Abstract
Objective
Long noncoding (lnc)RNAs regulate multiple biological processes including cancer. Oral squamous cell carcinoma (OSCC) is a common malignancy with poor prognosis. We aimed to identify the function of lncRNA HOXA10 antisense RNA (HOXA10-AS) and its clinical significance.
Methods
We used differential expression analysis to identify aberrantly expressed lncRNAs associated with OSCC. We identified key genes related to HOXA10-AS and their biological functions using bioinformatics tools and functional enrichment analyses. We predicted the function of HOXA10-AS using gene set enrichment and variation analyses and analyzed proliferation markers at the mRNA and protein levels. Finally, we silenced HOXA10-AS using antisense oligonucleotide and assessed proliferation ability using a cell counting kit (CCK8) and clone formation assays.
Results
In total, 506 aberrantly expressed lncRNAs were identified. HOXA10-AS was identified as a risk factor for OSCC and its expression was positively associated with tumor grade. We identified hub genes involved in regulating proliferation and predicted that HOXA10-AS is associated with an active cell cycle and increased proliferation. Silencing HOXA10-AS decreased proliferation in OSCC cell lines.
Conclusions
HOXA10-AS is involved in cell proliferation and silencing it decreases proliferation. Thus, HOXA10-AS could serve as prognostic biomarker and therapeutic target for OSCC.
Keywords: Long noncoding RNA, HOXA10-antisense RNA, oral squamous cell carcinoma, proliferation, bioinformatics, cell cycle regulation
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common malignancies of the oral and maxillofacial region and it has a poor prognosis.1 Its incidence accounts for 2% to 4% of all malignant tumors and about 80% of malignant tumors in the head and neck. There are about 350,000 new cases worldwide each year and the incidence is increasing.2 OSCC is characterized by high malignancy, strong invasion capability, and high lymphatic metastasis, all of which lead to the poor prognosis.1 Despite improvements in tumor diagnosis and treatment, surgical excision, postoperative radiotherapy, and chemotherapy remain the mainstream methods of treatment, and prognosis of OSCC patients has not significantly improved.3 The reasons for high mortality of OSCC include late diagnosis, the lack of suitable markers by which to predict prognosis, and lack of specific therapeutic targets.2 Therefore, exploration of key genes in development and progression of OSCC is essential and urgently needed to discover new diagnostic markers and therapeutic targets.
Long noncoding RNAs (lncRNAs) are a type of RNA more than 200 nucleotides (nt) in length that lack the ability to encode proteins.4 The lncRNAs take part in many biological processes, including regulation of gene expression, cell cycle, and cell differentiation.5 Previous studies have suggested that abnormal expression of lncRNAs is involved in the tumorigenesis of many cancers, including OSCC.1 Therefore, we aimed to identify the biological function of dysregulated lncRNA HOXA10-AS (HOXA10 antisense RNA) and uncover the underlying molecular characteristic of HOXA10-AS in OSCC tumorigenesis.
Materials and methods
Ethical approval
The data used in this study were from The Cancer Gene Atlas (TCGA) program; consent and ethics-related documents can be found here: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/history/policies. Further ethical approval by our institution was deemed unnecessary.
Data acquisition and preprocess
The FPKM (fragments per kilobase per million reads mapped) expression matrix and clinical follow-up information and protein expression matrix were downloaded from the UCSC Xena database (https://xenabrowser.net/). The immunohistochemistry stained slides of normal oral mucosa and OSCC were from The Human Protein Atlas (HPA; https://www.proteinatlas.org/). Patients whose anatomic neoplasm subdivisions were from the oral cavity (including oral tongue, buccal mucosa, alveolar ridge, floor of mouth, lip, hard palate, and oral cavity) were selected for our studies; patients without clinical information or with incomplete details in the database were excluded.
Bioinformatic analysis
The R package “edgR” (https://www.R-project.org/) was used for differentially expressed gene (DEG) analysis, and the cut-off values for differentially expressed lncRNAs were set as follows: false discovery rate (FDR) <0.01 and |log2FC| >1, where FC is fold change. The R packages “survival” and “survminer” were used for Cox proportional hazards model construction and Kaplan-Meier (KM) plot visualization. The R package “WGCNA” (weighted gene co-expression network analysis) was conducted to identify genes closely related to HOXA10-AS.6 Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed in DAVID (https://david.ncifcrf.gov/). P-values <0.05 were regarded as significant.7,8 The online database STRING (https://string-db.org/) was used to identify relationships between key genes. A protein–protein interaction (PPI) network was created using the online database STRING (https://string-db.org/cgi/input.pl) and visualized using Cytoscape software (version 3.7.1; https://cytoscape.org/). Patients were grouped into two groups according to HOXA10-AS expression: low-HOXA10-AS expression group (lowest 20% of patients) and high-HXOA10-AS expression group (highest 20% of patients). The R package “clusterProfiler” was used for gene set enrichment analysis (GSEA). The R package “GSVA” was used for gene set variation analysis (GSVA). Commonly used markers for cell cycle process and cell proliferation activity were assessed between the low-HOXA10-AS and high-HOXA10-AS groups from mRNA and protein levels.
Cell culture and transfection
The CAL27 and SCC9 OSCC cell lines were obtained from Shanghai Cell Bank (Chinese Academy of Sciences, Shanghai, China). The two cell lines are well characterized and derived from tongue. Cells were cultured in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and 1% penicillin-streptomycin, at 37°C in humidified air containing 5% CO2. Specific antisense oligonucleotide (ASO) for HOXA10-AS was purchased from RiboBio (Guangdong, China). CAL27 and SCC9 cells cultured in 6-well plates were transfected with 50 nM ASO-HOXA10-AS and ASO controls according to the manufacturer’s instructions.
Cell viability proliferation assay
The CAL27 and SCC9 cells were cultured in 96-well plates at a density of 5 × 103 per well. After 24 hours of incubation, Cell Counting Kit-8 (CCK8, MEC, Shanghai, China) assays were used to measure cell viability. The CAL27 and SCC9 cells were cultured, 1000 cells per well, in 6-well plates for 14 days. The cell clones were washed with PBS, fixed in methanol, and stained with 0.1% crystal violet; colonies that contained more than 50 cells were counted.
Quantitative PCR analysis
Total RNA was isolated from cells using RNAiso Plus reagent (Takara Biotechnology Co. Ltd., Dalian, China). Complementary DNA was synthesized using a PrimeScript RT reagent kit with a genomic DNA Eraser (Perfect Real Time; Takara Biotechnology Co. Ltd.). Quantitative (q)PCR was conducted with a SYBR Premix Ex Taq II kit (Takara Biotechnology Co. Ltd.). The levels of mRNA were normalized to the levels of U6 mRNA. All qPCR procedures, including the design of the primers, validation of PCR conditions, and quantification, were performed according to Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines.
Statistical analysis
All statistical analyses were conducted in R software (version R x64 3.6.1; https://www.R-project.org/). All experiments were performed at least three times, and the data (bar graphs) are presented as means ± standard deviations (SD). One-way ANOVA was performed for multiple group comparisons, and mean values of different groups were compared using the Student–Newman–Keuls (SNK) test. P < 0.05 was considered to denote significant differences.
Results
HOXA10-AS is dysregulated and related to prognosis
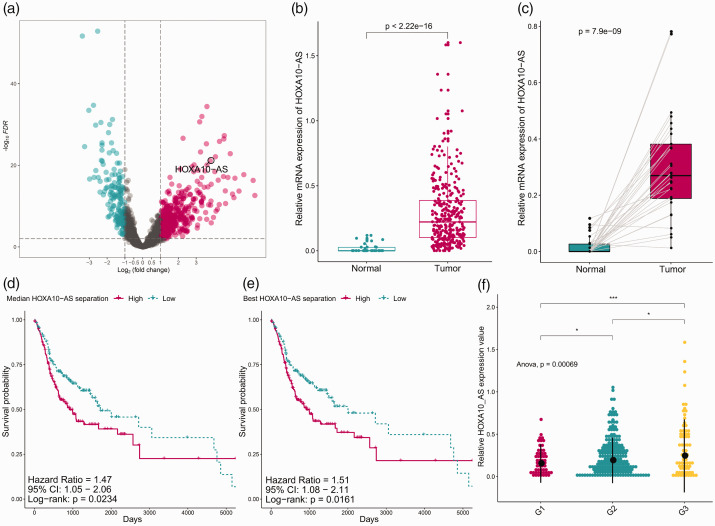
Using the criteria of |log2FC| >1 and FDR <0.01, we obtained 506 differentially expressed lncRNAs. The differentially expressed lncRNAs are presented in a volcano plot (Figure 1A). From the DEG results, we found that HOXA10-AS was aberrantly upregulated in OSCC, with significantly increased (P < 0.001) expression in tumor samples (Figure 1B and C). Expression of HOXA10-AS was associated with histological tumor grades and overall survival status of patients. Higher expression of HOXA10-AS was associated with higher tumor grade (P < 0.001), shorter survival time (P < 0.05), and poorer prognosis (P < 0.05), suggesting that upregulation of HOXA10-AS was a risk factor (Figure 1D–F).
Figure 1.
HOXA10-AS expression and its clinical significance. (a) Volcano plot showed differentially expressed lncRNAs between normal and OSCC samples. (b) Boxplots indicated that lncRNA HOXA10-AS was upregulated. (c) Pairwise boxplots indicated that HOXA10-AS was highly expressed in tumor samples. (d) Analysis of relationship between HOXA10-AS expression and grades. (e, f) Kaplan–Meier curves show overall survival time between HOXA10-AS low- and high-expression groups. HOXA10-AS, HOXA10 antisense RNA; lncRNA, long noncoding RNA; OSCC, oral squamous cell carcinoma; FDR, false discovery rate.
HOXA10-AS related module and key genes
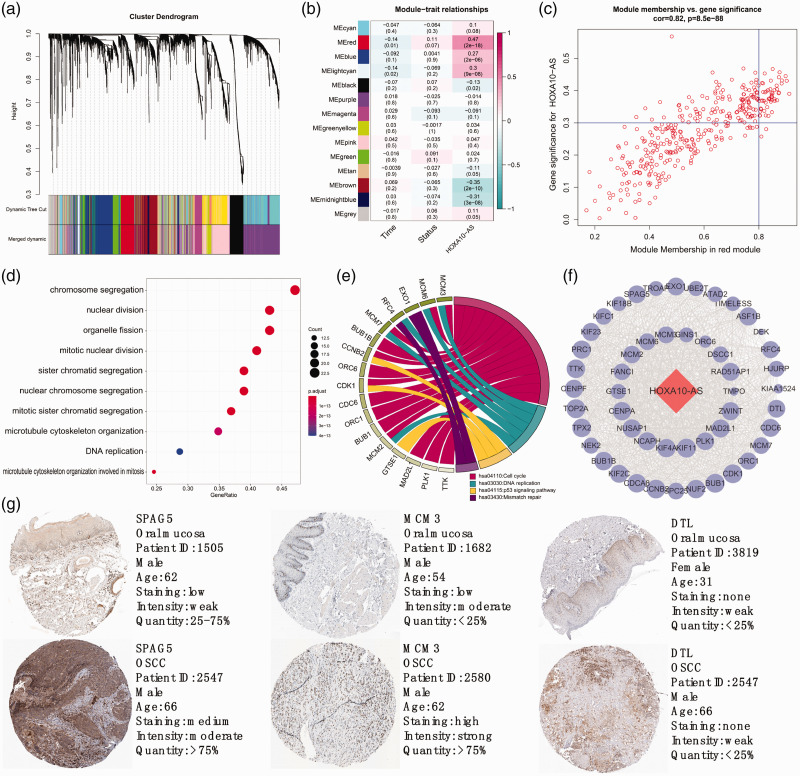
A total of 5028 genes were identified by the WGCNA package as being closely related to HOXA10-AS, and 14 modules were identified (Figure 2A). Further analysis revealed that the red module (a set of genes with similar functions or expression patterns) was the most significant module (P < 0.001) related to HOXA10-AS expression (Figure 2B). Fifty key genes were identified that played critical role in the red module (Figure 2C). Results showed that these 50 key genes were almost all involved in cell proliferation–related terms (Figure 2D) and similar results were observed for KEGG pathway analysis (Figure 2E). These results strongly suggested that HOXA10-AS plays an essential role in OSCC proliferation and cell cycle regulation. The PPI network revealed that the 50 key genes were closely related to each other, indicating they may function together (Figure 2F). We chose the top five key genes to assess their status between normal tissue and tumor samples. However, two of the five were not recorded in the HPA database (Figure 2G). We found that SPAG5 and MCM3 were significantly upregulated in OSCC samples but DTL did not differ between OSCC and control samples.
Figure 2.
Identification of key regulating module and biological functions of HOXA10-AS. (a) Samples were clustered into modules; modules are the branches of the clustering tree. (b) Correlation between module eigengenes and HOXA10-AS expression. (c) Scatterplot of genes in the red module. (d, e). GO and KEGG enrichment analyses for key genes related to HOXA10-AS. (f) PPI network revealed relationships among the key genes. (g) Validation of expression of the top hub genes in the red module. The protein expression of SPAG5 and MCM3 were upregulated in OSCC, whereas expression of DTL was unchanged. HOXA10-AS, HOXA10 antisense RNA; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein interaction.
HOXA10-AS promotes cell proliferation
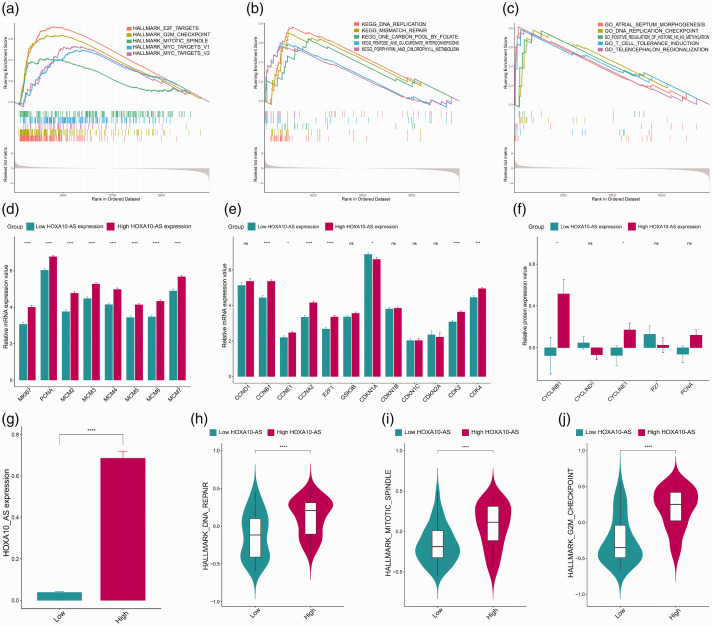
GSEA was applied to identify the deregulated gene sets that were enriched in the high-HOXA10-AS expression group. Many cell proliferation-related gene sets were enriched in the high-HOXA10-AS group, including cell cycle regulation and DNA replication (Figure 3A–C). These results indicated that HOXA10-AS was closely associated with cell proliferation. We detected commonly used markers of cell proliferation and cell cycle at the transcriptional and translational levels. We detected mRNA expression of cell proliferation markers (Figure 3D) and protein expression (Figure 3F); all markers were significantly highly expressed in the high-HOXA10-AS group compared with the low-HOXA10-AS group, indicating that HOXA10-AS could promote cell proliferation. The commonly used cell cycle promotion markers showed similar results (Figure 3E and 3F). GSVA was used to detect cell proliferation activity in the high-HOXA10-AS expression group. We found that the high-HOXA10-AS expression group (Figure 3G) had higher proliferation activity, including higher G2/M checkpoint activity (Figure 3H), higher mitotic spindle score (Figure 3I), and higher DNA repair rate (Figure 3J).
Figure 3.
HOXA10-AS is involved in regulation of OSCC cell proliferation. (a–c) Gene set enrichment analysis revealed the gene sets of hallmarks and KEGG biological processes enriched in patients with high expression of HOXA10-AS. (d, e) mRNA expression of common cell proliferation and cell cycle markers. (f) Protein expression of cell proliferation and cell cycle markers. (g) Expression differences between patients with high and low expression of HOXA10-AS. (h–j) Gene set variation analysis revealed proliferation scores. Data were presented as mean ± standard error (SE); *P < 0.05, **P < 0.01, ***P < 0.001. HOXA10-AS, HOXA10 antisense RNA; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Silencing HOXA10-AS decreases cell proliferation
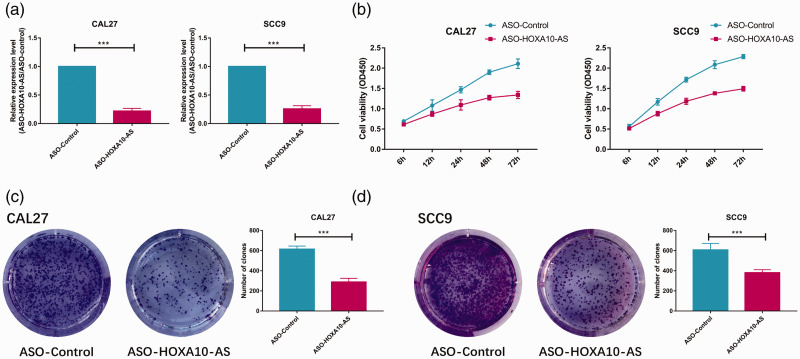
We used ASO (antisense oligonucleotide) to silence HOXA10-AS in CAL27 and SCC9 cells. The expression of HOXA10-AS was significantly silenced (P < 0.001) in ASO-HOXA10-AS compared with ASO-control (Figure 4A). In the CCK8 assay, cell viability was inhibited in a time-dependent manner after silencing (P < 0.001) compared with that of the control group (Figure 4B). Similar results were seen in the clone formation assay. The number of clones in the ASO-HOXA10-AS group was less (P < 0.001) than that in the control group (Figure 4C and D).
Figure 4.
Silencing HOXA10-AS decreases OSCC proliferation ability. (a) HOXA10-AS was silenced by ASO in OSCC cell lines CAL27 and SCC9, and qPCR analysis of the HOXA10-AS expression levels were conducted. (b) CAL27 and SCC9 cells were transfected with ASO-control and ASO-HOXA10-AS; 72 hours later, cell growth curves were measured by CCK8 at the indicated time points. (c, d) Colony formation ability of CAL27 and SCC9 was inhibited when HOXA10-AS was silenced. Data are presented as mean ± standard deviation (SD) (n = 3) *P < 0.05, **P < 0.01, ***P < 0.001.HOXA10-AS, HOXA10 antisense RNA; OSCC, oral squamous cell carcinoma; ASO, antisense oligonucleotide; qPCR, quantitative PCR; CCK8, Cell Counting Kit-8.
Discussion
LncRNAs are essential regulators associated with many diseases, including OSCC. For example, Zhang et al.9 reported that lncRNAs are involved in immune regulation of periodontitis, Chen et al.10 found that several lncRNAs contribute to development of pericarditis, and Qin et al.11 found that the lncRNA Xist is closely related to plexiform arteriopathy. OSCC, being the eighth most common malignancy worldwide, is known for its high mortality and short survival time, and its pathogenesis is poorly understood.12,13 In our study, lncRNA HOXA10-AS was upregulated in patients with OSCC and significantly related to overall survival as well as histological grade in OSCC. Analysis using the WGCNA package revealed that the red module (a set of genes with similar functions or expression patterns) was significantly associated with HOXA10-AS and demonstrated that key genes in this module were involved in cell proliferation. Using GSEA and GSVA, we identified the key function of HOXA10-AS, as shown by marker genes in cell proliferation. We confirmed the results by experimental validation.
The lncRNA studied here, HOXA10-AS, is also known as HOXA10-AS antisense RNA. Only two studies to date have discussed its oncogenic effects.14,15 Dong et al. investigated its functional role in glioma. Consistent with our results, they found that HOXA10-AS was upregulated in glioma tissues and cell lines. Expression of HOXA10-AS was also significantly related to glioma grade, and analysis of HOXA10-AS knockdown confirmed its positive effects on cell proliferation.15 In a study by Sheng et al.,14 elevated expression of HOXA10-AS was found in lung adenocarcinoma tissues, and upregulation of HOXA10-AS in lung adenocarcinoma cells enhanced its proliferation. These studies confirm our findings related to this lncRNA in OSCC. Many lncRNAs have been found to play vital roles by modulating downstream pathways in the tumorigenesis of OSCC, particularly in cell proliferation. The increased expression of lncRNA BANCR could enhance proliferation of OSCC cells via the MAPK signaling pathway.16 A study conducted by Liu et al.17 revealed that the lncRNA ANRIL enhanced the proliferation ability of OSCC by regulating the transforming growth factor (TGF)-β/Smad pathway. Overall, our findings were consistent with earlier studies and identified a new mechanism for HOXA10-AS in regulation of cell proliferation.
There are some limitations in this research. The in-depth mechanism of HOXA10-AS regulation network remains to be elucidated, and detailed relationships among HOXA10-AS, genomic features, and the tumor microenvironment have not been explored or experimentally validated. In future studies, we will investigate the association of HOXA10-AS with other carcinoma features such as infiltrating immunocytes, tumor stemness, SNP, copy number variation, and methylation. In addition, tumor xenografts in athymic nude mice are an effective method by which to investigate the role of lncRNAs or genes in vivo. Because of the short duration of short interfering (si)RNA transfection, which can last only 1 or 2 weeks, we could not explore the role of HOXA10-AS comprehensively in this study. In future studies, we will focus on the mechanism and effect of lncRNA HOXA10-AS in vivo. Further validation of these results is needed using in vivo assays and deeper cellular functional experiments.
In summary, we identified aberrantly expressed lncRNAs in OSCC and found that lncRNA HOXA10-AS plays an essential role in cell proliferation to promote OSCC tumorigenesis. Silencing HOXA10-AS can decrease cell proliferation.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 81972002).
ORCID iD
References
- 1.Rivera C, Oliveira AK, Costa RAP, et al. Prognostic biomarkers in oral squamous cell carcinoma: a systematic review. Oral Oncol 2017; 72: 38–47. DOI: 10.1016/j.oraloncology.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7–34. DOI: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Marur S, Forastiere AA. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc 2016; 91: 386–396. DOI: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 4.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016; 17: 47–62. DOI: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 5.Xing L, Zhang X, Chen A. Prognostic 4-lncRNA-based risk model predicts survival time of patients with head and neck squamous cell carcinoma. Oncol Lett 2019; 18: 3304–3316. DOI: 10.3892/ol.2019.10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Feng H, Li Z, et al. Application of weighted gene co-expression network analysis to identify key modules and hub genes in oral squamous cell carcinoma tumorigenesis. Onco Targets Ther 2018; 11: 6001–6021. DOI: 10.2147/OTT.S171791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing L, Zhang X, Feng H, et al. Silencing FOXO1 attenuates dexamethasone-induced apoptosis in osteoblastic MC3T3-E1 cells. Biochem Biophys Res Commun 2019; 513: 1019–1026. DOI: 10.1016/j.bbrc.2019.04.112. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Feng H, Du J, et al. Aspirin promotes apoptosis and inhibits proliferation by blocking G0/G1 into S phase in rheumatoid arthritis fibroblast-like synoviocytes via downregulation of JAK/STAT3 and NF-kappaB signaling pathway. Int J Mol Med 2018; 42: 3135–3148. DOI: 10.3892/ijmm.2018.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Ren L, Yan X, et al. Identification of immune-related lncRNAs in periodontitis reveals regulation network of gene-lncRNA-pathway-immunocyte. Int Immunopharmacol 2020; 84: 106600 DOI: 10.1016/j.intimp.2020.106600. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Sun F, Zhang Y, et al. Comprehensive molecular characterization of circRNA-associated ceRNA network in constrictive pericarditis. Ann Transl Med 2020; 8: 549. DOI: 10.21037/atm-20-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin S, Predescu DN, Patel M, et al. Sex differences in the proliferation of pulmonary artery endothelial cells: implications for plexiform arteriopathy. J Cell Sci 2020; 133: jcs237776. DOI: 10.1242/jcs.237776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amit M, Yen TC, Liao CT, et al. Improvement in survival of patients with oral cavity squamous cell carcinoma: an international collaborative study. Cancer 2013; 119: 4242–4248. DOI: 10.1002/cncr.28357. [DOI] [PubMed] [Google Scholar]
- 13.Xing L, Zhang X, Tong D. Systematic profile analysis of prognostic alternative messenger RNA splicing signatures and splicing factors in head and neck squamous cell carcinoma. DNA Cell Biol 2019; 38: 627–638. DOI: 10.1089/dna.2019.4644. [DOI] [PubMed] [Google Scholar]
- 14.Sheng K, Lu J, Zhao H. ELK1-induced upregulation of lncRNA HOXA10-AS promotes lung adenocarcinoma progression by increasing Wnt/beta-catenin signaling. Biochem Biophys Res Commun 2018; 501: 612–618. DOI: 10.1016/j.bbrc.2018.04.224. [DOI] [PubMed] [Google Scholar]
- 15.Dong CY, Cui J, Li DH, et al. HOXA10AS: a novel oncogenic long noncoding RNA in glioma. Oncol Rep 2018; 40: 2573–2583. DOI: 10.3892/or.2018.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao C, Kong F, Zhang S, et al. Long non-coding RNA BANCR promotes proliferation and migration in oral squamous cell carcinoma via MAPK signaling pathway. J Oral Pathol Med 2019; ▪: ▪. DOI: 10.1111/jop.12968. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Ning SB, Fu S, et al. Effects of lncRNA ANRIL on proliferation and apoptosis of oral squamous cell carcinoma cells by regulating TGF-beta/Smad pathway. Eur Rev Med Pharmacol Sci 2019; 23: 6194–6201. DOI: 10.26355/eurrev_201907_18435. [DOI] [PubMed] [Google Scholar]