||
| Abbott Architect | IgG | Recombinant full length nucleocapsid protein | ||||
| Euroimmun AG | IgA | IgG | S1-Domain of Spike protein including receptor-binding domain | |||
| Vircell S.L.U. | IgG | Recombinant antigens from Spike glycoprotein and nucleocapsid protein | ||||
| Mikrogen GmbH | IgG | Recombinant full length nucleocapsid protein | ||||
| in-house ELISA | IgA | IgG | IgM | Full length spike glycoprotein | ||
| Genspeed Biotech | IgG | Receptor Binding Domain / Full length spike glycoprotein / Nucleoprotein | ||||
Characteristics of different SARS-CoV-2 assays used in this study.
3. Results
For diagnostic tests, the determination of the parameters sensitivity and specificity are usually obtained by comparison with a so-called "gold standard" assay. Since such a clear-cut standard diagnostic test for the characterization of SARS-CoV-2-specific antibodies is still lacking, the evaluation of performance parameters remains challenging. In analogy to previous studies [12,13] we have therefore defined the immunoglobulin status of the 110 patient sera used in this evaluation as follows: Serum was regarded as SARS-CoV-2 positive if at least three out of five of the commercially available assays yielded a positive IgG signal for the respective sample (Epitope diagnostics, Abbott Architect, Euroimmun, Vircell, Mikrogen).
When applying these criteria we have designated 51 patient samples as positive, and 59 samples as negative based on the presence of IgG antibodies, respectively. Subsequently, we have tested the serum samples in parallel for the presence of immunoglobulins specific for the respective SARS-CoV-2 antigens according to instructions of the assay manufacturers (see also Table 1).
To estimate a potential diagnostic benefit that might arise through the detection of additional immunoglobulin classes directed against SARS-CoV-2 antigens, we had to expand the immunoglobulin status of the serum samples used in this study for a second analysis. For example, if both assays that detect the presence of IgA turned out positive (Euroimmun, in-house ELISA) in absence of a detectable IgG-signal, the patient sample was also regarded positive. In addition, if IgM was detected by the in-house ELISA in the absence of other immunoglobulin classes, the sample was also regarded positive.
For the assays that utilize only IgG to determine an induced immune response to SARS-CoV-2 antigens (Epitope diagnostics, Abbott Architect, Vircell, Mikrogen, Genspeed), we achieved sensitivity values of 98, 94, 98, 100 and 100 %, respectively. The according specificity values that we have obtained were 78, 100, 83, 97 and 93 %. A summary of the sensitivity and specificity values obtained during the course of this study can be found in Table 2 .
Table 2.
| Assay Manufacturer | True positive | False positive | True negative | False negative | Sensitivity | Specificity | ||
|---|---|---|---|---|---|---|---|---|
| Epitope diagnostics Inc. | 50 | 13 | 46 | 1 | 0,98 | 0,78 | ||
| Abbott Architect | 48 | 0 | 59 | 3 | 0,94 | 1,00 | ||
| Euroimmun AG | 47 | 1 | 58 | 4 | 0,92 | 0,98 | ||
| Vircell S.L.U. | 50 | 10 | 49 | 1 | 0,98 | 0,83 | ||
| Mikrogen GmbH | 51 | 2 | 57 | 0 | 1,00 | 0,97 | ||
| in-house ELISA | 49 | 4 | 55 | 2 | 0,96 | 0,93 | ||
| Genspeed Biotech | 51 | 4 | 55 | 0 | 1,00 | 0,93 |
Sensitivity and specificity values for SARS-CoV-2 assays based on the detection of IgG.
The Euroimmun ELISA that we have used offers two immunoglobulin classes for antigen detection (IgA and IgG). When only considering the results of the IgG-based detection, sensitivity and specificity values of 92 and 98 % could be achieved. When IgA detection was included in the course of the analysis, these values for the Euroimmun ELISA increased to 96 and 98 %, respectively. With our in-house ELISA we have the capability to detect all three immunoglobulin classes for SARS-CoV-2 antigen detection in parallel (IgA, IgG and IgM). Again, for reason of comparability we have made a first analysis also based solely on IgG-detection and obtained sensitivity and specificity values of 96 and 93 %. Naturally, these performance parameters increased for the in-house ELISA when we also included the detection of the additional immunoglobulins to 97 and 96 %, respectively. The data including the results obtained by using additional immunoglobulin classes for the Euroimmun and the in-house ELISA are summarized in Table 3 . However, one has to bear in mind that a diagnostic question aiming at the detection of all immunoglobulin classes in a patient’s serum would inevitably lead to a decrease of the performance parameters of the commercial assays solely based on IgG-detection.
Table 3.
| Assay Manufacturer | True positive | False positive | True negative | False negative | Sensitivity | Specificity | ||
|---|---|---|---|---|---|---|---|---|
| Euroimmun AG | 50 | 1 | 57 | 2 | 0,96 | 0,98 | ||
| in-house ELISA | 57 | 2 | 49 | 2 | 0,97 | 0,96 |
Sensitivity and specificity values for SARS-CoV-2 assays utilizing the detection of IgA / IgG (Euroimmun AG) and IgA / IgG / IgM (in-house ELISA).
Another observation that we have made is that in 5 samples analyzed with the Genspeed assay, antibodies exclusively to either the nucleocapsid protein (3 samples) or the full length spike glycoprotein (2 samples) have been detected. This raises the possibility, that a patient is developing an immune response that might very well differ substantially between individuals and assays detecting only a single antigen could lack diagnostic sensitivity. Therefore, the utilization of additional antigens in parallel for COVID-19 detection is another feature that we would certainly encourage for future testing regimes [10,14].
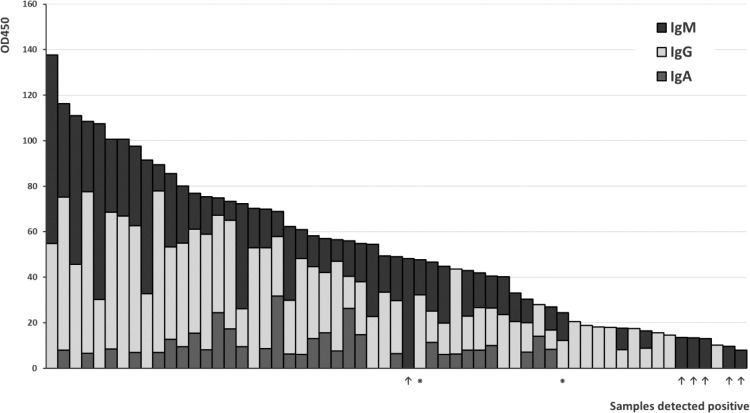
In Fig. 1 the data obtained from the serum samples analyzed with the in-house ELISA for all three immunoglobulin classes is summarized. For each sample, the measured signal intensities with values higher than the LoD threshold are plotted on the Y-axis. As clearly visible, the prominent immune response is usually provided by IgG followed by IgM class antibodies. IgA antibodies represent only a minor fraction in this respect. In most cases, we have detected multiple immunoglobulin classes in the samples identified as positive in the course of this study. In 14 samples we demonstrated the presence of only one immunoglobulin class (8 IgG, 6 IgM). In 16 samples we detected the expression of two immunoglobulin classes in parallel (2 IgA/IgG, 14 IgG/IgM), and in the majority of the samples (i.e. 29) all three immunoglobulin classes (IgA/IgG/IgM) were present.
Fig. 1.
Signal intensities for immunoglobulin classes A, G and M as obtained with the in-house ELISA. Samples designated false positive according to the criteria described in the Results section are indicated with an asterisk (⁕). Samples that are lacking detectable IgG antibodies and tested positive for IgM with our in-house ELISA are labeled with an arrow (↑).
The distribution of the signal intensities of the immunoglobulin data as obtained with our in-house ELISA test is shown as a boxplot diagram in Fig. 2 . The number of samples for analysis and the respective median values are IgA = 31 (8,41), IgG = 53 (23,6) and IgM = 49 (15,92) [15]. While antibodies of the IgA class represent only a minor fraction found in the patient samples analyzed in our study, IgG and IgM antibodies are predominant, although at highly varying concentrations.
Fig. 2.
Distribution of the signal intensities of the immunoglobulin data as obtained with the in-house ELISA. The box indicates the interquartile range (IQR), the whiskers the range of values that are within 1.5x IQR and the horizontal lines indicate the respective median.
4. Discussion
Especially in the face of an ongoing pandemic, the rapid development and production of diagnostic kits and devices is a matter of highest importance. It usually requires about 1–2 weeks before immunoglobulins specific for SARS-CoV-2 antigens can be detected in the circulation of a patient after the first viral particles have been encountered by the immune system. The initial immunoglobulins produced during an immune response consist mainly of IgM molecules, whereas IgG antibodies determine the SARS-CoV-2 seroprevalence in the general population (i.e. the long lasting immuno-reactivity usually attributed to herd immunity). Recently published studies indicate that the majority of patients exhibit seroconversion approximately 2–3 weeks after first symptoms were observed and that IgM was shown to decline already about 4 weeks after onset of symptoms whereas IgG levels remained elevated for at least several weeks [[16], [17], [18]].
The aim of this study was to evaluate the performance of an in-house ELISA designed to detect all three immunoglobulin classes (i.e., IgA, IgG and IgM) with the underlying idea to utilize also the early immunoreactive response to an SARS-CoV-2 infection for diagnostic purposes. We have compared our test to five commercially available serological assays for SARS-CoV-2 as well as a rapid test based on a microfluidic-chip setup. Comparable results were obtained for the commercial assays only detecting IgG. However, the detection of IgM antibodies in case of the in-house ELISA allowed for the identification of six additional samples solely positive for the presence of IgM that would have been missed otherwise.
In conclusion, the SARS-CoV-2 serological assays evaluated in the course of this study demonstrated sufficient sensitivity and specificity values for a robust identification of individuals with a previous SARS-CoV-2 infection. However, we have demonstrated that relying only on the detection of the IgG antibody titer for diagnostic purposes led to false negative results in several cases, most likely with samples obtained during the rather early phases of an immune response. We have shown, that the inclusion of additional immunoglobulin classes further increases the assay sensitivity and should be considered a valuable asset during the selection process of a diagnostic assay, a notion also argued already by others [19,20]. In addition, also the parallel analysis of more than one SARS-CoV-2 antigen for serological characterization – as demonstrated by the Genspeed COVID-19 assay – might proof beneficial for increasing diagnostic performance.
Credit Author Statement
CW established the in-house ELISA, performed experiments and wrote the manuscript. All authors designed the study, analyzed the data and contributed in writing the manuscript.
Funding
This study was partially supported by the MED-CALL program of the Medical Faculty, Johannes Kepler University Linz (Linz, Austria) to CW and the Emergency-Call Covid-19 (Austrian Research Promotion Agency FFG 880919) to DB.
Conflicts of Interest Statement
CW has been an employee of Greiner Bio-One Diagnostics GmbH from 2009 – 2016. During this time, he was member of the project team that developed the Genspeed assay platform. There is currently no professional affiliation of CW with Genspeed Biotech GmbH. The Division of Pathophysiology is serving as a reference laboratory for Genspeed Biotech GmbH within the Emergency-Call Covid-19 (FFG 880919). All other authors declare no competing interests.
Acknowledgments
The authors would like to thank the laboratory teams of the Austrian Red Cross, Blood Transfusion Service for Upper Austria, Linz, Austria and the Department of Pathology and Microbiology, Institute of Pathology and Molecular Pathology, Kepler University Hospital, Linz, Austria, for excellent technical assistance in performing all the analyses with the commercial available SARS-CoV-2 ELISA kits. Genspeed Biotech GmbH has offered the components and reagents for validation of the microfluidic chip assay free of charge.
References
- 1.Zhu N. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siordia J.A. Epidemiology and clinical features of COVID-19. A review of current literature. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104357). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman V.M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2431. doi: 10.2807/1560-7917.ES.2020.25.3.2000045). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weidner L. Quantification of SARS-CoV-2 antibodies with eight commercially available immunoassays. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104540). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104480). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Clinical performance of different SARS-CoV-2 IgG antibody tests. J. Med. Virol. 2020 doi: 10.1002/jmv.26145). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloch E.M. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Invest. 2020;130:2757–2765. doi: 10.1172/JCI138745). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roback J.D., Guarner J. Convalescent plasma to treat COVID-19. JAMA. 2020;323:1561. doi: 10.1001/jama.2020.4940). [DOI] [PubMed] [Google Scholar]
- 9.Dhama K., Sharun K., Tiwari R., Dadar M., Malik Y.S., Singh K.P., Chaicumpa W. COVID-19, an emerging coronavirus infection. Advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum. Vaccin. Immunother. 2020;16:1232–1238. doi: 10.1080/21645515.2020.1735227). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gattinger P. Antibodies in serum of convalescent patients following mild COVID-19 do not always prevent virus receptor binding. Allergy. 2020 doi: 10.1111/all.14523). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg. Microbes Infect. 2020;9:469–473. doi: 10.1080/22221751.2020.1732837). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krüttgen A., Cornelissen C.G., Dreher M., Hornef M., Imöhl M., Kleines M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104394). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Ory F., Guisasola M.-E.-E., Balfagón P., Sanz J.C. Comparison of commercial methods of immunoblot, ELISA, and chemiluminescent immunoassay for detecting type-specific herpes simplex viruses-1 and -2 IgG. J. Clin. Lab. Anal. 2018:32. doi: 10.1002/jcla.22203). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Assis R.R. Analysis of SARS-CoV-2 antibodies in COVID-19 convalescent plasma using a coronavirus antigen microarray . bioRxiv : the preprint server for biology. 2020 doi: 10.1101/2020.04.15.043364). [DOI] [Google Scholar]
- 15.Postma M., Goedhart J. PlotsOfData—a web app for visualizing data together with their summaries. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000202). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao A.T., Gao C., Zhang S. Profile of specific antibodies to SARS-CoV-2. The first report. J. Infect. 2020;81:147–178. doi: 10.1016/j.jinf.2020.03.012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W. Evaluation of Nucleocapsid and Spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.00461-20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espejo A.P., Akgun Y., Al Mana A.F., Tjendra Y., Millan N.C., Gomez-Fernandez C., Cray C. Review of current advances in serologic testing for COVID-19. Am. J. Clin. Pathol. 2020 doi: 10.1093/ajcp/aqaa112). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chau C.H., Strope J.D., Figg W.D. COVID-19 clinical diagnostics and testing technology. Pharmacotherapy. 2020 doi: 10.1002/PHAR.2439). [DOI] [PMC free article] [PubMed] [Google Scholar]