Abstract
Rationale
Non-typeable Haemophilus influenzae (NTHi) is a common inhabitant of the human nasopharynx and upper airways that can cause opportunistic infections of the airway mucosa including bronchopulmonary infections in patients with chronic obstructive pulmonary disease (COPD). It is clear that opportunistic infections contribute significantly to inflammatory exacerbations of COPD; however, there remains much to be learned regarding specific host and microbial determinants of persistence and/or clearance in this context.
Methods
In this study, we used a recently described ferret model for COPD, in which animals undergo chronic long-term exposure to cigarette smoke, to define host–pathogen interactions during COPD-related NTHi infections.
Results
NTHi bacteria colonised the lungs of smoke-exposed animals to a greater extent than controls, and elicited acute host inflammation and neutrophilic influx and activation, along with a significant increase in airway resistance and a decrease in inspiratory capacity consistent with inflammatory exacerbation; notably, these findings were not observed in air-exposed control animals. NTHi bacteria persisted within multicellular biofilm communities within the airway lumen, as evidenced by immunofluorescent detection of bacterial aggregates encased within a sialylated matrix as is typical of NTHi biofilms and differential bacterial gene expression consistent with the biofilm mode of growth.
Conclusions
Based on these results, we conclude that acute infection with NTHi initiates inflammatory exacerbation of COPD disease. The data also support the widely held hypothesis that NTHi bacteria persist within multicellular biofilm communities in the lungs of patients with COPD.
Short abstract
Infection of smoke-exposed ferrets with COPD results in mucus obstruction and respiratory symptoms as in patients, and the bacteria are in a distinct mode of growth consistent with biofilms https://bit.ly/3euXpbQ
Lessons for clinicians
Patients with chronic obstructive pulmonary disease (COPD) experience a progressive decline in lung function over time that is accelerated by chronic and acute phases of infection, known as exacerbations. Bacterial exacerbations lay a significant burden on COPD patients, contributing significantly to morbidity and mortality. It is important to understand the complex host–microbe interactions at play during these exacerbations and improve modelling of the development of disease post exacerbation to better guide treatment. In this study, we provide clear evidence that non-typeable Haemophilus influenzae, a common opportunist in patients with COPD, forms mature biofilm structures within the lungs of smoke-exposed ferrets shortly after infection, concomitant with the development of key components of an inflammatory exacerbation event, including a decline in airway function, an increase in proteolytic and pro-inflammatory activity, and thickening of airway walls. The significance of this work is in identifying bacterial biofilms as a therapeutic target in COPD during exacerbations.
Introduction
Chronic obstructive pulmonary disease (COPD) is among the leading causes of death worldwide [1]. COPD is a collective term for progressive lung disorders that include emphysema, chronic bronchitis, and irreversible airway damage [1–3]. All presentations of COPD result in decreased lung function that progressively worsens over time [1] and can be accelerated by inflammatory exacerbations [4, 5]. Patients with COPD are frequently colonised throughout the upper and lower airways with opportunists that normally reside within the nasopharyngeal microbiome, including non-typeable Haemophilus influenzae (NTHi) [6–10]. NTHi is a gram-negative coccobacillus that is highly adapted to its normal niche in the human upper airways [6, 10]. In patients with impaired mucociliary defences, NTHi can cause opportunistic mucosal infections that can be chronic in nature [10, 11]. NTHi is known to reside within multicellular biofilm communities on the airway mucosa during some opportunistic infections, such as otitis media, and there are data to suggest that biofilms may also have relevance in COPD-related infections [10].
Smoke-exposed ferrets display many analogous phenotypes seen in the human COPD lung, including emphysema, bronchitis, and immunomodulation [12–15]. In addition, this animal model readily displays mucus hypersecretion, a phenotype absent from other commonly used COPD models, which is a major contributing factor to the pathogenesis of the disease [16–18]. The results utilising this novel model show that NTHi elicits symptoms consistent with a COPD exacerbation event, and also confirm that NTHi bacteria reside within biofilm communities within the lung. These findings confirm long-standing clinical observations regarding NTHi persistence in vivo. These findings provide important insights guiding development of future treatments for infection in COPD, and efforts on clearance of bacterial biofilms in particular could provide significant benefit.
Methods
Animals and cigarette smoke exposure
Ferrets (Mustela putorius furo) were acquired from Marshall BioResources (North Rose, NY, USA) and randomly assigned to experimental groups exposed to whole cigarette smoke or ambient air (control). Ferrets were administered cigarette smoke and monitored as described previously [12].
Strains and bacterial preparation
NTHi 86-028NP and NTHi 7P49H1 [13–16] were cultured on brain heart infusion agar (Difco) supplemented with nicotinamide adenine dinucleotide (10 µg·mL−1, Sigma; St Louis, MO, USA) and hemin (10 µg·mL−1, ICN Biochemical; Costa Mesa, CA, USA) (sBHI). Bacteria were harvested from the surface of overnight plate cultures and resuspended in PBS to a bacterial density of ∼108 CFU·mL−1 to generate the inoculum.
Ferret infections
Ferrets (Mustela putorius furo) were acquired from Marshall BioResources and randomly assigned to experimental groups exposed to whole cigarette smoke or ambient air (control). Ferrets were restrained in customised nose-only exposure tubes with a 24-port plenum connected to mainstream smoke output and were exposed to 60 min of smoke from 3R4F research cigarettes, twice daily for a period of 6 months. Cigarette smoke was generated by an automated cigarette smoke generator (InExpose model, SCIREQ, Montreal, Canada), with a 24-cigarette autoloading carousel. An in-line gas analyser for oxygen, carbon monoxide, and particulate matter provided real-time estimates of cigarette smoke intensity exposure. Animals were monitored continuously, and analytics demonstrated particulate matter (200 μg·L−1 of total particulate matter) and carbon monoxide levels (∼1%–3%) typical of other animal smoke exposure systems [12].
Bronchoalveolar lavage fluid collection
To collect bronchoalveolar lavage fluid (BALF), ferrets were anaesthetised as previously described and a bronchoscope inserted down the trachea to the lobar bronchi [12]. For females, 1 mL of PBS was injected into the lungs, after which, as much fluid as possible was re-collected. Males received 2 mL of PBS. BALF was collected pre-infection and 3 days and 7 days post-infection.
Bacterial load was assessed by plate-count. Briefly, BALF fluids were serially diluted and plated on sBHI containing 3 µg·mL−1 of vancomycin and incubated at 37°C overnight. Separate aliquots of BALF were used for quantitative PCR on the Applied-Biosystems 7500 System (Applied-Biosystems, Foster City, CA, USA), as an additional measure of bacterial counts. NTHi-specific probes were used; these are listed below in table 1. The reaction mix and cycling conditions were performed as described previously [17]. A standard curve of NTHi genomic DNA (gDNA) serially diluted from 20 000 pg to 0.2 pg was used for each run.
TABLE 1.
Primers and probes used for quantitative PCR bacterial enumeration
| Target gene | Primer/probe | Sequence (5′ to 3′) | Amplicon size bp |
| fucP | FucP F | GCCGCTTCTGAGGCTGG | 68 |
| FucP R | AACGACATTACCAATCCGATGG | ||
| FucP Probe | 6FAM-TCCATTACTGTTTGAAATAC–QSY | ||
| hpd3 | Hpd3 F | GGTTAAATATGCCGATGGTGTTTG | 151 |
| Hpd3 R | TGCATCTTTACGCACGGTGTA | ||
| Hpd3 Probe | VIC–TTGTGTACACTCCGTTGGTAAAAGAACTTGCAC-QSY | ||
| omp26 | Omp26 F | ACCGCACTTGCTTTAGGTATT | 105 |
| Omp26 R | GCGATCTGGGTGATGTTGAA | ||
| Omp26 Probe | JOE-TTGCTTCAGGCTATGCTTCCGCT-BHQ |
Micro computed tomography of ferrets and quantification of lung airways changes
All ferrets underwent micro computed tomography (µCT) imaging prior to infection and at 3 days and 7 days post-infection, using established procedures that have recently been described in detail [18]. Imaging was gated for inspiratory phase of respiration. Ferrets were placed under anaesthesia using 5% isoflurane, after which the lungs were imaged using MiLabs µCT (High-Resolution U-CTHR). Ferrets were imaged under ultra-focused magnification in gated scan mode with a 55-kV X-ray and 0.19-mA current. µCT images were reconstructed using MiLabs software (MiLabs, Utrecht, Netherlands) and then visualised using pMOD (PMOD Technologies, Zurich, Switzerland). Luminal diameter and bronchial wall thickness were measured in apical, medial, and caudal lobes of each lung.
Flexivent spirometry
Lung function in ferrets was determined utilising the Flexivent ventilator system, using module 5 according to the manufacturer's instructions (SCIREQ). Ferrets were sedated prior to intubation and then connected to the ventilator system. After collection of baseline parameters, resistance, compliance, elastance, inspiratory capacity, and pressure–volume loops were measured.
Myeloperoxidase levels
Myeloperoxidase (MPO) activity present in BALF and in lung homogenate was measured using the MPO activity assay kit (Abcam, Cambridge, UK) according to the manufacturer's instructions.
Zymography
A 7.5% polyacrylamide separating gel with 0.1% gelatin (Bio-rad, Hercules, CA, USA) was cast and allowed to solidify for 45 min. Samples were loaded into wells with 4× loading buffer (200 mM Tris-Cl (pH 6.8), 400 mM dithiothreitol (DTT), 8% SDS, 0.4% bromophenol blue, 40% glycerol) and electrophoresed (50 V for 30 min), then immersed in 50 mL of 2.5% Triton X-100, incubated with gentle shaking at room temperature for 30 min and washed three times with distilled water. The gel was then placed in approximately 200 mL of 50 mM Tris (pH 8.0) and 5 mM CaCl2 and shaken gently for at least 12 h at 37°C, stained for 1 h in 0.05% Coomassie Brilliant Blue R-250 in 45% methanol/10% acetic acid, destained in 45% methanol/10% acetic acid, washed with dH2O, and imaged. Relative protein activity was determined via densitometry using ImageJ Fiji [18].
Immunofluorescent staining
Ferret lung tissue was sectioned via cryostat (5 µm/section) and fixed onto glass slides. NTHi bacteria were stained using polyclonal rabbit antiserum and goat anti-rabbit IgG Alexa 488 secondary antibody conjugate. Cover slips were mounted using Prolong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) (Thermo Fisher, Waltham, MA, USA). Confocal laser scanning microscopic analyses were performed using a Nikon A1R TE2000 inverted microscope (Nikon, Tokyo, Japan).
NTHi sialylated moieties in biofilm matrix were stained utilising specific lectin conjugates, essentially as described previously [19]. Maackia amurensis lectin (MAA) Texas red conjugated specific for Neu5Ac α(2,3) galactose, and Sambucus nigra lectin (SNA) Texas red specific for Neu5Ac α(2,6)Gal/GalNAc were diluted to a final concentration of 100 µg·mL−1 in 0.01 M phosphate, 0.15 M NaCl, 0.05 M sodium azide buffer according to the manufacturer's instructions.
To image neutrophil extracellular traps (NETs), slides were stained with anti-citrullinated histone H3 (Abcam) and goat anti-rabbit Alexa 647 secondary antibody conjugate (Thermo Fisher). All lung images were taken from day 7 post-infection lung tissue sections.
Reverse transcriptase quantitative PCR of Haemophilus influenzae biofilm-associated genes
Bacterial RNA was extracted using the Monarch total RNA miniprep kit (New England Biolabs, Ipswich, MA, USA) following the manufacturer's guidelines. Reverse transcriptase quantitative PCR (RT-qPCR) was performed using the Applied-Biosystems 7500 System, and oligonucleotide probes specific for pdgX, luxS, dps, hktE, and omp26 (table 2). The omp26 transcript was chosen as an endogenous control given that its expression does not vary between planktonic or biofilm mode of growth (S1). The reaction mix used was the NEB Luna Universal following the manufacturer's directions for cycling conditions (New England Biolabs). All samples were run in duplicate. Transcript measures were normalised relative to omp26 levels from the same sample. Relative quantification of gene expression was determined using the comparative CT method (2ΔΔCT).
TABLE 2.
Primers and probes used for reverse transcriptase quantitative PCR gene expression analysis
| Target gene | Primer/probe | Sequence (5′ to 3′) | Amplicon size bp |
| pdgX | PdgX F | GCACTCGTCAGGGTGATAAA | 109 |
| PdgX R | GATGAGCAAGTTGGAGTGAATG | ||
| PdgX Probe | 6FAM-TGATCGTGTTCTCATTACCGGGCG-QSY | ||
| luxS | LuxS F | GAAGTGTCTGAGGCTTGGTTAG | 103 |
| LuxS R | CCGTATAGCTTCCGCATTGATAG | ||
| LuxS Probe | 6FAM-AGGTGTACAAGATCAAGCTTCTATTCCCG-QSY | ||
| dps | Dps F | GGGCTACCACTGGAACATTA | 104 |
| Dps R | GTTCAGCCACCTCATCTACTC | ||
| Dps Probe | 6FAM-TGCGTGTAATGCAAAGAAGTTTACGCC-QSY | ||
| hktE | HktE F | CAACAACCAGACTTTGCTGAAC | 104 |
| HktE R | ACGAGGTTGGCTGAAGTAATC | ||
| HktE Probe | 6FAM-TCGTATCAACGGAGATGCAGCACA-3IABkFQ | ||
| omp26 | Omp26 F | ACCGCACTTGCTTTAGGTATT | 104 |
| Omp26 R | GCGATCTGGGTGATGTTGAA | ||
| Omp26 Probe | JOE-TTGCTTCAGGCTATGCTTCCGCT-BHQ |
Statistics
Data was analysed by the one-way ANOVA with Tukey's multiple comparisons test. All bar graphs represent the mean and error bars represent sem. All ferret experiments were repeated at least three times. Data from multiple independent animal experiments were averaged together. Animal numbers in each group are denoted in the figure legends. p-values ≤0.05 were considered statistically significant.
Results
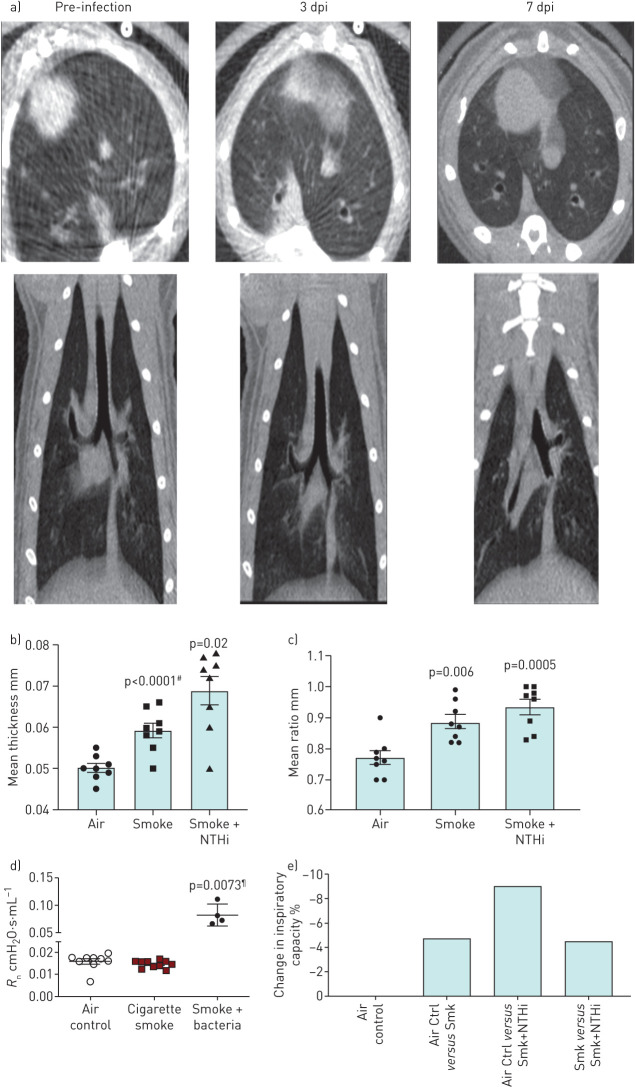
To investigate whether NTHi infection induced a COPD bacterial exacerbation event, µCT imaging, airway functional analysis, and histopathology were utilised. Airway infections in COPD cause structural changes, which include airway wall thickening from intraluminal inflammation and mucus expression. µCT analysis revealed that NTHi infection resulted in morphological changes within the lung. Evidence of airway wall thickening and classical “tree in bud” opacities was found, which are indicative of airway obstruction due to impaction within the bronchioles [20]. These airway changes were detectable by day 3 post-infection and remained present at day 7 post-infection (figure 1a). Quantification of these morphological changes showed that airway wall thickness and the ratio of thickness to diameter are significantly increased post-infection (figure 1b).
FIGURE 1.
Non-typeable Haemophilus influenzae (NTHi) infection induces significant morphological and functional decline in lung function in smoke-exposed ferret model of chronic obstructive pulmonary disease (COPD) indicative of an exacerbation event. Representative micro computed tomography (µCT) images of smoke-exposed ferret lungs infected with NTHi with quantification of airway walls. a) Axial view and posterior views of smoke-exposed ferret lungs prior to infection and post-infection at days 3 and 7. By day 3 post-infection (dpi) there is radiological evidence of consolidation (ground glass infiltrates), infection, and bronchial impaction, which is indicative of mucus build up in the bronchioles. b) Airway wall thickness and ratio of wall thickness to airway diameter of air controls, uninfected smoke-exposed ferrets, and infected smoke-exposed ferrets. #: comparison to air; ¶: comparison to smoke alone. Compared to air controls, smoke-exposed infected ferrets had significantly thicker airway walls and ratio of wall thickness to airway diameter indicating increased blockage and constriction. Mean±sem. n=8. c) Airway functional analysis of ferret airways. Central airway resistance is significantly increased in infected smoke-exposed animals compared to both smoke only and air controls as determined by one-way ANOVA. Mean±sem. n=4–10. Measurement of percent change in inspiratory capacity in pre- and post-infected smokers shows a synergistic effect between smoke exposure and bacterial infection.
Forced oscillatory spirometry was used to measure a variety of airway function parameters: tissue damping, central and single compartment airway resistance, and inspiratory capacity. Airway functional analysis did reveal a trend of worsening airway function in infected animals as indicated by multiple parameters, including airway resistance, tissue damping, and inspiratory capacity (figure 1c). Central airway resistance was significantly increased in smoke-exposed animals infected with NTHi, and infected animals displayed a synergistic effect with smoke resulting in a further decrease in inspiratory capacity.
Taken together, these data indicate that NTHi infection induces noticeable inflammation, airway wall thickening, and bronchiole impaction more than that caused by smoking alone, which contribute to a detectable decline in airway function, primarily by increasing central airway resistance and reducing inspiratory capacity.
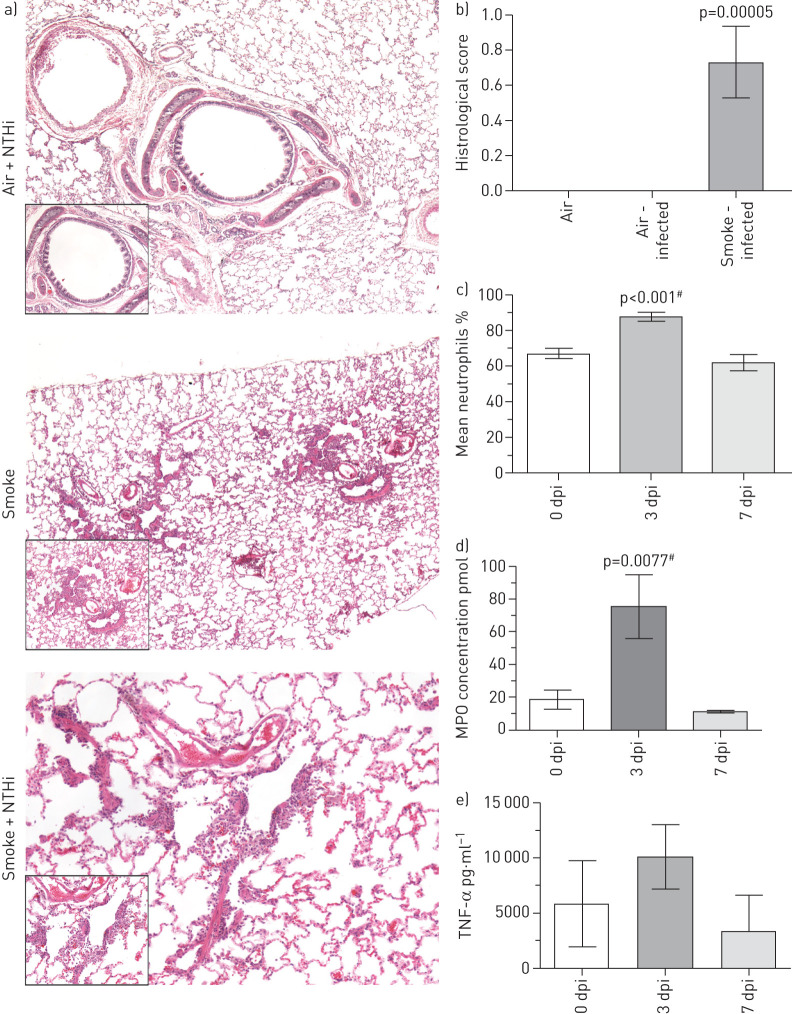
To gain further insight into the host response to NTHi infection in COPD, the inflammatory changes and immune cell influx post-infection with NTHi were compared by histopathological analysis of lung tissue sections. Infected smoke-exposed animals displayed significant neutrophilia and histological evidence of airway damage, particularly widening of the air spaces (figure 2a). By comparison, infected air control animals displayed healthy airways with no neutrophilia. NTHi infection resulted in a significant increase in mean neutrophil percent and counts present in the BALF that peaked at day 3 post-infection and returned to normal levels by day 7 (figure 2c). Lung macrophages and eosinophil levels were not significantly different post-infection. Additionally, MPO concentration present in the lungs was significantly increased at day 3 post-infection compared to pre-infection, which coincided with the neutrophil influx, indicating these infiltrating neutrophils were active (figure 2d).
FIGURE 2.
Smoke-exposed animals infected with non-typeable Haemophilus influenzae (NTHi) display hallmark immunological responses associated with chronic obstructive pulmonary disease (COPD) infection and exacerbation. a) Haematoxylin and eosin staining of ferret lungs. All large images are taken at 4×, while zoomed in sections were taken at 10×, except for the NTHi+Smoke, where the zoom is 10× and 20×, respectively. All lung sections were taken at day 7 post-infection (dpi). b) Quantification of histological scoring of stained lung sections. Mean±sem, n=4–9. Histological scores compared by one-way ANOVA. c) Mean percentage of neutrophils from total cell counts from smoke-exposed animals infected with either strain of NTHi at days 0, 3, and 7 post-infection. There is a significant increase in the percentage of the total cell counts that neutrophils take up by day 3 post-infection. This increased percentage decreases back to baseline levels by day 7 post-infection, which correlates with both mean neutrophil count and myeloperoxidase (MPO) concentration levels, which also decrease by this time. Mean±sem (n=10). d) Concentration of MPO present within bronchoalveolar lavage fluid (BALF) collected from smoke-exposed ferrets infected with H. influenzae 7P49H1. e) Tumour necrosis factor-α (TNF-α) levels present in ferrets infected with 7P49H1. Mean±sem (n=8). By day 3 post-infection there is a significant increase in the amount of MPO present within BALF as determined by one-way ANOVA. There was no significant difference between MPO concentration at day 0 and day 7 post-infection. These data coincide with persistence data for this strain where at 3 days post-infection smoke-exposed ferrets still maintain high CFUs of NTHi. In addition, these data indicate that a significant acute neutrophil-mediated inflammation is induced by NTHi. Mean±sem (n=4). #: versus day 0.
Additionally, there was an increase in the pro-inflammatory cytokine tumour necrosis factor-α (TNF-α) (figure 2e). The increase in TNF-α peaks at day 3 post-infection before returning to pre-infection levels later in the course of infection.
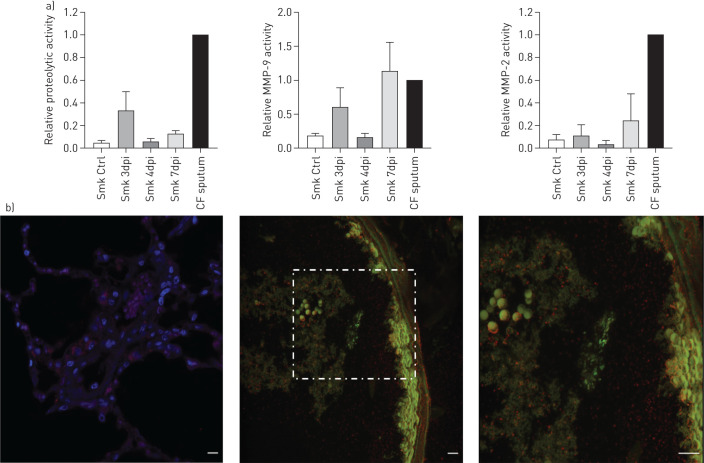
Using gel zymography, the overall proteolytic activity and the activity of two matrix metallopeptidases (MMPs), MMP-2 and MMP-9, were determined. Both MMPs are gelatinases/collagenases known to be increased in COPD patients. Cystic fibrosis (CF) sputum was used as a positive control given the highly active protease environment present in CF. Smoke exposure induced a small amount of proteolytic activity within the lungs, while bacterial infection further enhanced the proteolytic activity present in smoke-exposed lungs. Of the two MMPs, MMP-9 was induced at higher levels following bacterial infection than MMP-2 (figure 3a). Overall, these data indicate that NTHi infection in smoke-exposed ferrets induces significant pro-inflammatory, potentially damaging responses, similar to those seen in COPD patients experiencing NTHi infection or exacerbation.
FIGURE 3.
Non-typeable Haemophilus influenzae (NTHi) infection induces significant increases in proteolytic activity and neutrophil extracellular trap (NET) formation, which mimics what is seen in COPD patients and may contribute to airway damage and remodelling. a) Densitometric quantification of gel zymography of relative overall proteolytic activity, relative matrix metallopeptidase (MMP)-9 activity, and relative MMP-2 activity. The positive control used was cystic fibrosis (CF) sputum, represented by the well-labelled CF. Zymography showcasing proteolytic activity present in ferret bronchoalveolar lavage fluid (BALF). Smoke-exposed only animals harbour a baseline level of proteolytic activity that is higher than air controls. Infection seems to further induce proteolytic activity in the lungs. By comparison, infected smoke-exposed animals display more activity, as evidenced by an increase in the bands. This activity is most pronounced at day 3 post-infection and seems to wane during the following days. b) Immunofluorescent staining shows NETs surrounding NTHi within the lungs in the smoke-exposed animals only. Nuclei were stained with DAPI (blue) and bacteria were stained with anti-NTHi polyclonal antibodies (green), while NETs were stained using anti-Histone H3 Citrulline R2, R8, R17 (red). Images were taken at 60× magnification, while the right most panel is a 2× zoom of the region of interest indicated by the white box. dpi: day post-infection. Scale bars: 50 μm.
We also performed immunofluorescent microscopic analysis to compare the formation of NETs within ferret lungs. NETs were clearly visible within the lumen of the lungs of smoke-exposed animals following infection of a good portion of the lung environment, particularly around NTHi biofilms (figure 3b); this is consistent with prior work from our laboratory in other presentations of chronic NTHi infection [21]. NETs are generally ineffective at clearance of NTHi and may be contributing to the development of a pro-inflammatory lung environment and decline in airway function [21].
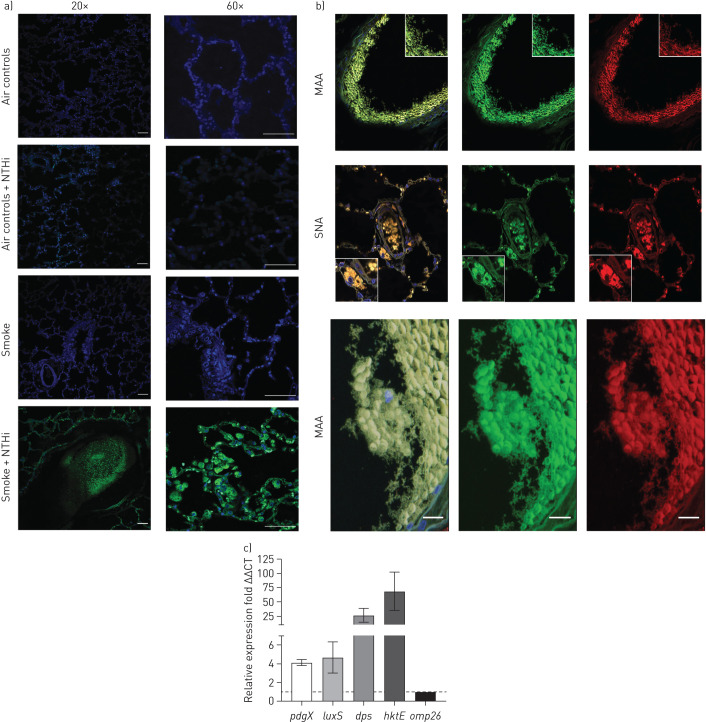
We next addressed the long-standing hypothesis that NTHi bacteria persist within biofilms within the airways during COPD-related infections [12, 22, 23]. Specifically, it has been demonstrated that NTHi can form adherent biofilms on the apical surfaces of airway epithelial cells, and the in vivo expression profile of NTHi shows an increase in biofilm-related genes, such as pdgX [22, 23]. In smoke-exposed ferret lungs, we observed abundant NTHi bacteria within multicellular aggregates on the mucosal surfaces, both in the lung parenchyma and air space, which is consistent with biofilm formation (figure 4a); these communities were reactive with lectins that are specific for the NTHi biofilm matrix. Images showing the staining of MAA and SNA alone are present in supplementary figure S2. Additionally, NTHi bacteria were observed adherent to the epithelial cell surface in smoke-exposed animals; the bacterial biofilms are thickest at the cell surface. In contrast, air control animals infected with NTHi lacked these bacterial aggregates and displayed only individual NTHi bacteria or small focal aggregates within the lungs.
FIGURE 4.
a) Confocal immunofluorescent imaging of ferret lungs. Smoke exposure enables non-typeable Haemophilus influenzae (NTHi) to form aggregates within the lungs. Nuclei were stained with DAPI (blue) and bacteria were stained with anti-NTHi polyclonal antibodies conjugated to Alexa 488 (green). Infected smoke-exposed animals exhibit bacterial aggregates both in the airway and lung parenchyma. By contrast, infected air control animals displayed few punctate NTHi in the lungs. Both uninfected smoke and air controls displayed no NTHi. Scale bars=50 µm. b) Confocal imaging of sialic acid lectin staining of NTHi biofilms. Nuclei were stained with DAPI (blue), bacteria were stained with anti-NTHi polyclonal antibodies conjugated to Alexa 488 (green), sialic α(2,3) galactose was stained with Maackia amurensis lectin (MAA) conjugated to Texas red (red), and Neu5Acα(2,6) Gal/GalNAc was stained with Sambucus nigra lectin (SNA) conjugated to Texas red (red). Images were taken at 20× zoom. Sialic acid staining overlaps with regions stained for NTHi suggesting biofilm formation is occurring within the airways during exacerbation; this is indicated by the yellow in the composite images. c) Relative expression of NTHi biofilm genes determined by reverse transcriptase quantitative PCR (RT-qPCR) in infected smoke-exposed animals. Three genes associated with NTHi biofilm growth were all significantly upregulated in comparison to the housekeeping gene omp26. Of interest, dps, a DNA-binding protein associated with resistance to environmental stress was very highly upregulated. hktE, the gene encoding for NTHi catalase, was also very highly upregulated, giving indications that NTHi was growing in a highly oxidatively stressful environment in the COPD lung. All samples were run in triplicate. Mean±sem. n=4. Scale bars: 50 μm.
To further define the NTHi communities observed in lung tissues, lectin staining utilising sialic acid-binding lectins (MAA and SNA) to test for carbohydrate linkages that have been associated with the NTHi biofilm matrix was performed. From the confocal images we observed significant colocalisation of NTHi extracellular matrix components (sialic acid) with NTHi bacterial aggregates. Indeed, both NTHi biofilm-specific sialic acid linkages stained for colocalised with NTHi within the airways of infected smoke-exposed ferrets, providing evidence that these aggregates are collections of NTHi within a biofilm matrix (figure 4b). In contrast, air controls lacked NTHi aggregates and displayed no evidence of bacterial biofilm components.
Gene expression profiles of NTHi bacteria in vivo were obtained using RT-qPCR. Expression of several NTHi genes associated with biofilm growth and resistance to environmental stress (pdgX, luxS, dps, and hktE) was measured and normalised relative to a constitutively expressed gene (omp26). Aggregates within infected smoke-exposed ferrets express a genetic profile indicative of biofilm formation. All genes examined were expressed at least four-fold higher than the control gene, omp26. Of these, hktE was the most highly expressed with a median increase of expression around 68-fold. It is important to note that pdgX is only expressed when NTHi is growing in a stationary biofilm, and not planktonically (figure 4c). Thus, the observation that pdgX is highly expressed is a good indication that these aggregates present in the smoke-exposed animals are true biofilms. Expression of stress factors such as dps and hktE support the conclusion that the COPD lung environment includes significant oxidative stress.
Overall, these data show that NTHi infection causes lung damage and functional decline in smoke-exposed ferrets, much as occurs in patients with COPD. This damage is noticeable shortly after infection, supporting the conclusion that infection initiates an inflammatory exacerbation event in this animal model. Notably, we have also obtained conclusive evidence that NTHi resides within biofilm communities encased in a chemically distinct matrix and with transcript profiles consistent with the biofilm, but not planktonic, phase of growth. Defining the host and bacterial factors that contribute to bacterial persistence (or clearance) in this model system will be an important avenue for future work.
Discussion
Changes occurring in the lungs of patients with COPD include chronic inflammation, extracellular matrix abnormalities, excessive mucus viscosity, reduced mucociliary clearance, and immune and epithelial barrier dysfunctions mediated by smoke exposure [6, 24]. Prior work has established that the smoke-exposed ferret model recapitulates human COPD symptoms, and of note this model emulates pathophysiologies not represented in other models, such as chronic bronchitis and mucosal abnormalities, including a reduction in mucociliary clearance [12, 25]. These pathophysiologies are key components of COPD, particularly the mucus phenotypes, which can have significant impacts on host–microbe interactions. For example, it is well documented that a reduction in mucociliary clearance induced by smoke exposure heightens sensitivity to bacterial infection and promotes chronic bacterial infection within the lungs [26]. Bacterial infection drastically reduces airway function and promotes airway damage in COPD patients; however, an understanding of the dynamics of this process and the complex bacterial–host interactions that take place are not fully understood [2, 14, 27]. However, it is well documented that repeat acute exacerbations increase the risk of further exacerbation events and are an important contributor to the degradation of a patient's quality of life, ballooning healthcare costs, and patient mortality and morbidity. These events are associated with heightened bacterial loads within the lungs, underpinning the concept that bacteria are a prime aetiological agent of exacerbations [28–31]. Changes in bacterial load, acquiring a new, perhaps more virulent strain of a common COPD pathogen, changes in the localisation of bacteria within the lungs, or combinations of these seem to be triggering events of COPD exacerbations [16, 32]. There may be some degree of variation in virulence among NTHi lineages, as it has been noted that NTHi isolated from patients undergoing an exacerbation induced significantly more inflammation and neutrophil recruitment than strains isolated from patients in stable COPD within a mouse model of COPD [32].
While it is clear the exacerbations in COPD are linked to bacterial infections, the host–microbe interplay occurring during the exacerbation event and the bacteria-dependent factors that are important for influencing severity are not fully understood. Thus, utilising a physiologically relevant model for COPD we hoped to make a contribution towards that question.
In smoke-exposed animals, NTHi infection caused morphological changes within the lung, that included thickening of the airway walls, consolidation, mucus accumulation, and bronchiole impaction. Concurrently, infection resulted in a significant increase in central airway resistance and a decline in inspiratory capacity. Together, these data suggest that NTHi infection elicits significant host responses consistent with an inflammatory exacerbation event.
Infection of smoke-exposed animals elicited airway neutrophilia, an increase in pro-inflammatory TNF-α and proteolytic activity. Airway neutrophilia is a common feature of COPD and other chronic inflammatory lung diseases, such as cystic fibrosis [33, 34]. Additionally, this feature is associated with disease severity, airway obstruction, and forced expiratory volume in 1 s decline in COPD. These neutrophils migrating into the lung will release a variety of active enzymes from their granules, such as MPO and neutrophil elastase, and produce NETs, which can degrade components of the extracellular matrix, contributing to the progression of disease [35–39]. NTHi infection stimulated the influx of activated neutrophils into the lungs, as well as increased the levels of active neutrophil enzymes, MPO, MMP-2, and MMP-9. Skewing the balance of protease/anti-protease activity towards the damaging, pro-inflammatory protease axis is a central component of the progression of COPD disease. NETs were seen surrounding NTHi biofilms as has been reported in other chronic infection models [19, 21, 40]. NETs have been known to increase the mucus viscosity by contributing to the increase in extracellular DNA within the mucus [40, 41]; thus, the formation of NETs in vivo may complicate rather than help resolve opportunistic NTHi infection. As the ferret model recapitulates many clinical features of humans with COPD, this model represents an ideal platform for translational studies regarding COPD-related infections.
These infection studies also provide new insights into the state of growth and metabolism of NTHi within the COPD lung. Utilising immunofluorescent staining, NTHi aggregates surrounded by their associated biofilm components were observed in the smoke-exposed animals. Also increased transcript levels for factors associated with biofilm mode of growth (pdgX, luxS, dps, and hktE) were observed. These data strongly support the conclusion that NTHi resides within biofilm communities in the context of COPD. Certainly, future goals will include longer term infection studies to mimic the long-term persistence seen in COPD.
Given that NTHi can persist well after the initial exacerbation period, it is reasonable to assume that these persistent population are residing in biofilms, protecting them from the chronically inflammatory environment and antibiotic treatment [8]. Thus, these biofilm resident populations are able to remain within the lung for extended periods of time inducing low-grade chronic inflammation around their local area and contributing to further lung damage and loss of function over time. There are a variety of strategies for targeting bacterial biofilms ranging from the use of nanoparticles capable of penetrating the biofilm matrix, mucolytics to break apart the biofilm, enzymes to digest the biofilm, or other compounds targeting bacterial processes, such as quorum signalling [42, 43]. A combination of antibiofilm and antibiotic treatment in patients undergoing or recovering from a bacterial exacerbation may help effectively clear these biofilm populations. Managing these bacterial biofilms could contribute to a reduction in airway inflammation, promote healthier airway function, and slow the progression and/or severity of COPD. Antibiofilm research is an important field with the potential to improve outcomes for COPD or other chronic NTHi infections.
The duration of NTHi persistence in vivo in our infection studies merits specific additional comment. Most of the existing animal models for NTHi infection involve acute infections of relatively short duration; it has only been with the chinchilla model for otitis media infections that chronic infections involving biofilms have been possible [17, 19, 44, 45]. Further work to define the bacterial determinants of colonisation and persistence in this context will be of particular importance.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
FIGURE S1 00200-2020.figureS1 (121.1KB, tif)
FIGURE S2 00200-2020.figureS2 (2MB, tif)
Support statement
This study received financial support from National Institutes of Health (NIH) grants R21 AI133445, R35 HL135816 and P30 DK072482. B.C. Hunt was supported by a trainee fellowship on NIH T32 HL134640. Funding information for this article has been deposited with the Crossref Funder Registry.
Footnotes
Author contributions: B.C. Hunt, A. Gaggar, S.M. Rowe, S.V. Raju and W.E. Swords conceived and designed the experiments. B.C. Hunt, S. Stanford, X. Xu and J. Li performed the experiments. B.C. Hunt, A. Gaggar, S.V. Raju and W.E. Swords analysed and interpreted the data. B.C. Hunt and W.E. Swords wrote the manuscript.
This article has supplementary material available from openres.ersjournals.com
Conflict of interest: B.C. Hunt has nothing to disclose.
Conflict of interest: D. Stanford has nothing to disclose.
Conflict of interest: X. Xu has nothing to disclose.
Conflict of interest: J. Li has nothing to disclose.
Conflict of interest: A. Gaggar has nothing to disclose.
Conflict of interest: S.M. Rowe reports grants, personal fees and nonfinancial support from Synedgen/Synspira during the conduct of the study; grants and personal fees from Novartis, grants and personal fees from Bayer, grants from Translate Bio, nonfinancial support from Proteostasis, grants, personal fees and nonfinancial support from Galapagos/Abbvie, grants, personal fees and other from Synedgen/Synspira, grants from Eloxx, grants from Celtaxsys, grants, personal fees, nonfinancial support and other from Vertex Pharmaceuticals Inc., personal fees from Renovion, grants and personal fees from Arrowhead, grants and other from Ionis, grants from AstraZeneca, outside the submitted work.
Conflict of interest: S.V. Raju has nothing to disclose.
Conflict of interest: W.E. Swords reports grants from Merck outside the submitted work.
References
- 1.Hassett DJ, Borchers MT, Panos RJ. Chronic obstructive pulmonary disease (COPD): evaluation from clinical, immunological and bacterial pathogenesis perspectives. J Microbiol 2014; 52: 211–226. doi: 10.1007/s12275-014-4068-2 [DOI] [PubMed] [Google Scholar]
- 2.Zhou X, Li Q, Zhou X. Exacerbation of chronic obstructive pulmonary disease. Cell Biochem Biophys 2015; 73: 349–355. doi: 10.1007/s12013-015-0605-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forey BA, Thornton AJ, Lee PN. Systematic review with meta-analysis of the epidemiological evidence relating smoking to COPD, chronic bronchitis and emphysema. BMC Pulm Med 2011; 11: 36. doi: 10.1186/1471-2466-11-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim V, Criner GJ. The chronic bronchitis phenotype in chronic obstructive pulmonary disease: features and implications. Curr Opin Pulm Med 2015; 21: 133–141. doi: 10.1097/MCP.0000000000000145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim V, Criner GJ. Chronic bronchitis and chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2013; 187: 228–237. doi: 10.1164/rccm.201210-1843CI [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su YC, Jalalvand F, Thegerstrom J, et al. . The interplay between immune response and bacterial infection in COPD: focus upon non-typeable Haemophilus influenzae. Front Immunol 2018; 9: 2530. doi: 10.3389/fimmu.2018.02530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leung JM, Tiew PY, Mac Aogain M, et al. . The role of acute and chronic respiratory colonization and infections in the pathogenesis of COPD. Respirology 2017; 22: 634–650. doi: 10.1111/resp.13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parameswaran GI, Murphy TF. Chronic obstructive pulmonary disease. Drugs Aging 2009; 26: 985–995. doi: 10.2165/11315700-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 9.Mammen MJ, Sethi S. COPD and the microbiome. Respirology 2016; 21: 590–599. doi: 10.1111/resp.12732 [DOI] [PubMed] [Google Scholar]
- 10.Clementi CF, Murphy TF. Non-typeable Haemophilus influenzae invasion and persistence in the human respiratory tract. Front Cell Infect Microbiol 2011; 1: 1. doi: 10.3389/fcimb.2011.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stämpfli MR, Anderson GP. How cigarette smoke skews immune responses to promote infection, lung disease and cancer. Nat Rev Immunol 2009; 9: 377–384. doi: 10.1038/nri2530 [DOI] [PubMed] [Google Scholar]
- 12.Raju SV, Kim H, Byzek SA, et al. . A ferret model of COPD-related chronic bronchitis. JCI Insight 2016; 1: e87536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison A, Dyer DW, Gillaspy A, et al. . Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J Bacteriol 2005; 187: 4627–4636. doi: 10.1128/JB.187.13.4627-4636.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakaletz LO, Tallan BM, Hoepf T, et al. . Frequency of fimbriation of nontypable Haemophilus influenzae and its ability to adhere to chinchilla and human respiratory epithelium. Infect Immun 1988; 56: 331–335. doi: 10.1128/IAI.56.2.331-335.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Xie J, Patel M, et al. . Nontypeable Haemophilus influenzae genetic islands associated with chronic pulmonary infection. PLoS One 2012; 7: e44730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sethi S, Evans N, Grant BJB, et al. . New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med 2002; 347: 465–471. doi: 10.1056/NEJMoa012561 [DOI] [PubMed] [Google Scholar]
- 17.de Gier C, Pickering JL, Richmond PC, et al. . Duplex quantitative PCR assay for detection of Haemophilus influenzae that distinguishes fucose- and protein D-negative strains. J Clin Microbiol 2016; 54: 2380–2383. doi: 10.1128/JCM.00982-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanford D, Kim H, Bodduluri S, et al. . Airway remodeling in ferrets with cigarette smoke induced COPD using microCT imaging. Am J Physiol Lung Cell Mol Physiol 2020; in press [DOI: 10.1152/ajplung.00328.2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schindelin J, Arganda-Carreras I, Frise E, et al. . Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurcisek J, Greiner L, Watanabe H, et al. . Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun 2005; 73: 3210–3218. doi: 10.1128/IAI.73.6.3210-3218.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimon G, Yonit W-W, Gabriel I, et al. . The ‘tree-in-bud’ pattern on chest CT: radiologic and microbiologic correlation. Lung 2015; 193: 823–829. doi: 10.1007/s00408-015-9759-x [DOI] [PubMed] [Google Scholar]
- 22.Hong W, Juneau RA, Pang B, et al. . Survival of bacterial biofilms within neutrophil extracellular traps promotes nontypeable Haemophilus influenzae persistence in the chinchilla model for otitis media. JIN 2009; 1: 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy TF, Kirkham C, Sethi S, et al. . Expression of a peroxiredoxin-glutaredoxin by Haemophilus influenzae in biofilms and during human respiratory tract infection. FEMS Immunol Med Microbiol 2005; 44: 81–89. doi: 10.1016/j.femsim.2004.12.008 [DOI] [PubMed] [Google Scholar]
- 24.Starner TD, Zhang N, Kim G, et al. . Haemophilus influenzae forms biofilms on airway epithelia: implications in cystic fibrosis. Am J Respir Crit Care Med 2006; 174: 213–220. doi: 10.1164/rccm.200509-1459OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhat TA, Panzica L, Kalathil SG, et al. . Immune dysfunction in patients with chronic obstructive pulmonary disease. Ann Am Thorac Soc 2015; 12: Suppl. 2, S169–S175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghorani V, Boskabady MH, Khazdair MR, et al. . Experimental animal models for COPD: a methodological review. Tob Induc Dis 2017; 15: 25. doi: 10.1186/s12971-017-0130-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagaitkar J, Demuth DR, Scott DA. Tobacco use increases susceptibility to bacterial infection. Tob Induc Dis 2008; 4: 12. doi: 10.1186/1617-9625-4-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurst JR, Vestbo J, Anzueto A, et al. . Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med 2010; 363: 1128–1138. doi: 10.1056/NEJMoa0909883 [DOI] [PubMed] [Google Scholar]
- 29.Wedzicha JA, Singh R, Mackay AJ. Acute COPD exacerbations. Clin Chest Med 2014; 35: 157–163. doi: 10.1016/j.ccm.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 30.Barnes PJ, Burney PGJ, Silverman EK, et al. . Chronic obstructive pulmonary disease. Nat Rev Dis Primers 2015; 1: 15076. doi: 10.1038/nrdp.2015.76 [DOI] [PubMed] [Google Scholar]
- 31.Garcha DS, Thurston SJ, Patel AR, et al. . Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax 2012; 67: 1075–1080. doi: 10.1136/thoraxjnl-2012-201924 [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson TMA, Patel IS, Wilks M, et al. . Airway bacterial load and FEV1 decline in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2003; 167: 1090–1095. doi: 10.1164/rccm.200210-1179OC [DOI] [PubMed] [Google Scholar]
- 33.Chin CL, Manzel LJ, Lehman EE, et al. . Haemophilus influenzae from patients with chronic obstructive pulmonary disease exacerbation induce more inflammation than colonizers. Am J Respir Crit Care Med 2005; 172: 85–91. doi: 10.1164/rccm.200412-1687OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laval J, Ralhan A, Hartl D. Neutrophils in cystic fibrosis. Biol Chem 2016; 397: 485–496. doi: 10.1515/hsz-2015-0271 [DOI] [PubMed] [Google Scholar]
- 35.Jasper AE, McIver WJ, Sapey E, et al. . Understanding the role of neutrophils in chronic inflammatory airway disease. F1000Res 2019; 8: F1000 Faculty Rev-557. doi: 10.12688/f1000research.18411.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tetley TD. New perspectives on basic mechanisms in lung disease. 6. Proteinase imbalance: its role in lung disease. Thorax 1993; 48: 560–565. doi: 10.1136/thx.48.5.560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crisford H, Sapey E, Stockley RA. Proteinase 3; a potential target in chronic obstructive pulmonary disease and other chronic inflammatory diseases. Respir Res 2018; 19: 180. doi: 10.1186/s12931-018-0883-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genschmer KR, Russell DW, Lal C, et al. . Activated PMN exosomes: pathogenic entities causing matrix destruction and disease in the lung. Cell 2019; 176: 113–126. doi: 10.1016/j.cell.2018.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirkham PA, Barnes PJ. Oxidative stress in COPD. Chest 2013; 144: 266–273. doi: 10.1378/chest.12-2664 [DOI] [PubMed] [Google Scholar]
- 40.Sethi S, Mallia P, Johnston SL. New paradigms in the pathogenesis of chronic obstructive pulmonary disease II. Proc Am Thorac Soc 2009; 6: 532–534. doi: 10.1513/pats.200905-025DS [DOI] [PubMed] [Google Scholar]
- 41.Papayannopoulos V. Neutrophils facing biofilms: the battle of the barriers. Cell Host Microbe 2019; 25: 477–479. doi: 10.1016/j.chom.2019.03.014 [DOI] [PubMed] [Google Scholar]
- 42.Papayannopoulos V, Staab D, Zychlinsky A. Neutrophil elastase enhances sputum solubilization in cystic fibrosis patients receiving DNase therapy. PLoS One 2011; 6: e28526. doi: 10.1371/journal.pone.0028526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy R, Tiwari M, Donelli G, et al. . Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence 2018; 9: 522–554. doi: 10.1080/21505594.2017.1313372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batoni G, Maisetta G, Esin S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim Biophys Acta 2016; 1858: 1044–1060. doi: 10.1016/j.bbamem.2015.10.013 [DOI] [PubMed] [Google Scholar]
- 45.Bakaletz LO. Chinchilla as a robust, reproducible and polymicrobial model of otitis media and its prevention. Expert Rev Vaccines 2009; 8: 1063–1082. doi: 10.1586/erv.09.63 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
FIGURE S1 00200-2020.figureS1 (121.1KB, tif)
FIGURE S2 00200-2020.figureS2 (2MB, tif)