Abstract
Background
Human metapneumovirus (HMPV) is an important aetiologic agent of respiratory tract infection (RTI). This study aimed to describe its genetic diversity and clinical impact in patients attended at a tertiary university hospital in Barcelona from the 2014-2015 to the 2016-2017 seasons, focusing on the emerging duplications in G gene and their structural properties.
Methods
Laboratory-confirmed HMPV were characterised based on partial-coding F and G gene sequences with MEGA.v6.0. Computational analysis of disorder propensity, aggregation propensity and glycosylation sites in viral G predicted protein sequence were carried out. Clinical data was retrospectively reviewed and further associated to virological features.
Results
HMPV prevalence was 3%. The 180- and 111-nucleotide duplications occurred in A2c lineage G protein increased in prevalence throughout the study, in addition to short genetic changes observed in other HMPV lineages. The A2c G protein without duplications was calculated to protrude over F protein in 23% of cases and increased to a 39% and a 46% with the 111- and 180-nucleotide duplications, respectively. Children did not seem to be more affected by these mutant viruses, but there was a strong association of these variants to LRTI in adults.
Discussion
HMPV presents a high genetic diversity in all lineages. Novel variants carrying duplications might present an evolutionary advantage due to an improved steric shielding, which would have been responsible for the reported increasing prevalence and the association to LRTI in adults.
Keywords: Human metapneumovirus, duplication, steric shielding, epidemiology, genetic diversity, clinical impact
1. INTRODUCTION
Human metapneumovirus (HMPV) is an important aetiologic agent of upper and lower respiratory tract infections (URTI and LRTI) in children and adults [1].
HMPV belongs to the Pneumoviridae family together with human respiratory syncytial virus (HRSV) [2], causing an indistinguishable symptomatology [1]. HMPV is an enveloped, lineal, negative-sense, single-stranded RNA virus, classified into HMPV-A and HMPV-B genotypes and subdivided into subgenotypes A1, A2 (A2a, A2b and A2c lineages) [1,3,4], B1 and B2 (B2a and B2b lineages) [5].
The fusion (F) and the attachment (G) proteins are the major envelope glycoproteins. F protein is the major cross-protective antigenic determinant and is highly conserved between genotypes (88%) [4]. Hence, it is the main target for most vaccine strategies under development [6]. Differently, G protein is weakly immunogenic [7], with 28% genetic divergence between genotypes and 74-82% intra-genotype [4]. In addition, 180- and 111-nucleotide duplications have been recently described into G protein’s ectodomain [[8], [9], [10], [11]].
The aims of this study were to describe circulation pattern, genetic diversity and clinical impact of HMPV in paediatric and adult population attended at a tertiary university hospital in Barcelona from the 2014-2015 to the 2016-2017 seasons, focusing on the emergence and spread of variants carrying these two nucleotide duplications.
2. MATERIALS AND METHODS
2.1. Sample collection
From October/2014 to May/2017, respiratory specimens (nasopharyngeal aspirates, nasal and pharyngeal swabs, bronchoaspirates, bronchoalveolar, bronchoselective and tracheal washes and sputums) were received for the laboratory-confirmation of respiratory viruses from children and adults attended at the Hospital Universitari Vall d’Hebron with suspicion of respiratory tract infection (RTI). Institutional Review Board approval (PR(AG)161/2016) was obtained from the hospital’s Clinical Research Ethics Committee.
2.2. Respiratory viruses’ detection
During the HRSV and influenza epidemics, rapid tests were performed for a fast diagnosis, which were based on immunocromatography (Alere BinaxNOW® Influenza A&B/RSV, Alere, USA), immunofluorescence (Sofia RSV FIA, Quidel, USA) or real-time RT-PCR (GenXpert Flu/RSV XC, Cepheid, USA). Samples received out of HRSV/FLUV epidemics or negative for rapid tests were analysed by immunofluorescence (D3 Ultra 8™ DFA Respiratory Virus, Diagnostic HYBRIDS, USA) or mainly by real-time RT-PCR (Anyplex II RV16, Seegene, Korea, until 2015; Allplex Respiratory Panels 1-3, Seegene, Korea, since 2015). Total nucleic acids were extracted using NucliSense easyMAG (BioMérieux, Marcy l’Etoile) and kept at -80 °C.
2.3. HMPV phylogenetic analysis
Both partial F and G genes were retrospectively sequenced from all HMPV laboratory-confirmed samples. The amplification was performed using the One-Step RT-PCR kit (Qiagen, Hilden, Germany), conditions in Table 1 . PCR products were purified using Exo-SAP-IT (USB, Affymetrix Inc., Cleveland, USA) and sequenced by the ABI Prism BigDye Terminator v3.1 (Applied Biosystems, Carlsbad, USA) on the ABI PRISM 3130xl Genetic Analyzer (Thermo Fisher Scientific, Waltham, USA). Nucleotide sequences were edited and assembled using MEGA v6.0 [12]. A collapse to haplotypes was done with ALTER server [13]. The best fit substitution model was determined by MEGA v6.0, and the lowest Bayesian information criterion score model was used and evaluated with 1,000 bootstrap resamplings.
Table 1.
Primers and PCR conditions.
| Gene | Primers | CAN97-83 |
PCR conditions | Fragment length (bp) | Reference | |
|---|---|---|---|---|---|---|
| Initial position | Final position | |||||
| G | GF: GAGAACATTCGRRCRATAGAYA |
6,247 | 6,268 | 50ºC×30’ – 95 °C×15’ – 45x (95 °C×30” – 59 °C×30” – 72 °C×1’) – 72ºc×10’ | 924 | Ludewick HP et al., 2005 |
| GR: AGATAGACATTRACAGTGGATT |
7,149 | 7,170 | ||||
| F | FF: CAATGCWGGRATAACACCAGC |
3,693 | 3,713 | 50ºC×30’ – 95 °C×15’ – 45x (95 °C×30” – 55 °C×30” – 72 °C×1’) – 72 °C×10’ | 745 | Designed for this study |
| FR: ATTGAAYTGATCYTCAGGAAAC |
4,416 | 4,437 | ||||
2.4. Computational analysis, generation of unfolded ensembles and geometric analyses of G predicted protein sequence
The propensity to adopt disordered conformations of three G sequences with and without nucleotide duplications was analysed using the MetaDisorder server [14], their propensity to aggregate using the Pasta 2.0 server [15], and the prediction of potential N- and O-glycosylation sites using NetNGlyc 1.0 [16] and NetOglyc 4.0 [17] servers, respectively.
Ensembles consisting of 2,000 unfolded conformations were generated for each of the three G sequences using the ProtSA server [18]. The PDB file of each conformation was analysed to compute the distance between the N atom of the first extracellular residue (Asn52) and the more distant atom, as well as the radius of gyration of the particular conformation.
2.5. Clinical features
Demographic (age and sex) and clinical features (URTI/LRTI, co-morbidities, co-infections, antibiotic use, need, type and length of respiratory support, length of hospital stay, ICU admission or exitus) of HMPV-laboratory confirmed cases were retrospectively reviewed from medical records and related to viral features. Patients included in the demographic study were those with clinical presentation of URTI or LRTI, whilst patients with other symptoms rather than respiratory were excluded from the study. For the severity study, only patients hospitalised due to LRTI were included, and exclusion criteria were those cases with other symptoms rather than LRTI and hospitalisation due to other clinical reasons even though the patient manifested RTI.
2.6. Statistical analysis
Data were analysed with R software v3.5.1. For categorical data, Chi-squared or Fisher’s exact test were performed. For numerical variables, t student, Mann-Withney, ANOVA or Kruskall-Wallis tests were performed according to the need. Statistical significance was taken at the p-value <0.05.
3. RESULTS
3.1. HMPV epidemiology
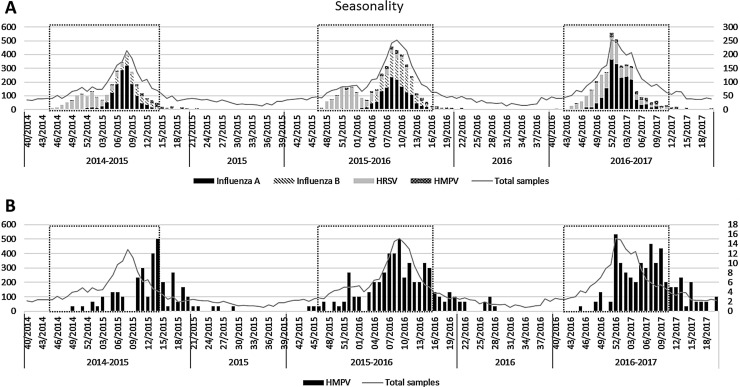
A total of 20,132 samples of 14,769 patients were tested, of which 9,370 (47%) were laboratory-confirmed for at least one respiratory virus. Though being in a similar period, HRSV and influenza epidemics varied between seasons (Fig. 1 A), being stablished during weeks 45/2014-13/2015 in the first season, 47/2015-16/2016 in the second season and 43/2016-11/2017 in the third season. Overall these 14,769 patients, 11,185 (76%) samples were collected during all 3 consecutive HRSV/influenza epidemics and 3,584 (24%) out of these epidemics (Supplementary Table 1). Regarding the methods for detection, from all these 14,769 patients, 3,316 samples (22%) were tested with the immunofluorescence assay, while 8,483 (57%) were tested with the real-time RT-PCR multiplex assays. Importantly, 656 (4%) samples which tested negative for the immunofluorescence assay were re-tested with the real-time RT-PCR multiplex assays. The remaining samples had been tested with a rapid test. The use of immunofluorescence and PCR-based assays were changing throughout the period of study, decreasing for immunofluorescence assays and increasing for molecular methods (59% vs 27% in 2014-2015; 15% vs 57% in 2015-2016 and <1% vs 74% in 2016-2017, respectively).
Fig. 1.
Seasonality. The X-axis represent the timeline of the study, the left Y-axis represents the number of total samples received and the right Y-axis represent the number of samples of each specific virus. HRSV/influenza epidemics period are framed in a black dotted box. A) shows the seasonality of HRSV, Influenza A virus, Influenza B virus and HMPV. B) shows the seasonality of HMPV.
HMPV was laboratory-confirmed in 423 (2%) respiratory samples (nasopharyngeal swabs or aspirates, bronchoaspirates, bronchoalveolar washes or tracheal aspirates) from 407 (3%) patients, 221 (3%) of them occurring in the paediatric population and 186 (3%) in adults, showing an important prevalence in the adult population, higher than in children in the 2015-2016 season (Table 2 ). This prevalence varied according to whether it was during HRSV/influenza epidemics or not (Supplementary Table 1). During the HRSV/influenza epidemics, 82-95% of HMPV laboratory-confirmed samples previously had a negative result in the rapid tests for HRSV and influenza. Over the 3 seasons, the proportion of HMPV laboratory-confirmed samples tested by immunofluorescence decreased while those tested by PCR-based assays increased (61%-11%-0% vs 47%-91%-100%, respectively and by season).
Table 2.
Demographic data of total patients and patients with HMPV.
| Season | Total patients |
Patients with HMPV |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Pediatric | Adult | Total | Pediatric | Adult | Total | ||||
| 2014 - 2015 | 1,939 | 1,744 | 3,683 | 54 | 3% | 36 | 2% | 90 | 2% |
| 2015 | 194 | 547 | 741 | 3 | 2% | 2 | <1% | 5 | 1% |
| 2015 - 2016 | 2,591 | 2,209 | 4,800 | 64 | 2% | 87 | 4% | 151 | 3% |
| 2016 | 222 | 564 | 786 | 10 | 5% | 0 | 0% | 10 | 1% |
| 2016 - 2017 | 2,832 | 1,927 | 4,759 | 90 | 3% | 61 | 3% | 151 | 3% |
| Overall | 7,778 | 6,991 | 14,769 | 221 | 3% | 186 | 3% | 407 | 3% |
75/407 (18%) HMPV cases presented co-infections with 88 respiratory viruses: rhinovirus (26, 30%), adenovirus (15, 17%), human bocavirus (15, 17%), enterovirus (11, 13%), HRSV (6, 7%), human coronavirus (HCoV) 229E (5, 6%), HCoV OC43 (5, 6%), HCoV NL63 (2, 2%), human parainfluenza 3 (1, 1%), influenza A (1, 1%) and B (1, 1%).
Weekly distribution of HMPV showed a higher circulation from February to April in the first two seasons, but started at mid December in 2016-2017 season (Fig. 1B). The peaks of incidence of the first two seasons were in March, but the last season presented a pattern with two peaks.
3.2. Genetic characterisation of viral strains
Phylogenetic analyses of HMPV F and G sequences from 387 strains revealed that HMPV-A (201/387, 52%) and HMPV-B (185/387, 48%) co-circulated, one HMPV-A/B was observed. The remaining 20/407 (5%) HMPV could not be characterised due to low RNA quality or low viral load. HMPV-B (61%) predominated during the 2014-2015 season and HMPV-A (62%) during 2015-2016. No difference in circulation was observed during the 2016-2017 season (Table 3 ).
Table 3.
Distribution of genotypes and lineages throughout the study period.
| Genetic group |
Season |
Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2014-2015 | 2015 | 2015-2016 | 2016 | 2016-2017 | ||||||||
| A1 | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| A2a | 9 | 10% | 1 | 20% | 0 | 0% | 0 | 0% | 1 | 1% | 11 | 3% |
| A2b | 12 | 14% | 0 | 0% | 22 | 16% | 1 | 10% | 2 | 1% | 37 | 10% |
| A2c | 13 | 15% | 0 | 0% | 65 | 46% | 8 | 80% | 67 | 47% | 153 | 40% |
| A2cwt | 11 | 85% | 0 | 0% | 46 | 71% | 6 | 75% | 31 | 46% | 94 | 61% |
| A2c180dup | 2 | 15% | 0 | 0% | 18 | 28% | 1 | 12.5% | 25 | 37% | 46 | 30% |
| A2c111dup | 0 | 0% | 0 | 0% | 1 | 1% | 1 | 12.5% | 11 | 17% | 13 | 9% |
| B1 | 37 | 42% | 4 | 80% | 22 | 16% | 1 | 10% | 42 | 29% | 106 | 27% |
| B2 | 17 | 19% | 0 | 0% | 31 | 22% | 0 | 0% | 31 | 22% | 79 | 20% |
| B2a | 5 | 29% | 0 | 0% | 2 | 6% | 0 | 0% | 3 | 10% | 10 | 13% |
| B2b | 12 | 71% | 0 | 0% | 29 | 94% | 0 | 0% | 28 | 90% | 69 | 87% |
| Total | 88 | 100% | 5 | 100% | 140 | 100% | 10 | 100% | 143 | 100% | 386 | 100% |
A2cwt: A2c strains without any duplication
A2c180dup: A2c strains carrying the 180-nucleotide duplication
A2c111dup: A2c strains carrying the 111-nucleotide duplication
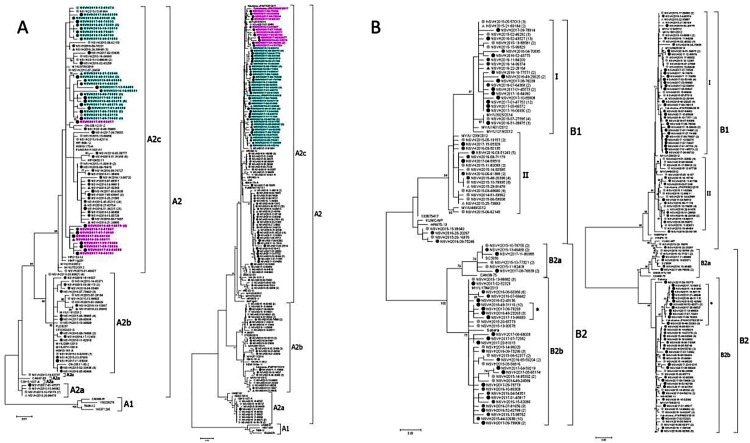
Phylogenetic analyses of F (382, 94%) and G (365, 90%) sequences (Fig. 2 ) showed congruent results. Overall, 11 (3%) samples belonged to A2a, 37 (10%) to A2b, 153 (40%) to A2c, 106 (27%) to B1 and 79 (20%) to B2 (Table 3).
Fig. 2.
Phylogenetic trees. Phylogenetic trees of F and G sequences from (A) HMPV-A and (B) HMPV-B, respectively. The analyses were performed on G sequences from nucleotide position 6340–6891 in reference to CAN97-83 strain (accession number AY297749) for HMPV-A and 6319-6921 CAN98-75 (accession number AY297748) for HMPV-B. On F sequences, nucleotide positions were 3846-4287 in reference to CAN97-83 strain and 3843-4284 in reference to CAN98-75. All 4 phylogenetic trees were inferred by using the maximum likelihood method, based on the Kimura 2-parameter for the F gene and General Time Reversible for the G gene. Numbers at the tree branch nodes represent the measure of support calculated by the bootstrap method (1000 replicates); only those exceeding 70% are shown. Sequences are marked with solid circles or triangles depending on whether they were collected during the seasonal or the interseasonal period (2014-2015 and 2015 in light grey, 2015-2016 and 2016 in dark grey and 2016-2017 in black). A2c sequences with duplications in the G protein have their name in bold turquoise for the 180-nucleotide duplication and in bold pink for the 111-nucleotide duplications.
Genetic characterisation of A2 G revealed that A2a and A2c sequences generally had a length of 220 aa, and A2b of 218 due to premature stop codons. Genetic characterisation of 153 A2c strains revealed the presence of the novel 180- (A2c180dup; 46; 30%) and 111-nucleotide (A2c111dup; 13; 9%) duplications into G ectodomain with increasing prevalence (Table 3). While all A2c180dup clustered together, two subgroups could be observed in the F phylogenetic tree (Fig. 2). Differently, A2c111dup G clustered into 2 groups but their F genes clustered together (except NSVH2017-09-82477).
B1 G clustered into two phylogenetic groups (I and II), differing in the acquisition of a premature stop codon in the 232 aa (relative to KU375606) in all strains belonging to group II (Fig. 2), but one (NSVH2015-12-87728). In addition, two sequences presented deletions of one (NSVH2017-04-59510) or two aa (NSVH2016-03-50135). Genetic characterisation of 10 B2a and 69 B2b G sequences revealed the acquisition of short duplications. Whereas B2a group, represented by CAN98-75 strain (AY297748), presented the aa KE in the 160-161 positions, B2b group presented 1 or 2 duplications (KEKE and KEKEKE). Also, a group differed from the rest of B2b group presenting an R after only one KE, which could probably be a mutation of the duplication of K (Fig. 2).
The sequences of the present study were submitted to GenBank (MN617398-MN617753).
3.3. Structural biology of G protein
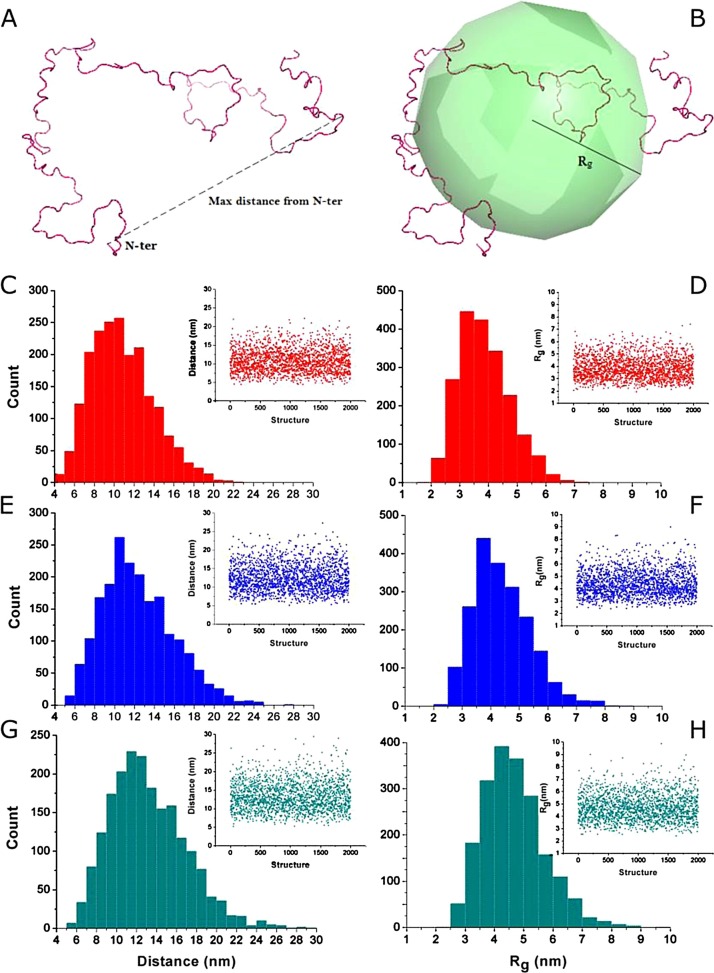
The ectodomain ensemble of the non-glycosylated form of G protein of NSVH2015-06-62150 (A2cwt) was simulated [[18], [19], [20]], resulting in a composition of conformations with maximal length (Dmax) of 4.5-22.2 nm and radii of gyration (Rg) of 1.9-7.4 nm (Fig. 3 ).
Fig. 3.
Maximum distance and radius of gyration analyses of the disordered ensembles of G protein. A) Representation of the maximum distance measured between the N atom of the first extracellular residue (Asn52) and the more distant atom to this. B) Depiction of the radius of gyration calculated for an individual conformation. C, E, G) Histograms of the maximum distances measured in the disordered ensemble for the wild type, 111- and 180-nucleotide duplications variants of the G protein, respectively. Insets at the right-hand part of each panel depict scatter-plots of maximum distances versus structure. D, F, H) Histograms of the radius of gyration of structures in the disordered ensemble for the wild type, 111- and 180-nucleotide duplications variants of the G protein, respectively. Similarly, insets at the right-hand part of each panel show scatter-plots of radius of gyration versus structure of each ensemble.
The MetaDisorder server predicted that both duplications were as fully disordered as A2cwt, with no self-aggregation segments. Also, the glycosylation pattern showed a similar distribution of the numerous O-glycosylation sites (Fig. 4 ), with 23-26 additional O-glycosylation sites in A2c180dup and 12-13 in A2c111dup.
Fig. 4.
Glycosylation pattern of HMPV-A G protein. Multiple alignment of deduced G amino acid sequences of HMPV-A. Only positions reflecting an amino acid change or a putative N- or O-glycosylated site are shown, amino acids were numerated following the reference sequence CAN99-81 (accession number AY574224). Amino acid positions 40 and 41 correspond to the transmembrane domain, while the remaining positions correspond to the ectodomain. The 180-nucleotide duplications in the ectodomain are framed in a turquoise box, the 111-nucleotide duplications are in a pink box. N- and O-glycosylated sites are marked in purple.
Once verified that duplications did not differ from A2cwt, 2000-conformation unfolded ensembles were generated for NSVH2017-09-78834 (A2c111dup) and NSVH2015-19-63118 (A2c180dup). A2c111dup increased size to Dmax 5.1-27.3 nm and Rg 2.4-9.0 nm, while A2c180dup increased Dmax 5.2-29.4 nm and Rg 2.4-9.9 nm (Fig. 3).
The pre-fusion conformation of the F trimer is calculated to protrude 13 nm [21]. According to the distance distributions of the three ensembles, the actual fraction of G protein’s ectodomain protruding more than 13 nm from the membrane amounts to 23% in the A2cwt, and it increases to 39% in A2c111dup and 46% and A2c180dup.
3.4. Clinical impact of human metapneumovirus
Due to the absence of clinical information (2), non-amplification (20) or manifestation of other syndromes rather than URTI or LRTI (20), clinical features of 203 paediatric and 162 adult cases were finally studied (Table 4 ).
Table 4.
Clinical features of patients infected with HMPV.
| Factor | Pediatric cases |
Adult cases |
||||
|---|---|---|---|---|---|---|
| URTI (n = 42) | LRTI (n = 161) | p | URTI (n = 77) | LRTI (n = 85) | p | |
| Season* | 0.908 | 0.020 | ||||
| 2014-2015 | 10 (25.6 %) | 42 (26.1 %) | 14 (18.4 %) | 19 (22.6 %) | ||
| 2015-2016 | 10 (25.6 %) | 44 (27.3 %) | 46 (60.5 %) | 33 (39.3 %) | ||
| 2016 | 1 (2.56 %) | 9 (5.59 %) | 0 (0.00 %) | 0 (0.00 %) | ||
| 2016-2017 | 18 (46.2 %) | 66 (41.0 %) | 16 (21.1 %) | 32 (38.1 %) | ||
| Age | 2.18 [0.88;6.41] | 1.17 [0.49;2.61] | 0.011 | 66.8 [53.7;77.1] | 74.7 [61.3;82.8] | 0.001 |
| Age group | 0.001 | 0.031 | ||||
| 0-2 years old | 19 (45.2 %) | 95 (59.0 %) | 0 (0.00 %) | 0 (0.00 %) | ||
| 2-5 years old | 10 (23.8 %) | 52 (32.3 %) | 0 (0.00 %) | 0 (0.00 %) | ||
| 5-15 years old | 11 (26.2 %) | 14 (8.70 %) | 0 (0.00 %) | 0 (0.00 %) | ||
| 15-64 years old | 2 (4.76 %) | 0 (0.00 %) | 34 (43.6%) | 23 (27.4%) | ||
| >64 years old | 0 (0.00 %) | 0 (0.00 %) | 44 (56.4%) | 61 (72.6%) | ||
| Sex | 1.000 | 0.197 | ||||
| Female | 18 (42.9 %) | 67 (41.6 %) | 45 (58.4 %) | 40 (47.1 %) | ||
| Male | 24 (57.1 %) | 94 (58.4%) | 32 (41.6%) | 45 (52.9 %) | ||
| Subgenotype | 0.763 | 0.052 | ||||
| A/B | 0 (0.00 %) | 0 (0.00 %) | 0 (0.00 %) | 1 (1.18 %) | ||
| A2 | 24 (57.1 %) | 83 (51.6 %) | 37 (48.1 %) | 46 (54.1 %) | ||
| B1 | 11 (26.2 %) | 44 (27.3 %) | 17 (22.1 %) | 26 (30.6 %) | ||
| B2 | 7 (16.7 %) | 34 (21.1 %) | 23 (29.9 %) | 12 (14.1 %) | ||
| Sublineage | 0.032 | 0.246 | ||||
| A2a | 5 (11.9%) | 2 (1.2%) | 1 (1.3 %) | 2 (2.4 %) | ||
| A2b | 6 (14.3%) | 14 (8.7%) | 7 (9.1 %) | 7 (8.3 %) | ||
| A2c | 13 (31.0%) | 67 (41.6 %) | 29 (37.7 %) | 37 (44.0 %) | ||
| B1 | 11 (26.2) | 44 (27.3%) | 17 (22.1%) | 26 (31.0%) | ||
| B2a | 0 (0.00 %) | 5 (3.1%) | 3 (3.9 %) | 2 (2.4 %) | ||
| B2b | 7 (16.7%) | 29 (18.0%) | 20 (26.0 %) | 10 (11.9%) | ||
| Duplication | 0.768 | 0.048 | ||||
| 111 | 1 (2.4 %) | 5 (3.1%) | 1 (1.3 %) | 6 (7.1 %) | ||
| 180 | 6 (14.3%) | 16 (9.9 %) | 7 (9.1 %) | 15 (17.6 %) | ||
| no | 35 (83.3%) | 140 (87.0%) | 69 (89.6%) | 64 (75.3%) | ||
| Comorbidities | 0.046 | 0.937 | ||||
| Yes | 13 (31.0 %) | 80 (49.7 %) | 63 (81.8 %) | 71 (83.5 %) | ||
| Non | 29 (69.0 %) | 81 (50.3 %) | 14 (18.2 %) | 14 (16.5 %) | ||
| Respiratory comorbidities | <0.001 | 0.799 | ||||
| Asthma | 1 (2.38 %) | 32 (19.9 %) | 0 (0.00 %) | 0 (0.00 %) | ||
| Pneumopathy | 0 (0.00 %) | 20 (12.42%) | 0 (0.00 %) | 0 (0.00 %) | ||
| EPOC | 0 (0.00 %) | 0 (0.00 %) | 15 (19.5 %) | 19 (22.4 %) | ||
| Non | 41 (97.6 %) | 109 (67.7 %) | 62 (80.5 %) | 66 (77.6 %) | ||
| Cardiopathy | 0.532 | <0.001 | ||||
| Yes | 2 (4.76 %) | 14 (8.70 %) | 11 (14.3 %) | 35 (41.2 %) | ||
| Non | 40 (95.2 %) | 147 (91.3 %) | 66 (85.7 %) | 50 (58.8 %) | ||
| Oncohematology | 0.276 | 0.022 | ||||
| Yes | 4 (9.52 %) | 8 (4.97 %) | 31 (40.3 %) | 19 (22.4 %) | ||
| Non | 38 (90.5 %) | 153 (95.0 %) | 46 (59.7 %) | 66 (77.6 %) | ||
| Immunodepression | 0.008 | 0.203 | ||||
| Immunodeficiency | 1 (2.38 %) | 2 (1.24 %) | 23 (29.9 %) | 17 (20.0 %) | ||
| TPH | 3 (7.14 %) | 0 (0.00 %) | 0 (0.00 %) | 0 (0.00 %) | ||
| Non | 38 (90.5 %) | 159 (98.8 %) | 54 (70.1 %) | 68 (80.0 %) | ||
| Diabetes mellitus | . | 0.114 | ||||
| Yes | 0 (0.00 %) | 0 (0.00 %) | 12 (15.6 %) | 23 (27.1 %) | ||
| Non | 42 (100 %) | 161 (100 %) | 65 (84.4 %) | 62 (72.9 %) | ||
| Prematurity | 0.422 | . | ||||
| Yes | 3 (7.14 %) | 21 (13.0 %) | 0 (0.00 %) | 0 (0.00 %) | ||
| Non | 39 (92.9 %) | 140 (87.0 %) | 77 (100 %) | 85 (100 %) | ||
| Chronic kidney disease | . | 0.221 | ||||
| Yes | 0 (0.00 %) | 0 (0.00 %) | 9 (11.7 %) | 17 (20.0 %) | ||
| Non | 42 (100 %) | 161 (100 %) | 68 (88.3 %) | 68 (80.0 %) | ||
| Bacteria co-infection | 0.244 | 0.005 | ||||
| Yes | 4 (9.52 %) | 7 (4.35 %) | 10 (13.0 %) | 28 (32.9 %) | ||
| Non | 38 (90.5 %) | 154 (95.7 %) | 67 (87.0 %) | 57 (67.1 %) | ||
| Viral co-infection | 0.384 | 1000 | ||||
| Yes | 11 (26.2 %) | 30 (18.6 %) | 5 (6.49 %) | 5 (5.88 %) | ||
| Non | 31 (73.8 %) | 131 (81.4 %) | 72 (93.5 %) | 80 (94.1 %) | ||
| Antibiotic | <0.001 | <0.001 | ||||
| Yes | 7 (16.7 %) | 85 (52.8 %) | 42 (54.5 %) | 79 (92.9 %) | ||
| Non | 35 (83.3 %) | 76 (47.2 %) | 35 (45.5 %) | 6 (7.06 %) | ||
| Duplication vs the rest | 0.743 | 0.018 | ||||
| A2c w/ duplication | 7 (22.6 %) | 21 (17.9 %) | 8 (13.3 %) | 21 (36.2 %) | ||
| Other types | 24 (77.4 %) | 96 (82.1 %) | 69 (86.7 %) | 64 (63.8 %) | ||
| A2c sublineage | 0.202 | 0.034 | ||||
| A2c w/ duplication | 7 (53.8 %) | 21 (31.3 %) | 8 (10.4 %) | 21 (24.7 %) | ||
| A2c w/o duplication | 6 (46.2 %) | 46 (68.7 %) | 21 (89.6 %) | 16 (75.3 %) | ||
The median age of paediatric patients presenting LRTI (1.17; IQR 0.49-2.61) was significantly lower (p 0.011) than those with URTI (2.18; IQR 0.88-6.41). A2c lineage was more associated to LRTI (p 0.032) than other lineages, but A2c with duplications were not associated with a higher risk of LRTI compared to other strains (p 0.743 and 0.202).
The median age of adult patients presenting LRTI (74.7; IQR 61.3-82.8) was significantly higher (p 0.001) than those with URTI (66.8; IQR 53.7-77.1), each year of age increasing 1.03 times the odds of having LRTI. No lineages or subgenotypes were more associated to LRTI (p 0.052 and 0.246). Cardiopathy was associated to LRTI (OR 4.2, IC 95% 2-9.43, p < 0.001). A2c strains with duplications were significantly more associated to LRTI, compared to all other strains (OR 2.83, IC 95% 1.17-6.84, p 0.018) or to A2cwt (OR 3.45, IC 95% 1.22-9.77, p 0.034).
For the severity study, only patients hospitalised due to LRTI (176) were considered, being 116 (66%) paediatric (Table 5 ) and 60 (34%) adult patients (Table 6 ). Children infected with A2 were more likely to be admitted in the ICU (OR 5.14, IC 95% 1.06-24.95, p 0.031). No other variables were found to be significant.
Table 5.
Severity of HMPV disease in paediatric patients. HFNC = high flux nasal cannula; CO = conventional oxygenotherapy (low flow nasal cannula or oxygen mask); MV = mechanical ventilation.
| Pediatric patients | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Factor | Subgenotypes | Duplications vs other strains | A2c sublineage | |||||||
| A2 | B1 | B2 | p | A2c w/ duplication | Other strains | p | A2c w/ duplication | A2c w/o duplication | p | |
| Length hospital stay | 4.50 [3.00;7.75] | 5.00 [4.00;7.00] | 5.50 [3.00;7.25] | 0.880 | 5.00 [4.00;11.00] | 5.00 [3.00;7.00] | 0.199 | 5.00 [4.00;11.0] | 4.00 [3.00;6.00] | 0.166 |
| Respiratory support | 1.000 | 0.297 | 0.406 | |||||||
| Yes | 47 (81.0 %) | 27 (79.4 %) | 19 (79.2 %) | 14 (93.3%) | 79 (78.2%) | 14 (93.3 %) | 25 (78.1 %) | |||
| Non | 11 (19.0 %) | 7 (20.6 %) | 5 (20.8 %) | 1 (6.67%) | 22 (21.8%) | 1 (6.67 %) | 7 (21.9 %) | |||
| Type of respiratory support | 0.520 | 0.140 | 0.391 | |||||||
| HFNC | 13 (22.4 %) | 7 (20.6 %) | 7 (29.2 %) | 3 (20.0%) | 24 (23.8%) | 3 (20.0 %) | 7 (21.9 %) | |||
| CO | 28 (48.3 %) | 20 (58.8 %) | 10 (41.7 %) | 8 (53.3%) | 50 (49.5%) | 8 (53.3 %) | 16 (50.0 %) | |||
| MV | 6 (10.3 %) | 0 (0.00 %) | 2 (8.33 %) | 3 (20.0%) | 5 (5.00%) | 3 (20.0 %) | 2 (6.25 %) | |||
| Non | 11 (19.0 %) | 7 (20.6 %) | 5 (20.8 %) | 1 (6.67%) | 22 (21.8%) | 1 (6.67 %) | 7 (21.9 %) | |||
| Length respiratory support | 3.00 [2.00;5.75] | 4.00 [2.00;5.75] | 3.00 [2.00;6.00] | 0.874 | 3.00 [3.00;9.00] | 4.00 [2.00;5.00] | 0.208 | 3.00 [3.00;9.00] | 3.00 [2.00;4.25] | 0.122 |
| ICU admission | 0.031 | 0.633 | 1.000 | |||||||
| Yes | 9 (15.5 %) | 0 (0.00 %) | 2 (8.33 %) | 2 (13.3%) | 9 (8.9%) | 2 (13.3 %) | 5 (15.6 %) | |||
| Non | 49 (84.5 %) | 34 (100 %) | 22 (91.7 %) | 13 (86.7%) | 92 (91.1%) | 13 (86.7 %) | 27 (84.4 %) | |||
| Length ICU stay | 6.00 [4.00;7.00] | . [.;.] | 13.0 [9.00;17.0] | 0.340 | 9.00 [7.50;10.50] | 5.00 [4.00;7.00] | 0.340 | 9.00 [7.50;10.5] | 4.00 [4.00;6.00] | 0.155 |
| Exitus | . | . | . | |||||||
| Yes | 0 (0 %) | 0 (0 %) | 0 (0 %) | 0 (0 %) | 0 (0 %) | 0 (0%) | 0 (0%) | |||
| Non | 58 (100 %) | 34 (100 %) | 24 (100 %) | 15 (100%) | 101 (100%) | 15 (100 %) | 32 (100 %) | |||
Table 6.
Severity of HMPV disease in adult patients. HFNC = high flux nasal cannula; CO = conventional oxygenotherapy (low flow nasal cannula or oxygen mask); MV = mechanical ventilation
| Adult patients | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Factor | Subgenotypes | Duplications vs other strains | A2c sublineage | |||||||
| A2 | B1 | B2 | p | A2c w/ duplication | Other strains | p | A2c w/ duplication | A2c w/o duplication | p | |
| Length hospital stay | 4.00 [2.00;9.00] | 9.00 [2.00;12.0] | 4.00 [2.00;7.25] | 0.327 | 4.00 [2.00;9.00] | 6.00 [2.00;10.50] | 0.442 | 4.00 [2.00;9.00] | 2.50 [2.00;7.50] | 0.787 |
| Respiratory support | 0.655 | 0.639 | 0.596 | |||||||
| Yes | 30 (90.9 %) | 15 (88.2 %) | 8 (80.0 %) | 11 (84.6%) | 42 (89.4%) | 11 (84.6 %) | 13 (92.9 %) | |||
| Non | 3 (9.09 %) | 2 (11.8 %) | 2 (20.0 %) | 2 (15.4%) | 5 (10.6%) | 2 (15.4 %) | 1 (7.14 %) | |||
| Type of respiratory support | 0.145 | 0.780 | 0.450 | |||||||
| HFNC | 4 (12.1 %) | 1 (5.88 %) | 1 (10.0 %) | 2 (15.4%) | 4 (8.5%) | 2 (15.4 %) | 1 (7.14 %) | |||
| CO | 25 (75.8 %) | 9 (52.9 %) | 6 (60.0 %) | 8 (61.5%) | 32 (68.1%) | 8 (61.5 %) | 12 (85.7 %) | |||
| MV | 1 (3.03 %) | 5 (29.4 %) | 1 (10.0 %) | 1 (7.69%) | 6 (12.8%) | 1 (7.69 %) | 0 (0.00 %) | |||
| Non | 3 (9.09 %) | 2 (11.8 %) | 2 (20.0 %) | 2 (15.4%) | 5 (10.6%) | 2 (15.4 %) | 1 (7.14 %) | |||
| Length respiratory support | 2.00 [1.00;5.00] | 3.00 [1.00;10.0] | 1.00 [1.00;5.50] | 0.304 | 1.00 [1.00;4.00] | 2.00 [1.00;6.00] | 0.449 | 1.00 [1.00;4.00] | 1.00 [1.00;4.25] | 0.938 |
| ICU admission | 0.151 | 0.182 | 1.000 | |||||||
| Yes | 2 (6.06 %) | 4 (23.5 %) | 2 (20.0 %) | 0 (0%) | 8 (17%) | 0 (0.00 %) | 1 (7.14 %) | |||
| Non | 31 (93.9 %) | 13 (76.5%) | 8 (80.0 %) | 13 (100%) | 39 (83%) | 13 (100 %) | 13 (92.9 %) | |||
| Length ICU stay | 9.50 [6.75;12.2] | 11.0 [5.00;21.0] | 10.5 [8.75;12.2] | 0.939 | . [.;.] | 10.5 [5.5;15.25] | . | . [.;.] | 15.0 [15.0;15.0] | . |
| Exitus | 0.096 | 1.000 | 1.000 | |||||||
| Yes | 2 (6.06 %) | 1 (5.88 %) | 3 (30.0 %) | 1 (7.69%) | 5 (10.6%) | 1 (7.69 %) | 1 (7.14 %) | |||
| Non | 31 (93.9 %) | 16 (94.1 %) | 7 (70.0 %) | 12 (92.3%) | 42 (89.4%) | 12 (92.3 %) | 13 (92.9 %) | |||
4. DISCUSSION
This study reports recent data on prevalence, genetic diversity, structural biology of G protein and clinical features of HMPV in Barcelona, Spain.
The positivity rate of HMPV was similar to recent reports [4,22,23]. Interestingly, the prevalence in adults was similar or even higher than in children, which emphasizes the importance of HMPV in adults. HMPV prevalence increased throughout the three seasons, probably due to the higher implementation of molecular methods, though there might be an underestimation, as a large number of positive samples for HRSV and influenza by rapid assay were not tested for other respiratory viruses. Most co-infections were with rhinoviruses, adenoviruses and bocaviruses, as previously reported [23,24].
HMPV presented a clear seasonality, as previously described [2,6,24]. Interestingly, the last season presented a different pattern, showing two different peaks in one epidemic season without changes among circulating genotypes. Interestingly, the prevalence of HMPV was higher out of the HRSV/influenza epidemics in the first season but did not vary in the second and third seasons. This could be due to the higher implementation of PCR-based assays in detriment to the use of immunofluorescence assays. Moreover, the great majority of HMPV laboratory-confirmed samples during these epidemics were previously tested by rapid assays and had a negative result, which would suggest that there might be many more samples that would be positive for HMPV but are not tested due to a HRSV or influenza positive result when HMPV circulation is coincidental with influenza epidemics. Thus, HMPV prevalence could be underestimated due to the lack of search of this virus when samples are HRSV or influenza laboratory-confirmed during the epidemics.
Genetic characterisation revealed that both genotypes co-circulated with a shift in predominance, as expected [2]. However, there was an unpredicted co-predominance in the third season, which could be due to either an intermediate alternation of genotypes or the emergence of HMPV-A viruses with new antigenic features that would evade the immunity created on the previous season.
Congruent classification of both F and G genes was expected, as no genetic recombination has been described for HMPV. All subgenotypes were detected except A1, suggesting it has extinguished and been replaced by A2, according to previous studies [3]. According to the data of the present study, A2c lineage appears to be replacing A2a and A2b. Moreover, A2c strains with duplications might be replacing A2cwt in the near future, as they might present an improved mechanism of immune evasion. In fact, a group in Japan observed that A2c111dup had totally replaced the rest of A2 strains [25], including A2c180dup. Interestingly, our group has observed how both A2c111dup and A2c180dup have replaced together the rest of HMPV-A viruses, being the latter more prevalent [26].
Different lengths of G protein have been observed due to premature stop codons, as previously described [3]. A2b and A2c lineages included viruses with G proteins of 218 and 220 aa respectively; and two different genetic groups (I and II) could be distinguished within B1 subgenotype, with a difference of 10 aa in length, which might evolve into novel lineages. Also, nucleotide duplications can lengthen the G aa sequence, such as long duplications in A2c, and short duplications in B2. For B2 viruses, KE duplications or KER variants should be monitored next seasons to reveal whether they confer an evolutionary advantage. The deletions observed seem not to have been fixed in the viral population.
Once these A2c111dup and A2c180dup were described, one of the aims was to study their structural properties. G has a heavily glycosylated pattern [21], enhanced by the emergence of duplications that increase the number of potential glycosylation sites. Although it is a very disordered protein and seems to have numerous random conformations, a composition of these conformations could be done. This prediction suggests that both A2c180dup and A2c111dup proteins protrude more than A2cwt. This finding supports the hypothesis of Leyrat [21], who suggested that G protein had a shielding function towards F protein, masking its antigenic epitopes, and at the same time validates the hypothesis that these novel long duplications would enhance this immune evasion mechanism, as it would hide more efficiently F epitopes [8].
Sequences of the newly described A2c lineage [3,4] were compared to sequences of the previously described A2b1 and A2b2 sublineages [27,28] and clustered together; that is to say, A2b and A2c lineages are exactly the same as A2b1 and A2b2, respectively. This misunderstanding between the genetic classification used in several articles highlights the urgent need of an official classification, as well as universal criteria to define new genotypes or lineages.
Furthermore, clinical impact was also assessed. As in literature [2], LRTI is more common in children under 2 and adults over 65. Moreover, adults have an increase of 1.03 times the odds of suffering LRTI every passing year. The presence of chronic medical conditions as cardiopathy, more frequent in the elderly, may be responsible for this, so HMPV should be tightly surveilled in these cases. Comorbidities are also associated with LRTI in children, especially respiratory comorbidities and immunodepression. In this study, prematurity and cardiopathies were not associated with a major risk of developing LRTI in children, in opposite to previous studies [[29], [30], [31], [32]].
Paediatric and adult patients underwent more antibiotic treatment when manifesting LRTI than URTI. However, only 8% of children and 30% of adults treated with antibiotics had a positive bacterial culture. Hence, over-antibiotic prescription is still reported.
Regarding infections by A2c, children seemed to be as affected by A2c with duplications as by A2cwt or other lineages, as it is probably a primary infection. Instead, A2c with duplications were more associated with LRTI in adults than A2cwt or other lineages. Although adults should have an efficient immune response [6], they have 3.45 times more odds of manifesting LRTI when infected by A2c with duplication than by A2cwt. This suggests that it might be acting as a primary infection, which supports the hypothesis of G protein’s steric shielding over F protein. HMPV is known for the many immune evasion strategies it has, so this could be a new mechanism developed in recent years [33,34]. Whether strains with duplication cause more severe disease could be demonstrated neither in children nor in adults.
The increasing prevalence of viral variants carrying a duplication into the ectodomain of the G protein throughout the study period, the association of A2c111dup and A2c180dup with more severe disease in adults, and the prediction of an enhancing steric shielding of the G protein masking antigenic epitopes of the F protein suggest that these duplications might confer an evolutionary advantage contributing to the immune evasion during the infection. This mechanism would be similar to that described for other viruses which have been reported to evade the immune response due to the glycosylation they present in their envelopes [35]. Given that F protein is the main target for most vaccine strategies currently under development, the fact that it could be masked by G should be taken into account.
Declaration of Competing Interest
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
This study was supported by the Spanish Ministry of Economy and Competitiveness (grants BFU2016-78232-P), Instituto de Salud Carlos III and by the European Regional Development Fund, through the Interreg V-A programme: POCTEFA 2014-2020 (grant Pirepred EFA086/15). It was also co-financed by the European Development Regional Fund (ERDF) "A way to achieve Europe", Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0003). We also would like to acknowledge the Statistics and Bioinformatics Unit (UEB) in Vall d’Hebron Research Institute (VHIR) for the statistical analyses.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104590.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Panda S., Mohakud N.K., Pena L., Kumar S. Human metapneumovirus: Review of an important respiratory pathogen. Int. J. Infect. Dis. 2014;25:45–52. doi: 10.1016/j.ijid.2014.03.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shafagati N., Williams J. Human metapneumovirus - what we know now. F1000Research. 2018;7:135. doi: 10.12688/f1000research.12625.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jagušić M., Slović A., Ljubin-Sternak S., Mlinarić-Galinović G., Forčić D. Genetic diversity of human metapneumovirus in hospitalized children with acute respiratory infections in Croatia. J. Med. Virol. 2017;89:1885–1893. doi: 10.1002/jmv.24884. [DOI] [PubMed] [Google Scholar]
- 4.Chow W.Z., Chan Y.F., Oong X.Y., Ng L.J., Nor’E S.S., Ng K.T., Chan K.G., Hanafi N.S., Pang Y.K., Kamarulzaman A., Tee K.K. Genetic diversity, seasonality and transmission network of human metapneumovirus: identification of a unique sub-lineage of the fusion and attachment genes. Sci. Rep. 2016;6:27730. doi: 10.1038/srep27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr M.J., Waters A., Fenwick F., Toms G.L., Hall W.W., O’Kelly E. Molecular epidemiology of human metapneumovirus in Ireland. J. Med. Virol. 2008;80:510–516. doi: 10.1002/jmv. [DOI] [PubMed] [Google Scholar]
- 6.Kumar P., Srivastava M. Prophylactic and therapeutic approaches for human metapneumovirus. Virus Dis. 2018;29:434–444. doi: 10.1007/s13337-018-0498-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papenburg J., Carbonneau J., Isabel S., Bergeron M.G., Williams J.V., De Serres G., Hamelin M.-È., Boivin G. Genetic diversity and molecular evolution of the major human metapneumovirus surface glycoproteins over a decade. J. Clin. Virol. 2013;58:541–547. doi: 10.1016/j.jcv.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 8.Piñana M., Vila J., Gimferrer L., Valls M., Andrés C., Codina M.G.M.G., Ramón J., Martín M.C.M.C., Fuentes F., Saiz R., Alcubilla P., Rodrigo C., Pumarola T., Antón A. Novel human metapneumovirus with a 180-nucleotide duplication in the G gene. Future Microbiol. 2017;12:565–571. doi: 10.2217/fmb-2016-0211. [DOI] [PubMed] [Google Scholar]
- 9.Piñana M., Andrés C., Gimferrer L., Codina M.G., Fuentes F., del M., Martín C., Rubio S., Alcubilla P., Isern A., Osuna M., Pumarola T., Antón A. A novel human metapneumovirus carrying a 111-nucleotide duplication within the G gene detected at a tertiary university hospital in Catalonia since the 2015-2016 season. 20th ESCV Annu. Meet. 2017 p. 52. [Google Scholar]
- 10.Saikusa M., Kawakami C., Nao N., Takeda M., Usuku S., Sasao T., Nishimoto K., Toyozawa T. 180-nucleotide duplication in the G gene of human metapneumovirus A2b subgroup strains circulating in Yokohama city, Japan, since 2014. Front. Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saikusa M., Nao N., Kawakami C., Usuku S., Sasao T., Toyozawa T., Takeda M., Okubo I. A novel 111-nucleotide duplication in the G gene of human metapneumovirus. Microbiol. Immunol. 2017;61:507–512. doi: 10.1111/1348-0421.12543. [DOI] [PubMed] [Google Scholar]
- 12.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glez-Peña D., Gómez-Blanco D., Reboiro-Jato M., Fdez-Riverola F., Posada D. ALTER: Program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010;38:14–18. doi: 10.1093/nar/gkq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozlowski L.P., Bujnicki J.M. MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics. 2012;13:1. doi: 10.1186/1471-2105-13-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh I., Seno F., Tosatto S.C.E., Trovato A. PASTA 2.0: An improved server for protein aggregation prediction. Nucleic Acids Res. 2014;42:301–307. doi: 10.1093/nar/gku399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta R., Jung E., Brunak S. 2004. Prediction of N-glycosylation sites in human proteins, Prep. [Google Scholar]
- 17.Steentoft C., Vakhrushev S.Y., Joshi H.J., Kong Y., Vester-Christensen M.B., Schjoldager K.T.-B.G., Lavrsen K., Dabelsteen S., Pedersen N.B., Marcos-Silva L., Gupta R., Paul Bennett E., Mandel U., Brunak S., Wandall H.H., Levery S.B., Clausen H. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estrada J., Bernadó P., Blackledge M., Sancho J. ProtSA: A web application for calculating sequence specific protein solvent accessibilities in the unfolded ensemble. BMC Bioinformatics. 2009;10:1–8. doi: 10.1186/1471-2105-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernado P., Blanchard L., Timmins P., Marion D., Ruigrok R.W.H., Blackledge M. A structural model for unfolded proteins from residual dipolar couplings and small-angle x-ray scattering. Proc. Natl. Acad. Sci. 2005;102:17002–17007. doi: 10.1073/pnas.0506202102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernadó P., Blackledge M., Sancho J. Sequence-specific solvent accessibilities of protein residues in unfolded protein ensembles. Biophys. J. 2006;91:4536–4543. doi: 10.1529/biophysj.106.087528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leyrat C., Paesen G., Charleston J., Renner M., Grimes J. Structural insights into the human metapneumovirus glycoprotein ectodomain. J. Virol. 2014;88:11611–11616. doi: 10.1128/JVI.01726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reiche J., Jacobsen S., Neubauer K., Hafemann S., Nitsche A., Milde J., Wolff T., Schweiger B. Human metapneumovirus: Insights from a ten-year molecular and epidemiological analysis in Germany. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J.Y., Yun K.W., Lim J.W., Lee M.K., Lim I.S., Choi E.S. Clinical and genetic features of human metapneumovirus infections in children. Pediatr. Int. 2015:22–26. doi: 10.1111/ped.12782. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L., Liu W., Liu D., Chen D., Tan W., Qiu S., Xu D., Li X., Liu T., Zhou R. Epidemiological and clinical features of human metapneumovirus in hospitalised paediatric patients with acute respiratory illness: a cross- sectional study in Southern China, from 2013 to 2016. BMJ Open. 2018;8:6–12. doi: 10.1136/bmjopen-2017-019308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saikusa M., Nao N., Kawakami C., Usuku S., Tanaka N., Tahara M., Takeda M., Okubo I. Predominant detection of the subgroup A2b human metapneumovirus strain with 111-nucleotide duplication in Yokohama City, Japan in 2018. Jpn. J. Infect. Dis. 2019:350–352. doi: 10.7883/yoken.jjid.2019.124. [DOI] [PubMed] [Google Scholar]
- 26.Piñana M., Andrés C., Gimferrer L., Codina M.G., del M., Martín C., Esperalba J., Fuentes F., Rubio S., Alcubilla P., Pumarola T., Anton A. Human metapneumovirus: are the new duplications within the G gene responsible for doubling its prevalence? 21st Congr. Eur. Soc. Clin. Virol. 2018 [Google Scholar]
- 27.Regev L., Meningher T., Hindiyeh M., Mendelson E., Mandelboim M. Increase human metapneumovirus mediated morbidity following pandemic influenza infection. PLoS One. 2012;7:2–7. doi: 10.1371/journal.pone.0034750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neemuchwala A., Duvvuri V.R., Marchand-Austin A., Li A., Gubbay J.B. Human metapneumovirus prevalence and molecular epidemiology in respiratory outbreaks in Ontario, Canada. J. Med. Virol. 2015;87:269–274. doi: 10.1002/jmv.24024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuster J.E., Khuri-Bulos N., Faouri S., Shehabi A., Johnson M., Wang L., Fonnesbeck C., Williams J.V., Halasa N. Human Metapneumovirus Infection in Jordanian Children. Pediatr. Infect. Dis. J. 2015;34:1335–1341. doi: 10.1097/inf.0000000000000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pancham K., Sami I., Perez G.F., Huseni S., Kurdi B., Rose M.C., Rodriguez-Martinez C.E., Nino G. Human metapneumovirus infection is associated with severe respiratory disease in preschool children with history of prematurity. Pediatr. Neonatol. 2016;57:27–34. doi: 10.1016/j.pedneo.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papenburg J., Hamelin M.È., Ouhoummane N., Carbonneau J., Ouakki M., Raymond F., Robitaille L., Corbeil J., Caouette G., Frenette L., De Serres G., Boivin G. Comparison of risk factors for human metapneumovirus and respiratory syncytial virus disease severity in young children. J. Infect. Dis. 2012;206:178–189. doi: 10.1093/infdis/jis333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis C.R., Stockmann C., Pavia A.T., Byington C.L., Blaschke A.J., Hersh A.L., Thorell E.A., Korgenski K., Daly J., Ampofo K. Incidence, morbidity, and costs of human metapneumovirus infection in hospitalized children. J. Pediatric Infect. Dis. Soc. 2016;5:303–311. doi: 10.1093/jpids/piv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Céspedes P.F., Palavecino C.E., Kalergis A.M., Bueno S.M. Modulation of host immunity by the human metapneumovirus. Clin. Microbiol. Rev. 2016;29:795–818. doi: 10.1128/CMR.00081-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolli D., Bao X., Casola A. Human metapneumovirus antagonism of innate immune responses. Viruses. 2012;4:3551–3571. doi: 10.3390/v4123551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook J.D., Lee J.E. The Secret Life of Viral Entry Glycoproteins: Moonlighting in Immune Evasion. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.