Abstract
Background
Fabry disease (α-galactosidase deficiency) is an X-linked genetic disease caused by a variety of pathogenic GLA variants. The phenotypic heterogeneity is considerable, with two major forms, classic and later-onset disease, but adjudication of clinical phenotype is currently lacking for many variants. We aimed to determine consensus phenotypic classification for previously unclassified GLA variants from the GLA-specific fabry-database.org database.
Methods
A Fabry disease genotype–phenotype workgroup developed a five-stage iterative system based on expert clinical assessment, published literature and clinical evidence of pathogenicity using a 2-point scoring system based on clinical hallmarks of classic disease. Kaplan–Meier (KM) analysis of severe clinical event-free survival was used as final validation. Results were compared with those from web-based disease databases and in silico pathogenicity prediction programmes.
Results
Final consensus on classifications of ‘pathogenic’ was achieved for 32 of 33 GLA variants (26 ‘classic’ phenotype, 171 males; 6 ‘later-onset’ phenotype, 57 males). One variant remained of uncertain significance. KM curves were similar for the known fabry-database.org database phenotypes and when workgroup consensus classifications were added, and the curves retained the same separation between ‘classic’ and ‘later-onset’ phenotypes.
Conclusion
The iterative system implemented by a Fabry disease genotype–phenotype workgroup achieved phenotypic classifications for variants that were previously unclassified. Clinical pathogenicity associated with a particular GLA variant defined in affected males appears to have predictive value and also generally correlates with risk for affected females. The newly established classifications can be of benefit to the clinical care of Fabry patients harbouring these variants.
Keywords: expert opinion, Fabry disease, genotype-phenotype correlations, variants, genetics
Introduction
Fabry disease (OMIM #301500) is a rare, progressive, X-linked inherited, multisystemic, lysosomal disorder. It is caused by genetic variations in GLA (HUGO Gene Nomenclature Committee ID: 4296; Gene Entrez: 2717; NCBI reference sequence: NM_000169.2), which encodes α-galactosidase (α-Gal, Enzyme Commission number: EC 3.2.1.22; UniProt ID: P06280).1 Molecular studies of GLA variants have identified a remarkable variety of variants underlying the phenotypic heterogeneity of this genetic disorder.2–18 At present, nearly 1000 different GLA variants have been reported (table 1). Most of the variants are ‘private’ and confined to individual pedigrees with possible variability in phenotypic expression due to phenotype-modifying factors (eg, genetic background, epigenetics and environmental conditions).19 20
Table 1.
Types of GLA variants reported in the Human Gene Mutation Database
| GLA variant types | Number | % of total |
| Missense (pathogenic with classic or later-onset phenotypes, or benign with no or unspecific, Fabry disease-unrelated phenotype) | 605 | 61.0 |
| Nonsense (pathogenic with classic phenotype) | 84 | 8.5 |
| Splicing substitutions (pathogenic with classic or later-onset phenotypes) | 49 | 4.9 |
| Frameshift (pathogenic with classic phenotype)* | 253 | 25.5 |
| Small deletions† | 140 | 14.1 |
| Gross deletions | 39 | 3.9 |
| Small insertions/duplications | 43 | 4.3 |
| Gross insertions/duplications | 8 | 0.8 |
| Small indels | 16 | 1.6 |
| Complex rearrangements (including inversions) | 7 | 0.7 |
| Total | 991 | 100.0 |
*Small and gross are arbitrarily defined as 20 or less and more than 20 nucleotides, respectively.
†Some small deletions occur in-frame and do not necessarily lead to premature termination of mRNA translation.
GLA variants can be disease-causing (pathogenic or likely pathogenic), of uncertain significance, likely benign, or benign.1 21–23 In hemizygous males with pathogenic GLA variants associated with the classic phenotype, clinical symptoms typically appear in childhood (eg, diffuse angiokeratomas, cornea verticillata, dysaesthesia in the extremities). α-Gal activity is severely deficient (<1%) or absent, and marked accumulation of globotriaosylceramide (GL-3, Gb3) and globotriaosylsphingosine (lyso-GL-3) generally leads to multiorgan failure and reduced life expectancy. In contrast, patients with pathogenic GLA variants associated with non-classic, later-onset phenotypes have varying levels of residual α-Gal activity, variable age of symptom onset and fewer clinical manifestations of Fabry disease, often confined to the heart. Phenotypic presentations are even more variable in heterozygous female patients due to both the nature and type of the GLA variant and the X-chromosome inactivation profiles in the various organs.24
Consensus recommendations for the interpretation of variant pathogenicity have been published by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) and are based on criteria using a process incorporating several lines of evidence (eg, scientific and medical literature, disease databases, in silico prediction programmes, functional data).23 In parallel, due to the broad heterogeneity in the natural history of Fabry patients, there has been increasing interest among experts in the Fabry disease scientific community for establishing potential associations between GLA genotypes and phenotypic presentations (eg, International Fabry Disease Genotype-Phenotype Database (dbFGP, http://dbfgp.org/dbFgp/fabry/), fabry-database.org GLA variant database25) in order to optimise clinical management. However, many GLA variants remain absent from or unclassified in those gene-specific databases.
With over 6500 patients (44% males) enrolled, the Fabry Registry has the potential to have several individuals with the same genotype, offering a unique opportunity for assessment of their phenotypic characteristics. Based on the presence or absence of clinical characteristics associated with the classic versus the later-onset Fabry phenotype, the multispecialty Fabry disease genotype–phenotype workgroup developed a novel Fabry phenotype consensus classification system (which we present in this paper) to facilitate the classification of GLA variants reported to the Fabry Registry. This approach was used to assign phenotype classifications to variants that had remained unassigned in the fabry-database.org database, provided that the required Fabry Registry data were available for at least four male patients harbouring a specific GLA variant.
Methods
Fabry disease genotype–phenotype workgroup
The international genotype–phenotype workgroup was composed of nine established experts in Fabry disease with extensive experience in patients’ clinical care and knowledge of GLA variants, including personally observed clinical expressions (all authors of the present paper). They were all Fabry Registry Advisory Board members representing four subspecialties (genetics (n=4), nephrology (n=3), neurology (n=1) and paediatrics (n=1)) from major referral centres in seven different countries. The workgroup met during Registry Advisory Board meetings and had telephone conference meetings, with further exchange of information via email.
Fabry Registry
Deidentified patient natural history data from the Fabry Registry (NCT00196742, sponsor: Sanofi Genzyme) were used in the analyses. The Fabry Registry is a multicentre, international, longitudinal, observational programme that was initiated in 2001 and designed to track the natural history and treatment outcomes of patients with Fabry disease. Patient and investigator participation is voluntary. Recommended schedules of clinical assessments are available but treating physicians determine the actual frequency of assessments according to a patient’s individualised need for medical care and routine follow-up.
Consensus procedures
A five-stage iterative system was implemented to reach a final consensus on the criteria to classify the phenotypes associated with GLA variants that were not classified in the fabry-database.org database.25 This database for clinicians and researchers contains published GLA variants in patients with Fabry disease and assessment of the variants’ phenotypes, as reported in the literature, and had been operational for several years prior to initiation of the current research project (the dbFGP became operational in 2018). It was constructed and is maintained by an independent research group led by Dr H Sakuraba (Department of Clinical Genetics, Meiji Pharmaceutical University, Tokyo, Japan). Standardised Human Genome Variation Society nomenclature was used to describe the GLA variants26 and, as recommended, standard terminology was used for variant classification, that is, ‘pathogenic’ (clinically differentiated as ‘classic’ or ‘later-onset’ phenotype), ‘likely pathogenic’ (‘classic’ or ‘later-onset’ phenotype), ‘uncertain significance’, ‘likely benign’ and ‘benign’.23 The process was limited to phenotypes in male patients, given the multifactorial phenotypic variability in female patients with a specific pathogenic GLA variant.
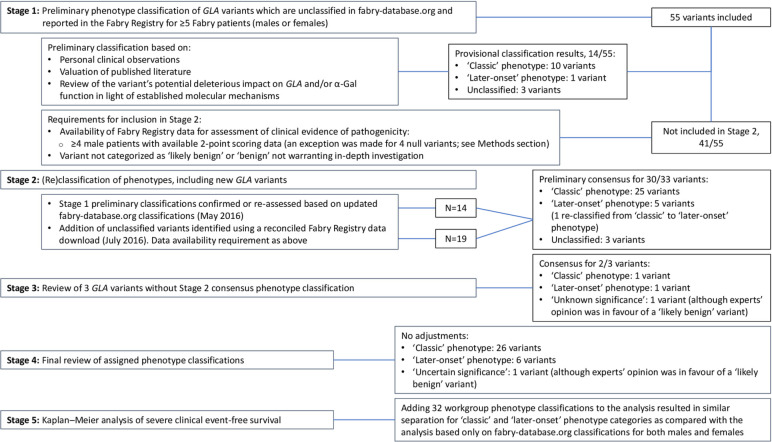
The five stages of the workgroup’s classification system are outlined in figure 1 together with an overview of GLA variant inclusion and exclusion. In Stage 1, individual workgroup members gave preliminary expert clinical assessment of phenotype classification for GLA variants that are unclassified in the fabry-database.org database and occurred in ≥5 Fabry patients (males and females) enrolled in the Fabry Registry. The preliminary assessment was based on personal clinical observations, valuation of published literature and review of the variant’s potential deleterious impact on GLA and/or α-Gal function in light of established molecular mechanisms. Variants qualified for inclusion in Stage 2 if ≥4 male patients having a particular variant had Fabry Registry 2-point scoring data available for assessment of clinical evidence of pathogenicity (see below). Variants categorised as ‘likely benign’ or ‘benign’ not warranting in-depth investigation were excluded. In Stage 2, preliminary Stage 1 phenotype classifications were confirmed or reassessed based on updated fabry-database.org classifications (May 2016) and GLA variant 2-point scores. Unclassified variants, which were identified using a reconciled Fabry Registry data download (July 2016), were added to the process in Stage 2 if they met the data availability criterion. Some null variants that were present in ≥4 male patients, with <4 patients having 2-point scoring data available, were included in the process as the deleterious impact of these variants dominated the expert’s clinical phenotype overall assessment as ‘classic’.
Figure 1.
The five stages of the workgroup’s classification system and an overview of GLA variants inclusion and exclusion.
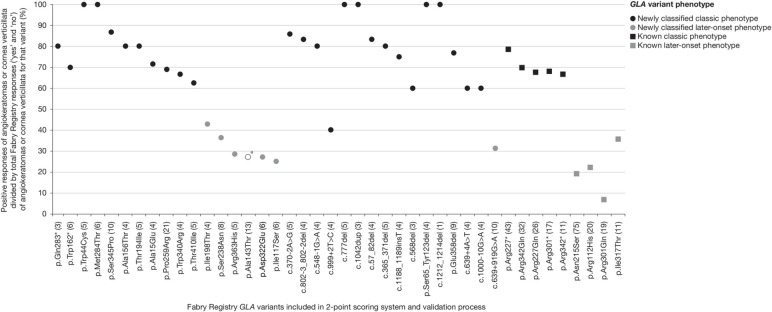
The assessment of clinical evidence of pathogenicity used a GLA variant 2-point scoring system based on absence/presence of hallmark symptoms of the classic phenotype of Fabry disease (ie, diffuse angiokeratomas and cornea verticillata) as recorded in the Fabry Registry (figure 2 and online supplementary methods). GLA variant 2-point scores were calculated as follows: the total number of positive responses for angiokeratomas or cornea verticillata for males having the GLA variant divided by the total number of responses for angiokeratomas or cornea verticillata status for that variant as a percentage (see online supplementary methods for an example of calculation). Two-point scores could range from 0% (indicative of probability of lower level of pathogenicity, that is, ‘later-onset’ variant or non-pathogenicity) to 100% (highest level of probability of pathogenicity and ‘classic’ phenotype). The 2-point scoring system was successfully tested for its precision in distinguishing classic from later-onset Fabry phenotypes. Variants known as being associated with the classic phenotype (n=5) or a later-onset phenotype (n=4) and (relatively) frequently occurring in male patients enrolled in the Fabry Registry (total of 129 and 125 males with usable 2-point scoring responses, respectively) were included in the method validation process and were appropriately separated (figure 2). In Stage 3, the workgroup members performed a detailed review of GLA variants that had not reached a consensus phenotype classification during Stage 2, and Stage 4 encompassed a final review of all assigned phenotype classifications. In Stage 5, Kaplan–Meier (KM) analysis of severe clinical event-free survival was performed using natural history clinical data from adult male patients enrolled in the Fabry Registry and, for further validation, of data from female patients (see also ‘Statistical considerations’).
Figure 2.
GLA variant 2-point scoring validation and results. aGenetic variant of unknown significance (experts’ opinion in favour of a likely benign GLA variant; further research required). *Translation termination codon. Numbers in brackets indicate the number of males who had usable 2-point scoring responses (angiokeratomas or cornea verticillata) recorded in the Fabry Registry per variant.
jmedgenet-2019-106467supp001.pdf (284.7KB, pdf)
As recommended in the ACMG/AMP guidelines for interpretation of variant pathogenicity,23 our final phenotype classifications were compared with results obtained from established web-based Fabry disease-specific (ie, dbFGP) and broader genetic variants databases (ClinVar database and Leiden Open Variation Database (LOVD)), and from in silico programmes commonly used for prediction of pathogenicity of missense variants (ie, Polymorphism Phenotyping V.2 (PolyPhen-2), Scale-Invariant Feature Transform (SIFT) and MutationTaster). The Exome Aggregation Consortium (ExAC) database was used to retrieve the frequencies of variants in the general population, if available.
Statistical considerations
In the KM analysis of severe clinical event-free survival (time to median (50%) event-free survival), data were analysed by fabry-database.org pathogenicity available for GLA variants (with ‘classic’ or ‘later-onset’ phenotype) or unclassified variants, and by combining phenotype classifications from this database with the newly established workgroup consensus classifications. A severe clinical event was defined as a composite event including renal (kidney transplant, chronic dialysis), cardiac (significant cardiac procedures, arrhythmia, angina pectoris, congestive heart failure) and cerebrovascular events (stroke) and death. Statistical analyses were performed using SAS statistical software V.9.2 (SAS Institute, Cary, North Carolina, USA). The log-rank test was used to compare the survival distributions for variants with ‘classic’ and ‘later-onset’ phenotypes. An alpha level of 0.05 was used as the criterion for statistical significance.
Results
Stage 1: Preliminary phenotype classification of 55 GLA variants
A provisional consensus of ‘pathogenic’ was achieved for 11 of 55 (20%) variants (10 with the ‘classic’ phenotype and 1 (p.Arg363His) with a ‘later-onset’ phenotype) (online supplementary table 1). Three variants remained unclassified at this stage. Forty-one variants were excluded from further evaluation (see Methods section).
Stage 2: (Re)classification of phenotypes, including new GLA variants
Thirty-three GLA variants, including 19 variants added to the process using a reconciled Fabry Registry data download, qualified for assessment in Stage 2 (table 2, figure 1). These variants were found at a mean frequency of 7.6 males per variant and were from a total of 252 adult male patients with Fabry disease. Preliminary consensus of ‘pathogenic’ was achieved for 30 of 33 GLA variants, including 25 variants with the ‘classic’ phenotype (2-point scores ranging from 60% to 100%) and five with a ‘later-onset’ phenotype (2-point scores ranging from 25% to 43%) (table 2). The p.Ser238Asn (11 males) missense variant, initially classified as ‘pathogenic’ with the ‘classic’ phenotype during Stage 1, was reclassified as ‘pathogenic’ with a ‘later-onset’ phenotype, taking the 2-point score of 36% into account. The reclassification of this variant merits caution in clinical application of the classification.
Table 2.
Workgroup GLA variant phenotype classification consensus results; Stages 1–4
| Fabry Registry GLA variants (n=33) |
Variant type | No. of males with genotype | No. of males with genotype and usable clinical data* | GLA variant 2-point score (%) | GLA variant phenotype consensus† | ||
| Provisional | Final | ||||||
| Stage 1 | Stage 2 | Stages 3 and 4 | |||||
| p.Trp44Cys | Missense | 7 | 5 | 100 | Classic | Classic | Classic |
| p.Met284Thr | Missense | 6 | 6 | 100 | – | Classic‡ | Classic |
| c.777del | Frameshift | 6 | 5 | 100 | – | Classic‡ | Classic |
| c.1042dup | Frameshift | 5 | 3§ | 100 | – | Classic‡ | Classic |
| p.Ser65_Tyr123del | Large deletion | 4 | 4 | 100 | – | Classic‡ | Classic |
| c.1212_1214del | Small deletion | 7 | 1§ | 100 | – | Classic‡ | Classic |
| p.Ser345Pro | Missense | 11 | 10 | 87 | Unclassified | Classic | Classic |
| c.370-2A>G | Splice site | 5 | 5 | 86 | – | Classic‡ | Classic |
| c.802–3_802-2del | Splice site | 5 | 4 | 83 | – | Classic‡ | Classic |
| c.57_82del | Frameshift | 4 | 4 | 83 | – | Classic‡ | Classic |
| p.Gln283* | Nonsense | 4 | 3§ | 80 | Classic | Classic | Classic |
| p.Ala156Thr | Missense | 5 | 4 | 80 | Classic | Classic | Classic |
| p.Thr194Ile | Missense | 5 | 5 | 80 | Classic | Classic | Classic |
| c.548–1G>A | Splice site | 4 | 4 | 80 | – | Classic‡ | Classic |
| c.365_371del | Frameshift | 6 | 5 | 80 | – | Classic‡ | Classic |
| p.Glu358del | Small deletion | 13 | 9 | 77 | Classic | Classic | Classic |
| c.1188_1189insT | Frameshift | 5 | 4 | 75 | Classic | Classic | Classic |
| p.Ala15Glu | Missense | 6 | 4 | 71 | – | Classic‡ | Classic |
| p.Trp162* | Nonsense | 7 | 6 | 70 | Classic | Classic | Classic |
| p.Pro259Arg | Missense | 28 | 21 | 69 | Classic | Classic | Classic |
| p.Trp340Arg | Missense | 4 | 4 | 67 | – | Classic‡ | Classic |
| p.Thr410Ile | Missense | 5 | 5 | 63 | Classic | Classic | Classic |
| c.568del | Frameshift | 4 | 3§ | 60 | – | Classic‡ | Classic |
| c.639+4A>T | Intronic | 6 | 4 | 60 | – | Classic‡ | Classic |
| c.1000–10G>A | Intronic | 4 | 4 | 60 | – | Classic‡ | Classic |
| p.Ile198Thr | Missense | 8 | 4 | 43 | – | Later-onset‡ | Later-onset |
| c.999+2T>C | Splice site | 5 | 4 | 40 | – | Unclassified‡ | Classic |
| p.Ser238Asn | Missense | 11 | 8 | 36 | Classic | Later-onset | Later-onset |
| c.639+919G>A | Intronic | 13 | 10 | 31 | – | Later-onset‡ | Later-onset |
| p.Arg363His | Missense | 12 | 5 | 29 | Later-onset | Later-onset | Later-onset |
| p.Asp322Glu | Missense | 7 | 6 | 27 | Unclassified | Unclassified | Later-onset |
| p.Ala143Thr | Missense | 24 | 13 | 27 | Unclassified | Unclassified | GVUS¶ |
| p.Ile117Ser | Missense | 6 | 6 | 25 | – | Later-onset‡ | Later-onset |
*Usable data on angiokeratomas or cornea verticillata status available in the Fabry Registry.
†Phenotypes associated with ‘pathogenic’ GLA variants include ‘classic’ and ‘later-onset’ phenotypes.
‡Novel GLA variants (n=19) resulting from using a more recent Fabry Registry reconciled data download (July 2016) added during Stage 2 of the process (hence the 'dash' in the Stage one column).
§Null variant present in ≥4 male patients with <4 patients having 2-point scoring data available. Included in the process as the deleterious impact of the variant dominated the expert’s overall clinical phenotype assessment as ‘classic’.
¶Genetic variant of unknown significance; experts’ opinion in favour of a likely benign GLA variant.
Stage 3: Review of GLA variants without consensus phenotype classification
During Stage 2, preliminary phenotype consensus was not reached for three variants (p.Asp322Glu, c.999+2T>C and p.Ala143Thr) (table 2). A further review of the data by the workgroup resulted in consensus for two of these three variants.
The p.Asp322Glu variant was originally reported in the literature as a pathogenic variant with a classic phenotype, with no clinical information provided.27 Using an artificial substrate, residual enzyme activity of 1.5% of mean values in normal subjects has recently been reported,28 but the low 2-point score of 27% (6 males) led to a workgroup classification as ‘pathogenic’ variant with a ‘later-onset’ phenotype.
The c.999+2T>C splice site variant is an intronic variant at a consensus site of splicing,29 which are known to invariably cause aberrant splicing.18 Despite having a 2-point score of 40% (4 males), workgroup consensus was that the classification was a ‘pathogenic’ variant with the ‘classic’ phenotype, as described originally (no clinical information was provided in that report).18
For p.Ala143Thr (2-point score of 27%, 13 males), the classification could not be established in the present analysis; although experts’ opinion was in favour of a ‘likely benign’ variant, additional research is required (see also Discussion section).
Stage 4: Final review of assigned phenotype classifications
Final review of the 32 of 33 (97%) GLA variants that achieved a consensus classification of ‘pathogenic’ (‘classic’ phenotype: n=26, 171 males; ‘later-onset’ phenotype: n=6, 57 males) did not lead to adjustments (table 2). The p.Ala143Thr missense variant remained of uncertain significance (GVUS).
Stage 5: Kaplan–Meier analysis of severe clinical event-free survival
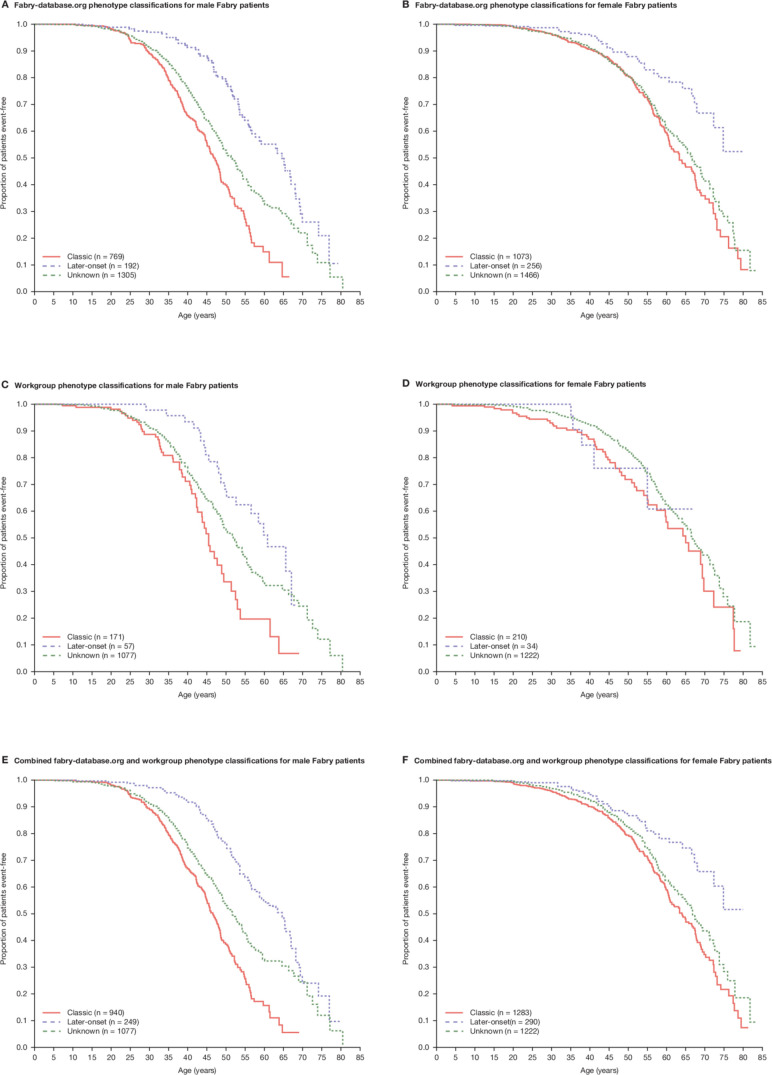
For males, the KM analysis included 2266 adult Fabry patients. Using the workgroup’s 32 consensus male classifications, the GLA variants of 228 of 1305 males (17.5%) in the Fabry Registry, previously unclassified in fabry-database.org (middle curve in figure 3A), could be classified as ‘pathogenic’ with the ‘classic’ phenotype (n=171) or the ‘later-onset’ phenotype (n=57) (figure 3C). Males with pathogenic GLA variants with a ‘classic’ phenotype reported first severe events at a younger age than those with variants associated with ‘later-onset’ phenotypes. Median age at first severe event (preinclusion (figure 3A) and postinclusion (figure 3E) of consensus classifications, respectively) was 46.8 (n=769) and 46.6 years (n=940) for males with variants associated with ‘classic’ phenotypes, 64.6 (n=192) and 64.6 years (n=249) for males with variants with ‘later-onset’ phenotypes and 51.5 (n=1305) and 51.7 years (n=1077) for males with unclassified variants.
Figure 3.
Kaplan–Meier estimates of median age at first reported natural history severe clinical event: male Fabry patients by (A) fabry-database.org phenotype classification, (C) workgroup classification and (E) fabry-database.org and workgroup classifications; female Fabry patients by (B) fabry-database.org phenotype classification, (D) workgroup classification and (F) fabry-database.org and workgroup classifications. Patients with the p.Ala143Thr variant were excluded from the analysis.
For females, KM analysis included 2795 patients. Using the male consensus classifications, the GLA variants of 244 of 1466 females (16.6%) in the Fabry Registry, previously unclassified (middle curve in figure 3B), could be classified as ‘pathogenic’ with ‘classic’ (n=210) or ‘later-onset’ phenotypes (n=34) (figure 3D). The difference between ages at first report of severe events for females with variants with ‘classic’ or ‘later-onset’ phenotypes was less pronounced as compared with males. Median age at first severe event (preinclusion (figure 3B) and postinclusion (figure 3F) of consensus classifications, respectively) was 63.4 (n=1073) and 64.0 years (n=1283) for females with ‘pathogenic’ variants with ‘classic’ phenotypes, whereas the survival function did not reach 50% in both analyses (median time could not be computed) for those with variants with ‘later-onset’ phenotypes (n=256 and n=290, respectively). Median age at first severe event (preinclusion and postinclusion of consensus classifications) was 66.7 (n=1466) and 66.8 years (n=1222), respectively, for females with unclassified variants.
For both males and females, adding the workgroup phenotype classifications to the analysis resulted in similar separation for patients within the ‘classic’ and ‘later-onset’ phenotype categories (figure 3E, F, respectively) as compared with the analysis based only on fabry-database.org classifications (figure 3A, B, respectively) (log-rank test p values<0.0001). This observation further validated our classification method.
Comparison of final classifications with other types of variant evidence of pathogenicity
The workgroup phenotype classifications were compared with results obtained from disease databases and results retrieved from in silico programmes. The latter were restricted to missense variants (n=15), as null variants (nonsense, frameshift, splice site and deletions) are considered very strong evidence of pathogenicity.23 All null variants included in the research had indeed all been classified as ‘pathogenic’ with the ‘classic’ phenotype by the workgroup. The results of the comparison are shown in the online supplementary table 2. Notable observations, taking the aggregate data into account, included the following.
Of the missense variants classified as ‘pathogenic’ with a ‘later-onset’ phenotype, two of five had a discrepant ‘classic’ phenotype classification in the dbFGP database (p.Asp322Glu and p.Ile117Ser). These discordant classifications were primarily based on results of published in vitro expression studies.30 In ClinVar, results of assessments of pathogenicity were available for only 7 of 15 missense variants and most were based on reports from clinical testing labs without supporting information. The p.Trp44Cys variant was listed as being of ‘uncertain significance’ and the p.Trp340Arg and p.Thr410Ile variants as ‘likely pathogenic’, whereas the workgroup classifications were ‘pathogenic’ with the ‘classic’ phenotype (dbFGP database: ‘classic’). In silico predictions supported pathogenicity. In LOVD, 11 missense variants were listed, but classifications were not available. The in silico programmes predicted non-pathogenicity for the p.Arg363His variant, whereas the workgroup and dbFGP classified this variant as ‘pathogenic’ with a ‘later-onset’ phenotype. A discrepant MutationTaster prediction of ‘polymorphism’ was found for the p.Ala15Glu missense variant. The p.Ala143Thr variant remained of ‘uncertain significance’ since the workgroup opinion was in favour of a ‘likely benign’ variant and the results from disease databases and in silico programmes were inconclusive.
Discussion
A novel, five-stage iterative system for consensus phenotype classification of GLA variants that had been reported to the Fabry Registry but had remained unclassified in the fabry-database.org database was developed. The system is based on expert clinical assessment of the Fabry disease genotype–phenotype workgroup members, evaluation of published literature, clinical evidence of variant pathogenicity in the Fabry Registry (2-point scoring system), and severe clinical event-free survival analyses. Final workgroup consensus classification of ‘pathogenic’ was achieved for 32 of 33 GLA variants, including 26 classifications as ‘pathogenic’ with a ‘classic’ phenotype and 6 with the ‘later-onset’ phenotype. The p.Ala143Thr missense variant remained of uncertain significance.
Ascertaining genotype–phenotype associations in Fabry disease is complicated by the rarity of the disease, the striking allelic heterogeneity with nearly 1000 known GLA variants, the variation in clinical expressivity and the lack of published clinical data that could support adjudications. Consequently, phenotype classification is currently lacking for many GLA variants. Although progress has been made in elucidating the natural history of Fabry disease, a better understanding of genotype–phenotype relationships is needed to predict disease progression, improve the definition of interventional studies, set expectations regarding therapeutic outcomes and assist in the selection of modality for various current and future treatments for Fabry disease.31 32 Published therapeutic recommendations highlight the importance of an individualised approach to the care of Fabry patients reflecting, among other things, genotype, phenotype and gender.22 Therefore, accurate phenotype classification of GLA variants, including precise clinical differentiation between the ‘classic’ and ‘later-onset’ pathogenic phenotypes, and reclassification once new supporting data become available, can be of benefit to the clinical care of patients suffering from this rare disorder. For example, we found that for 171 male patients enrolled in the Fabry Registry, their previously unclassified GLA variants could be classified as ‘pathogenic’ with the ‘classic’ phenotype. With this knowledge, caregivers could tailor their clinical management strategy to the needs of the patients with this severe, progressive phenotype. Available enzyme replacement therapies (ERTs) include agalsidase beta (1 mg/kg body weight every 2 weeks (EOW)) and agalsidase alfa (0.2 mg/kg EOW). Chaperone therapy is restricted to patients with amenable variants, for which pathogenicity should have been documented.22
Currently, there is a lack of reference studies in which patients with pathogenic variants with ‘classic’ and ‘later-onset’ phenotypes are analysed separately. This is important, as the disease generally progresses at different rates, and some organ involvement may be delayed or absent in ‘later-onset’ phenotypes of Fabry disease. A recent study stratified ERT-treated male and female patients with ‘pathogenic’ variants by ‘classic’ and ‘later-onset’ phenotypes based on presence/absence of characteristic Fabry disease symptoms and, in males, residual enzyme activity. Not surprisingly, that study found that phenotype (ie, ‘classic’ or ‘later-onset’) is among the strong predictors of the risk to develop clinically important events and of decline in renal function.21
There remains doubt as to whether the p.Ala143Thr missense variant should be considered pathogenic or not, with many lines of evidence in favour of a ‘likely benign’ variant.33 34 Much of the debate concerning this ‘likely benign’ variant has arisen as a result of screening programmes which have identified p.Ala143Thr at a relatively high frequency (allele frequencies, ExAC: European population, 0.001271; overall population, 0.0007748; that is, at frequencies higher than frequencies of 0.0007502 and 0.0004331, respectively, reported for the p.Ser126Gly variant which was classified as ‘likely benign’ in the dbFGP database), with conclusions of pathogenicity based on unspecific clinical symptoms in individuals rather than tissue biopsies.33 34 A retrospective study has shown that most males with the p.Ala143Thr variant present with significantly high residual α-Gal activity, normal lyso-GL-3 levels and absence of renal and cardiac involvement, leading those authors to conclude that it is a ‘likely benign’ variant,33 whereas another study showed cardiac symptoms in 6 of 10 patients with this variant but in association with normal α-Gal activity levels.35 It has been hypothesised that, in the pedigree of the patient originally described by Anderson,36 a second yet undiscovered variant of GLA may actually lead to manifestations of Fabry disease rather than p.Ala143Thr itself. Contributing to the diagnostic dilemma of this genotype, there is some evidence that unspecific signs and symptoms described are unrelated to Fabry disease based on biopsies in individuals with the p.Ala143Thr variant.37 Therefore, further research needs to detail the medical history, clinical examination, biological workup and biomarkers of Fabry disease (ie, lyso-GL-3) as well as tissue biopsy data when possible, obtained in individuals with this variant who develop unspecific symptoms before claiming these are Fabry disease-related.38
Given the X-linked inheritance of Fabry disease, we suggest assessing GLA variant pathogenicity in male patients if sufficient numbers of patients with the relevant clinical data exist, because of the multifactorial phenotypic variability in females with the GLA variant. Heterozygous females harbouring a pathogenic variant are at risk of developing the same form of phenotype as observed in males who have the variant; however, many will develop only incomplete phenotypes and some may remain asymptomatic. Comparison of severe clinical event-free survival for patients enrolled in the Fabry Registry based on existing phenotype classifications with the survival of patients with the previously unclassified GLA variants added to the analysis showed similar survival distributions for ‘pathogenic’ variants with ‘classic’ or ‘later-onset’ phenotypes for each of the sexes. This may suggest that the proportions of patients with ‘classic’ and ‘later-onset’ phenotypes in the cohorts with unclassified phenotypes approximate the proportions in the overall male and female cohorts with established phenotypes. Thus, this method based on severe clinical events observed in male and female patients provided further validation of the workgroup’s phenotype classification efforts.
We also observed that females with variants associated with ‘later-onset’ phenotypes may not have enough severe events to randomise them into clinical trials, whereas in male patients with these variants, more severe events were documented.
As recommended by the ACMG/AMP,23 our final classifications were compared with other lines of evidence of pathogenicity of the studied GLA variants. In silico programmes have not been clinically validated for prediction of GLA variant pathogenicity, are not able to discriminate between subclasses of predictions of ‘pathogenic’ (ie, ‘classic’ vs ‘later-onset’ phenotypes), can only be used for prediction of pathogenicity of missense or splice-site variants and tend to have low specificity. We found only one discrepancy (p.Arg363His missense variant) between the combined prediction from three programmes and phenotype classification from the workgroup and the dbFGP database. There were two discrepancies between our classification of ‘pathogenic’ with a ‘later-onset’ phenotype and classifications available in the dbFGP database. However, the dbFGP classification of those two variants relied only on results from in vitro expression studies,30 39 in the absence of clinical reports. Although such studies supportive of a damaging effect are recognised as strong evidence of pathogenicity,23 the results should be interpreted with caution, as expression studies may not reflect the complexities of the in vivo environment. One of the variants that was classified differently is the p.Asp322Glu variant. Based on diffuse angiokeratoma and cornea verticillata data, workgroup consensus reached at the end of 2016 was that this ‘pathogenic’ variant is associated with a ‘later-onset’ phenotype. This is strengthened by a recent paper indicating the absence of significant renal involvement in this variant.28 GLA variant information available in the disease databases ClinVar and LOVD was generally limited, and supporting information was particularly lacking in the LOVD. To expand the body of evidence, researchers, geneticists and clinicians are encouraged to share information on GLA variants by submitting complete clinical data to existing major databases and publishing their molecular findings together with precise clinical descriptions.
We recognise several limitations to our GLA variant phenotype classification system. There may have been patient selection bias towards enrolment of more severely affected patients in the Fabry Registry, and 2-point scoring data (angiokeratomas, cornea verticillata) were not available for all enrolled patients, indicating incomplete recording of clinical assessments. Diffuse or widespread angiokeratomas are considered a hallmark of classic Fabry disease but the presence of a more limited number of ‘red spots’ on the skin is not pathognomonic and may have accounted for increased scoring for ‘later-onset’ variants or the p.Ala143Thr variant. Similarly, the differential diagnosis of Fabry disease cornea verticillata includes a phenocopy that may be drug-induced and the result of amiodarone intake in a subset of patients with Fabry disease with cardiac arrhythmia.1 The GLA variant sample size used for testing the precision of the 2-point scoring system was small, and the age of the patients (although all were adult males) is not accounted for in the 2-point scoring process. Furthermore, the fabry-database.org database relies on curation and quality of the published literature.25 Last, the analysis did not include biochemical parameters performed by a single central reference lab (enzyme activity, lyso-GL-3) and no in vitro GLA variant expression studies could be performed.
In conclusion, our validated approach for clinical phenotyping based on clinical data from Fabry patients enrolled in the Fabry Registry was able to predict the phenotypic manifestations for GLA variants that had no prior phenotype classification documented in the fabry-database.org database. Clinical pathogenicity associated with a particular GLA variant defined in affected males appears to have predictive value and generally correlates with risk for affected females. The newly established phenotype classifications can be of benefit to the clinical care of Fabry patients harbouring these GLA variants.
Electronic database information
URLs for Fabry disease-specific variant databases, broader genetic variants databases and in silico programmes referred to in this article are as follows:
ClinVar database, https://www.ncbi.nlm.nih.gov/clinvar/ (last accessed February 2019).
ExAC Database (gene: GLA), http://exac.broadinstitute.org/gene/ENSG00000102393 (last accessed February 2019).
Fabry-database.org GLA variant database, http://fabry-database.org/mutants (last accessed February 2019).
Human Gene Mutation Database (gene: GLA), http://www.hgmd.cf.ac.uk/ac/gene.php?gene=GLA (last accessed December 2019).
International Fabry Disease Genotype–Phenotype Database (dbFGP), http://dbfgp.org/dbFgp/fabry/ (last accessed February 2019).
Leiden Open Variation Database, http://www.lovd.nl/3.0/home (last accessed February 2019).
MutationTaster, http://www.mutationtaster.org/ (last accessed February 2019).
Polymorphism Phenotyping v2, http://genetics.bwh.harvard.edu/pph2/ (last accessed February 2019).
SIFT, http://provean.jcvi.org/genome_submit_2.php (last accessed February 2019).
Acknowledgments
The authors would like to thank the patients who agreed to participate in the Fabry Registry and Badari Gudivada (Sanofi Genzyme) for statistical programming support.
Footnotes
Contributors: DPG, JPO, DGB, HWY, RJH, JP, CW, WRW and DGW are members of the Fabry disease genotype–phenotype workgroup. DPG drafted the article. All authors have critically reviewed the manuscript for important intellectual content and are responsible for all content and editorial decisions. All authors approved the final version of the manuscript and received no honoraria related to the development of this publication.
Funding: This research was funded by Sanofi Genzyme, the sponsor of the Fabry Registry. The authors received editorial/writing support in the preparation of this manuscript from Tom Rouwette of Excerpta Medica, funded by Sanofi Genzyme, and Hans Ebels of Sanofi Genzyme. DPG was supported by the Plan National Maladies Rares of the French Ministry of Health.
Disclaimer: The funder had a role in Fabry Registry data collection and analysis.
Competing interests: DPG is a consultant for Sanofi Genzyme and Takeda/Shire, was an investigator in clinical trials sponsored by Amicus Therapeutics and Sanofi Genzyme and has received speaker honoraria and travel support from Amicus Therapeutics, Sanofi Genzyme and Takeda/Shire. JPO has received consulting honoraria and unrestricted research grants and funding for research projects from Sanofi Genzyme, has received speaker honoraria from Sanofi Genzyme and Takeda/Shire and has received conference and travel support from Amicus Therapeutics, Sanofi Genzyme and Takeda/Shire. DGB has received speaker honoraria from Amicus Therapeutics, Sanofi Genzyme and Takeda/Shire. HWY has received honoraria from Sanofi Genzyme. RJH has received consulting honoraria from Sanofi Genzyme, Amicus Therapeutics, Protalix Corporation and Takeda/Shire, was an investigator in clinical trials sponsored by Amicus Therapeutics, Sanofi Genzyme and Takeda/Shire and received research funding from Sanofi Genzyme, Protalix Corporation and Amicus Therapeutics; these activities have been monitored and found to be in compliance with the conflict of interest policies at Cincinnati Children’s Hospital Medical Center. RL is an employee of Sanofi Genzyme. JP has received honoraria and travel support from Amicus Therapeutics, Protalix Corporation, Sanofi Genzyme and Takeda/Shire. CW has received research support from Sanofi Genzyme and is a consultant for Actelion Pharmaceuticals, Protalix Corporation, Boehringer Ingelheim GmbH and Sanofi Genzyme. WRW consults for Sanofi Genzyme and was an investigator in clinical studies and trials sponsored by Amicus Therapeutics, Protalix Corporation, Sanofi Genzyme and Takeda/Shire and has received research funding from Sanofi Genzyme, Amicus Therapeutics and Takeda/Shire; these activities are monitored and are in compliance with the conflict of interest policies at Emory University School of Medicine. DGW received consulting honoraria from Amicus Therapeutics, Sanofi Genzyme, Actelion Pharmaceuticals, AVROBIO, Freeline Therapeutics and Protalix Biotherapeutics and has received research funding from Amicus Therapeutics and Sanofi Genzyme. DPG, JPO, DGB, HWY, RJH, JP and CW are Regional or International Fabry Registry Board members and have received Fabry Registry Board honoraria.
Patient consent for publication: Not required.
Ethics approval: The Fabry Registry protocol, informed consent form and any locally required authorisation documents are reviewed and approved by the local fully constituted Institutional Review Board or Independent Ethics Committee unless the site provides the Registry with documentation that approval is not required or has been waived.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available on reasonable request. Qualified researchers may request access to patient-level data and related study documents including the clinical study report, study protocol with any amendments, blank case report form, statistical analysis plan and dataset specifications. Patient-level data will be anonymised and study documents will be redacted to protect the privacy of trial participants. Further details on Sanofi’s data sharing criteria, eligible studies and process for requesting access can be found at: https://www.clinicalstudydatarequest.com.
References
- 1. Germain DP. Fabry disease. Orphanet J Rare Dis 2010;5:30 10.1186/1750-1172-5-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ashley GA, Shabbeer J, Yasuda M, Eng CM, Desnick RJ. Fabry disease: twenty novel alpha-galactosidase A mutations causing the classical phenotype. J Hum Genet 2001;46:192–6. 10.1007/s100380170088 [DOI] [PubMed] [Google Scholar]
- 3. Ashton-Prolla P, Tong B, Shabbeer J, Astrin KH, Eng CM, Desnick RJ. Fabry disease: twenty-two novel mutations in the alpha-galactosidase A gene and genotype/phenotype correlations in severely and mildly affected hemizygotes and heterozygotes. J Investig Med 2000;48:227–35. [PubMed] [Google Scholar]
- 4. Davies JP, Eng CM, Hill JA, Malcolm S, MacDermot K, Winchester B, Desnick RJ. Fabry disease: fourteen α-galactosidase A mutations in unrelated families from the United Kingdom and other European countries. Eur J Hum Genet 1996;4:219–24. 10.1159/000472202 [DOI] [PubMed] [Google Scholar]
- 5. Eng CM, Ashley GA, Burgert TS, Enriquez AL, D’Souza M, Desnick RJ. Fabry disease: thirty-five mutations in the alpha-galactosidase A gene in patients with classic and variant phenotypes. Mol Med 1997;3:174–82. 10.1007/BF03401671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eng CM, Niehaus DJ, Enriquez AL, Burgert TS, Ludman MD, Desnick RJ. Fabry disease: twenty-three mutations including sense and antisense CpG alterations and identification of a deletional hot-spot in the alpha-galactosidase A gene. Hum Mol Genet 1994;3:1795–9. 10.1093/hmg/3.10.1795 [DOI] [PubMed] [Google Scholar]
- 7. Germain DP, Biasotto M, Tosi M, Meo T, Kahn A, Poenaru L. Fluorescence-assisted mismatch analysis (FAMA) for exhaustive screening of the alpha-galactosidase A gene and detection of carriers in Fabry disease. Hum Genet 1996;98:719–26. 10.1007/s004390050292 [DOI] [PubMed] [Google Scholar]
- 8. Germain DP. A new phenotype of Fabry disease with intermediate severity between the classical form and the cardiac variant. Contrib Nephrol 2001;136:234–40. 10.1159/000060194 [DOI] [PubMed] [Google Scholar]
- 9. Germain DP, Poenaru L. Fabry disease: identification of novel alpha-galactosidase A mutations and molecular carrier detection by use of fluorescent chemical cleavage of mismatches. Biochem Biophys Res Commun 1999;257:708–13. 10.1006/bbrc.1999.0310 [DOI] [PubMed] [Google Scholar]
- 10. Germain DP, Shabbeer J, Cotigny S, Desnick RJ. Fabry disease: twenty novel alpha-galactosidase A mutations and genotype-phenotype correlations in classical and variant phenotypes. Mol Med 2002;8:306–12. 10.1007/BF03402156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ishii S, Kase R, Sakuraba H, Suzuki Y. Characterization of a mutant alpha-galactosidase gene product for the late-onset cardiac form of Fabry disease. Biochem Biophys Res Commun 1993;197:1585–9. 10.1006/bbrc.1993.2659 [DOI] [PubMed] [Google Scholar]
- 12. Ishii S, Nakao S, Minamikawa-Tachino R, Desnick RJ, Fan J-Q. Alternative splicing in the alpha-galactosidase A gene: increased exon inclusion results in the Fabry cardiac phenotype. Am J Hum Genet 2002;70:994–1002. 10.1086/339431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ishii S, Sakuraba H, Suzuki Y. Point mutations in the upstream region of the alpha-galactosidase A gene exon 6 in an atypical variant of Fabry disease. Hum Genet 1992;89:29–32. 10.1007/BF00207037 [DOI] [PubMed] [Google Scholar]
- 14. Ploos van Amstel JK, Jansen RP, de Jong JG, Hamel BC, Wevers RA. Six novel mutations in the alpha-galactosidase A gene in families with Fabry disease. Hum Mol Genet 1994;3:503–5. 10.1093/hmg/3.3.503 [DOI] [PubMed] [Google Scholar]
- 15. Sakuraba H, Oshima A, Fukuhara Y, Shimmoto M, Nagao Y, Bishop DF, Desnick RJ, Suzuki Y. Identification of point mutations in the alpha-galactosidase A gene in classical and atypical hemizygotes with Fabry disease. Am J Hum Genet 1990;47:784–9. [PMC free article] [PubMed] [Google Scholar]
- 16. Shabbeer J, Yasuda M, Luca E, Desnick RJ. Fabry disease: 45 novel mutations in the alpha-galactosidase A gene causing the classical phenotype. Mol Genet Metab 2002;76:23–30. 10.1016/S1096-7192(02)00012-4 [DOI] [PubMed] [Google Scholar]
- 17. Shabbeer J, Yasuda M, Benson SD, Desnick RJ. Fabry disease: identification of 50 novel alpha-galactosidase A mutations causing the classic phenotype and three-dimensional structural analysis of 29 missense mutations. Hum Genomics 2006;2:297–309. 10.1186/1479-7364-2-5-297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Topaloglu AK, Ashley GA, Tong B, Shabbeer J, Astrin KH, Eng CM, Desnick RJ. Twenty novel mutations in the α-galactosidase A gene causing Fabry disease. Mol Med 1999;5:806–11. 10.1007/BF03401993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cammarata G, Fatuzzo P, Rodolico MS, Colomba P, Sicurella L, Iemolo F, Zizzo C, Alessandro R, Bartolotta C, Duro G, Monte I. High variability of Fabry disease manifestations in an extended Italian family. Biomed Res Int 2015;2015:504784 10.1155/2015/504784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rigoldi M, Concolino D, Morrone A, Pieruzzi F, Ravaglia R, Furlan F, Santus F, Strisciuglio P, Torti G, Parini R. Intrafamilial phenotypic variability in four families with Anderson-Fabry disease. Clin Genet 2014;86:258–63. 10.1111/cge.12261 [DOI] [PubMed] [Google Scholar]
- 21. Arends M, Wanner C, Hughes D, Mehta A, Oder D, Watkinson OT, Elliott PM, Linthorst GE, Wijburg FA, Biegstraaten M, Hollak CE. Characterization of classical and nonclassical Fabry disease: a multicenter study. JASN 2017;28:1631–41. 10.1681/ASN.2016090964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ortiz A, Germain DP, Desnick RJ, Politei J, Mauer M, Burlina A, Eng C, Hopkin RJ, Laney D, Linhart A, Waldek S, Wallace E, Weidemann F, Wilcox WR. Fabry disease revisited: management and treatment recommendations for adult patients. Mol Genet Metab 2018;123:416–27. 10.1016/j.ymgme.2018.02.014 [DOI] [PubMed] [Google Scholar]
- 23. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Echevarria L, Benistan K, Toussaint A, Dubourg O, Hagege AA, Eladari D, Jabbour F, Beldjord C, De Mazancourt P, Germain DP. X-chromosome inactivation in female patients with Fabry disease. Clin Genet 2016;89:44–54. 10.1111/cge.12613 [DOI] [PubMed] [Google Scholar]
- 25. Saito S, Ohno K, Sakuraba H. Fabry-database.org: database of the clinical phenotypes, genotypes and mutant α-galactosidase A structures in Fabry disease. J Hum Genet 2011;56:467–8. 10.1038/jhg.2011.31 [DOI] [PubMed] [Google Scholar]
- 26. den Dunnen JT. Sequence variant descriptions: HGVS nomenclature and Mutalyzer. Curr Protoc Hum Genet 2016;90:7.13.1–19. 10.1002/cphg.2 [DOI] [PubMed] [Google Scholar]
- 27. Lee BH, Heo SH, Kim G-H, Park J-Y, Kim W-S, Kang D-H, Choe KH, Kim W-H, Yang SH, Yoo H-W. Mutations of the GLA gene in Korean patients with Fabry disease and frequency of the E66Q allele as a functional variant in Korean newborns. J Hum Genet 2010;55:512–7. 10.1038/jhg.2010.58 [DOI] [PubMed] [Google Scholar]
- 28. Adalsteinsdottir B, Palsson R, Desnick RJ, Gardarsdottir M, Teekakirikul P, Maron M, Appelbaum E, Neisius U, Maron BJ, Burke MA, Chen B, Pagant S, Madsen CV, Danielsen R, Arngrimsson R, Feldt-Rasmussen U, Seidman JG, Seidman CE, Gunnarsson GT. Fabry disease in families with hypertrophic cardiomyopathy: clinical manifestations in the classic and later-onset phenotypes. Circ Cardiovasc Genet 2017;10:e001639 10.1161/CIRCGENETICS.116.001639 [DOI] [PubMed] [Google Scholar]
- 29. Breathnach R, Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem 1981;50:349–83. 10.1146/annurev.bi.50.070181.002025 [DOI] [PubMed] [Google Scholar]
- 30. Benjamin ER, Della Valle MC, Wu X, Katz E, Pruthi F, Bond S, Bronfin B, Williams H, Yu J, Bichet DG, Germain DP, Giugliani R, Hughes D, Schiffmann R, Wilcox WR, Desnick RJ, Kirk J, Barth J, Barlow C, Valenzano KJ, Castelli J, Lockhart DJ. The validation of pharmacogenetics for the identification of Fabry patients to be treated with migalastat. Genet Med 2017;19:430–8. 10.1038/gim.2016.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Germain DP. Fabry disease: the need to stratify patient populations to better understand the outcome of enzyme replacement therapy. Clin Ther 2007;29:S17–8. 10.1016/S0149-2918(07)80122-6 [DOI] [PubMed] [Google Scholar]
- 32. Wanner C, Arad M, Baron R, Burlina A, Elliott PM, Feldt-Rasmussen U, Fomin VV, Germain DP, Hughes DA, Jovanovic A, Kantola I, Linhart A, Mignani R, Monserrat L, Namdar M, Nowak A, Oliveira J-P, Ortiz A, Pieroni M, Spada M, Tylki-Szymańska A, Tøndel C, Viana-Baptista M, Weidemann F, Hilz MJ. European expert consensus statement on therapeutic goals in Fabry disease. Mol Genet Metab 2018;124:189–203. 10.1016/j.ymgme.2018.06.004 [DOI] [PubMed] [Google Scholar]
- 33. Lenders M, Weidemann F, Kurschat C, Canaan-Kühl S, Duning T, Stypmann J, Schmitz B, Reiermann S, Krämer J, Blaschke D, Wanner C, Brand S-M, Brand E. Alpha-galactosidase A p.A143T, a non-Fabry disease-causing variant. Orphanet J Rare Dis 2016;11:54 10.1186/s13023-016-0441-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Terryn W, Vanholder R, Hemelsoet D, Leroy BP, Van Biesen W, De Schoenmakere G, Wuyts B, Claes K, De Backer J, De Paepe G, Fogo A, Praet M, Poppe B. Questioning the Pathogenic Role of the GLA p.Ala143Thr "Mutation" in Fabry Disease: Implications for Screening Studies and ERT. JIMD Rep 2013;8:101–8. 10.1007/8904_2012_167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De Brabander I, Yperzeele L, Ceuterick-De Groote C, Brouns R, Baker R, Belachew S, Delbecq J, De Keulenaer G, Dethy S, Eyskens F, Fumal A, Hemelsoet D, Hughes D, Jeangette S, Nuytten D, Redondo P, Sadzot B, Sindic C, Sheorajpanday R, Thijs V, Van Broeckhoven C, De Deyn PP. Phenotypical characterization of α-galactosidase A gene mutations identified in a large Fabry disease screening program in stroke in the young. Clin Neurol Neurosurg 2013;115:1088–93. 10.1016/j.clineuro.2012.11.003 [DOI] [PubMed] [Google Scholar]
- 36. Gaggl M, El-Hadi S, Aigner C, Sunder-Plassmann G. The renal history of Fabry disease. G Ital Nefrol 2016;33:1–4. [PubMed] [Google Scholar]
- 37. Smid BE, Hollak CE, Poorthuis BJ, van den Bergh Weerman MA, Florquin S, Kok WE, Lekanne Deprez RH, Timmermans J, Linthorst GE. Diagnostic dilemmas in Fabry disease: a case series study on GLA mutations of unknown clinical significance. Clin Genet 2015;88:161–6. 10.1111/cge.12449 [DOI] [PubMed] [Google Scholar]
- 38. Germain DP, Jurca-Simina IE. Principles of human genetics and Mendelian inheritance : Burlina AP, Neurometabolic hereditary diseases of adults. Cham, Switzerland: Springer, 2018: 1–28. 10.1007/978-3-319-76148-0 [DOI] [Google Scholar]
- 39. Lukas J, Giese A-K, Markoff A, Grittner U, Kolodny E, Mascher H, Lackner KJ, Meyer W, Wree P, Saviouk V, Rolfs A. Functional characterisation of alpha-galactosidase A mutations as a basis for a new classification system in Fabry disease. PLoS Genet 2013;9:e1003632 10.1371/journal.pgen.1003632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jmedgenet-2019-106467supp001.pdf (284.7KB, pdf)