Abstract
The Envelope (E) protein in SARS Coronavirus (CoV) is a small structural protein, incorporated as part of the envelope. A major fraction of the protein has been known to be associated with the host membranes, particularly organelles related to intracellular trafficking, prompting CoV packaging and propagation. Studies have elucidated the central hydrophobic transmembrane domain of the E protein being responsible for much of the viroporin activity in favor of the virus. However, newer insights into the organizational principles at the membranous compartments within the host cells suggest further complexity of the system. The lesser hydrophobic Carboxylic-terminal of the protein harbors interesting amino acid sequences- suggesting at the prevalence of membrane-directed amyloidogenic properties that remains mostly elusive. These highly conserved segments indicate at several potential membrane-associated functional roles that can redefine our comprehensive understanding of the protein. This should prompt further studies in designing and characterizing of effective targeted therapeutic measures.
Keywords: SARS CoV E protein, Membrane, Structure, Covid-19, Amyloidogenesis
Abbreviations: CoV, Coronavirus; SARS, Severe Acute Respiratory Syndrome; CoV-2, Coronavirus-2; MERS-CoV, Middle East respiratory syndrome Coronavirus; ORFs, open reading frames; S, spike; E, envelope; M, membrane protein; N, nucleocapsid; ACE2, angiotensin-converting enzyme 2; RNA, Ribonucleic Acid; ER, Endoplasmic Reticulum; ERGIC, Endoplasmic Reticulum-Golgi intermediate compartment; TMD, transmembrane domain; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; Asp, Aspartate; PBD, PDZ-binding domain; IBV, Infectious bronchitis virus; MHV, Mouse hepatitis virus; NMR, Nuclear Magnetic Resonance; mRNA, Messenger RNA; Cys, Cystein; cDNA, complementary Deoxyribonucleic Acid; LLPS, liquid-liquid phase separation; IDRs, intrinsically disordered regions; LCDs, low complexity domains; BLAST, basic local alignment search tool; MEGA, Molecular Evolutionary Genetics Analysis; Pro, Proline; HMA, 5-(N, N-hexamethylene)amiloride; Val, Valine; Leu, Leucine;; Bcl-xL, B-cell lymphoma-extra-large.
Graphical abstract
1. Introduction
The recent outbreak of the deadly Severe Acute Respiratory Syndrome (SARS) Coronavirus-2 (CoV-2), has become a global threat reaching a total cumulative count of over million cases worldwide [1]. World Health Organization declared Coronavirus Disease 19 (COVID-19) a pandemic on March 11, 2020 [2]. Since the past two decades, this is the third major outbreak of the novel coronavirus family after SARS [3] and MERS-CoV [4] in 2003 and 2012, respectively. The repeated global threat from this family of viruses indicates their invincible nature owing to their highly evolving characteristics and plasticity. The efficiency of the virus to remain unrecognized within the host cell and rapid hijacking of the cellular machinery has been crucial for its virulence. This has necessitated pre-emptive prediction and an early diagnosis and response system to help us curb with this genus of efficient pathogens that often spread into the masses much before the appearance of the symptoms in the “super spreaders.” [5] The need of the hour demands developments of vaccines and therapeutic agents for efficient prophylactic treatment. However, diagnoses and treatment of viral diseases are particularly challenging, given the complex biology of viral particles once inside the host machinery [6]. Therefore, decoding of the cellular machinery inside the host cell may be crucial in determining newer alternative targets for drug designing.
The CoVs genome comprise of 6–10 open reading frames (ORFs) involved in encoding 16 non- structural proteins (NSP 1–16) and four structural proteins viz. spike (S), envelope (E), membrane protein (M) and the nucleocapsid (N) along with nine putative accessory factors [[7], [8], [9]]. While the NSP1–16 forms the replicase/ replication transcriptase complex prompting viral multiplication in the host cells, the structural proteins- S, E, M, and N are mainly involved in mediating host-pathogen interactions including viral packaging crucial for its propagation within the host [10]. Over the years several studies have been focusing on the efficiency of the viral proteins in mediating the virulence, hence serving as potential therapeutic targets [11,12]. However, a majority of the virulence factor is attributed to the host-membrane interactions [13]. This makes the lipid interacting interface of viral proteins as unique functional domains that can provide useful insight into our overall understanding. Knowledge of the functional interface of the host membrane interactions might provide us with a fresh perspective in defining newer therapeutic targets. Among the membrane proteins, the E protein is of particular interest, given its functional role in compromising the integrity of host membranes [14,15], manipulating them into forming favorable membranous compartments to virus particle generating factories.
The viral structural proteins have provided essential cues for therapeutic strategies [[16], [17], [18]]. The E protein, in particular, had been a less recognized component that has now been known for contributing significantly to the virulence factor within the host body. CoVs, deficient of E protein has been known to display lower viral titer, immature, and inefficient progenies [19,20]. Therefore, the E protein has been suggested to be essential for CoV viability and has the potential to be targeted for effective therapeutics development against the recent SARS-CoV-2 outbreak. It is the smallest (8.4–12 kDa size) transmembrane structural protein of CoV, with a highly variable structural propensity [21]. The CoV E protein has been extensively studied for its ability to alter the host cell membrane permeability by forming oligomeric cation-selective ion channels [22]. Parallel studies have proposed a major role in viral morphogenesis, especially during assembly and budding [23]. Apart from this, the amino acid sequence of the protein suggests several other potential purposes that are yet to be defined and characterized that might, in turn, aid in the development of effective therapeutic measures. The lack of understanding of the functional interface of virus-host interactions has restricted the characterization of the E protein as a potential target for therapeutic intervention [17,21]. The E protein can possibly adopt multiple topologies during infection [24]. The highly conserved sequences present at the C-terminal region of the protein suggest at a lesser-known amyloidogenic propensity [25]. Numerous other reports conclude that the E protein acts in co-ordination with other intracellular proteins and alters the activity of those proteins.
Identification and characterization of the transient host membrane-associated conformers of the E protein at different stages of protein synthesis, folding, and final packaging should enable us to gain a clear understanding of the viral system. In the present review of the available literature, we aim to define and delineate specific functional segments of the E protein at the functional interface of the host membranes (including Golgi/Endoplasmic Reticulum membranes). This should summarize the interspersed pieces of evidence to evaluate the immense functional potency of the CoV-2 E protein that can provide a novel perspective for therapeutic intervention.
2. The role of lipids in CoV infection
The involvement of host membrane lipids in the infectious cycle is shared by enveloped viruses and non-enveloped viruses [26]. Apart from taking advantage of cellular lipids that are usually located inside cells, viruses induce global metabolic changes on infected cells, leading to the rearrangement of the lipid dynamics to facilitate viral multiplication [27]. In some cases, these alterations produce the reorganization of intracellular membranes of the host cell, building an adequate microenvironment for viral replication. All these findings highlight the intimate connections between viruses and host lipid interactions.
Typical of the genus, the initial steps of SARS-CoVs entry include the attachment of the virus particle to a cellular receptor, angiotensin-converting enzyme 2 (ACE2- specific for SARS-CoV) located on the host plasma membrane surface [28]. The viral genome has to take entry into the host cell to reach the replication sites. Different lipids found either on the plasma membrane and/or the endosomal membranes can contribute to these processes by enabling receptor clustering, virus internalization, or membrane fusion.
The host spectrum/tropism of individual CoVs have been known to be primarily determined by the S protein [29]. The functional aspects of the S protein have been known to mediate receptor binding and virus-cell fusion that occur at either the interface of the plasma membrane or within the endosomal vesicles [30]. The mechanism of membrane fusion of SARS-CoV-1 mostly belongs to the type I fusion system, where S protein of SARS-CoV-1 comprises of a number of membrane binding regions in the S2 domain identified as fusogenic peptide or FPs by several researchers [[31], [32], [33]]. Recently Bhattacharjya et al. have shown that one of these fusion peptides adopts β-sheet conformation either in the presence of phosphatidylcholine or phosphatidylcholine/cholesterol comprising membrane models with the help of circular dichroism spectroscopy [34]. Moreover, it has also been shown that the peptide undergoes oligomerization with increasing concentration of cholesterol in the membrane. In addition to receptor binding, the action of host cell-specific proteases can likely cleave the S protein, in turn, regulating the CoV infection and host- or tissue-tropism [35].
Once inside the host cellular environment, positive-stranded RNA viruses such as CoV compartmentalize their replication machinery efficiently within the host membranous compartments to evade detection by host pattern recognition receptors [36]. Such compartments are formed for the benefit of the virus by taking advantage of cellular pathways and lipid-modifying enzymes [27]. After transcription within the rough Endoplasmic Reticulum (ER), the viral membrane proteins associate with the Endoplasmic Reticulum-Golgi intermediate compartment (or the ERGIC) before being folded and promoting the budding off of the viral particle by hijacking the cellular secretory mechanism [37]. A major portion of the viral E protein has been known to remain localized at the site of intracellular trafficking, viz. the ERGIC system, participating in CoV assembly and budding [38,39]. Thus, much of the lipid components of the virus particles are comparable to these host organellar membranes being rich in phosphatidylcholines, phosphatidylethanolamines and a small fraction of phosphatidylinositols [40]. This has provided direct evidence that the host membranes can serve as crucial functional domains that are indispensable for viral propagation. Thus, mechanistic insight into the functional interface of host-membranes is particularly essential to obtain a comprehensive understanding of the target conformers for effective therapeutic strategies.
3. The host membrane interaction interface of E protein
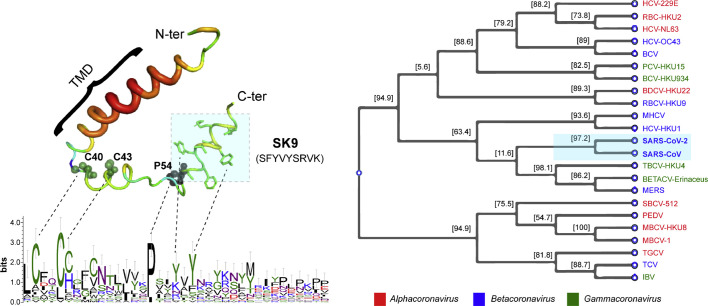
E protein of CoV is an integral membrane protein about 76–109 residues long and has been known to be most dynamic at the membranous functional interface [41]. The primary structural elements of the protein make it particularly interesting that delve into its possible role in viral propagation. Interestingly, despite the ever-evolving nature of the virion particles, the primary sequence of the protein remains mostly conserved. The SARS-CoV-2 E protein bears about 97% sequence similarity with that of the SARS-CoV. This alternatively directs into understanding the uniqueness of the sequence that is conserved to mediate some crucial functional roles of the protein (Fig. 1 ).
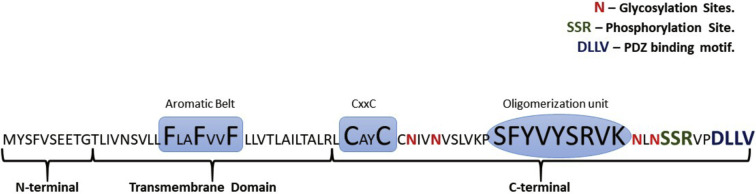
Fig. 1.
The amino acid sequence of the SARS-CoV-2 E Protein, with defined N-terminal, TMD and C-terminal. Conserved sequence motifs are FxxFxxF, CxxC, YVYSRVK. The figure specifically highlights the unique residual composition from the C-terminus that hint at the immense possible functional roles of this segment.
The primary amino acid sequence reveals a very short N-terminal hydrophilic sequence of about ten amino acids [42,43]. This is followed by a stretch of hydrophobic sequence that serves as the transmembrane domain (TMD). This twenty-eight residue long stretch has been known for the membrane-induced topologies of the protein that have been extensively defined and studied [44]. The absence of a canonical cleaved signal sequence suggests equal likelihood for the protein to be a type II (with its C-terminal targeted to the ER lumen) or type III (with its N-terminal targeted to the ER lumen) membrane protein. Therefore, the topology of the CoV E protein is highly debated in the literature and remains largely inconclusive. Table 1 summarizes the contradictory topology predicted for some of the widely studied CoV strains. The predicted topologies of the SARS CoV-2 E protein using prediction servers have been represented in Table 2 . However, further complicating matters, the topology models devised using prediction programs are mostly inconsistent with the previous experimental observations. Two membrane-associated topologies have been demonstrated for E protein (Fig. 2 )- hairpin or transmembrane [24,45]. In a report by Arbely et al. (2004), the protein was shown to adopt a highly unusual topology, comprising of a short transmembrane helical hairpin conformation that inverses about a previously unidentified pseudo-center of symmetry [46]. This hairpin structure could deform lipid bilayers and cause fragmentation [46,47].
Table 1.
Proposed topologies of CoV E proteins based on in vivo investigations.
| System | Protein condition | TOPOLOGY |
Reference | ||
|---|---|---|---|---|---|
| N-ter | C-ter | ||||
| TGEV E | Cell | Surface of Non-permeabilized infected cells | Cyto | Exo | [141] |
| IBV E | Cell | Radiolabeled | Golgi lumen | Cyto | [108] |
| MHV E | Cell | FLAG-tagged protein in the infected cell | Cyto | Cyto | [130,131] |
| SARS CoV E | Cell | N- or C- terminally FLAG-tagged | Cyto | Cyto | [45] |
| Expressed protein found to have N-linked oligosaccharides at the C-ter. | Cyto | ER lumen | |||
| Untagged protein expressed in the infected cell | Intracellular lumen (ERGIC) | Cyto | [38] | ||
Table 2.
List of predicted topologies of SARS CoV-2 Envelope protein using in silico prediction servers.
| Prediction server | N-ter (1−10) | HD (11–38) | C-ter (39–75) |
|---|---|---|---|
| TMHMM | Cyto | Transmembrane | lumen |
| MEMSAT | Lumen | Transmembrane | Lumen |
| PHOBIUS | Cyto | Transmembrane | Lumen |
| TMPred | Lumen | Transmembrane | Cyto |
Fig. 2.
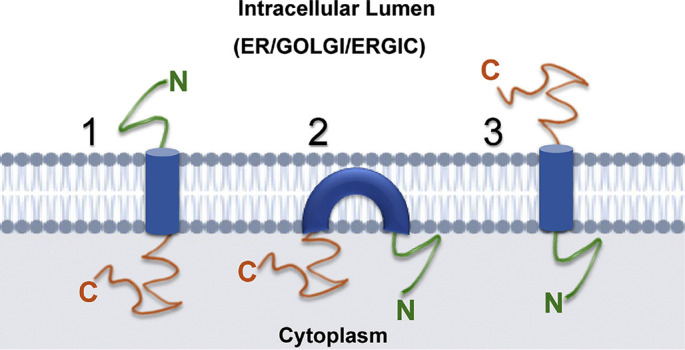
Experimentally evaluated and virtually predicted topologies of the CoV E protein. Orientation 1 represents a typical type III membrane protein; while 2 shows a membrane hairpin structure, 3 represents a type II membrane protein topology.
It has been suggested that post-translational modifications and several intermediate stages of maturation in the ER and Golgi environment can modulate the overall conformation of the molecule in response to the physiochemical properties of the membrane [[48], [49], [50]]. The specific lipid composition of the particular lipid bilayers of the ER-Golgi protein synthesis-packing machinery of the cell defines divergent physicochemical properties at the membranous interface. The lipid packing density has been known to underlie significant features of the resulting membrane- inducing characteristic local curvatures, surface charge, phase behavior, elasticity, hydrophobicity, degree of hydration, etc. [51] Membrane phase behavior in a cell has been known to permit the transient concentration of specific proteins and lipids into dynamic nanoscopic domains [52]. The level of palmitoylation may moderate the relative proportion of the membrane-associated conformers of the protein that again, in turn, can modulate the local membrane curvatures, facilitating further favorable association of the protein conformers [53].
Substitutions of the hydrophobic amino acid residues in the TMD of the E protein with charged amino acids have been shown to alter the migrating properties of the E protein in SDS-PAGE [44]. This, therefore, suggested changes in the overall conformation and membrane association of the mutant forms in comparison to the wild type E protein.
The TMD is followed by a lesser hydrophobic C-terminus with a fraction incorporated as part of the envelope in the virion particle (Fig. 1). The C-terminus, in particular, has been known to be associated with the redirection to the host ER and Golgi complex membranes that underlies some of the major functional domains of the protein [54,55]. Comprising of more than 50% of the entire protein sequence, this segment has been known to influence the topology in the host cell. There are several conflicting theories that either suggests a luminal localization of the C-terminal when associated with the ERGIC system or directed towards the cytoplasm [38,45]. The several experimental and in-silico prediction studies have been mostly inconclusive with respect to its localization and hence functional domain in the different strains of CoV. Nevertheless, the amino acid sequence of the C-terminal harbors some of the answers to the many known functional attribute of the protein sequence. Interestingly, it comprises of the conserved “FYxY” peptide sequence (where x = any amino acid) that can be correlated to its high amyloidogenic propensity [15,25]. Also, another highly conserved Cysteine containing motif- “CxxC” can enable disulfide isomerization to prompt membrane-directed conformational changes [56]. Apart from these highly conserved sequences throughout the genus, there are distinct potent glycosylation sites along the stretch that can serve as chaperone interacting motifs to help in the protein folding and/or aid in trafficking along with the cellular machinery [57]. Glycosylation of particular asparagine residues (Asn 45, Asn 48, Asn 64, and Asn 68) in the SARS-CoV has been shown to be crucial in maintaining the protein-oligomerization events associated with the host membranes [58]. Conversely, maintaining the glycosylation events have been suggested to be critical to maintaining the broad functional roles of the protein [59,60]. Further terminally, there is a distinct phosphorylation domain comprising of the “S67SR69” motif. Additionally, a PDZ-binding domain (PBM) in the C-terminal plays an essential role in host cell modifications necessary for viral infection and pathogenesis (Fig. 1) [61,62]. Mutation studies with the PBM have established its importance for viral infection, making it a significant factor of research for therapeutic and vaccine designing. [61,62]
4. The E protein as a functional viroporin
The primary structural features of the E protein have striking similarities with the viroporin class of proteins [[63], [64], [65]]. Viroporins ideally comprise of about 60–120 amino acids, with a distinct hydrophobic transmembrane domain. They are known for their interactions with membrane surfaces, often resulting in the expansion of the lipid bilayer [65]. Viroporins participate in several viral functions, including the prompting of viral particle release from host cells [66]. Although not essential for viral replication, viroporins modulate the host cellular machinery to make it favorable for viral propagation. They affect host cellular functions, including membrane vesicularization, glycoprotein tracking, and membrane permeability. The transmembrane domain could form hydrophilic pores in the membranes of virus-infected cells by oligomerization [63,67]. These hydrophilic channels would allow low molecular weight hydrophilic molecules to cross the membrane barrier, leading to the disruption of membrane potential, collapse of ionic gradients, and release of essential compounds from the cell.
Several studies with SARS CoV E protein have demonstrated the structural and functional similarities with viroporins. It has been demonstrated that the SARS-CoV E protein could enhance the membrane permeability of bacterial cells to o-nitrophenyl-β-d-galactopyranoside and hygromycin B, suggesting that the protein may function as a viroporin [68]. Similar observations were reported on mouse hepatitis virus E protein [64]. The E protein, as a viroporin, is present in low copy numbers in the virus particle, but has been implicated in membrane scission being mostly associated with the ERGIC and cis Golgi- consistent with a predominant role as a mediator of virus assembly and release at this location [69]. The lipid content at these locations may also enhance virus budding [70].
5. Highjacking of the host protein synthesis machinery
The CoV E protein interactions with host cell tight-junction proteins provide for direct evidence of a plausible role in manipulating the synthesis and release of newer viral particles [61]. Several studies show that the E protein is mainly localized to the perinuclear regions of the cells. However, the exact subcellular localization has been an issue of debate in the current literature [71]. Clear ER localization of the CoV IBV E protein was observed at early time points in a time-course experiment using an overexpression system [72]. Alternatively, expression of the Flag-tagged SARS-CoV in cells stably expressing the T7 RNA polymerase, Buchholz et al., showed that the protein exhibits typical Golgi localization patterns [73]. Nevertheless, the viral packaging within the host cellular environment and eventual propagation has attracted much attention over the years.
According to the current understanding, upon the S protein-mediated successful receptor binding, membrane fusion, and/or internalization of the virion, the viral genome is introduced into the host cytoplasm [30]. Once deposited, the positive sense RNA strand is directly translated into polyproteins that are later cleaved and processed to establish a replication/transcription complex on ER membranes [74]. These specialized membrane compartments are formed by manipulating the protein-synthesis, packaging, and distribution system of the cell involving the ER-ERGIC-Golgi complex. This helps the viral replication machinery to evade detection by host pattern recognition receptors (Fig. 2). Both viral and hijacked host proteins are used in this process, taking advantage of cellular pathways and lipid-modifying enzymes to the benefit of the virus, sequestering newly formed RNAs away from host immune sensors [75,76]. This eventually allows the virus to manipulate the normal secretory pathway of the host cellular machinery to transport and deliver the virus protein cargo at the site of final packaging, and eventual budding [69].
The virulence of the strain is, in fact, determined by the efficiency in their mechanism to concentrate the RNA synthesis machinery and subsequent packaging within these specialized compartments that are, in turn, modulated by the associated structural proteins [77]. Among the structural proteins, the E protein has been particularly intriguing either alone or in collaboration with the M protein [71]. Interestingly, studies have shown that overexpression of the E protein is sufficient to induce the packaging and assembly of the CoV particles upon efficient manipulation of the host cellular machinery [78]. Rottier et al. suggested that the E protein has no genuine structural function in the virion envelope itself where it occupies frequent, regular positions within the lattice built by M protein [79]. However, its primary role lies in the functional interface of the host packaging and assembling membranes (Fig. 3 ). The frequent but strategic positions within the lattice hint at a possible morphogenetic function to generate the required membrane curvature for the final viral particle formation (Fig. 3, inset).
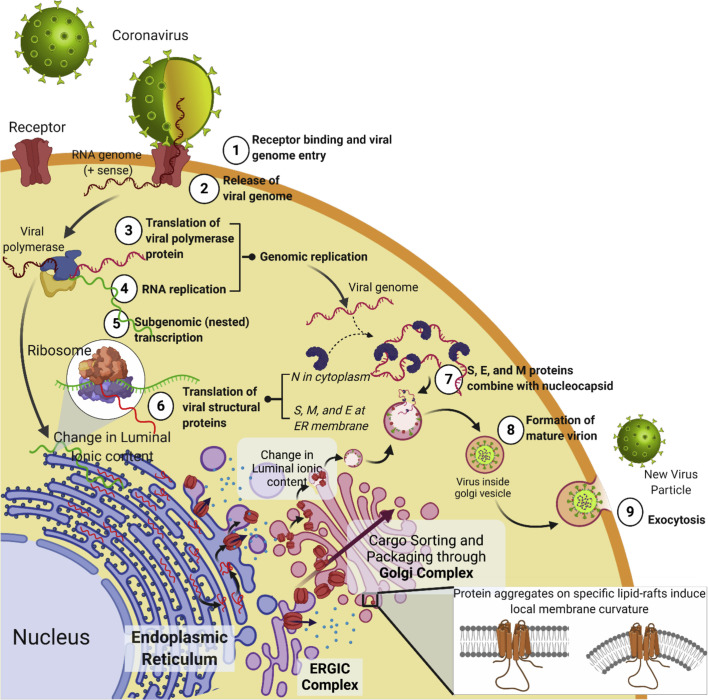
Fig. 3.
E protein-host membrane interactions. The stepwise functional interactions of the E protein with the host membranous interfaces in hijacking the cellular protein synthesis machinery to prompt viral propagation. The entry step of virus requires receptor binding followed by cell fusion that allows the entry of the viral genome into the host cellular environment. Once inside, the viral replication machinery is consolidated within the membranous compartments of host intracellular-trafficking system including the ER-ERGIC-Golgi complexes. Being in a protective environment of the host organelles, the viral structural proteins are transcribed, translated and eventually hijack the entire machinery to the benefit of the virus. The membrane-directed interactions prompt dynamic changes in the membranes of the secretory organelles, resulting in the eventual packaging and release or budding off of the new viral particles from the host cell. The figure was prepared with https://biorender.com.
Studies have endowed the C-terminal to be associated with the redirection of the E protein to the ER, and Golgi complex for assembly, budding, and intracellular trafficking of infectious virions [55]. Interestingly, sequence analyses of the C-terminal residues suggest the prevalence of some degenerate ER export signal sequences. These are closely comparable to the most common FxxxFxxxF [80] and the [R/K](x)[R/K] [81] dibasic motifs that have been known to be responsible for the export of glycosyltransferases. The last 6-residue “R103DKLYS108” stretch from the IBV E protein terminal has been defined to serve as the ER retention signal [72]. The site-directed mutagenesis of this composite lysine residue (K105) to glutamine resulted in the accumulation of E in the Golgi apparatus. [56].
Maturing through the Golgi complex, the E protein is expected to impart some very crucial functions with respect to vesicularization events, by inducing local curvatures at the site of membrane-directed accumulation (Fig. 3). Two principle mechanisms have been described for moving and delivering cargo proteins through the secretory pathway of the Golgi complex, the cisternal maturation, and the formation of mega-vesicles [82,83]. Despite the extensive studies performed, experimental evidence for both the systems remains inadequate and far from being elusive. During CoV infection, virions have been observed in large vesicle depots resembling mega-vesicles derived from Golgi/ERGIC membranes, indicating that remodeling of the Golgi complex may be crucial for virion trafficking [84].
Understanding the structural transitions that the E protein undergoes, from after its synthesis to its association with the ERGIC membrane complex, would greatly facilitate the characterization of potential targets. We expect the protein to undergo stepwise dynamic structural transitions with specific roles bound to the membranous component. This gradual transition is dependent equally on both the respective membrane composition along with the conformational changes in the protein as it is translated, modified, and matures along with the ER-Golgi network system. Previous studies performed by Knoops et al., revealed a single network of interconnected ER-derived membranes, with distinct membranous structures upon infection with the SARS-CoV [85]. Further knowledge of the membrane composition of the specific compartments of the ERGIC complex should provide useful insight into the native environments of the membrane that dictates the overall dynamics of interaction. Studies have demonstrated the virus-induced membrane rearrangements involve dynamic events such as membrane fusion and fission to prompt viral propagation by hijacking the entire cellular machinery. However, progress in our in-depth understanding of this whole machinery has been stymied by the inability to track the functional interface of the protein at the atomic-level resolution in real-time. Identifying these intermediate functional conformers could broaden our chances of finding the best target motif for therapeutic design.
6. Unregulated ion-channel formation by the transmembrane domain
The CoV E protein has been known for its possible pro-apoptotic viroporin activity majorly by mobilizing calcium ions, prompting an overall change in the ionic environment of the homeostatic cell [86,87]. The virus particles, generally, hijack the cellular homeostatic machinery of the host cell by compromising the membrane integrity that facilitates its regulatory mechanisms within the environment of the host cells. It has been suggested that the E protein can directly disrupt membranes through the formation of ion channels upon membrane-directed oligomerization events mediated upon the insertion of the TMD. In this context, the TMD has received much interest owing to their membrane-spanning attribute. Molecular dynamic simulation studies with synthetic peptides have revealed that the TMD could form ion channels by homo-oligomerization into stable dimers, trimers, and pentamers [88]. Extensive studies performed in Prof. Torres's laboratory have particularly focused on the structural and functional relationship of the E proteins from the SARS CoV, IBV, and MHV [89]. These studies have provided useful insight into understanding the role of post-translational modification in the functional oligomerization of the TMD. [90] Additionally, in silico prediction studies based on high-resolution NMR spectroscopic studies provided useful insight into understanding the probable conformers that define the TMD induced ion channels. Hence, these studies directed the defining of therapeutics targeting the resultant ion channel pores [91].
The hollow structure of the oligomers allows the unregulated efflux of ions from the cellular compartments across the hydrophilic interior of the pore, modulating the membrane potential, altering the ionic environment of the lumen. Alterations in ion concentration would promote translation of viral mRNAs, given the translation of mRNAs from many cytolytic animal viruses is reasonably resistant to high sodium concentrations [63,67]. Since the secretory pathway of the host cellular machinery is highly sensitive to perturbations in ionic strength, viral propagation can trigger a detrimental apoptotic cascade.
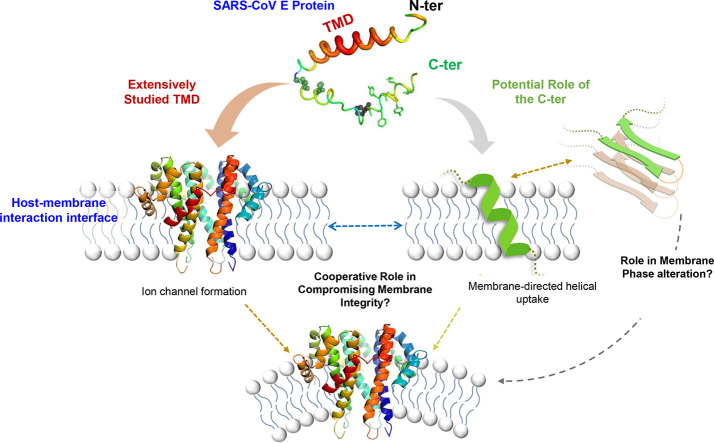
7. Membrane-directed amyloidogenic propensity of the C-terminal domain of the E protein
Unregulated ion channel formation upon E protein oligomerization in the host membrane is believed to be one of the primary processes underlying the viral pathophysiology [92]. As discussed previously, much of the studies have been focusing on the TMD for the high molecular weight conformers (multimers) of the protein upon host membrane interactions [93,94]. However, interestingly, the immediate adjacent segment (towards the C-terminal) hint, at least in part, at a possible role in the membrane directed oligomerization propensity of the molecule. Upon inspection into the C-terminal segment of the protein, we can see that it is marked by the presence of unique motifs that can prompt much of the membrane-directed conformational changes of the protein molecule. The primary sequence of the C-terminal region reveals possible functional characteristics of the protein that are lesser known in literature. Interestingly, the sequence is distinctive of amyloidogenic proteins and peptides that have been studied in the literature for their membrane-directed indispensable functions. High-resolution structural models of amyloid oligomers have been obtained in the context of membrane interactions in neurodegenerative diseases [95]. Membrane-induced oligomerization prompts membrane compromise, often leading to vesicularization into smaller lipid aggregates [[96], [97], [98]]. Studies with amyloid proteins have hinted at the possibilities of a change in the local curvature being induced by the membrane-associated high molecular weight conformers of the proteins (Fig. 3, inset) [98]. Several reports have suggested specific lipid domains to serve as templates for protein anchorage. This is followed by rearrangement of the protein conformers resulting in aggregates that form “wedges” within the bilayer to facilitate curvature change [99,100]. Such protein aggregates are driven mostly by the hydrophobic interactions that relocate within the acyl chain region of the lipid bilayer. These interactions result in a change in the lipid packing density and hence the overall membrane integrity [101].
The structural similarity of the C-terminal segment of the E protein to amyloid proteomics indicates at analogous purposes that can be complementary to or function in parallel to its viroporin activities. The oligomerization properties of E protein have been defined previously [25]. E protein expressed in bacterial and mammalian cell systems under reducing conditions was shown to exist as monomers. However, upon change to non-reducing conditions, they formed homodimers and homotrimers [68]. The existing literature has focused on the phenomenon of TMD multimerization associated with the ion channel formation that lacks mechanistic details on the initiation. However, site-directed mutagenesis studies revealed that the two-cysteine residues, immediately adjacent to the TMD in the C-terminal, were essential for the oligomerization, leading to the induction of membrane permeability. The TMD is immediately followed by a “CxxC” redox motif, which is highly conserved among the coronaviridae family of viruses. This motif is very crucial for several reasons. The oxidized CxxC motif can maintain the structural topology of the transmembrane region as well as that of its contiguous cytoplasmic domain, inclusive of glycosylation sites, involved in signaling and protein-protein interaction [102]. A probable activation of a thiol in this motif can trigger the participation of the other Cys residues in the formation of the inter-subunit disulfide bond. The activated thiol will then attack the disulfide and cause its isomerization into a disulfide isomer within the motif. This may lead to the refolding of the transmembrane region and may activate its fusogenic potential [103,104]. Interestingly, a similar CxxC motif is present as a part of the C-terminal of S protein sequence. Bioinformatic studies have suggested at the possibilities of forming inter-protein disulphide bridges that might indicate at a crossover or cooperation between these two structural proteins in the viral membrane interactions [41]. The formation of a disulfide bond may also play a crucial role in the oligomerization of the E protein, forming stable dimers, trimers, and pentamers depending on its functional requirement [105]. Thus even though the TMD spans the lipid bilayer, the CxxC motif could serve as an essential key to defining the membrane-associated oligomerization events- providing newer targets for pre-emptive therapeutic intervention.
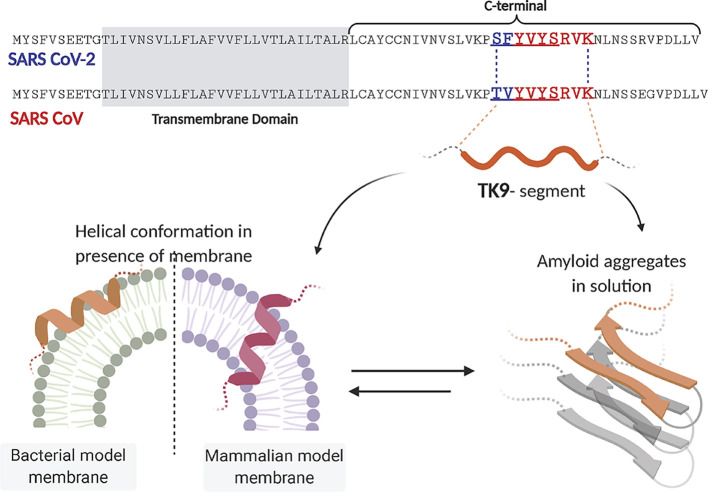
Apart from the consequential involvement of this segment in the viroporin function, the amyloidogenic segment of the E proteins can function independently at the membranous interface. Further downstream, the C-terminal harbors the “FYxY” composite peptide sequence (Fig. 4 ) that has been known for a high propensity of amyloid formation [106,107]. Interestingly, a self-assembling short peptide segment with the composite sequence has been studied to have a membrane-mediated function. Bhunia and co-workers studied a 9-residue long peptide stretch (TK9, T55VYVYSRVK63) in SARS CoV E protein (Fig. 4) for its physical changes in the presence of both zwitterionic and negatively charged model membrane micelles [15]. The TK9 peptide segment was shown to adopt amyloid fibrillary structures in solution (Fig. 4) [25]. Further studies with TK9 provided evidence for its aggregation propensity that closely resembles amyloid proteins. Intriguingly, the composite 5-residue peptide motif- T55VYV58 in TK9 was found to serve as the critical sequence motif that may be critical for the beta-sheet formation [25]. However, in vitro measures demonstrated the differential orientation of the TK9 peptide with respect to the membranous environment: correlating well with the structure and function at the membrane-protein interactions interface. This atomic-resolution NMR based structural elucidation in the presence of membrane mimics have improved understanding of the molecular mechanism of CoV infection under physiological conditions. Studies demonstrated the helical propensity of this particular segment from the C-terminal region (Fig. 4) of the protein to impart host specificity to the composite virus. The peptide's affinity was further manifested by its pronounced membrane-integrity disruption ability towards the mammalian compared to the bacterial membrane mimic- implicated in the viral pathogenesis. TK9 penetrated deeper into the acyl region of the neutral micelles mimicking the mammalian membrane and its intracellular organelles as opposed to the superficial conformation when in association with the bacterial model membrane mimics (Fig. 4). This provided evidence for the host membrane-associated conformational change in the protein segments that is specific to a particular lipid composition (species specificity). This study alternatively offered useful insight into the fact that the composite lipid molecules play particularly important roles in the interaction dynamics and conformational uptake. These segment-based studies highlighted the exclusive function of the amyloidogenic segment of the E protein that acts independently of the TMD-associated oligomerization events. Similarly, another short 9-residue peptide, NK9 (N45IVNVSLVK53) further upstream, was identified and characterized, being marked by sheet breaker-hydrophobic residues, like Ile, Val, and Leu [25]. The experimental intervention into these segments can provide useful insight into the topology of the protein, its membrane-bound orientation, and eventual functional conformation uptake.
Fig. 4.
The amyloidogenic propensity of the C-terminal segment of the SARS CoV E-protein that have a definite functional role at the membrane-interacting interface. The sequence comparison between the CoV-2 and CoV E protein C-terminal, highlights the conserved amyloidogenic segment, with a difference in only a few residues. The underlined segment in either protein can account for the fibrillation propensity. Previous studies by Bhunia et al. have highlighted the functional significance of the TK9 segment (colored residues) in SARS-CoV. The peptide-based studies have demonstrated the amyloidogenic aggregates in solution. Alternatively, the segment has been studied for the differential membrane-directed functioning. The peptide undergoes a helical conformation when in association with membranous environments. Comparison between bacterial and mammalian model membrane mimicking systems showed the different orientation of the peptide. This suggested the specific functional role of this peptide segment and its membrane-directed structural change. Preliminary studies with the TY5 peptide segment (underlined) had also shown the significance of the “FYxY” sequence, characteristic of amyloid proteins, and peptides [15,25]. The figure was prepared with https://biorender.com.
These preliminary studies have provided useful links to the immense possibilities of the functional membrane-directed conformers of the E protein, which remains largely unexplored. Nevertheless, epitope mapping for these interfaces can serve as critical targets for effective therapeutic intervention. Preliminary sequence homology analyses performed to compare the E protein primary sequence of SARS CoV to that of the novel CoV-2, revealed a minimal difference with the “FYxY” motif remaining conserved (Fig. 1). The first Threonine and the second Valine from the TK9 sequence have only been substituted by a Serine and Phenylalanine residues, respectively (Fig. 4). This might indicate at an unperturbed oligomerization potency of the segment. This conserved amyloidogenic sequence indicates a very crucial functional role of the segment that contributes significantly to the virulence. The fact that hydrophobic interactions can provide stability in localization within the acyl-chain layer of the lipid bilayers leaves us with some obvious open questions. Does the C-terminal induce the initial docking of the protein to the host membranes? Do these hydrophobic interactions prompt the downstream oligomerization events to the formation of the membrane-associated multimers? Alternatively, does the lipid-composition specificity of the C-terminal determine the conformational uptake and the eventual functional topology?
These questions are vital for a comprehensive understanding of viral propagation. Interestingly, hydrophobic protection of these segments within the acyl-chain region can serve as open templates for downstream aggregation. This can result in membrane-associated aggregates that, in analogy to the amyloidogenic proteins, can modulate the overall membrane integrity. Thus, it is essential to study the interface of interaction between the E protein and the host membrane, either as whole or directed to particular functional peptide segments.
8. Membrane remodeling for the final viral particle packaging and budding
One of the major functional aspects of CoV E protein has been associated with virus assembly and its subsequent release for propagation in the host physiology. Indirect immunofluorescence microscopy showed that E protein is localized to the Golgi complex in cells transiently expressing IBV E [108]. When co-expressed with IBV M, both from cDNA and in IBV infection, the two proteins are colocalized in Golgi membranes, near the CoV budding site [109]. Thus, even though IBV E is present at low levels in virions, it is apparently expressed at high levels in infected cells near the site of virus assembly. The subcellular localization of the SARS CoV E protein was analyzed using the sera from immunized mice by ELISA and immunofluorescence using cells infected with recombinant (rSARS-CoV) and viruses lacking the E gene (rSARS-CoV-∆E) as a negative control [38].
Several parallel studies have endowed the C-terminal to be associated with the redirection of the E protein to the ER, and Golgi complex for the intracellular trafficking of infectious virions [55]. Experimental studies have shown that the C-terminal domain of the E protein, in fact, plays a crucial role in virus budding [110,111]. The deletion of the domain resulted in the free distribution of the mutant protein and a dysfunctional viral assembly. Alternatively, mutations introduced into the cytoplasmic tail of MHV E protein by targeted RNA recombination resulted in elongated virions, further indicating at the critical involvement of the domain in the budding mechanism [72].
The difference in the lipid composition along the secretory pathway might, in fact, have a direct role on the E protein topology. Moving from the ER to the Golgi and finally the plasma membrane, there is a definite gradient in the concentration of specific lipid components that induce a unique physical property of the lipid-bilayers in terms of thickness and rigidity [40]. The ER presents a lower fraction of Cholesterol or Sphingolipids that allows maintenance of a relatively fluid lipid compartments for the remodeling associated with the protein association, sorting and accumulation [112]. This fluidity can have a direct role in the gradual compartmentalization of the ER-associated membranes prompting viral particle formations.
The host membrane-mediated oligomerization of the E protein may be responsible for much of the membrane compromise in favor of the viral propagation. The membrane-associated aggregates can compromise the overall lipid bilayer integrity [113,114]. The hydrophobic interactions involving the lipid acyl regions can result in increased fluidity of the membrane surface, prompting phase separation of the lipid domains. Biomolecular phase separation is suggested to drive the organization of cellular organelles involved in cellular packaging and compartmentalization [115]. Differential distribution of the membrane proteins and their site of interaction with specific lipid rafts, help determine the local curvature and hence characterize propensity to bud off from the surface [112,116]. The membranous compartments of ER and the Golgi complex presents with a fair fraction of zwitterionic and neutral lipids- i.e., phosphatidylcholine and phosphatidylethanolamines [112]. In vitro studies by Khattari et al. have demonstrated that the membrane-directed topology of the SARS CoV E protein imposes a direct constraint on the lipid-bilayer thickness and the acyl-chain ordering [117]. Their studies suggested that the increasing protein concentrations in the organellar membrane along the secretory pathway (greater protein/lipid concentrations), the TMD of the SARS CoV E protein induces bilayer lipid arrangements. Specific inter-molecular interactions with the lipid molecules and intra-chain hydrogen bonds and salt-bridge interactions allow the uptake of a topology that traverses across the membrane bilayer. However, the exact trans-membrane topology of the protein was highly debated and remains elusive [23]. Nevertheless, these intra- and intermolecular interactions can be considered to have the ability to rearrange and induce direct morphological changes in the host membranes. The final viral packaging requires assembling of the replicated RNA strands and the other viral proteins before the final membrane budding.
The putative transmembrane domains of the E protein can also serve a ‘catalytic’ function in the membrane packaging [118]. Cross-talk between distant protein molecules can, in fact, serve as a “zipper” to close the ‘neck’ of the viral particle as it pinches off from the membrane in the terminal phase of budding [8,119].
Despite the immense possibilities, much of the progress in our understanding of the actual mechanism of action is still hypothetical and far from being elusive. It is, therefore, essential to study the interface of interaction between the E protein and the replication complex. Particularly, the membrane-mediated aggregation, that compromises the local curvature, inducing the final scission.
9. Sequence homology and the possible conservation of the specific functional domains
CoV gained particular notoriety when the SARS CoV outbreak shook the world around 2002–2003. The aftermath leads to the identification of many newer family members. Soon after that, studies were in full swing all around the globe to determine the mechanism of viral propagation. Several parallel studies provided useful insight into the understanding of the structural and functional uniqueness of the viral proteins and RNA in their search for finding the ideal target for therapeutic intervention. Almost immediately after SARS, the endemic spread of the MERS-CoV in 2012 reignited the urge to gain in-depth insights into the system all over the world for designing effective therapeutic measures. The recent outbreak of the novel COVID-19 soon turned into a pandemic, threatening health standards globally. Lu et al. first published the genomic characterization of COVID-19 and also reported it to be significantly divergent from SARS-CoV, claiming it to be a new human-infecting β-coronavirus [120].
A comparative analysis of small membrane/ envelope protein of different coronaviruses could provide an interpretation of the molecular events that could advance these viruses from developing acute infections to one producing a pandemic. An analysis of the sequence conservation can also help in identifying the regions required in the typical functions of membrane interaction, oligomerization, localization in the host cell, and viral infection. Such information is of immediate significance and would contribute to vaccine designing and facilitate the evaluation of vaccine candidate immunogenicity. Additionally, this would help in reflecting the potential effects of mutational events as the virus is transmitted through the human population. Recent studies have revealed that the homology with a CoV strain isolated from pangolin was ~99%. This indicated that while SARS-CoV-2 have been known to evolve from the bat CoV, pangolins might have helped as an intermediate host [121]. Nevertheless, further research is required to justify these theories.
We performed a BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) search for the E protein and selected the sequences from the different genus of the coronaviridae family. Homology analyses and sequence alignment were conducted using the MEGA software. The Muscle program of the MEGA software was used to perform multiple sequence alignment, and a phylogenetic tree was created by using a maximum likelihood approach (Fig. 5 ) [122].
Fig. 5.
The conserved sequence analyses of the SARS-CoV-2 E protein with other members of the family. The left panel shows a multiple sequence alignment of several different CoV E proteins. The highly conserved Cysteine and Proline residues are labeled and represented with a sphere. The amyloidogenic SK9 fragment and TMD are highlighted in the figure. The multiple sequence alignment of proteins were carried out using ClustalW2 at the European Bioinformatics Institutes server, and WebLogo 3 was used to generate the figure. The right panel represents a Phylogenetic tree, based on amino acid sequences for E proteins of the CoV family. The tree was built using Mega X software based on maximum likelihood. Multiple sequence alignment was performed using the clustal method. The following full length amino acid sequences were used: Human coronavirus 229E (HCV-229E), Human coronavirus NL63 (HCV-NL63), Miniopterus bat coronavirus 1 (MBCV-1), Miniopterus bat coronavirus HKU8 (MBCV-HKU8), Porcine epidemic diarrhea virus (PEDV), Rhinolophus bat coronavirus HKU2 (RBC-HKU2), Scotophilus bat coronavirus 512 (SBCV-512), Murine hepatitis virus strain JHM (MHCV), Bovine coronavirus (BCV), Human coronavirus OC43 (HCV-OC43), Betacoronavirus Erinaceus/VMC/DEU/2012 (BetaCV-Erinaceus), Human coronavirus HKU1 (HCV-HKU1), Middle East respiratory syndrome-related coronavirus (MERS), Rousettus bat coronavirus HKU9 (RBCV-HKU9), Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Severe acute respiratory syndrome-related coronavirus (SARS-CoV), Tylonycteris bat CoV HKU4 (TBCV-HKU4), Infectious bronchitis virus (IBV), Turkey coronavirus (TCV), Bottlenose dolphin coronavirus HKU22 (BDCV-HKU22), Bulbul coronavirus HKU11–934 (BCV-HKU-934), Porcine coronavirus HKU15 (PCV-HKU15). Genus of the viruses is colour coded in the figure.
Predictive tools such as TMHMM [123], Memsat [124], Phobius [125], and the hydrophobic moment plot method [126] of Eisenberg and co-workers allowed the calculation of the transmembrane regions. Much has been defined to correlate the structural conformation of the TMD with its ion channel formation. However, sequence analyses of the C-terminal have revealed crucial information that has been out of the limelight for several years. Despite the fact that the C-terminal has shown the potential to underlie much of the protein's dynamic functionality associated with membrane budding and scission, it has received much lesser attention.
Several secondary structure prediction programs predicted two β-strands within the SARS-CoV E protein tail sequence, including the N45IVNVSLVK53 and the T55VYVYSRVK63. The predicted β-strands fit the criteria for forming a β-hairpin, which is a simple structural motif with two β-strands linked by a short loop of two to five amino acid residues. Interestingly, a highly conserved proline residue, Pro 54, resides between the predicted β-strands- responsible for the β-coil-β motif [54]. The antiparallel β-strands form hydrogen bonds to stabilize the hairpin structure. Previous reports with the synthetic peptide fragment of E (i.e., I46-S60), encompassing the predicted β-hairpin segment, was found to produce ~100% β-structure and was completely resistant to Hydrogen–deuterium exchange in Fourier transform experiments. Alternatively, the study showed that titration with the drug molecule, HMA (5-(N, N-hexamethylene) amiloride) resulted in large perturbations of the Val 49 and Leu 65 residues, which are far apart in the sequence. The structural model proposed by Jaume Torres's group, therefore, suggested that these two residues were coming spatially close in the membrane-associated pentamer and might belong to different monomers. Comparison with mutation-based data hypothesized this β structure to be in dynamic equilibrium with an α-helical intermediate form. In fact, a delicate balance between these two forms may alter the membrane-dynamics and subsequent processes in the infected cell (e.g., membrane scission, binding to protein partners, or E protein localization). Disturbing this predicted β-strand region along with the substitution of the conserved proline residues had resulted in the disruption of the cytoplasmic Golgi complex signal, consequently changing the subcellular localization of E protein [55].
Taken together, the results support the hypothesis that the three-dimensional structure of E protein and not the primary sequence dictates much of its function. The sum of amino acid substitutions per site from between sequences is presented in Fig. 5. Analyses were performed using the Poisson correction model. The estimated value of the shape parameter for the discrete Gamma Distribution is ~15.1, suggesting that the occurrence of the conserved amino acid residues among the coronavirus family E protein is not entirely random. However, upon close inspection with other E protein sequences of different species of the same genus, the value decreased to ~6.8. Substitution patterns and rates were estimated under the Jones-Taylor-Thornton (1992) model (+G) [127,128]. The model evolutionary rate differences among sites were obtained using a discrete Gamma distribution (10 categories, [+G]). All the ambiguous positions were eliminated for each of the sequence pairs (pairwise deletion option). Evolutionary analyses were conducted in MEGA X [129].
Collectively, the data reveals that sites observed to be constant among sequences might have crucial biological significance correlated to their function. Besides, deleterious mutations are much more likely to be found in population-level data. This indicates that sites that would predictably be considered as ‘invariable’ at the phylogenetic level might have transient polymorphisms at the population level. In the analyses of the E protein sequences from the cornaviridae family, altering the number of gamma categories did not have any noticeable effect on the estimate of the substitution rate.
10. E protein- a potential therapeutic target
CoV E protein has received lesser attention when compared to the other structural proteins (like S and M) majorly owing to a very low copy number in the viral envelope [42,130,131]. It is the smallest and yet has been the most puzzling of the major structural proteins. Earlier deletion based studies had proven that the viral life cycle endures the absence of E protein, implying that other viral genes could counterbalance for its loss [132]. However, recent evidence collected with recombinant CoVs missing E protein, exhibit considerably lowered viral titers, crippled viral development, or produce progeny incapable of further propagation. [10,11] Several parallel studies have highlighted the crucial role played by the E and M proteins in the propagation of the viral genome into the host cell and virulence. Remarkably, studies have reported that viruses produced by the deletion of the SARS-CoV E gene can be attenuated in at least three different animal models that conferred protection against SARS-CoV pathogenesis [133,134]. But, the lack of complete information and a limited number of findings have prevented an understanding of the exact mechanism underlying the definite functional role of E protein in viral infection. Nevertheless, these have provided the basis of understanding that the CoV E might be involved in several aspects of the viral replication cycle, including host cell responses such as apoptosis, inflammation, and even autophagy in association with other nonstructural viral proteins [135]. Soon after the identification, studies have been in full swing all around the globe to determine the mechanism of the viroporin action. Several parallel studies from all over provided useful insight into understanding the structural and functional uniqueness of the viral protein in their search for finding the absolute target for therapeutic intervention.
Extensive studies performed in Prof. Torres's laboratory provide useful insight into the structural conformation of the ion channel pores created by E protein that seems to be a very attractive target for therapeutic intervention [91]. Their studies have prompted the designing and characterization of specific inhibitors for these channels that would perturb the primary functional attribute of the protein. Hexamethylene amiloride has been studied to have blocked this E protein-associated ion channel activity in the mammalian cells expressing SARS-CoV E protein [136]. However, viroporin proteins, like the SARS-CoV E protein, can exhibit a multitude of diverse functions unrelated to their ion-channel properties that are yet to be defined. The dynamic functional interfaces of the protein have been underestimated for its functionality in the propagation. This study can be correlated with earlier docking screening by Gupta et al., where more than 4000 phytochemicals were evaluated on the same protein, and three of those, i.e., belachinal, macaflavanone E, and vibsanol B were found to be particularly promising. [130] Based on the experimental evidences, it is worth mentioning that the two hits, i.e., glycyrrhizic acid and cepharanthine, are supporting their significant activity against the SARS viruses. [130]
More recently, Anatoly Chernyshev has established the potential of the SARS-CoV-2 E protein as a pharmaceutical target [94,137]. These studies have defined 36 approved drugs, which theoretically could block the E protein-induced ion channel and eventually hinder the virus's life cycle. [138] However, these studies have been based on in silico approaches, and hence much of the drug candidates have to be subjected to further experimental investigations.
Considering the lengthy procedure of new drug development, the existing strategy of drug repurposing has turned into one of the preferred solutions for the immediate treatment of SARS-CoV-2 affected individuals [139]. Long-term drug development objectives for the pharmaceutical industry incorporate the identification of inhibitors targeted at the replication or infection routes associated with SARS-CoV-2 or other allied coronavirus infections [140].
11. Conclusions
Knowledge of the coronavirus structural proteins and their mechanisms of viral action can be crucial in defining alternative therapeutic targets. Despite the extensive research efforts worldwide, a definite cure to target CoV propagation is still elusive. Effective therapeutic intervention into viral systems without compromising the host cell integrity requires an in-depth understanding of the specific functional interface. Especially, with the ever-evolving viral components, as seen in the COVID-19 specimen, immense multidimensional research is the need for the hour. Among the structural proteins, the E protein have not been a convincing choice for therapeutic targeting, despite the evidence for a crucial viroporin activity. Much of the knowledge has remained restricted to only its ion channel formation ability. Although this might be serving as a significant functionality, it is definitely not the comprehensive representation of the immense potential that the protein is possibly capable of undertaking. We believe, in-depth understanding of its functional interface with the host-membranes can undermine newer possibilities and functional attributes that have not received the deserved attention so far. Attempts to determine the crucial protein domains associated with the host membranes are innovative in terms of targeted therapeutic designing. Several studies have been gradually exemplifying the protein's essential role in viral packaging and propagation. The conserved sequences in the C-terminal region directly prompt the understanding of evolutionarily crucial functional roles of this segment that is yet to be conclusively defined. The knowledge is incomplete, essential insight into the protein's functional interface should provide newer insight into understanding and structure-function correlation. This should enable the effective designing of targeted therapeutics. The entire portrayals of biologics with characters suitable for inhibiting several key CoV proteins could serve as a basis for drug development.
Declaration of Competing Interest
The authors declare no competing financial interests.
Acknowledgements
This research was partly supported by Science and Engineering Research Board (File No. EMR/2017/003457 to AB), Government of India, partly by the Department of Biotechnology (BT/PR29978/MED/30/2037/2018 to AB) Govt. of India and partly by Bose Institute intramural extramural research fund (R/16/19/1615 to AB). SM and DB thank UGC, Govt. of India for providing Senior Research Fellowship.
References
- 1.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16(10):1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sohrabi C., Alsafi Z., O’Neill N., Khan M., Kerwan A., Al-Jabir A., Iosifidis C., Agha R. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., K. F. To, Lui S.F., Szeto C.C., Chung S., Sung J.J. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 4.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., Wang B., Wu B., Shang S., Zhang Y., Shi C. Characterizing super-spreading in microblog: an epidemic-based information propagation model. Physica A. 2016;463:202–218. doi: 10.1016/j.physa.2016.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burbelo P.D., Iadarola M.J., Chaturvedi A. Emerging technologies for the detection of viral infections. Futur. Virol. 2019;14(1):39–49. doi: 10.2217/fvl-2018-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham R.L., Sparks J.S., Eckerle L.D., Sims A.C., Denison M.R. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133(1):88–100. doi: 10.1016/j.virusres.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mortola E., Roy P. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 2004;576(1–2):174–178. doi: 10.1016/j.febslet.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and Development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolokoltsov A.A., Saeed M.F., Freiberg A.N., Holbrook M.R., Davey R.A. Identification of novel cellular targets for therapeutic intervention against Ebola virus infection by siRNA screening. Drug Dev. Res. 2009;70(4):255–265. doi: 10.1002/ddr.20303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Armas-Rillo L., Valera M.S., Marrero-Hernandez S., Valenzuela-Fernandez A. Membrane dynamics associated with viral infection. Rev. Med. Virol. 2016;26(3):146–160. doi: 10.1002/rmv.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surya W., Li Y., Torres J. Structural model of the SARS coronavirus E channel in LMPG micelles. Biochim. Biophys. Acta Biomembr. 2018;1860(6):1309–1317. doi: 10.1016/j.bbamem.2018.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh A., Bhattacharyya D., Bhunia A. Structural insights of a self-assembling 9-residue peptide from the C-terminal tail of the SARS corona virus E-protein in DPC and SDS micelles: a combined high and low resolution spectroscopic study. Biochim. Biophys. Acta Biomembr. 2018;1860(2):335–346. doi: 10.1016/j.bbamem.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazzon M., Marsh M. Targeting viral entry as a strategy for broad-spectrum antivirals. F1000Res. 2019;8 doi: 10.12688/f1000research.19694.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5) doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortego J., Ceriani J.E., Patiño C., Plana J., Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. 2007;368(2):296–308. doi: 10.1016/j.virol.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortego J., Escors D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76(22):11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S., Medhi B. Drug targets for corona virus: a systematic review. Indian J. Pharm. 2020;52(1):56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson L., McKinlay C., Gage P., Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330(1):322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruch T.R., Machamer C.E. The coronavirus E protein: assembly and beyond. Viruses. 2012;4(3):363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruch T.R., Machamer C.E. A single polar residue and distinct membrane topologies impact the function of the infectious bronchitis coronavirus E protein. PLoS Pathog. 2012;8(5) doi: 10.1371/journal.ppat.1002674. (e1002674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh A., Pithadia A.S., Bhat J., Bera S., Midya A., Fierke C.A., Ramamoorthy A., Bhunia A. Self-assembly of a nine-residue amyloid-forming peptide fragment of SARS corona virus E-protein: mechanism of self aggregation and amyloid-inhibition of hIAPP. Biochemistry. 2015;54(13):2249–2261. doi: 10.1021/acs.biochem.5b00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono A. Viruses and lipids. Viruses. 2010;2(5):1236–1238. doi: 10.3390/v2051236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorizate M., Kräusslich H.G. Role of lipids in virus replication. Cold Spring Harb. Perspect. Biol. 2011;3(10):a004820. doi: 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahajan M., Bhattacharjya S. NMR structures and localization of the potential fusion peptides and the pre-transmembrane region of SARS-CoV: implications in membrane fusion. Biochim. et Biophys. Acta (BBA)-Biomembr. 2015;1848(2):721–730. doi: 10.1016/j.bbamem.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahajan M., Chatterjee D., Bhuvaneswari K., Pillay S., Bhattacharjya S. NMR structure and localization of a large fragment of the SARS-CoV fusion protein: implications in viral cell fusion. Biochim. et Biophys. Acta (BBA)-Biomembr. 2018;1860(2):407–415. doi: 10.1016/j.bbamem.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakraborty H., Bhattacharjya S. Mechanistic insights of host cell fusion of SARS-CoV-1 and SARS-CoV-2 from atomic resolution structure and membrane dynamics. Biophys. Chem. 2020;265:106438. doi: 10.1016/j.bpc.2020.106438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meher G., Bhattacharjya S., Chakraborty H. Membrane cholesterol modulates oligomeric status and peptide-membrane interaction of severe acute respiratory syndrome coronavirus fusion peptide. J. Phys. Chem. B. 2019;123(50):10654–10662. doi: 10.1021/acs.jpcb.9b08455. [DOI] [PubMed] [Google Scholar]
- 35.Li F. Structure, function, and evolution of coronavirus spike proteins. Ann. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frieman M., Heise M., Baric R. SARS coronavirus and innate immunity. Virus Res. 2008;133(1):101–112. doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romero-Brey I., Bartenschlager R. Endoplasmic reticulum: the favorite intracellular niche for viral replication and assembly. Viruses. 2016;8(6) doi: 10.3390/v8060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nieto-Torres J.L., Dediego M.L., Alvarez E., Jiménez-Guardeño J.M., Regla-Nava J.A., Llorente M., Kremer L., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Escriba P.V., Busquets X., Inokuchi J., Balogh G., Torok Z., Horvath I., Harwood J.L., Vigh L. Membrane lipid therapy: modulation of the cell membrane composition and structure as a molecular base for drug discovery and new disease treatment. Prog. Lipid Res. 2015;59:38–53. doi: 10.1016/j.plipres.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Wu Q., Zhang Y., Lü H., Wang J., He X., Liu Y., Ye C., Lin W., Hu J., Ji J., Xu J., Ye J., Hu Y., Chen W., Li S., Bi S., Yang H. The E protein is a multifunctional membrane protein of SARS-CoV. Genom. Proteome. Bioinforma. 2003;1(2):131–144. doi: 10.1016/S1672-0229(03)01017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo L., Hurst K.R., Masters P.S. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J. Virol. 2007;81(5):2249–2262. doi: 10.1128/JVI.01577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., Dediego M.L., Enjuanes L., Aguilella V.M. Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochim. Biophys. Acta. 2013;1828(9):2026–2031. doi: 10.1016/j.bbamem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y., Yuan Q., Torres J., Tam J.P., Liu D.X. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology. 2006;349(2):264–275. doi: 10.1016/j.virol.2006.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan Q., Liao Y., Torres J., Tam J.P., Liu D.X. Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells. FEBS Lett. 2006;580(13):3192–3200. doi: 10.1016/j.febslet.2006.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arbely E., Khattari Z., Brotons G., Akkawi M., Salditt T., Arkin I.T. A highly unusual palindromic transmembrane helical hairpin formed by SARS coronavirus E protein. J. Mol. Biol. 2004;341(3):769–779. doi: 10.1016/j.jmb.2004.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saleem M., Morlot S., Hohendahl A., Manzi J., Lenz M., Roux A. A balance between membrane elasticity and polymerization energy sets the shape of spherical clathrin coats. Nat. Commun. 2015;6:6249. doi: 10.1038/ncomms7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang S., Wang Y. Golgi structure formation, function, and post-translational modifications in mammalian cells. F1000Res. 2017;6:2050. doi: 10.12688/f1000research.11900.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braakman I., Hebert D.N. Protein folding in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2013;5(5):a013201. doi: 10.1101/cshperspect.a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellgaard L., McCaul N., Chatsisvili A., Braakman I. Co- and post-translational protein folding in the ER. Traffic. 2016;17(6):615–638. doi: 10.1111/tra.12392. [DOI] [PubMed] [Google Scholar]
- 51.Bassereau P., Jin R., Baumgart T., Deserno M., Dimova R., Frolov V.A., Bashkirov P.V., Grubmüller H., Jahn R., Risselada H.J., Johannes L., Kozlov M.M., Lipowsky R., Pucadyil T.J., Zeno W.F., Stachowiak J.C., Stamou D., Breuer A., Lauritsen L., Simon C., Sykes C., Voth G.A., Weikl T.R. The 2018 biomembrane curvature and remodeling roadmap. J. Phys. D. Appl. Phys. 2018;51(34) doi: 10.1088/1361-6463/aacb98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.King C., Sengupta P., Seo A.Y., Lippincott-Schwartz J. ER membranes exhibit phase behavior at sites of organelle contact. Proc. Natl. Acad. Sci. U. S. A. 2020;117(13):7225–7235. doi: 10.1073/pnas.1910854117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Escribá P.V., González-Ros J.M., Goñi F.M., Kinnunen P.K., Vigh L., Sánchez-Magraner L., Fernández A.M., Busquets X., Horváth I., Barceló-Coblijn G. Membranes: a meeting point for lipids, proteins and therapies. J. Cell. Mol. Med. 2008;12(3):829–875. doi: 10.1111/j.1582-4934.2008.00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Surya W., Claudine S., Torres J. Structure of a conserved Golgi complex-targeting signal in coronavirus envelope proteins. J. Biol. Chem. 2014;289(18):12535–12549. doi: 10.1074/jbc.M114.560094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen J.R., Lin L.D., Machamer C.E. Identification of a Golgi complex-targeting signal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus envelope protein. J. Virol. 2011;85(12):5794–5803. doi: 10.1128/JVI.00060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chivers P.T., Laboissière M.C., Raines R.T. The CXXC motif: imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996;15(11):2659–2667. [PMC free article] [PubMed] [Google Scholar]
- 57.Sriram V., Willard C.A., Liu J., Brutkiewicz R.R. Importance of N-linked glycosylation in the functional expression of murine CD1d1. Immunology. 2008;123(2):272–281. doi: 10.1111/j.1365-2567.2007.02696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Y., Lu K., Pfefferle S., Bertram S., Glowacka I., Drosten C., Pöhlmann S., Simmons G. A single asparagine-linked glycosylation site of the severe acute respiratory syndrome coronavirus spike glycoprotein facilitates inhibition by mannose-binding lectin through multiple mechanisms. J. Virol. 2010;84(17):8753–8764. doi: 10.1128/JVI.00554-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westerbeck J.W., Machamer C.E. A coronavirus E protein is present in two distinct pools with different effects on assembly and the secretory pathway. J. Virol. 2015;89(18):9313–9323. doi: 10.1128/JVI.01237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16(1):69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teoh K.T., Siu Y.L., Chan W.L., Schlüter M.A., Liu C.J., Peiris J.S., Bruzzone R., Margolis B., Nal B. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol. Biol. Cell. 2010;21(22):3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Javier R.T., Rice A.P. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J. Virol. 2011;85(22):11544–11556. doi: 10.1128/JVI.05410-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gonzalez M.E., Carrasco L. Viroporins. FEBS Lett. 2003;552(1):28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- 64.Madan V., García M.E.J., Sanz M.A., Carrasco L. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 2005;579(17):3607–3612. doi: 10.1016/j.febslet.2005.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scott C., Griffin S. Viroporins: structure, function and potential as antiviral targets. J. Gen. Virol. 2015;96(8):2000–2027. doi: 10.1099/vir.0.000201. [DOI] [PubMed] [Google Scholar]
- 66.Nieto-Torres J.L., Verdiá-Báguena C., Castaño-Rodriguez C., Aguilella V.M., Enjuanes L. Relevance of viroporin ion channel activity on viral replication and pathogenesis. Viruses. 2015;7(7):3552–3573. doi: 10.3390/v7072786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carrasco L. Modification of membrane permeability induced by animal viruses early in infection. Virology. 1981;113(2):623–629. doi: 10.1016/0042-6822(81)90190-2. [DOI] [PubMed] [Google Scholar]
- 68.Liao Y., Lescar J., Tam J.P., Liu D.X. Expression of SARS-coronavirus envelope protein in Escherichia coli cells alters membrane permeability. Biochem. Biophys. Res. Commun. 2004;325(1):374–380. doi: 10.1016/j.bbrc.2004.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alsaadi J., Jones I.M. Membrane binding proteins of coronaviruses. Futur. Virol. 2019;14(4):275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heaton N.S., Randall G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011;19(7):368–375. doi: 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corse E., Machamer C.E. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology. 2003;312(1):25–34. doi: 10.1016/S0042-6822(03)00175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lim K.P., Liu D.X. The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 2001;276(20):17515–17523. doi: 10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buchholz U.J., Finke S., Conzelmann K.K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 1999;73(1):251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reid C.R., Airo A.M., Hobman T.C. The virus-host interplay: biogenesis of +RNA replication complexes. Viruses. 2015;7(8):4385–4413. doi: 10.3390/v7082825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hagemeijer M.C., Rottier P.J., de Haan C.A. Biogenesis and dynamics of the coronavirus replicative structures. Viruses. 2012;4(11):3245–3269. doi: 10.3390/v4113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6(5):363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gale M., Jr., Tan S.L., Katze M.G. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 2000;64(2):239–280. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S., Bruzzone R., Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82(22):11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15(8):2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bermak J.C., Li M., Bullock C., Zhou Q.Y. Regulation of transport of the dopamine D1 receptor by a new membrane-associated ER protein. Nat. Cell Biol. 2001;3(5):492–498. doi: 10.1038/35074561. [DOI] [PubMed] [Google Scholar]
- 81.Giraudo C.G., Maccioni H.J. Endoplasmic reticulum export of glycosyltransferases depends on interaction of a cytoplasmic dibasic motif with Sar1. Mol. Biol. Cell. 2003;14(9):3753–3766. doi: 10.1091/mbc.E03-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Volchuk A., Amherdt M., Ravazzola M., Brügger B., Rivera V.M., Clackson T., Perrelet A., Söllner T.H., Rothman J.E., Orci L. Megavesicles implicated in the rapid transport of intracisternal aggregates across the Golgi stack. Cell. 2000;102(3):335–348. doi: 10.1016/s0092-8674(00)00039-8. [DOI] [PubMed] [Google Scholar]
- 83.Mironov A.A., Beznoussenko G.V., Nicoziani P., Martella O., Trucco A., Kweon H.S., Di Giandomenico D., Polishchuk R.S., Fusella A., Lupetti P., Berger E.G., Geerts W.J., Koster A.J., Burger K.N., Luini A. Small cargo proteins and large aggregates can traverse the Golgi by a common mechanism without leaving the lumen of cisternae. J. Cell Biol. 2001;155(7):1225–1238. doi: 10.1083/jcb.200108073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ulasli M., Verheije M.H., de Haan C.A., Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010;12(6):844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9) doi: 10.1371/journal.pbio.0060226. (e226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang C., Takeuchi K., Pinto L.H., Lamb R.A. Ion channel activity of influenza a virus M2 protein: characterization of the amantadine block. J. Virol. 1993;67(9):5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mould J.A., Paterson R.G., Takeda M., Ohigashi Y., Venkataraman P., Lamb R.A., Pinto L.H. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell. 2003;5(1):175–184. doi: 10.1016/s1534-5807(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 88.Torres J., Wang J., Parthasarathy K., Liu D.X. The transmembrane oligomers of coronavirus protein E. Biophys. J. 2005;88(2):1283–1290. doi: 10.1529/biophysj.104.051730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Torres J., Maheswari U., Parthasarathy K., Ng L., Liu D.X., Gong X. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007;16(9):2065–2071. doi: 10.1110/ps.062730007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lopez L.A., Riffle A.J., Pike S.L., Gardner D., Hogue B.G. Importance of conserved cysteine residues in the coronavirus envelope protein. J. Virol. 2008;82(6):3000–3010. doi: 10.1128/JVI.01914-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pervushin K., Tan E., Parthasarathy K., Lin X., Jiang F.L., Yu D., Vararattanavech A., Soong T.W., Liu D.X., Torres J. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5(7) doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Verdia-Baguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Torres J., Aguilella V.M., Enjuanes L. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432(2):485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.To, J, Surya W., Fung T.S., Li Y., Verdia-Baguena C., Queralt-Martin M., Aguilella V.M., Liu D.X., Torres J. Channel-inactivating mutations and their revertant mutants in the envelope protein of infectious bronchitis virus. J. Virol. 2017;91(5) doi: 10.1128/JVI.02158-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chernyshev A. 2020. Pharmaceutical Targeting the Envelope Protein of SARS-CoV-2: The Screening for Inhibitors in Approved Drugs. [Google Scholar]
- 95.Bhattacharyya D., Kumar R., Mehra S., Ghosh A., Maji S.K., Bhunia A. Multitude NMR studies of α-synuclein familial mutants: probing their differential aggregation propensities. Chem. Commun. (Camb.) 2018;54(29):3605–3608. doi: 10.1039/C7CC09597J. [DOI] [PubMed] [Google Scholar]
- 96.Stahelin R.V. Monitoring peripheral protein oligomerization on biological membranes. Methods Cell Biol. 2013;117:359–371. doi: 10.1016/B978-0-12-408143-7.00019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Azouz M., Cullin C., Lecomte S., Lafleur M. Membrane domain modulation of Aβ. Nanoscale. 2019;11(43):20857–20867. doi: 10.1039/c9nr06361g. [DOI] [PubMed] [Google Scholar]
- 98.Burke K.A., Yates E.A., Legleiter J. Biophysical insights into how surfaces, including lipid membranes, modulate protein aggregation related to neurodegeneration. Front. Neurol. 2013;4:17. doi: 10.3389/fneur.2013.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Daumke O., Lundmark R., Vallis Y., Martens S., Butler P.J., McMahon H.T. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449(7164):923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- 100.Plomann M., Wittmann J.G., Rudolph M.G. A hinge in the distal end of the PACSIN 2 F-BAR domain may contribute to membrane-curvature sensing. J. Mol. Biol. 2010;400(2):129–136. doi: 10.1016/j.jmb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 101.McMahon H.T., Boucrot E. Membrane curvature at a glance. J. Cell Sci. 2015;128(6):1065–1070. doi: 10.1242/jcs.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brazin K.N., Mallis R.J., Li C., Keskin D.B., Arthanari H., Gao Y., Wu S.L., Karger B.L., Wagner G., Reinherz E.L. Constitutively oxidized CXXC motifs within the CD3 heterodimeric ectodomains of the T cell receptor complex enforce the conformation of juxtaposed segments. J. Biol. Chem. 2014;289(27):18880–18892. doi: 10.1074/jbc.M114.574996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sjoberg M., Wallin M., Lindqvist B., Garoff H. Furin cleavage potentiates the membrane fusion-controlling intersubunit disulfide bond isomerization activity of leukemia virus Env. J. Virol. 2006;80(11):5540–5551. doi: 10.1128/JVI.01851-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li K., Zhang S., Kronqvist M., Wallin M., Ekstrom M., Derse D., Garoff H. Intersubunit disulfide isomerization controls membrane fusion of human T-cell leukemia virus Env. J. Virol. 2008;82(14):7135–7143. doi: 10.1128/JVI.00448-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li P.P., Nakanishi A., Clark S.W., Kasamatsu H. Formation of transitory intrachain and interchain disulfide bonds accompanies the folding and oligomerization of simian virus 40 Vp1 in the cytoplasm. Proc. Natl. Acad. Sci. U. S. A. 2002;99(3):1353–1358. doi: 10.1073/pnas.032668699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bemporad F., Taddei N., Stefani M., Chiti F. Assessing the role of aromatic residues in the amyloid aggregation of human muscle acylphosphatase. Protein Sci. 2006;15(4):862–870. doi: 10.1110/ps.051915806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gazit E. A possible role for pi-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002;16(1):77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 108.Corse E., Machamer C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000;74(9):4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lim K.P., Xu H.Y., Liu D.X. Physical interaction between the membrane (M) and envelope (E) proteins of the coronavirus avian infectious bronchitis virus (IBV) Adv. Exp. Med. Biol. 2001;494:595–602. doi: 10.1007/978-1-4615-1325-4_88. [DOI] [PubMed] [Google Scholar]
- 110.Fischer F., Stegen C.F., Masters P.S., Samsonoff W.A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 1998;72(10):7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baudoux P., Carrat C., Besnardeau L., Charley B., Laude H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 1998;72(11):8636–8643. doi: 10.1128/jvi.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Casares D., Escriba P.V., Rossello C.A. Membrane lipid composition: effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int. J. Mol. Sci. 2019;20(9) doi: 10.3390/ijms20092167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gorbenko G., Trusova V. Protein aggregation in a membrane environment. Adv. Protein Chem. Struct. Biol. 2011;84:113–142. doi: 10.1016/B978-0-12-386483-3.00002-1. [DOI] [PubMed] [Google Scholar]
- 114.Berterame N.M., Porro D., Ami D., Branduardi P. Protein aggregation and membrane lipid modifications under lactic acid stress in wild type and OPI1 deleted Saccharomyces cerevisiae strains. Microb. Cell Factories. 2016;15:39. doi: 10.1186/s12934-016-0438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yoshizawa T., Nozawa R.S., Jia T.Z., Saio T., Mori E. Biological phase separation: cell biology meets biophysics. Biophys. Rev. 2020;12(2):519–539. doi: 10.1007/s12551-020-00680-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Khattari Z., Brotons G., Akkawi M., Arbely E., Arkin I.T., Salditt T. SARS coronavirus E protein in phospholipid bilayers: an x-ray study. Biophys. J. 2006;90(6):2038–2050. doi: 10.1529/biophysj.105.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.van Geest M., Lolkema J.S. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol. Mol. Biol. Rev. 2000;64(1):13–33. doi: 10.1128/mmbr.64.1.13-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Den Boon J.A., Snijder E.J., Locker J.K., Horzinek M.C., Rottier P.J. Another triple-spanning envelope protein among intracellularly budding RNA viruses: the torovirus E protein. Virology. 1991;182(2):655–663. doi: 10.1016/0042-6822(91)90606-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lam T.T., Shum M.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S., Wei W., Cheung W.Y., Li W.J., Li L.F., Leung G.M., Holmes E.C., Hu Y.L., Guan Y. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 122.Chakrabarti S., Panchenko A.R. Coevolution in defining the functional specificity. Proteins. 2009;75(1):231–240. doi: 10.1002/prot.22239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 124.Jones D.T. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23(5):538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- 125.Käll L., Krogh A., Sonnhammer E.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 1984;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 127.Arenas M. Trends in substitution models of molecular evolution. Front. Genet. 2015;6:319. doi: 10.3389/fgene.2015.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 129.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Maeda J., Repass J.F., Maeda A., Makino S. Membrane topology of coronavirus E protein. Virology. 2001;281(2):163–169. doi: 10.1006/viro.2001.0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Raamsman M.J., Locker J.K., de Hooge A., de Vries A.A., Griffiths G., Vennema H., Rottier P.J. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 2000;74(5):2333–2342. doi: 10.1128/jvi.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xie Q., Cao Y., Su J., Wu J., Wu X., Wan C., He M., Ke C., Zhang B., Zhao W. Two deletion variants of Middle East respiratory syndrome coronavirus found in a patient with characteristic symptoms. Arch. Virol. 2017;162(8):2445–2449. doi: 10.1007/s00705-017-3361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69(4):635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., Enjuanes L. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M.L., Enjuanes L., Guo D. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One. 2008;3(10) doi: 10.1371/journal.pone.0003299. (e3299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wilson L., Gage P., Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353(2):294–306. doi: 10.1016/j.virol.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gupta M.K., Vemula S., Donde R., Gouda G., Behera L., Vadde R. Approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang C.H., Wang Y.F., Liu X.J., Lu J.H., Qian C.W., Wan Z.Y., Yan X.G., Zheng H.Y., Zhang M.Y., Xiong S., Li J.X., Qi S.Y. Antiviral activity of cepharanthine against severe acute respiratory syndrome coronavirus in vitro. Chin. Med. J. 2005;118(6):493–496. [PubMed] [Google Scholar]
- 139.Senanayake S.L. Drug repurposing strategies for COVID-19. Future Sci. 2020 doi: 10.4155/fdd-2020-0010. [DOI] [Google Scholar]
- 140.Abd El-Aziz T.M., Stockand J.D. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2) - an update on the status. Infect. Genet. Evol. 2020;83:104327. doi: 10.1016/j.meegid.2020.104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Godet M., L’Haridon R., Vautherot J.F., Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188(2):666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]