Abstract
The Covid-19 pandemic threatens to overwhelm healthcare systems around the world. The only current FDA approved treatment, which directly targets the virus, is the ProTide prodrug remdesivir. In its activated form, remdesivir prevents viral replication by inhibition of the essential RNA-dependent RNA polymerase. Like other ProTide prodrugs, remdesivir contains a chiral phosphorus center. The initial selection of the (SP)-diastereomer for remdesivir was reportedly due to the difficulty in producing the pure (RP)-diastereomer of the required precursor. However, the two currently known enzymes responsible for the initial activation step of remdesivir are each stereoselective and show differential tissue distribution. Given the ability of the Covid-19 virus to infect a wide array of tissue types, inclusion of the (RP)-diastereomer may be of clinical significance. To help overcome the challenge of obtaining the pure (RP)-diastereomer of remdesivir, we have developed a novel chemo-enzymatic strategy that utilizes a stereoselective variant of the phosphotriesterase from Pseudomonas diminuta to enable the facile isolation of the pure (RP)-diastereomer of the chiral precursor for the chemical synthesis of the (RP)-diastereomer of remdesivir.
Graphical Abstract
INTRODUCTION
The emergence of the Covid-19 pandemic has perhaps become the defining event of the 21st Century. As of July 1, 2020, there have been more than 10 million documented cases and more than 500,000 deaths worldwide (1). Consequently, there is an urgent need for the rapid development of effective antiviral therapies to relieve the ongoing burden on local healthcare systems. Covid-19 is caused by the SARS-CoV-2 RNA virus (2), and the RNA-dependent RNA polymerase from the virus is currently one of the most attractive targets for clinical therapy (3). The antiviral prodrug remdesivir was originally developed and tested against the Ebola virus. It has been shown to be a highly effective inhibitor of several RNA-dependent RNA polymerases from various corona viruses, including SARS-CoV-2 (4–8). The pre-existing safety and clinical data for remdesivir have facilitated current clinical trials and the United States FDA has granted approval of remdesivir for treatment of severe Covid-19 (9). Remdesivir is now the only approved treatment for Covid-19 that directly targets the virus.
Remdesivir is a phosphoramidate nucleotide prodrug (ProTide). The ProTides achieve higher efficacy with lower doses compared to their unmasked nucleoside counterparts by improving cellular availability and bypassing the rate-limiting phosphorylation step for nucleoside activation (10–12). The remdesivir prodrug contains a chiral phosphorus center with the current clinical compound being the (SP)-diastereomer (4, 7). However, the active form of the drug is the nucleotide triphosphate derivative of remdesivir, which causes inhibition of viral replication by delayed chain termination of the RNA-dependent RNA polymerase (6).
The activation mechanism of the ProTides is known to occur by enzymatic hydrolysis of the amino acid carboxylate ester, leading to spontaneous cyclization and hydrolysis (Scheme 1) (13–16). Subsequent hydrolysis of the phosphoramidate intermediate is catalyzed by a histidine triad protein (Hint1) releasing the monophosphate of remdesivir, which is then enzymatically phosphorylated to the ultimate triphosphate inhibitor (4, 13). The two enzymes that have been identified as the major contributors of the initial activation step are carboxylate esterase 1 (CES1) and cathepsin A (CatA) (13). CatA displays expression in a wide array of tissues and has a strong preference for hydrolysis of the (SP)-diastereomer of the prodrug. However, the expression levels of this enzyme vary widely among individual patients. Conversely, CES1 has been shown to have a strong preference for hydrolysis of the (RP)-diastereomer. The tissue distribution of this enzyme is more limited but it is the primary carboxylate esterase in some tissues. These differences have been utilized to target ProTides for specific tissues (17). However, in the case of Covid-19, viral infection of multiple tissues and organs, including lung, liver, vasculature, and kidney, has been implicated and thus a more widespread distribution of antiviral activity may be beneficial (2, 18, 19).
Scheme 1.
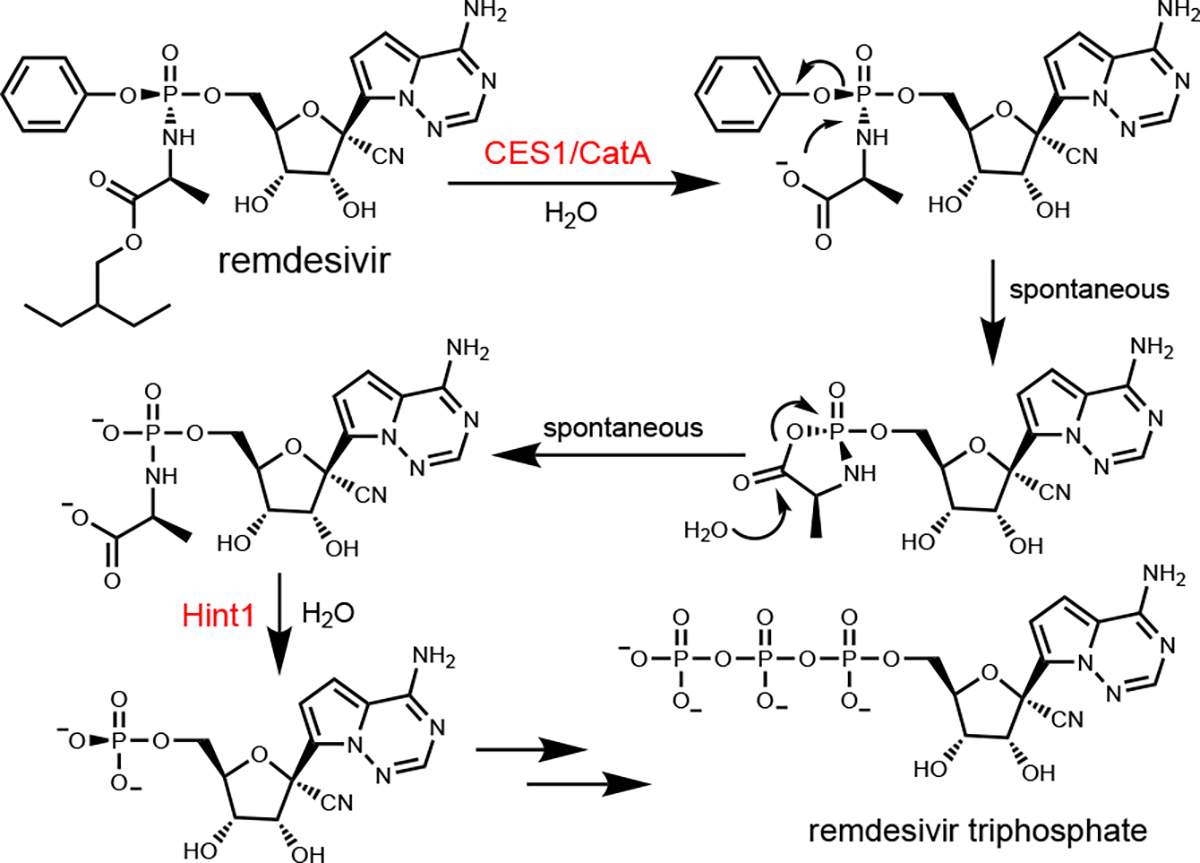
Biological activation of remdesivir.
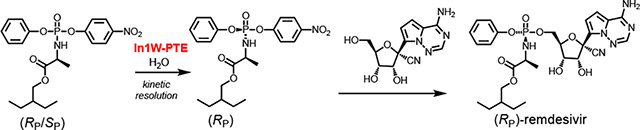
The initial selection of the (SP)-diastereomer of remedsivir was reported to be based on the facile crystallization of the (SP)-diastereomer of the p-nitrophenyl containing precursor (1)(7). In vitro testing of both the (SP)- and (RP)-diastereomers of remdesivir demonstrated that both compounds were effective antiviral inhibitors, and that the more effective inhibitor depended on the tissue-type being tested (7). These data suggest that in the treatment of Covid-19, the (RP)-diastereomer of remdesivir might be of some clinical importance. To enable the chemical preparation of the (RP)-diastereomer of remdesivir we have developed a chemo-enzymatic strategy that utilizes the stereoselective In1W variant of the phosphotriesterase from Pseudomonas diminuta (In1W-PTE) to facilitate the isolation of the (RP)-diastereomer of precursor 1 and subsequent coupling to the appropriate nucleoside analog (2) as illustrated in Scheme 2 (20).
Scheme 2.
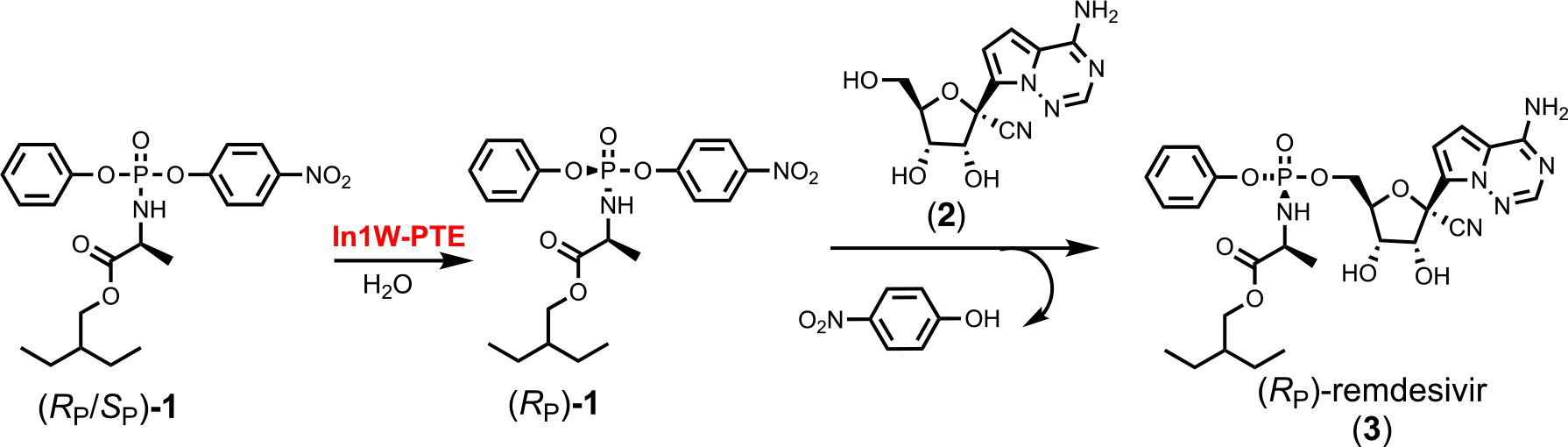
Chemo-enzymatic synthesis of (RP)-remdesivir.
MATERIALS and METHODS
Materials.
General laboratory chemicals and supplies were obtained from either Sigma Aldrich or VWR. Varients of phosphotriesterase (In1W-PTE and G60A-PTE) were expressed and purified as previously described, and then stored at −80 °C prior to use (21). The nucleoside analog for remdesivir (2) was purchasd from Carbosynth.
Synthesis of the Diastereomeric Precursor for Remdesivir (RP/SP-1).
The diastereomeric precursor of remdesivir (RP/SP-1) was synthesized as previously described with minor modifications (8). (S)-2-Ethylbutyl 2-aminopropanoate tosylate (1.14 g, 3.3 mmol, 1.1 equiv) was suspended in dichloromethane (10 mL) and the resulting mixture was cooled to −78 °C. Phenyl dichlorophosphate (0.52 mL, 3.3 mmol, 1.1 equiv, 95%) was then added, followed by the slow addition of triethylamine (0.92 mL, 6.6 mmol, 2.2 equiv). The resulting mixture was allowed to warm to room temperature and stirred for 3 h. The mixture was then cooled to 0 °C and 4-nitrophenol (0.42, 3.0 mmol, 1.0 equiv) was added, followed by the slow addition of triethylamine (0.46 mL, 3.3 mmol, 1.1 equiv). The resaction was allowed to warm to room temperature and stirred for 16 h. The reaction mixture was then concentrated under reduced pressure and the crude residue was subjected to silica gel chromatography and eluted with hexanes/ethyl acetate (7:1, 200 mL) and hexanes/ethyl acetate (3:1, 400 mL) to afford the final product (0.91 g, 61%) as a colorless semi-solid.
All NMR spectra were recorded on a Bruker 400 MHz NMR with an Advance III Console and 5 mm broad band probe. 1H NMR (400 MHz, CDCl3) δ 8.25–8.19 (m, 2H), 7.42–7.31 (m, 4H), 7.26–7.17 (m, 3H), 4.20 – 4.10 (m, 1H), 4.10–3.99 (m, 2H), 3.92–3.84 (m, 1H), 1.55–1.46 (m, 1H), 1.44–1.39 (m, 3H), 1.36–1.28 (m, 4H), 0.87 (t, J = 7.4 Hz, 6H). 31P NMR (160 MHz, CDCl3) δ – 3.20 (s), −3.25 (s).
High resolution mass spectrometry (ESI+); m/z [M+H]+ calc. for C21H28N2O7P: 451.1634; found: 451.1623.
Synthesis of (RP/SP)-Remdesivir (3).
To a 10-mL round bottom flask, under an argon atmosphere, the nucleoside analog component of remdesivir (compound 2) (9.0 mg, 0.031 mmol, 1.0 equiv) was dissolved in dry DMF (0.3 mL) and then 0.5 mL of dry THF was added. To the stirred solution at room temperature, t-butylmagnesium chloride (0.051 mL, 0.051 mmol, 1.6 equiv, 1.0 M in THF) was added, producing a white solid. After 30 min, (RP/SP)-1 (20 mg, 0.044 mmol, 1.4 equiv) dissolved in dry THF (0.7 mL) was added. The resulting mixture was stirred for 42 h and then concentrated under reduced pressure. The crude residue was subjected to silica gel chromatography using chloroform/methanol (12:1, 180 mL) to isolate (RP/SP)-remdesivir (10 mg, 54%).
1H NMR (400 MHz, CDCl3) δ 7.86 (s, 0.35H), 7.84 (s, 0.65H), 7.34–7.24 (m, 2H), 7.21–7.10 (m, 3H), 6.93–6.86 (m, 2H), 4.51 – 4.24 (m, 4H), 4.23–4.15 (m, 1H), 4.05–3.82 (m, 3H), 1.52–1.42 (m, 1H), 1.39–1.23 (m, 7H), 0.92–0.82 (m, 6H). 31P NMR (160 MHz, CDCl3) δ 3.52 (s), 3.47 (s).
HRMS (ESI+) m/z [M+H]+ calc. for C27H36N6O8P: 603.2332, found: 603.2320.
Synthesis of (RP)-Remdesivir (3).
To a 10-mL round bottom flask, under an argon atmosphere, compound 2 (9.0 mg, 0.031 mmol, 1.0 equiv) was dissolved in dry DMF (0.3 mL) and then 0.5 mL of dry THF was added. To the stirred solution at room temperature, t-butylmagnesium chloride (0.051 mL, 0.051 mmol, 1.6 equiv, 1.0 M in THF) was added, producing a white solid. After 30 min, (RP)-1 (20 mg, 0.044 mmol, 1.4 equiv) dissolved in dry THF (0.7 mL) was added. The resulting mixture was stirred for 42 h and then concentrated under reduced pressure. The crude residue was subjected to silica gel chromatography using chloroform/methanol (12:1, 180 mL) to obtain the (RP)-remdesivir product (7.0 mg, 38%).
1H NMR (400 MHz, CDCl3) δ 7.84 (s, 1H), 7.32–7.27 (m, 2H), 7.19–7.13 (m, 3H), 6.93 (d, J = 4.6 Hz, 1H), 6.90 (d, J = 4.6 Hz, 1H), 4.51 – 4.36 (m, 3H), 4.35–4.28 (m, 1H), 4.22 (t, J = 5.4 Hz, 1H), 4.02 (dd, J1 = 10.8 Hz, J2 = 5.8 Hz, 1H), 3.97 (dd, J1 = 10.8 Hz, J2 = 5.8 Hz, 1H), 3.90–3.80 (m, 1H), 1.52–1.42 (m, 1H), 1.38–1.22 (m, 7H) 0.87 (t, J = 7.4 Hz, 6H). 31P NMR (160 MHz, CDCl3) δ 3.52 (s).
HRMS (ESI+) m/z [M+H]+ calc. for C27H36N6O8P: 603.2332, found: 603.2319.
Determination of Stereoselective Hydrolysis of Remdesivir Precursor (RP/SP-1) by PTE.
Stock solutions of (RP/SP)-1 were made at ~2.0 mM in 60% MeOH. Assays were conducted in a final total volume of 1.0 mL containing 50 mM HEPES (pH 8.5), 20% MeOH and ~35 μM (RP/SP)-1. Reactions were monitored at 400 nm using a Molecular Devices SpectaMax 384 Plus spectrometer. The total concentration of (RP/SP)-1 was determined by the addition of 50 mM NaOH and the reaction followed to completion by the release of p-nitrophenol at 400 nm (E400 = 17 mM−1 cm−1) in a 1-cm plastic cuvette. Enzymatic reactions were initiated by the addition of appropriately diluted enzyme to allow for completion of the reaction in approximately 15 min. Enzyme variants were evaluated for clear biphasic hydrolysis or the selective hydrolysis of 50% of the total substrate sample.
Enzyme Reaction Kinetics.
Enzymatic reactions were conducted as above with 30 nM and 3.0 μM In1W-PTE, and 3.5 μM G60A-PTE at 25 °C (20). Reactions were followed to completion by the release of p-nitrophenol and the data converted to fraction hydrolyzed , plotted as a function of time and fit to a single exponential to determine according to equations 1–3, where is the absorbance at time , is the initial absorbance, is the final absorbance, is the magnitude of the exponential phase, is the exponential rate constant and is the concentration of enzyme used.
| (1) |
| (2) |
| (3) |
Isolation of Enantiomerically Pure (RP)-1.
The reaction was conducted in a volume of 1.0 L using 50 mM HEPES (pH 8.0), 30% MeOH, 100 μM (RP/SP)-1 and 3 × 0.22 mg In1W-PTE. (RP/SP)-1 (45 mg) was dissolved into 300 mL of methanol. Separately, 700 mL of a solution containing 50 mM HEPES (pH 8.0), and 0.22 mg In1W-PTE was prepared. The two solutions were mixed and the reaction allowed to proceed at room temperature with gentle stirring. The progress of the reaction was monitored by removal of 980 μL samples at various time points. The absorbance of the sample was measured at 400 nm and the progress of the reaction checked by the addition of 20 μL of 10 M NaOH to hydrolyze the remaining substrate. At 1 h the reaction was found to be 25% complete and a second aliquot of In1W-PTE was added. At 2 h the reaction was found to be 40% complete and a third aliquot of In1W-PTE was added and the reaction allowed to procced for an additional hour. The diastereomeric purity of the remaining substrate was evaluated by removal of 20 mL samples at 10, 25, 60, and 105 min. The methanol was removed from the samples by evaporation under reduced pressure and the reaction stopped by extraction of the remaining substrate into dichlormethane (3 × 10 mL). The organic layer was washed 3 times with 50 mL of 25 mM HEPES (pH 8.0) and dried over NaSO4. The solvent was removed and the residue dissolved in 700 μL of deuterated DMSO and the 31P NMR spectrum was recorded.
Once the reaction reached 50% completion, methanol was removed from the remaining solution by evaporation under reduced pressure. The pure (RP)-diastereomer of compound 1 was recovered by extraction in dichloromethane (3 × 100 mL). The organic layer was washed with copious amounts of 25 mM HEPES (pH 8.0) to remove the contaminating p-nitrophenol and then dried over NaSO4. The solvent was removed yielding 20 mg (97%) of the pure (RP)-diastereomer (ee >95%).
RESULTS and DISCUSSION
Previous work from this laboratory demonstrated that diastereomers of ProTide precursors can be isolated by kinetic resolution using engineered variants of PTE (20). The majority of known ProTides utilize an alanine residue esterified at the carboxylate group with methanol or isopropanol (10). Unlike these compounds, remdesivir is esterified with 2-ethyl butanol, which adds substantial bulk to the phosphorus moiety (7). Wild-type PTE favors substrates with smaller ester groups attached to the central phosphorus core. For example, there is a 15-fold reduction in activity for hydrolysis of diisopropyl p-nitrophenyl phosphate relative to the hydrolysis of diethyl p-nitrophenyl phosphate (22). The In1W-PTE variant has previously been shown to be much more tolerant of larger ester groups and to stereoselectively hydrolyze ProTide precursors (20). When In1W-PTE was tested with the (RP/SP)-1 precursor of remdesivir, it rapidly hydrolyzed half of the total sample, indicating robust catalytic activity and high stereoselectivity (Figure 1). Other known variants of PTE, including G60A, GGY, YT, BHR-30 and BHR-72, which show strong stereoselectivity with other compounds, were also tested for stereoselective hydrolysis of (RP/SP)-1 but none of these mutants were found to be suitable (22–24).
Figure 1.
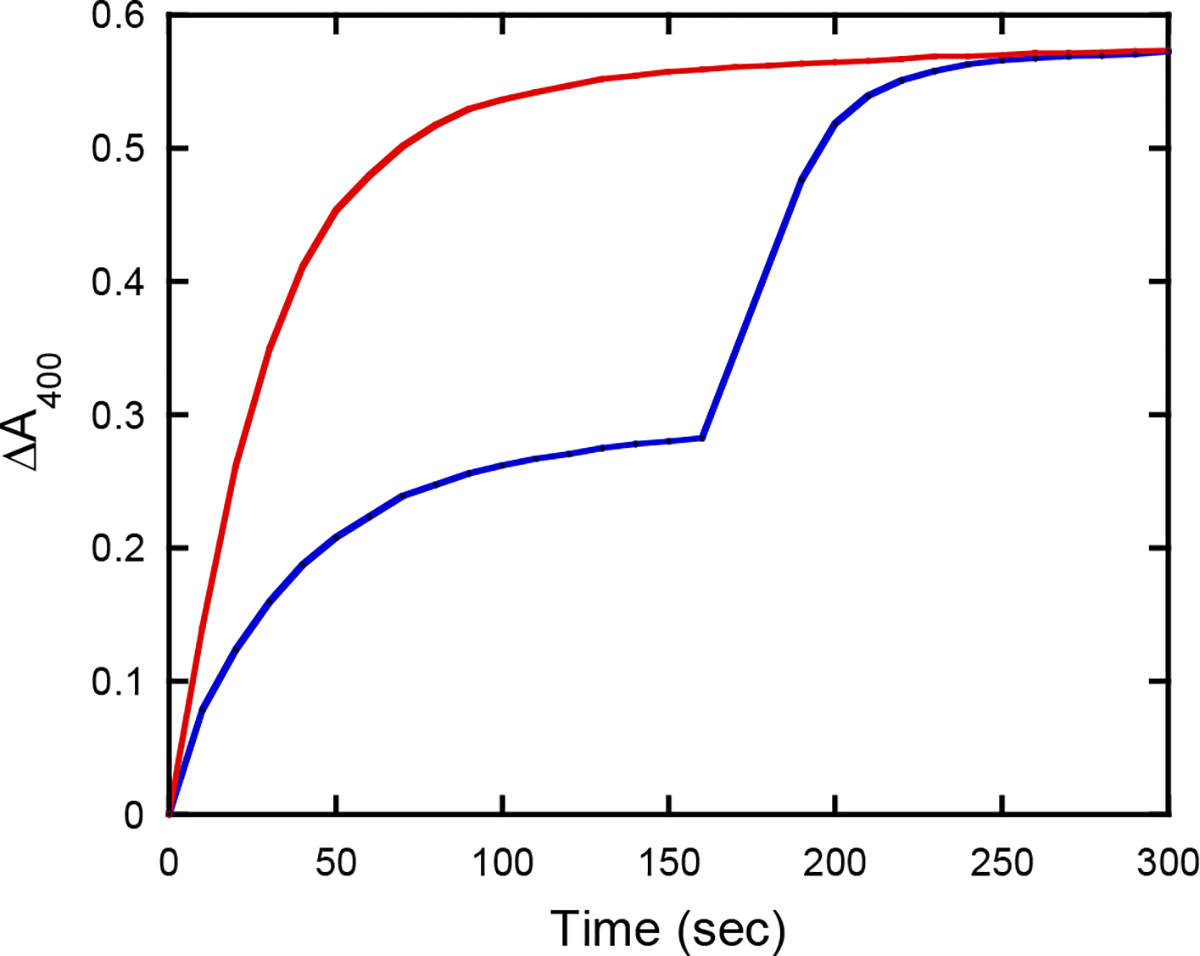
Chemical and enzymatic hydrolysis of (SP/RP)-1. Reactions contained 10 mM HEPES-KOH buffer (pH 8.5) and 20% MeOH. The red line represents the time course for the chemical hydrolysis of 34 μM (RP/SP)-1 by 50 mM NaOH. The blue line represents the time course for hydrolysis of a single isomer of (RP/SP)-1 catalyzed by the addition of 29 nM In1W-PTE to 34 μM (RP/SP)-1, followed by hydrolysis of the other isomer by the addition of 50 mM NaOH.
The two diastereomers of (RP/SP)-1 are easily distinguished from one another by 31P NMR spectroscopy (Figure 2a). It has previously been established that the (SP)-diastereomer of ProTide precursors appears downfield of the (RP)-diastereomer in multiple solvents (20, 25). To determine which diastereomer of compound 1 was preferentially hydrolyzed by In1W-PTE, the reaction was followed over time. As shown in Figure 2, over a time course of 3 h, the substrate composition goes from a 1:1 mixture of the (SP)- and (RP)-diastereomers to only the single (RP)-diastereomer being present, demonstrating the stereoselective hydrolysis of the (SP)-diastereomer by In1W-PTE.
Figure 2.
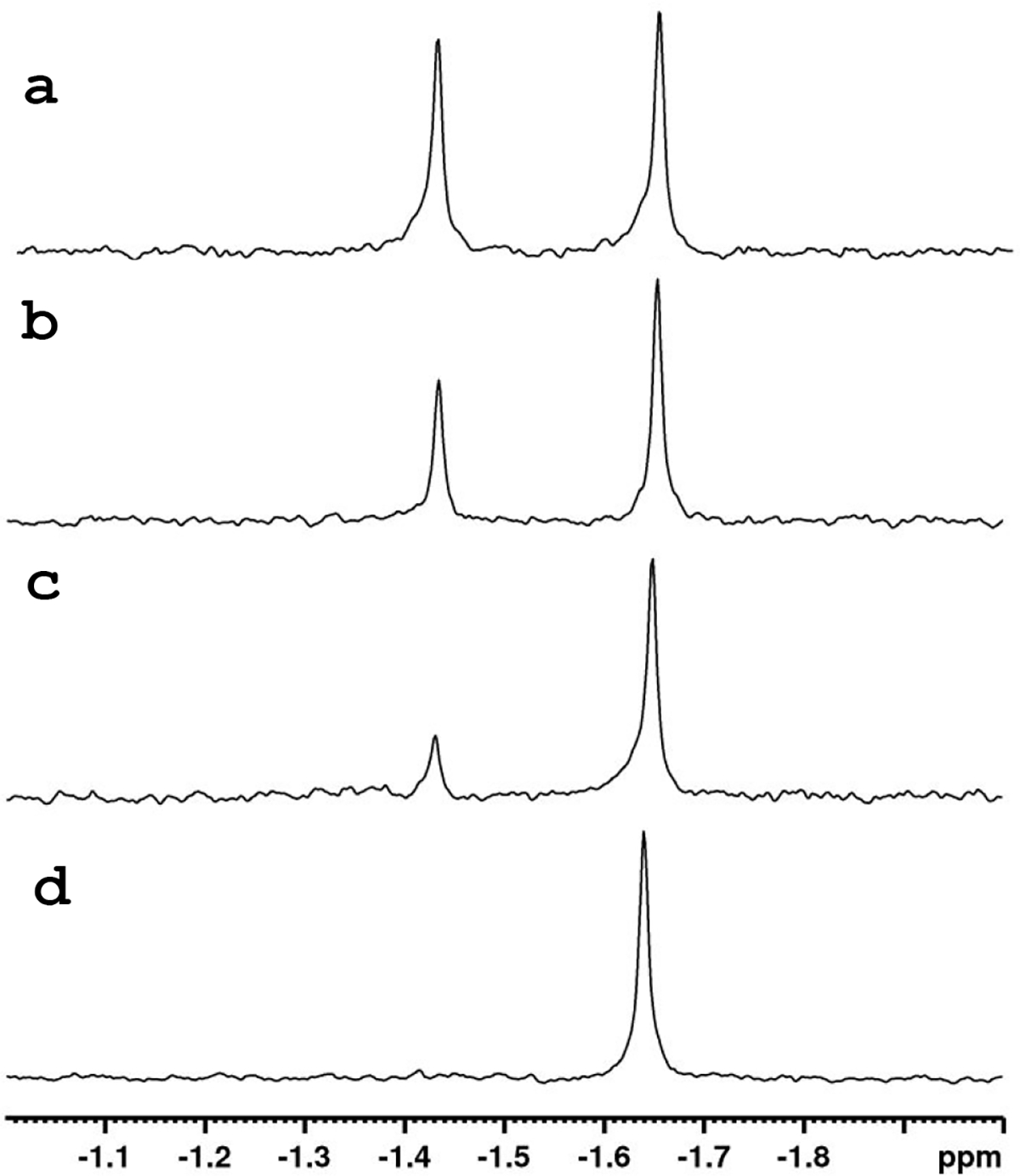
31P-NMR spectra of (RP/SP)-1 after hydrolysis catalyzed by In1W-PTE. The remaining substrate was recovered by extraction with DMSO of the reaction solution at various times (a) before the addition of enzyme, (b) 60 min; (c) 105 min; and (d) 190 min incubation.
In order to determine the extent of the stereoselectivity of PTE with compound 1, the enzymatic efficiency of In1W-PTE and G60A-PTE were measured using a total hydrolysis assay (26). This method allows for the measurement of for both diastereomers in a single assay by first hydrolyzing the preferred enantiomer with a low concentration of enzyme, and then subsequently hydrolyzing the second enantiomer by addition of a higher enzyme concentration. Kinetic measurements for the stereoselective hydrolysis of (RP/SP)-1 catalyzed by In1W exhibited a greater than 200-fold preference for hydrolysis of the (SP)-isomer relative to the (RP)diastereomer . Interestingly, the variant G60A, which showed a stereoselective preference for hydrolysis of the (RP)-isomer of the ProTide precursor with an isopropyl ester (20), did not show measurable stereoselective hydrolysis of compound 1, and the catalytic activity of (RP)-1 is reduced more than 10-fold compared to the precursor with an isopropyl ester.
Given the strong stereoselectivity of In1W-PTE, isolation of the (RP)-diastereomer was attempted on a larger scale. Approximately 41 mg of (RP/SP)-1 was dissolved in 30% methanol with 50 mM HEPES-KOH buffer (pH 8.0). The reaction was catalyzed by the addition of a total of 0.65 mg of In1W-PTE. The reaction was monitored by UV/Vis spectroscopy at 400 nm, and after the reaction had reached 50% completion, residual 1 was recovered by organic extraction using dichloromethane. From 41 mg of (RP/SP)-1, In1W-PTE enabled the recovery of 20 mg of the pure (RP)-diastereomer (97%) and contamination by the (SP)-diastereomer was below the detection limit as determined by 31P NMR spectroscopy (Figure 2d).
Synthesis of (RP/SP)- and (RP)-Remdesivir.
The isolated (RP)-1 was used as the precursor for the synthesis of remdesivir according to previously established methods (7, 20). The displacement of p-nitrophenyl from compound 1 proceeds via an SN2-like mechanism which results in an inversion of stereochemistry. From 20 mg of (RP)-1 isolated, 7 mg of product containing a single stereoisomer was recovered (Figure 3a). As a control, the same reaction was carried out using racemic 1. The racemate yielded (RP/SP)-remdesivir in a ratio of (65:35) according to the 31P NMR spectrum (Figure 3b). (SP)-and (RP)-remdesivir can be distinguished from one another by 31P and 1H NMR spectroscopy, which allows for the definitive identification of the product using the isolated (RP)-1 as being (RP)-remdesivir.
Figure 3.
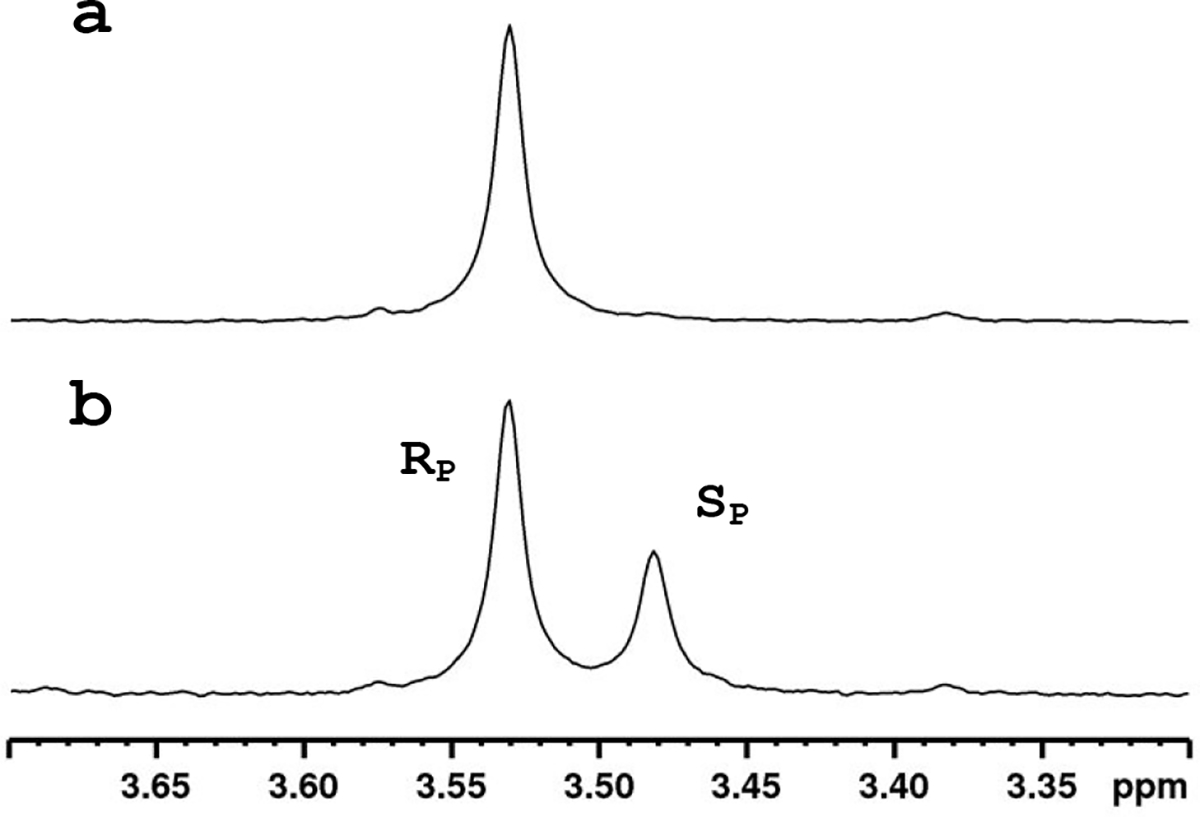
31P NMR spectra of remdesivir in methanol. (a) Spectrum of remdesivir synthesized from pure (RP)-1. ( b) Spectrum of remdesivir synthesized from (RP/SP)-1.
CONCLUSIONS
Given the ability of SARS-CoV-2 to infect multiple tissues, there is an urgent need for antiviral therapy that shows wide tissue distribution and cellular availability. Remdesivir has demonstrated strong efficacy for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2, and the use of the (RP)-diastereomer may be able to increase the tissue distribution and efficacy of treatment. Using the stereoselective In1W variant of PTE we have overcome this challenge and can readily synthesize the pure (RP)-diastereomer of remdesivir using a chemo-enzymatic strategy.
Funding Sources
This work was supported by grants from the National Institutes of Health (GM 116894) and the Robert A. Welch Foundation (A-0840).
Footnotes
Notes
Authors declare no competing financial interest.
REFERENCES
- [1].Johns Hopkins University (2020) CORONAVIRUS RESOURCE CENTER, https://coronavirus.jhu.edu/, Accessed: 7-1-2020.
- [2].Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, and Tan W (2020) A Novel Coronavirus from Patients with Pneumonia in China, 2019, New England Journal of Medicine 382, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang Q, Wu J, Wang H, Gao Y, Liu Q, Mu A, Ji W, Yan L, Zhu Y, Zhu C, Fang X, Yang X, Huang Y, Gao H, Liu F, Ge J, Sun Q, Yang X, Xu W, Liu Z, Yang H, Lou Z, Jiang B, Guddat LW, Gong P, and Rao Z (2020) Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase, Cell. 182, 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Eastman RT, Roth JS, Brimacombe KR, Simeonov A, Shen M, Patnaik S, and Hall MD (2020) Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19, ACS Cent. Sci. 6, 672–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, and Gotte M (2020) The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus, J. Biol. Chem. 295, 4773–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, and Gotte M (2020) Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency, J. Biol. Chem. 295, 6785–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Siegel D, Hui HC, Doerffler E, Clarke MO, Chun K, Zhang L, Neville S, Carra E, Lew W, Ross B, Wang Q, Wolfe L, Jordan R, Soloveva V, Knox J, Perry J, Perron M, Stray KM, Barauskas O, Feng JY, Xu Y, Lee G, Rheingold AL, Ray AS, Bannister R, Strickley R, Swaminathan S, Lee WA, Bavari S, Cihlar T, Lo MK, Warren TK, and Mackman RL (2017) Discovery and Synthesis of a Phosphoramidate Prodrug of a Pyrrolo[2,1-f][triazin-4-amino] Adenine C-Nucleoside (GS-5734) for the Treatment of Ebola and Emerging Viruses, J Med Chem 60, 1648–1661. [DOI] [PubMed] [Google Scholar]
- [8].Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, Larson N, Strickley R, Wells J, Stuthman KS, Van Tongeren SA, Garza NL, Donnelly G, Shurtleff AC, Retterer CJ, Gharaibeh D, Zamani R, Kenny T, Eaton BP, Grimes E, Welch LS, Gomba L, Wilhelmsen CL, Nichols DK, Nuss JE, Nagle ER, Kugelman JR, Palacios G, Doerffler E, Neville S, Carra E, Clarke MO, Zhang L, Lew W, Ross B, Wang Q, Chun K, Wolfe L, Babusis D, Park Y, Stray KM, Trancheva I, Feng JY, Barauskas O, Xu Y, Wong P, Braun MR, Flint M, McMullan LK, Chen SS, Fearns R, Swaminathan S, Mayers DL, Spiropoulou CF, Lee WA, Nichol ST, Cihlar T, and Bavari S (2016) Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys, Nature 531, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].US Food and Drug Administration (2020) Coronavirus (COVID-19) Update: FDA Issues Emergency Use Authorization for Potential COVID-19 Treatment, Washington DC. [Google Scholar]
- [10].Slusarczyk M, Serpi M, and Pertusati F (2018) Phosphoramidates and phosphonamidates (ProTides) with antiviral activity, Antivir. Chem. Chemother. 26, 2040206618775243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Murakami E, Wang T, Park Y, Hao J, Lepist EI, Babusis D, and Ray AS (2015) Implications of efficient hepatic delivery by tenofovir alafenamide (GS-7340) for hepatitis B virus therapy, Antimicrob. Agents Chemother. 59, 3563–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Slusarczyk M, Lopez MH, Balzarini J, Mason M, Jiang WG, Blagden S, Thompson E, Ghazaly E, and McGuigan C (2014) Application of ProTide technology to gemcitabine: a successful approach to overcome the key cancer resistance mechanisms leads to a new agent (NUC-1031) in clinical development, J. Med. Chem. 57, 1531–1542. [DOI] [PubMed] [Google Scholar]
- [13].Murakami E, Tolstykh T, Bao H, Niu C, Steuer HM, Bao D, Chang W, Espiritu C, Bansal S, Lam AM, Otto MJ, Sofia MJ, and Furman PA (2010) Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977, J. Biol. Chem. 285, 34337–34347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Birkus G, Wang R, Liu XH, Kutty N, MacArthur H, Cihlar T, Gibbs C, Swaminathan S, Lee W, and McDermott M (2007) Cathepsin A is the major hydrolase catalyzing the intracellular hydrolysis of the antiretroviral nucleotide phosphonoamidate prodrugs GS-7340 and GS-9131, Antimicrob. Agents Ch 51, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Derudas M, Carta D, Brancale A, Vanpouille C, Lisco A, Margolis L, Balzarini J, and McGuigan C (2009) The application of phosphoramidate protide technology to acyclovir confers anti-HIV inhibition, J. Med. Chem. 52, 5520–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Huang H, Fleming CD, Nishi K, Redinbo MR, and Hammock BD (2005) Stereoselective hydrolysis of pyrethroid-like fluorescent substrates by human and other mammalian liver carboxylesterases, Chem. Res. Toxicol. 18, 1371–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Feng JY, Wang T, Park Y, Babusis D, Birkus G, Xu Y, Voitenleitner C, Fenaux M, Yang H, Eng S, Tirunagari N, Kirschberg T, Cho A, and Ray AS (2018) Nucleotide Prodrug Containing a Nonproteinogenic Amino Acid To Improve Oral Delivery of a Hepatitis C Virus Treatment, Antimicrob Agents Chemother 62, e00620–18 DOI: 10.1128/AAC.00620-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Monteil V, Kwon H, Prado P, Hagelkruys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado Del Pozo C, Prosper F, Romero JP, Wirnsberger G, Zhang H, Slutsky AS, Conder R, Montserrat N, Mirazimi A, and Penninger JM (2020) Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2, Cell 181, 905–913 e907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhao B, Ni C, Gao R, Wang Y, Yang L, Wei J, Lv T, Liang J, Zhang Q, Xu W, Xie Y, Wang X, Yuan Z, Liang J, Zhang R, and Lin X (2020) Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids, Protein and Cell, DOI: 10.1007/s13238-020-00718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xiang DF, Bigley AN, Desormeaux E, Narindoshvili T, and Raushel FM (2019) Enzyme-Catalyzed Kinetic Resolution of Chiral Precursors to Antiviral Prodrugs, Biochemistry 58, 3204–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bigley AN, Xu C, Henderson TJ, Harvey SP, and Raushel FM (2013) Enzymatic neutralization of the chemical warfare agent VX: evolution of phosphotriesterase for phosphorothiolate hydrolysis, J. Am. Chem. Soc. 135, 10426–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen-Goodspeed M, Sogorb MA, Wu F, and Raushel FM (2001) Enhancement, relaxation, and reversal of the stereoselectivity for phosphotriesterase by rational evolution of active site residues, Biochemistry 40, 1332–1339. [DOI] [PubMed] [Google Scholar]
- [23].Bigley AN, Desormeaux E, Xiang DF, Bae SY, Harvey SP, and Raushel FM (2019) Overcoming the Challenges of Enzyme Evolution To Adapt Phosphotriesterase for V-Agent Decontamination, Biochemistry 58, 2039–2053. [DOI] [PubMed] [Google Scholar]
- [24].Tsai PC, Bigley A, Li Y, Ghanem E, Cadieux CL, Kasten SA, Reeves TE, Cerasoli DM, and Raushel FM (2010) Stereoselective hydrolysis of organophosphate nerve agents by the bacterial phosphotriesterase, Biochemistry 49, 7978–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ross BS, Reddy PG, Zhang HR, Rachakonda S, and Sofia MJ (2011) Synthesis of diastereomerically pure nucleotide phosphoramidates, J. Org. Chem. 76, 8311–8319. [DOI] [PubMed] [Google Scholar]
- [26].Chen-Goodspeed M, Sogorb MA, Wu F, Hong SB, and Raushel FM (2001) Structural determinants of the substrate and stereochemical specificity of phosphotriesterase, Biochemistry 40, 1325–1331. [DOI] [PubMed] [Google Scholar]


