Abstract
A contrasting genotype and allele frequency pattern between Africans and non-Africans in the Duffy (T-33C) locus is reported. Its near fixation in various populations suggest is no longer under natural selection, and that current distribution is possibly a relic of distant extreme selection combined with genetic drift during the out of Africa. We put this difference into the utility to infer the ancestral state of ambiguous loci in different populations.
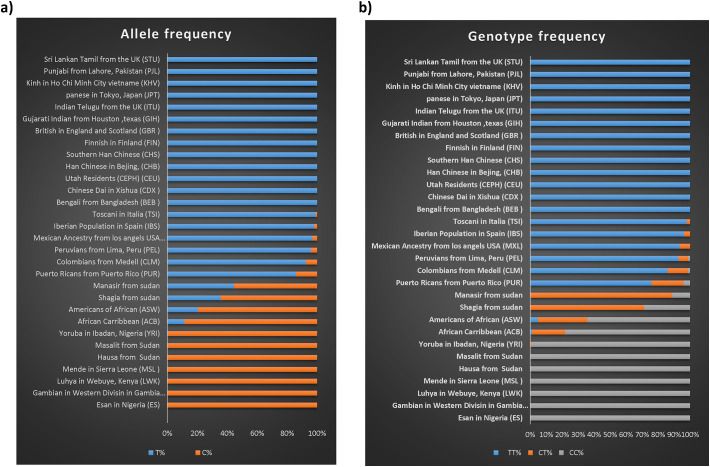
Differences in alleles frequency patterns between Africans and non-Africans have been documented and attributed to the large African population effective size (Watkins et al., 2001; Elhassan et al., 2014) genetic drift and possible selection episodes that might have occurred during or prior to the exodus from the continent. These differences may help shed light on hitherto hidden chapters and dangling questions on evolutionary history and the genetic basis of phenotypic variations (Tishkoff and Williams, 2002). Here we report on an interesting situation of a contrasting genotype and allele frequency pattern between Africans and non-Africans in the Duffy antigen gene. The pattern was originally observed in a 812 sample of 812 Sudanese from the MalariaGen genotype panel (Masalit & Hausa). Allele “C” was found to be most frequent among Africans almost reaching fixation (100%) in the majority of groups. The reverse was observed in non-Africans with the C allele at a frequency of 1% in Europeans (Iberians) and Amerindians (Mayan). In contrast, the T allele frequency was as high as (100%) in Europeans (Fig. 1a). The total genotype data of this locus comprises 3533 individuals from global populations, of which 2504 were from the 1000 genomes project (phase 3) (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes), and 217 genotypes from published data of Sudanese (Shaygia & Manasir) (Kempińska-Podhorodecka et al., 2012). Based on such allele and genotype frequencies, and accordingly we set to examine the likelihood of an interesting scenario related to human migrations. The polymorphism within the Duffy antigen gene (rs2814778) seems rather a relic of past extreme selection that conferred an adaptive advantage to humans in a specific environment during their exodus out of Africa, ~40KYB, with frequencies and impact aggravated thereafter by the sheer relatively small number of migrants under the influence of genetic drifts. Generally speaking, diseases are among the most recognizable forces exerting selection pressure, with the malaria parasite being a notable example. Malaria influence on the human genome has a history estimated to fall in a range of 10–15,000 years ago concurrent with the advent of agriculture (Kwiatkowski, 2005) which makes it much later to the first out of Africa episode. It has been widely documented that malaria was one of the factors determining population diversity manifested in atypical variants of hemoglobin, such as hemoglobin S (HbS), C (HBC) or E (HBE), as well as variability in the Duffy blood group system (Kempińska-Podhorodecka et al., 2012). DARC has also recently been associated with susceptibility to HIV infection and rate of progression to AIDS (He et al., 2008), and is shown to be prognostic for WBCs count and neutrophil count (benign ethnic neutropenia) in African Americans (Reich et al., 2009) as well as susceptibility to several other conditions capable of inducing equally significant selective pressure (de Carvalho and de Carvalho, 2011). Such contrasting pattern is unique and the utility of rs2814778 with its spectacular frequency distribution (Fig. 1a and b) is paramount. Past and present respiratory viral pandemics including the current Covid-19 and its relatively high mortality rates provides an intuitive example to a possible scenario that might have led to the current global frequency distribution.
Fig. 1.
a) The allele frequency pattern in the Duffy G298A, the “C” allele being more frequent in African populations with near fixation (100%) and the opposite is true for Europeans and non-Africans in general. b) The Genotype frequency of the global sample, exception is Shaygia and Manasir (from Sudan) seemingly consistent with an admixture between Africans and non-Africans in a population known to have such mixed ancestry.
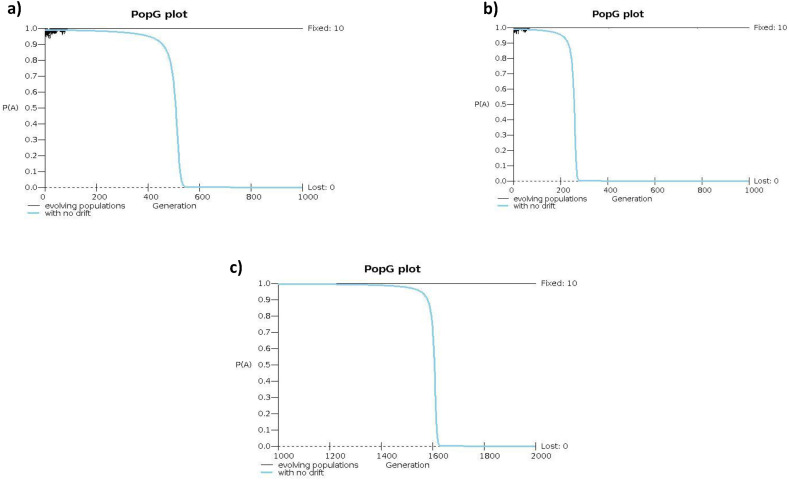
Single locus data can be quite informative in delineating population histories as opposed to multilocus or whole genomes, examples to quote but a few are the sickle cell mutation and the YAP insertion. It can help settle lingering quandaries, like the status of some mutations and traits in terms of ancestry, and whether they are indigenous to a particular population and geographical area or influenced by gene flow. The genetic population structure of the Masalit from Darfur of western Sudan and Hausa from Nigeria are interesting cases to consider. Both groups carry predominantly allele C of the (rs2814778), suggesting that either a) autosomes of Masalit as manifested in this locus had sustained no major gene flow from non-Africans chromosomes or b) gene flow has taken place but the introgression of the introduced allele has seized and disappeared due to a combined effect of drift and selection. Masalit according to Y chromosome data (Hassan et al., 2008) is around 71.9% E1b1b paternal haplogroup, mainly the V32 subclade, and approximately 6.3% also belong to the haplogroup J1 considered a par excellence marker of Afro-Asiatic ancestry and back migration. This might insinuate significant patrilineal gene flow from neighboring Afro-Asiatic-speaking populations, although it does not tally with the historical time frame of 500–1500 years for the migration of Arab speaking populations into the Sahel and a hypothesized subsequent admixture scenario (Bereir et al., 2007). To further consider other possible scenarios of an older back to Africa at a different time frame like the hypothetical introduction into Africa by Neolithic farmers of the Y haplogroup R1b common among Hausa in this case (40%) followed by selection episode and losing the accompanying T allele due to decreased fitness, we modeled similar scenarios using the program PopG (Felsenstein/Kuhner lab). The analysis shows that even with extreme selection/fitness values, complete fixation in the absence of drift required 300 generations (~7500 years), and ~ 10,000 years with less extreme fitness values seen in major human epidemics and outbreaks (Fig. 2a,b) at a frequency of 0.01 of the minor allele (T in this case). While a frequency of 0.001 which is more comparable to the present values will require 1400–1600 generations (~30–40,000 years) to reach fixation (Fig. 2c). A back to Africa scenario seems even less likely given the abundance of very old ancestral mitochondria and Y chromosome haplogroups with no or very low-frequency distribution out of Africa in Hausa and Masalit like A and B haplogroups (Hassan et al., 2008) 11. It has been previously shown by us that the Y chromosome can be a reliable marker for allele introgression into a population genome and in dating of such events (Bereir et al., 2007).
Fig. 2.
Simulation of allele frequency change over time for a scenario of allele T into Africa a) an initial frequency of 0.001 similar to the current frequencies and fitness of extreme selection at 1.4 a migration rate of 0.4 and population of 100 effective individuals, analysis shows that even with such high selection/fitness values, complete fixation without drift required 300 generations b) with more realistic fitness measures at 1.25 fixation and near fixation required 500 generations to fixation c) with a frequency of allele T approaching the present frequencies (0.0001) which is more comparable with the present values will require 1400–1600 generation to reach fixation.
1. Conclusion
The Duffy mutation (rs2814778) seems an insightful marker for African non-African ancestry and population admixture exemplified in admixture patterns in African Americans (Estalote et al., 2005 ) and some northern and east Africans. It would be interesting to use biomarkers such as Y-chromosome and/or mtDNA to quantify and inform on such admixture patterns of hypothesized mixed ancestry groups in continental sets and at the fringes of population interactions.
Credit author statement
Musab M. Ali Albsheer wrote the original draft, and analyzed the data; Muzamil Mahdi Abdel Hamid reviewed and edited the manuscript; Dominic Kwiatkowski Contributed genotype data and revised the manuscript; Ayman Hussien contributed to the data analysis, Muntaser Ibrahim conceptualized of the study and contributed to the analysis and wrote the final manuscript.
Declaration of Competing Interest
There are no conflicts of interest to disclose.
References
- Bereir Rihab E., Hassan Hisham Y., Salih Niven a, Underhill Peter a, Cavalli-Sforza Luigi L., Hussain Ayman a, Kwiatkowski Dominic, Ibrahim Muntaser E. Co-introgression of Y-chromosome Haplogroups and the sickle cell gene across Africa’s Sahel. Eur. J. Hum. Genet. 2007;15(11):1183–1185. doi: 10.1038/sj.ejhg.5201892. [DOI] [PubMed] [Google Scholar]
- de Carvalho Gledson Barbosa, de Carvalho Glauber Barbosa. Duffy blood group system and the malaria adaptation process in humans. Rev. Bras. Hematol. Hemoter. 2011;33(1):55–64. doi: 10.5581/1516-8484.20110016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhassan Nuha, EyoabIyasuGebremeskel Mohamed Ali Elnour, Isabirye Dan, Okello John, AymanHussien Dominic Kwiatksowski, JibrilHirbo Sara Tishkoff, Ibrahim Muntaser E. The episode of genetic drift defining the migration of humans out of Africa is derived from a large east African population size. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0097674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estalote Angela Cristina, Proto-siqueira Rodrigo, Araújo Wilson, Da Silva, Zago Marco Antonio, Marcos Palatnik. n.d. The mutation G298A → Ala100Thr on the coding sequence of the Duffy antigen/chemokine receptor gene in Non-Caucasian Brazilians. Genet. Mol. Res. 2005;4(2):166–173. [PubMed] [Google Scholar]
- Hassan Hisham Y., Underhill Peter a, Cavalli-Sforza Luca L., Ibrahim Muntaser E. Y-chromosome variation among Sudanese: restricted gene flow, concordance with language, geography, and history. Am. J. Phys. Anthropol. 2008;137(3):316–323. doi: 10.1002/ajpa.20876. [DOI] [PubMed] [Google Scholar]
- He Weijing Neil, Stuart Hemant Kulkarni, Wright Edward, Agan Brian K., Marconi Vincent C., Dolan Matthew J. Duffy antigen receptor for chemokines mediates trans -infection of hiv-1 from red blood cells to target cells and affects HIV-AIDS susceptibility. Cell Host Microbe. 2008;4(1):52–62. doi: 10.1016/j.chom.2008.06.002. July 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempińska-Podhorodecka Agnieszka, Knap Oktawian, Drozd Arleta, MariuszKaczmarczyk MiroslawParafiniuk, Parczewski Milosz, Ciechanowicz Andrzej. Analysis for genotyping duffy blood group in inhabitants of sudan, the fourth cataract of the Nile. Malar. J. 2012;11(April):115. doi: 10.1186/1475-2875-11-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski Dominic P. 2005. How Malaria Has Affected the Human Genome and What Human Genetics Can Teach Us about Malaria; pp. 171–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich David, Nalls Michael A., Kao W.H. Linda, Akylbekova Ermeg L., Tandon Arti, Mullikin James, Hsueh Wen-chi. Reduced neutrophil count in people of african descent is due to a regulatory variant in the duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5(1) doi: 10.1371/journal.pgen.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff Sarah a, Williams Scott M. Genetic analysis of African populations: human evolution and complex disease. Nat. Rev. Genet. 2002;3(8):611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- Watkins W.S., Ricker C.E., Bamshad M.J., Carroll M.L., Nguyen S.V., Batzer M.A., Harpending H.C., Rogers A.R., Jorde L.B. Patterns of ancestral human diversity: an analysis of alu-insertion and restriction-site polymorphisms. Am. J. Hum. Genet. 2001;68(3):738–752. doi: 10.1086/318793. [DOI] [PMC free article] [PubMed] [Google Scholar]