The landscape of mutations and clonal evolution trajectories in pediatric ALL reveal relapse-fated, drug-resistant subclones present at diagnosis and expanded by therapy.
Abstract
Relapse of acute lymphoblastic leukemia (ALL) remains a leading cause of childhood cancer-related death. Prior studies have shown clonal mutations at relapse often arise from relapse-fated subclones that exist at diagnosis. However, the genomic landscape, evolutionary trajectories, and mutational mechanisms driving relapse are incompletely understood. In an analysis of 92 cases of relapsed childhood ALL incorporating multimodal DNA and RNA sequencing, deep digital mutational tracking, and xenografting to formally define clonal structure, we identified 50 significant targets of mutation with distinct patterns of mutational acquisition or enrichment. CREBBP, NOTCH1, and RAS signaling mutations arose from diagnosis subclones, whereas variants in NCOR2, USH2A, and NT5C2 were exclusively observed at relapse. Evolutionary modeling and xenografting demonstrated that relapse-fated clones were minor (50%), major (27%), or multiclonal (18%) at diagnosis. Putative second leukemias, including those with lineage shift, were shown to most commonly represent relapse from an ancestral clone rather than a truly independent second primary leukemia. A subset of leukemias prone to repeated relapse exhibited hypermutation driven by at least three distinct mutational processes, resulting in heightened neoepitope burden and potential vulnerability to immunotherapy. Finally, relapse-driving sequence mutations were detected prior to relapse using droplet digital PCR at levels comparable with orthogonal approaches to monitor levels of measurable residual disease. These results provide a genomic framework to anticipate and circumvent relapse by earlier detection and targeting of relapse-fated clones.
Significance:
This study defines the landscape of mutations that preexist and arise after commencement of ALL therapy and shows that relapse may be propagated from ancestral, major, or minor clones at initial diagnosis. A subset of cases exhibits hypermutation that results in expression of neoepitopes that may be substrates for immunotherapeutic intervention.
See related video: https://vimeo.com/442838617
See related commentary by Ogawa, p. 21 .
See related article by S. Dobson et al .
This article is highlighted in the In This Issue feature, p. 5
Introduction
Relapsed acute lymphoblastic leukemia (ALL) is the second leading cause of cancer-related death in children (1). There are few targeted therapeutic approaches for relapsed ALL and outcome is frequently poor (2), even with the advent of immunotherapeutic approaches. ALL typically exhibits a relatively low burden of somatic mutations, which has allowed delineation of the nature and sequence of acquisition of genetic variants that drive treatment failure (3). These include inherited variants that are often associated with leukemia subtype (e.g., TP53 mutations and low hypodiploid ALL), founding chromosomal rearrangements (e.g., BCR-ABL1 and rearrangement of KMT2A), secondary genomic alterations (e.g., alteration of IKZF1), and somatic alterations that are enriched from minor clones or acquired after initiation of therapy (4). Mutations targeting signaling pathways, chromatin patterning, tumor suppression, and nucleoside metabolism are enriched at relapse (5). These can confer resistance to specific drugs, such as mutations in NT5C2 to thiopurines (4, 6–10) and mutations in the glucocorticoid receptor NR3C1 and acetyltransferase CREBBP to glucocorticoids (11), or confer sensitivity to targeted agents, such as RAS pathway mutations and MEK inhibition (12). Prior studies also suggest that “relapse-fated” clones commonly exist as minor clones at diagnosis; along with the predominant major clone, these originate from a common ancestral clone that undergoes divergent evolution (8).
The early identification and genetic characterization of relapse-fated clones offer the opportunity to improve treatment outcomes by anticipating relapse and adjusting therapy, or by targeting relapse-fated clones prior to the acquisition of additional mutations facilitating leukemic progression. However, prior genomic studies of relapsed ALL have typically been limited in cohort size and the extent of genomic analysis such that a rigorous analysis of the relapse driver mutations, formal delineation of clonal structure and disease progression, and deep sequencing to distinguish preexisting clones from acquired mutations has not been possible.
Results
Patterns of Relapse in ALL
Multiple tools were used to describe mutational landscape, clonal structure, and clonal evolution for 92 children with ALL and sequential diagnosis, remission and relapse samples treated on St Jude Total Therapy Studies (13, 14), results of which may be explored at https://stjuderesearch.org/site/data/relapsed-all (Supplementary Table S1). Somatic sequence variants detected at diagnosis and relapse were subjected to confirmatory capture-based sequencing at each time point to optimize estimation of mutant allele frequency (MAF) and time acquisition of relapse-associated mutations (Supplementary Fig. S1A–S1D). The deep sequencing identified subclonal somatic mutations in a subset of the remission samples (Supplementary Table S2), raising the possibility that mutational persistence early in therapy may predict relapse, as observed in acute myeloid leukemia (AML; ref. 15). However, comparative analysis of germline mutation burden in 12 cases from this relapsed cohort, with samples at days 27 to 49 and 20 cases that did not relapse, showed no correlation between mutational burden early in therapy and likelihood of relapse (Supplementary Data).
The burden of single-nucleotide variants (SNV), short insertions/deletions (indels), and copy number alterations (CNA) increased with disease progression (Supplementary Figs. S2A–S2D and S3A–S3C; Supplementary Tables S3–S7). Across the cohort, the majority of CNAs (60%) were preserved from diagnosis to relapse, whereas the majority of SNV/indels (74%) were acquired (Supplementary Table S8). Twenty-seven tumors from 18 patients were hypermutated (>85 mutations per sample, ∼1.3 mutations/Mb; Supplementary Fig. S4A–S4F), including 9 of 14 second relapses (64%), 6 of which were already hypermutated at first relapse. Apart from an increased mutational burden at early second relapse, no relationship was observed between mutation burden and time to relapse. CREBBP mutations (n = 15 cases) were associated with a longer time to relapse (mean 4.3 vs. 2.8 years, Student t test P = 0.019; Supplementary Table S9). Notably, 8 cases with outlier early relapse harbored combinations of alterations known to be involved in relapse development (Supplementary Data).
Frequently Mutated Genes and Pathways
A total of 4,509 genes harbored nonsilent sequence mutations at diagnosis (D) or relapse (R; Supplementary Tables S3 and S4). Clonal, nonsilent SNV/indels, or focal CNAs were acquired or selected for in 125 genes at first relapse in at least 2 cases and 28 genes in at least 3 cases. Among the recurrent mutated genes (≥2 cases), 38 were known cancer/leukemia genes (16) and 87 had not previously been described (Supplementary Table S10). Using GRIN (17), a model that incorporates analysis of multimodal genomic data (Methods; Supplementary Tables S11 and S12), 23 genes were significantly mutated (q < 0.1) at diagnosis and relapse, and 50 genes significantly enriched with mutations at R1 (by rising MAF or acquisition of mutation following diagnosis). Most (n = 20, 87%) of the D-R1 shared mutated genes, but only 14 (28%) R1 specific, were known targets of mutation in cancer/leukemia.
B-ALL relapses were enriched with mutations in Ras pathway (relapse 31.3% vs. diagnosis 17.9%) and epigenetic modifiers/regulators, including PRDM2 (n = 4), PHF19 (n = 3), TET3 (n = 3), and SIN3A (n = 3), 16 of which had not been reported in ALL (49.3% vs. 29.9%; Fig. 1A and B; Supplementary Figs. S5A, S5B and S6; Supplementary Table S13). Of 61 cases with signaling pathway mutations, 31 harbored at least one Ras pathway mutation at diagnosis, with 11 cases having multiple, commonly subclonal RAS pathway mutations at diagnosis (Supplementary Table S14). Seven cases showed convergence to one or two clonal Ras pathway mutations. In contrast, only three cases showed acquisition of new RAS pathway mutations at relapse. Thus, multiclonality of signaling pathway mutations is frequent at diagnosis in ALL, indicating that they are secondary lesions in leukemia evolution, and the observed mutational extinction and convergence to clonal dominance supports a selective advantage to RAS pathway mutations in many cases. In contrast, PI3K–AKT pathway mutations were common at diagnosis in T-ALL but were often lost at relapse, suggesting that inhibition of this pathway may not reduce likelihood of relapse.
Figure 1.
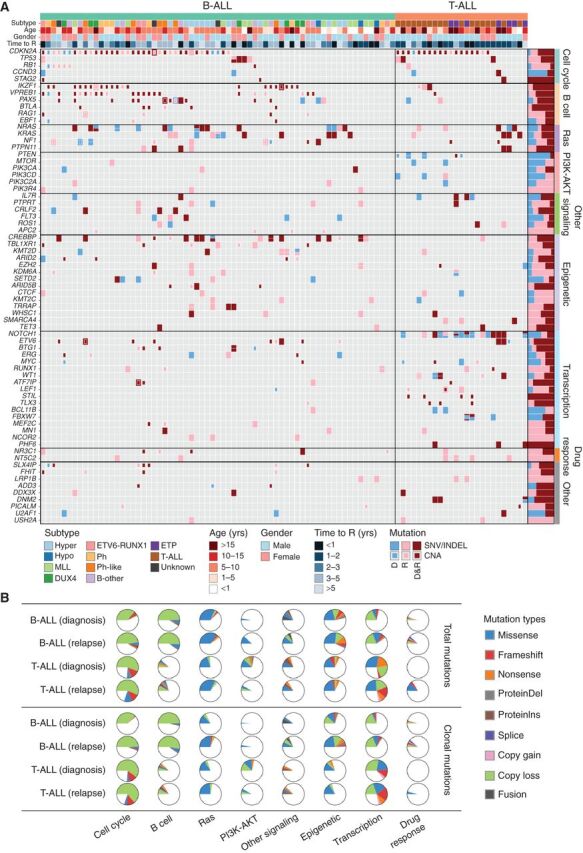
Somatic mutation spectrum in ALL at diagnosis and relapse. A, Nonsilent mutations in recurrently affected (≥3 cases) key genes (COSMIC Cancer Gene Census or reported leukemia relevant genes) in diagnosis (D) and first available relapse (R) sample per case. The B-ALL cases are grouped into well-defined disease subtypes, which include hyperdiploid (Hyper), hypodiploid (Hypo), KMT2A (MLL)-rearranged, DUX4-rearranged (DUX4), ETV6-RUNX1, BCR-ABL1 (Ph), Ph-like, and a group of other B-ALL subtypes including B-other, PAX5 P80R, and iAMP21 ALL. Mutations in the form of SNV/indels and focal CNAs are shown as rectangles with different sizes. Mutations observed only in D, only in R or shared by D and R are shown in blue, pink, and dark red colors, respectively. The prevalence for each gene mutation is shown in bar graph on the right. B, Distribution of recurrent mutations in key pathways. Top, all recurrent mutations; bottom, the clonal (MAF ≥ 30%) nonsilent mutations. Samples are divided into B-ALL (n = 67) and T-ALL (n = 25) and the mutation ratio in diagnosis and relapse stages is shown. Detailed mutation types are indicated by different colors.
Of the genes known to play a role in the development of ALL, 29 (including IKZF1, TP53, NR3C1, TBL1XR1, and PTPN11) showed universal enrichment at relapse, whereby mutations were always retained from diagnosis to relapse (i.e., were truncal variants) or subsequently emerged at relapse (Supplementary Table S15). Of these, six were never observed at diagnosis (NT5C2, LRP1B, USH2A, APC2, PIK3R4, and NCOR2). An additional 20 genes showed extinction of mutations present at diagnosis in only a single case (e.g., VPREB1, CREBBP, ETV6, and KDM6A). Several genes not previously reported to be mutated in ALL were notable for preservation or clonal selection of mutations from diagnosis to relapse, including PTPRT (n = 6), ROBO2 (n = 5), and TRRAP (n = 5), suggesting a role in promoting leukemogenesis and relapse (Supplementary Fig. S7A and S7B; Supplementary Tables S16 and S17). Mutations in the glucocorticoid receptor (NR3C1) and purine/pyrimidine synthesis pathway (NT5C2) were frequent in both B- and T-ALL at relapse, but often as subclonal events, suggesting that additional targeting of these drug-specific resistance-driving mutations may eradicate all relapse clones in many cases.
Integration of Mutational Landscapes with Clonal Structure
Using the rise and fall of CNAs and MAF of somatic SNV/indels, most tumor pairs [n = 79 D-R1/secondary tumor (S; 86%), and n = 12 R1-R2/S (92%)], showed clonal extinction and evolution of new clones in the subsequent tumor, indicating branching evolution. One quarter of the relapses (n = 28, including 2 second relapses) arose from the major clone (MAF > 30%) present at diagnosis or the previous relapse, and half of the relapses (n = 53, of which 6 were second relapses) developed from a minor clone (Fig. 2A and B; Supplementary Table S18). Of the 53 relapses arising from a minor clone, nine (17%) had relapse-enriched mutations already present in that subclone and 26 (49%) acquired new or additional relapse-enriched mutations following diagnosis. Nineteen tumors (18%), exhibited polyclonal evolution in which multiple diagnosis clones persisted at relapse.
Figure 2.
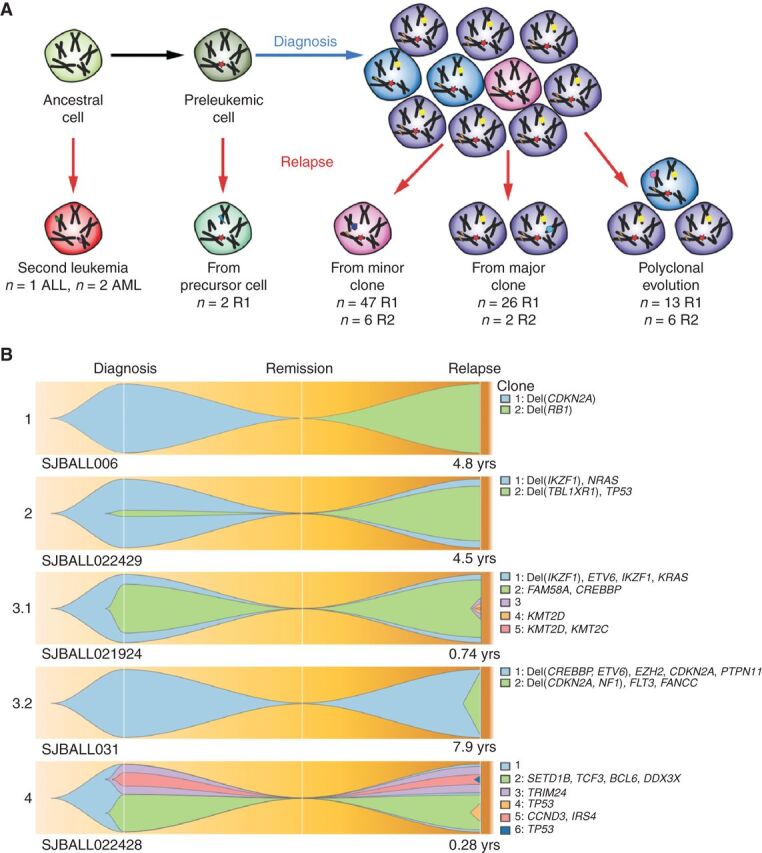
Patterns of relapse in ALL. A, Schematic overview of mechanisms of clonal evolution. Three patients developed a second primary tumor that was not clonally related to the previous tumor occurrence. Two patients developed a tumor that shared only one founding fusion between diagnosis and relapse, indicating the disease relapsed from a preleukemic cell. Further relapses arose through evolution from a minor clone, a major clone, or multiple clones. B, Fish plots of the clonal evolution models inferred from somatic mutations detected in diagnosis and relapse samples. MAF of the somatic mutations was used by the sciClone R package (14) to infer potential clonal clusters (shown in different colors) and visualized by Fishplot (ref. 13; see Methods). Four major clonal evolution models were observed: 1, relapse sample is a second primary leukemia with no somatic mutations shared with diagnosis; 2, a minor clone (somatic mutations' median MAF of the clone is less than 30%) in diagnosis develops into the major clone in relapse; 3, a major clone (somatic mutations' median MAF of the clone is greater than 30%) is preserved from diagnosis to relapse (3.1) or emerges as a major clone at relapse (3.2); 4, multiple subclones in diagnosis develop as multiple subclones in relapse. For each exemplary case, the time from diagnosis to relapse is indicated. Focal deletions and nonsilent somatic mutations on cancer genes (according to the COSMIC Cancer Gene Census and well-known leukemia relevant genes) for each inferred clone are shown on the right side.
We found that second relapses evolved more often in a polyclonal fashion (χ2P = 0.043) and with a shorter remission time than first relapses (average 1.5 years vs. 3 years, Student t test, P = 0.0044). Compared with first relapses, variants in second relapses were more often variants preserved from subclones (10% vs. 4%) or preserved at subclonal levels (11% vs. 2%) and less often acquired (69% vs. 79%), which reflects the polyclonal evolution model (χ2P < 2.2 × 10−16). Thus, initial evolution from diagnosis to relapse is characterized by mutational convergence, and commonly, emergence from a minor clone, but subsequent progression exhibits preservation of the initially selected clones and variants.
Second Primary Leukemia
Four first relapses (SJBALL006, SJTALL142, SJPHALL005, and SJPHALL022425) and one second relapse (SJTALL049) were fully discordant for all genetic alterations (SV, CNA, SNV/Indel, and antigen receptor rearrangements) or shared only a leukemia fusion (Supplementary Tables S19 and S20), suggesting distinct second leukemias masquerading as relapse. These scenarios are important to distinguish, as second leukemias may be curable with standard therapy and multiple primary tumors suggest heritable leukemia predisposition. Both tumors in SJBALL006 harbored MEF2D-BCL9 fusions, but with unique RNA and DNA breakpoints, and evidence of the second MEF2D-BCL9 fusion at low level in the primary sample (Supplementary Tables S20 and S21). In addition, there was a constitutional gain of the chromosome 1q neuroblastoma breakpoint (NBPF; ref. 18) region on chromosome 1q that contains MEF2D and BCL9, and discordant somatic complex genomic amplifications of the NBPF region at diagnosis and relapse (Fig. 3A–D; Supplementary Tables S6 and S22). Thus, while emergence of a tumor with the same fusion partners suggests relapse, this case represents germline structural variation promoting development of multiple tumors with distinct initiating fusions and different latencies of presentation.
Figure 3.
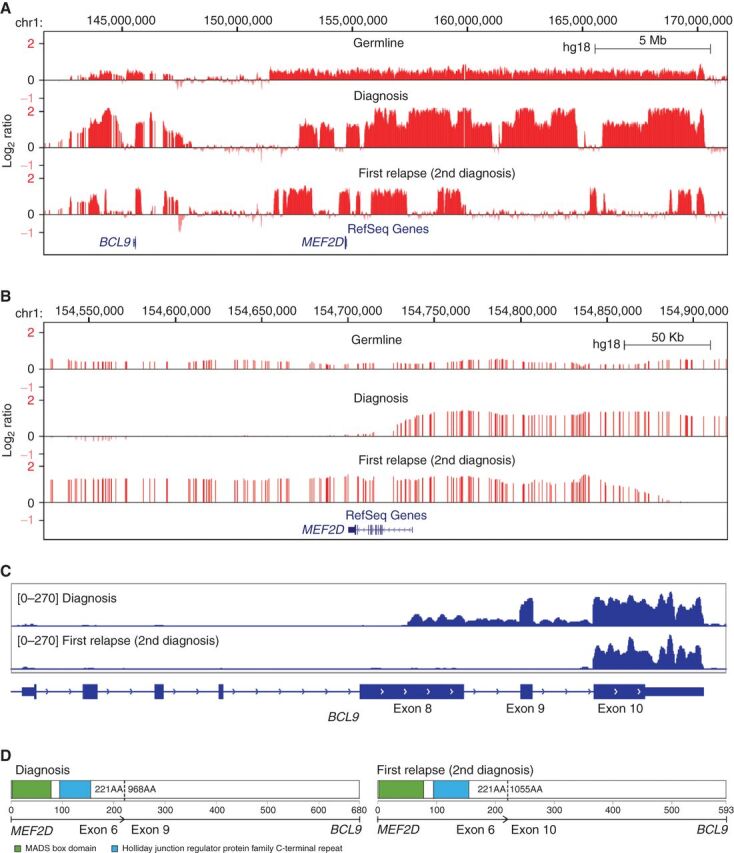
CNAs and MEF2D-BLC9 rearrangements in patient SJBALL006. Signal intensity from Affymetrix SNP6.0 microarrays was normalized to log2 ratio (>0 indicate copy gain; <0 indicate copy loss) and shown in the University of California, Santa Cruz Genome Browser in large (A) and focal scale (B) to show the distinct alteration patterns between diagnosis and “relapse” (second diagnosis) samples. Constitutional copy number gains were observed in the germline sample. C, RNA-seq depth on exons of BCL9 gene. The sequencing depth was scaled from 0- to 270-fold for both diagnosis samples. The uptick of expression of exon 9 and 10 was observed for first and second diagnosis samples, respectively, indicating different rearrangement breakpoints on BCL9, which was consistent with fusions called from RNA-seq. The RNA-seq library for the first diagnosis sample was total RNA, so the intronic region was covered by sequencing reads. D, Schematic visualization of MEF2D-BCL9 chimeric protein structure. Two fusion isoforms have been reported as the most recurrent MEF2D-BCL9 rearrangements (30).
BCR-ABL1 cases SJPHALL005 and SJPHALL022425 lacked shared nonsilent variants at diagnosis and relapse, raising the possibility of second leukemia rather than relapse. However, whole genome sequencing (WGS) of SJPHALL005 identified identical rearrangement breakpoints at diagnosis and relapse and 65 shared somatic noncoding SNVs, demonstrating relapse from a common, ancestral clone (Supplementary Table S20). SJTALL049 developed three tumors: STIL-TAL1 rearranged T-ALL at age 6 that relapsed at age 14 plus an independent BCR-ABL1–positive chronic myeloid leukemia (CML) at age 20. Breakpoint spanning PCR revealed unique STIL-TAL1 rearrangements at diagnosis and relapse, but WGS showed 19 shared noncoding SNVs. WGS showed no shared SNVs between either of the T-ALLs and the CML, indicating that the CML developed as a second primary tumor. Thus, completely genetically distinct second tumors masquerading as relapse of acute leukemia are rare, and even if complete nonsilent mutational discordance is present, they may arise from divergent evolution soon after leukemia initiation.
Three relapses were clinically considered second tumors based on a shift to myeloid lineage (SJTALL008, SJTALL124, and SJTALL164). However, these tumors revealed shared mutations between diagnosis and relapse, indicating a common clonal origin (Supplementary Data). This recapitulates the lineage plasticity that is independent of genetic variegation we have recently described in acute leukemia of ambiguous lineage (19) and highlights the importance of genomic analysis to accurately interpret the relationship of diagnosis and relapse samples.
Tracing the Evolution of Relapse
Bulk sequencing data may fail to unambiguously define a clonal evolution model, particularly for mutationally sparse samples and those exhibiting a continuum of variant MAFs. We performed limiting dilution xenografting and sequencing of eight matched diagnosis and relapse samples (Supplementary Table S1). Most (90.5% of 232) of the somatic mutations detected in primary samples were identified in at least one diagnosis or relapse xenograft (MAF ≥ 0.01; Supplementary Fig. S8A). Of the 22 mutations not captured in xenografts, 20 were observed only in the bulk diagnosis sample and not in the relapse sample, suggesting that such nonxenografted diagnosis mutations must be present in cells that lack clonal propagating potential and are less likely to initiate relapse.
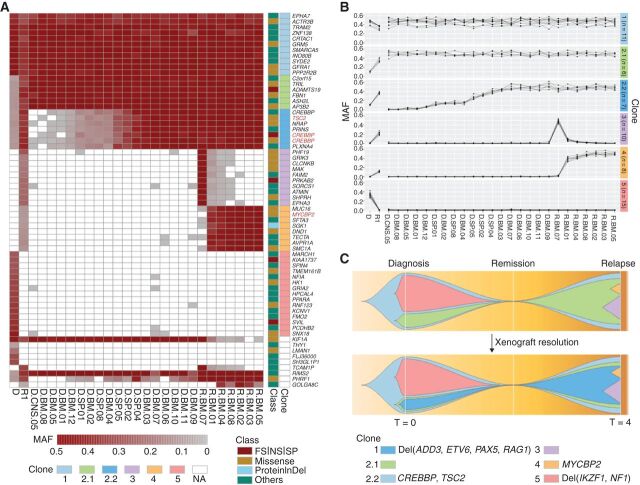
Genomic analysis including xenografting overcame the challenge of assigning mutations to individual subclones and enabled unambiguous delineation of clonal structure. Xenograft analysis of SJBALL036 (ETV6-RUNX1-like subtype) identified two linearly related clones (2.1 and 2.2) from mutations originally allocated to a common clone (Fig. 4A). Mutations in relapse-fated clone 2.1 were clonal in all xenografts, including those propagated from diagnosis, whereas the CREBBP mutations in clone 2.2 were observed in a subset of xenografts, indicating that subclone 2.2 arose from 2.1. In addition, xenografting demonstrated the selective advantage of clone 2.1 versus clone 5 that was lost at relapse and was not represented in any of the xenografts transplanted with the diagnosis sample. Furthermore, the xenografts derived from the relapse sample captured mutually exclusive variants, providing definitive evidence that clones 3 and 4 were unrelated, and represent branching evolution (Fig. 4B and C). Similarly, xenograft data of SJETV010, of which the second relapse sample is hypermutated (1,699 somatic mutations), resolved 13 clones following linear and branching patterns of evolution (Supplementary Fig. S8B and S8C). These data support the notion of branching evolution in ALL suggested by FISH and bulk sequencing analysis (20, 21), but now with mutational data enabling unambiguous clonal delineation. A subset of xenografts has also been utilized to demonstrate that relapse-fated clones may be detected at diagnosis that already exhibit resistance to therapy (22, 23).
Figure 4.
Integration of mutational landscape and xenografts resolves clonal structure in ALL. A, Somatic mutation spectrum of diagnosis (D), first relapse (R1), and xenografted leukemia samples. Leukemic cells from D (D.*.#) and R1 (R.*.#) from patient SJBALL036 were xenografted in mice and collected from bone marrow (*.BM.#), central nervous system (*.CNS.#), and spleen (*.SP.#). Cancer genes with nonsilent mutations are highlighted in red. FS, frameshift; NS, nonsense; SP, canonical splice site; proteinInDel, protein insertion/deletion. B, Delineation of clonal model from xenografted samples. MAF of SNV/indels were analyzed by sciClone (14) to infer clonal clusters. On the basis of the MAF in D and R1, clone 2.1 and 2.2 were indistinguishable. Xenograft data shows that clone 2.1 rises as a major clone (MAF = 0.5) in relapse alone or together with clone 2.2, indicating that 2.1 is the parental clone of 2.2. In addition, xenograft data showed variability in MAFs between clones 3 and 4, indicating that clones 3 and 4 were two distinct subclones. The clones are color-coded in the schema as in A. The number of somatic mutations in each clone is shown in parentheses. C, Fishplot of the leukemia evolution model. The top plot shows the original evolution model based on D and R1, and the bottom plot is the refined evolution model after incorporating the information from xenografted samples. The time (T) at diagnosis is defined as 0 and the first relapse was observed 4 years later. Nonsilent mutations and focal deletions (Del) affecting cancer genes are highlighted for each clone.
Tracing Mutation Acquisition Prior to Relapse
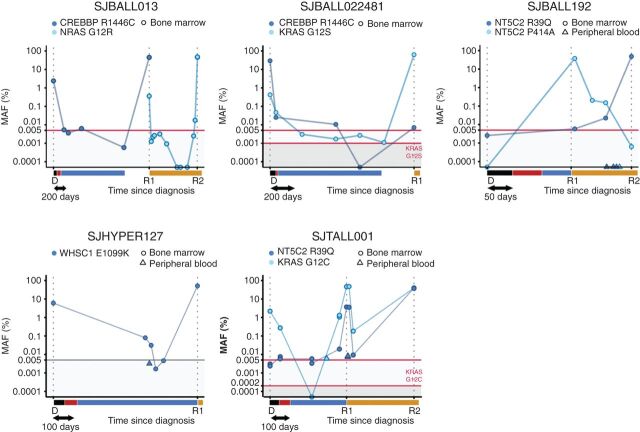
As xenografting identified resistance-driving mutations in relapse-fated subclones present at diagnosis, we sought to determine whether these mutations could be detected in patient samples obtained between diagnosis and relapse. We used droplet digital PCR (ddPCR) to track the emergence of relapse-specific mutations in CREBBP, NRAS, KRAS, NT5C2, and WHSC1 in 50 samples from 5 patients (Supplementary Table S23). With an input of 500 ng DNA a frequency of >0.005% (>7 copies) could be consistently detected (Supplementary Fig. S9A–S9D; Supplementary Table S24). ddPCR identified previously undetected minor clones in three tumor samples (NRAS G12R MAF = 0.4% in SJBALL013-R1, KRAS G12S MAF = 0.4% in SJBALL022481-D, NT5C2 R39Q MAF = 0.006% in SJBALL192-R1; Fig. 5), confirming the minor clone evolution model for these tumors. Difference in the temporal dynamics and occurrence of mutations in SJBALL022481 (CREBBP and KRAS), SJBALL192 (two NT5C2 mutations), and SJTALL001 (NT5C2 and KRAS) demonstrated clonal exclusivity of these mutations in each case. Despite complete remission by conventional minimal residual disease (MRD) testing, we detected tumor-specific mutations up to 534 days after the diagnosis (SJBALL022481, CREBBP R1446C), as well as 40 days prior to relapse (SJBALL013 NRAS G12R) in complete remission bone marrow samples. Moreover, in SJBALL022481, the KRAS G12S mutation was detectable at regular intervals during the 1,169 day period between diagnosis and relapse, even though the samples were deemed complete remission. Peripheral blood samples obtained from patients with B-lineage (SJBALL192, SJHYPER127) as well as T-lineage ALL (SJTALL001) with eventual bone marrow relapse were negative or contained much lower MAFs compared with the bone marrow. Thus, leukemic cells may persist in bone marrow and may be detected at low levels during complete remission, indicating the utility of this approach for disease monitoring and early relapse detection.
Figure 5.
ddPCR reveals mutations at low levels in intermediate complete remission samples. MAF of the indicated variants was determined in bone marrow (circle) and peripheral blood (triangle) samples for 5 patients. The time to diagnosis is scaled on the x-axis, with the treatment blocks indicated in black (induction), red (consolidation), blue (maintenance), and orange (relapse treatment). SJBALL192, SJHYPER127, and SJTALL001 relapsed during maintenance treatment. Detection limits are indicated with a red horizontal line and shaded background. Detection limits in gray were extrapolated from the other assays (i.e., not experimentally determined). The MAF at relapse of WHSC1 in SJHYPER127 was determined in our capture validation analysis as no DNA was available for ddPCR. The y-axis is in logarithmic scale.
Mutational Drivers of Hypermutation and Neoepitope Expression
Three percent of diagnoses, 17% of first relapse, and 64% of second relapse cases exhibited hypermutation. This was defined by an inflection at 85 mutations, approximately 1.3 mutations/Mb, a burden that was more conservative than the cutoff determined by the Segmented algorithm (ref. 24; Supplementary Fig. S4A). Hypermutation was observed in cases relapsing from minor or multiple clones of all 3 hypodiploid, 2 of 5 ETV6-RUNX1, 5 of 13 hyperdiploid, 1 Ph-like, 2 unclassified B-ALL, 1 ETP, and 4 T-ALL cases.
To understand the processes responsible for hypermutation, we used non-negative matrix factorization (NMF; ref. 25) and extracted four single-base substitution (SBS) signatures (Fig. 6A and B; Supplementary Fig. S10A and S10B). The high prevalence of hypermutation in second relapses suggests that hypermutation may be driven by treatment exposure. However, we did not uncover a mutational signature associated with treatment, such as temozolomide-associated signatures found in glioblastoma and melanoma (25). On the basis of the most prominent mutational signature (Supplementary Fig. S10B), we classified 17 hypermutated relapses from 13 patients into four groups (Fig. 6C). Two of these groups were characterized by well-established mutational processes. Group 2 (signature B mutations) resembles signatures associated with activity of the AID/APOBEC family of cytidine deaminases (26, 27). This mutational process is common in human cancer including ALL (25) and was postulated to occur in short bursts initiated by retrotransposon mobility (28). The mutational burden in this group was relatively low (99–156 acquired mutations) compared with the other three groups (group 1; 220–1,210, group 3; 860–1,218, group 4; 104–710). Group 3 cases have high contribution of signature C, which clusters with mismatch repair (MMR)–associated signatures, with highest similarity to SBS15 (Fig. 6B). Indeed, the three relapses in this group all had biallelic mutations in one of the MMR genes (see Supplementary Data) and had high levels of single-base insertions or deletions in simple repeats (Fig. 6D), a feature of MMR deficiency (29). Genetic alterations in the MMR pathway have been associated with resistance to drugs such as thiopurines in ALL (30), suggesting that this mechanism of hypermutation directly contributed to relapse in these cases.
Figure 6.
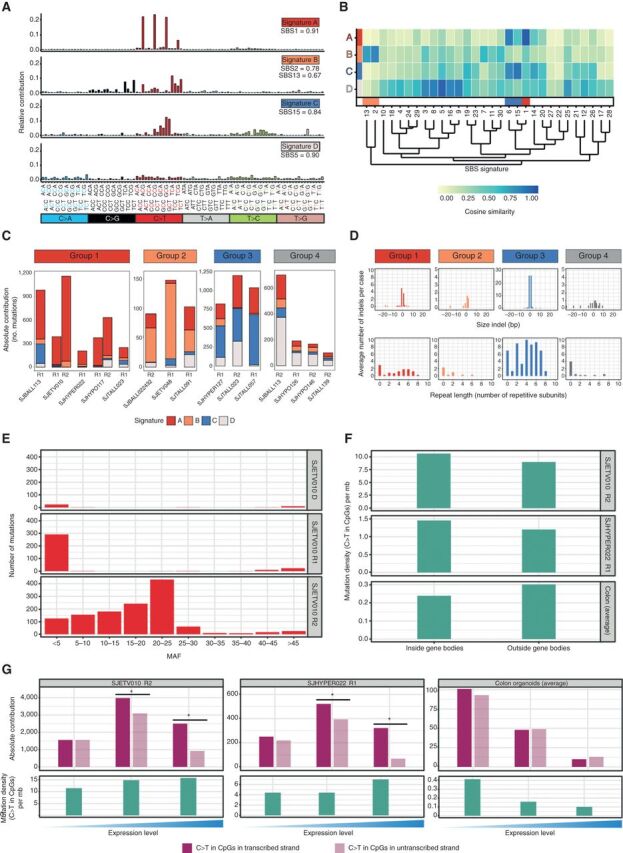
Mutational signature analysis of hypermutated relapses identifies multiple distinct mutational processes in hypermutation. A, Four mutational signatures identified in hypermutated ALL. Relative contribution of the different mutation types in their trinucleotide context, and cosine similarity values to reported COSMIC signatures are shown. B, Cosine similarity heatmap showing the hierarchical clustering of de novo signatures identified in this study with 30 known SBS signatures, including those associated with AID/APOBEC (orange bar), spontaneous deamination of meC (red bar), and mismatch repair (blue bar). C, Absolute contribution of each of the four signatures to the acquired mutations in 17 hypermutated relapse samples from 13 patients. Samples are grouped on the basis of the most prominent contributing signature. D, Average number and size of acquired indels in samples assigned to each group (top) and the number of repetitive subunits surrounding an inserted or deleted subunit (bottom). A value of 0 indicates that the indel is not located within a simple repeat. E, Total number of mutations (acquired and preserved) assigned with >95% confidence to signature A in the tumors of SJETV010, binned based on the mutation allele frequency (MAF). F, Density of C>T transitions in CpGs inside and outside gene bodies of two hypermutated relapses (SJETV010R2 and SJHYPER022R1) with high contribution of signature A mutations (top and middle) and healthy colon organoids with high contribution of SBS1 mutations (average of 3 organoids; bottom). G, Bar plots showing number of C>T transitions in CpGs on the transcribed and nontranscribed strand in relation to gene expression (top) and density of C>T transition in CpGs (bottom) in genes with no, low (<median) and high (≥median) expression (*, P < 0.05).
Two signatures could not be assigned to known mutational processes. Signature D (group 4) lacked a strong bias for a particular trinucleotide context and showed similarity to multiple mutational signatures. Signature A (group 1) resembled clock-like signature SBS1, which is a known consequence of a slow but progressive accumulation of C to T transitions at CpG sites owed to spontaneous deamination of methylated cytosines and is more apparent in cancers diagnosed at older age (29). Because the patients in our cohort are young, this process appears to be accelerated by an acquired imbalance between damage and repair. This was not caused by alterations in genes encoding regulators of DNA deamination. Interestingly, the signature A mutations in SJETV010 were subclonal (MAF < 0.5%) in the first relapse, but showed a much wider spread of allele frequencies in the second relapse, suggesting an ongoing endogenous mutational process initiated in a minor clone at first relapse (Fig. 6E). Recently, an SBS1-like signature (SBS74) has been reported that appears to be associated with MMR deficiency (31. Indeed, all hypermutated ALL relapses with MMR deficiency show signature A mutations (Fig. 6C; Supplementary Data).
To further compare the characteristics of signature A with the clock-like signature SBS1, we performed WGS of relapse and remission samples of SJETV010, SJHYPER022, and SJHYPO117, followed by somatic SNV calling and mutational signature extraction. In line with our findings in the coding regions, we identified highly concordant mutation densities, as well as composition and relative contribution of the four mutational signatures (Supplementary Fig. S10C–S10F). Signature SBS1 mutations have been described to occur throughout the genome and do not show strand asymmetry in transcribed regions (25, 32). We confirmed these observations using three recently published colon organoid samples (33), which are characterized by high prevalence of signature SBS1 mutations, mainly outside gene bodies. In contrast, we found that signature A mutation density was highest in gene bodies and showed strong transcription strand bias, particularly in genes with high expression in the respective samples (Fig. 6F and G; Supplementary Fig. S10G). We did not observe strand asymmetry associated with DNA replication for either signature A or signature SBS1 (Supplementary Fig. S10H). Transcriptional strand asymmetry can be the result of more efficient repair of the transcribed strand, or increased damage on the single-stranded, nontranscribed strand, two mechanisms that show opposite correlations with expression (34). Because signature A mutations are enriched in highly expressed genes, they may originate from transcription-coupled damage at the single-stranded, nontranscribed strand, as has been described for liver cancer (34).
The high prevalence of hypermutation in relapsed ALL suggested that this may result in increased generation of expressed neoepitopes that may be exploited by immunotherapeutic approaches to enhance antitumor responses of autologous T cells. We used WGS and RNA sequencing (RNA-seq) data to infer HLA class I types from each sample (35), predicted the binding affinities of all unique 8–12 amino acid peptides corresponding to SNVs and fusion proteins (36), and developed unweighted (UPAS) and weighted putative antigenicity scores (WPAS), the latter of which incorporates predicted sample-specific peptide: MHC binding with variant-specific expression. Although we observed variation in the number of fusion-encoded, predicted MHC-binding peptides across individual fusions (0–20, median = 1), fusion-encoded neoepitopes remained unchanged over time (Supplementary Table S21). In contrast, we observed that the number of predicted HLA-binding mutant peptides (≤500 nmol/L; ref. 37) per tumor rose with disease progression as a function of increased mutation burden, and thus particularly in hypermutated samples (P < 0.001, Supplementary Table S25; Supplementary Fig. S11A). In addition, the number of predicted MHC-binding peptides per tumor was significantly correlated with disease (B- and T-ALL), disease progression (D, R1, and R2), and signatures of hypermutation (Supplementary Fig. S11B). An expression-weighted antigenicity analysis comprising the subset of missense SNVs with expression data showed a significant effect of disease progression, with median WPAS lowest in R1 and highest in R2 variants (Supplementary Fig. S11C), and was particularly marked for known cancer genes (Supplementary Fig. S11D). These patterns may correspond to variations in immunologic pressure owed to, for instance, the distinct etiologies underlying B- and T-ALL and the successive use of immunosuppressive agents in treatment, respectively.
Discussion
Using multiple orthogonal approaches, we have described the patterns, dynamics, and drivers of clonal evolution in a large cohort of childhood relapsed ALL. These results have implications for the development of new approaches to monitor and treat ALL more effectively.
The scope of our study allowed us to identify relapsed enriched driver mutations more comprehensively than in prior studies and included newly identified targets of mutation as well as recurrent mutations in genes previously identified at relapse (11, 38, 39). We were able to identify distinct patterns of temporal acquisition across relapse-enriched targets of mutation, with mutations in genes such as CREBBP preserved from or acquired after diagnosis, and others in genes such as NT5C2 and USH2A only observed after initial therapy; importantly, these findings suggest a role for therapy in the induction of mutation, and/or a deleterious effect on initial leukemic fitness of these mutations (38). Mutations observed in different gene regulation pathways showed different frequencies of relapse-enriched genes between B- and T-ALL, indicating that distinct biological mechanisms drive the genetic alterations of the disease progression.
Tumors that evolved from, or retained multiple clones all had a short time to relapse, supporting a model in which early relapses are associated with dynamic clonal evolution and late relapses to a more inert pattern (40). These patterns of evolution, and description of the targets of mutation, have important implications for anticipation of relapse and modulation of therapy. Over half of relapses arise from a minor clone that commonly harbors, or acquires, relapse-enriched mutations that drive drug resistance. Analysis of this large cohort also enabled demonstration that relapse enriched drivers such as alterations of CREBBP, IKZF1, and NT5C2 are rarely lost if present at diagnosis or subsequently acquired, indicating that early detection may predict an increased likelihood of treatment failure. Moreover, in parallel studies using a subset of the xenografts described here, we have shown relapse-fated minor clones already exhibiting resistance to therapy are present at the time of diagnosis (22, 23). Thus, it is imperative that mutational profiling must now strive to achieve MRD levels of mutation detection at diagnosis or early in therapy, and we have shown the feasibility of this approach using ddPCR. An alternative approach is capture-based deep sequencing of regions of sequence and structural variation in ALL. Early detection will facilitate close monitoring and consideration of alternative treatment approaches such as intensification, immunotherapy, or novel drug approaches for drug-specific resistance (e.g., NT5C2 and thiopurines and CREBBP and corticosteroid resistance).
We show hypermutation is common at relapse, and driven by distinct mechanisms of mutation, including tumor-intrinsic processes, such as cytidine deaminase DNA editing activity (AID/APOBEC) as previously observed in diagnosis samples (41) and experimental models (42), or the acquisition of mutations that cause MMR deficiency, which may represent a mechanism of MMR-induced resistance to thiopurines in ALL (30). In addition, we identified a new SBS1-like signature (Signature A), characterized by transcriptional strand asymmetry and enrichment in expressed genes, caused by an unknown mutational mechanism that is acquired during leukemia progression. Importantly, the identification of hypermutation as a common phenomenon in relapsed ALL suggests that immunotherapeutic approaches intended to restore autoreactivity against neoantigen expression, such as checkpoint blockade, should be formally explored. Although long assumed to be poor targets for immunotherapy due to the relatively low mutation burden in comparison with other tumors (43), recent data have demonstrated that pediatric hematologic malignancies promote the generation of abundant and functional immune responses to tumor-specific neoepitopes despite the apparent inability of the immune system to control those tumors (44). In conjunction with those findings, our results suggest that not only does hypermutation drive the generation of HLA-restricted neoepitopes, but that these are expressed in an immunologically tolerized milieu that may be exploited with strategies to augment T-cell antitumor reactivity. It will now be of great interest to formally document the presence of autoreactive T-cell clones directed at neoepitopes induced by hypermutation, as we have described at diagnosis in ALL (44), and to formally test, in experimental models, whether immunomodulatory approaches can augment or restore antitumor reactivity in hypermutated ALL.
Methods
Subjects and Samples
The cohort included 92 children (31 female, 61 male) with relapsed B-progenitor (B-ALL, n = 67) or T-lineage [T-ALL, n = 25; including 7 early T-cell precursor ALL (ETP-ALL)] ALL patients treated on St. Jude Total Therapy Studies XI-XVI (median age at diagnosis 7.8 years; range, 1 month–18.7 years; Supplementary Table S1; refs. 45–47). The median time from diagnosis to first relapse was 2.7 years (range, 3 months–9.9 years). Sixteen cases developed a second relapse (range, 3 months–7 years). Relapse was very early (<1 year) in 17 patients, early (1–2 years) in 21 patients, and late (>5 years) in 14 patients. Nine cases developed a second tumor of different lineage, including 2 basal cell carcinomas, 1 B-cell lymphoma, 1 CML, and 5 AMLs. A total of 46 patients received bone marrow transplants at a median age of 12 years (range, 7 months–22 years), and all except one were allogeneic. Tumor samples with a blast percentage of less than 80% were flow sorted for the tumor population. Written informed consent was obtained from the patient and/or parent. The study was conducted in accordance with the Declaration of Helsinki, and was approved by the St. Jude Children's Research Hospital Institutional Review Board.
Genomic Analyses
DNA copy number aberrations were determined using SNP 6.0 microarrays (Affymetrix) in 307 samples from 92 patients (92 diagnosis, 84 relapse, 14 second relapse, 5 second tumor, and 91 germline samples; Supplementary Table S1). Data were analyzed using reference normalization (48) and circular binary segmentation (49).
We performed whole-exome sequencing (WES) on 276 samples from 92 cases (Supplementary Table S26) and WGS on 99 samples from 36 cases (Supplementary Table S27). Exomes were captured using the TruSeq Exome Library Prep Kit (67 Mb, 1 μg DNA input) or the Nextera Rapid Capture Expanded Exome Kit (62 Mb, 50 ng DNA input; Illumina). Paired-end sequencing was performed with the HiSeq 2500 sequencer (Illumina). The data were mapped to human reference genome hg19 and variant calling was performed using Bambino (50–52). All somatic SNVs and indels identified at diagnosis or relapse were validated using NimbleGen SeqCap Target Enrichment according to manufacturer's instructions (Roche NimbleGen) and resequenced using a HiSeq 2500 genome sequencer to a mean coverage >350× (250–500 ng DNA input; Supplementary Table S28). We performed transcriptome sequencing (RNA-seq) for TRIzol extracted RNA for 115 samples obtained from 66 cases (Supplementary Table S29). We used 1 μg RNA for library preparation with the TruSeq RNA Library Prep Kit v2 (Illumina) and 2 × 100 bp paired-end sequencing was performed on a HiSeq 2500 sequencer (Illumina). RNA-seq data were mapped to human reference genome hg19 using StrongArm and fusions were identified using CICERO (53) and FusionCatcher (54, 55).
B-ALL Subtyping Based on Gene Expression Profile from RNA-seq
Read counts for annotated genes (Ensembl Homo sapiens GRCh37 v75) were called by HTSeq (version 0.6.0; ref. 56) and processed by DESeq2 R package (57) to normalize gene expression into regularized log2 values (rlog). A subtype predication model was trained by Prediction Analysis of Microarrays based on a cohort of 309 samples from our previous studies (16, 53, 58), which consists of 8 B-ALL subtypes: IGH-DUX4 (n = 42), TCF3-PBX1 (n = 41), ETV6-RUNX1 (n = 42), hyperdiploid (n = 46), MEF2D-rearranged (n = 21), KMT2A (MLL)-rearranged (n = 44), BCR-ABL1 (n = 44), and ZNF384-rearranged (n = 27). The trained model was applied with 100 evenly divided thresholds (control selected feature genes from 5,000 to 50) and the probability score was averaged to predict subtypes for the enrolled RNA-seq samples.
Mutation Analysis and Clonal Modeling
Variants with a total coverage of <5 reads (combining WGS/WES and validation) were excluded. Variants with ≤2 variant reads were considered wild type; those with 3 to 8 variant reads only considered mutant when both WES/WGS and capture validation techniques identified the variant or the variant was called with higher coverage in other (tumor) samples of the same patient. A MAF of <30% was considered subclonal, 30% to 75% heterozygous, and MAF ≥75% homozygous.
MutSigCV (59) and the Genomic Random Interval Model (GRIN; ref. 17) analyses were performed to identify potential driver lesions (Supplementary Tables S11 and S12). MutSigCV is limited to analysis of sequence mutations, whereas GRIN incorporates multimodal genomic data including sequence and structural variants with robust adjustment for background mutation rate to identify significantly altered genes/regions, and unlike CNA-specific tools such as GISTIC (60), is not influenced by full chromosomal aneuploidies.
Two-dimensional MAF plots were used to visualize the relationship between sequential samples (4). We used sciClone (14) and manual curation incorporating xenograft data to assign variants to clones. Clonal evolution was visualized using ClonEvol (61) and fishplot (13). Noncoding variants were also considered to resolve the nature of clonal evolution in cases with presumed evolution from a major clone, which resulted in; required reclassification from evolution from a major to a minor clone in 5 of 27 cases. Retention of multiple clones was deemed polyclonal evolution, and relapse from an ancestral precursor where D/R tumors share only the founding translocation and up to two SNV/indels.
Xenografting
Diagnosis and/or relapse tumors of 8 cases [3 ETV6-RUNX1, 1 ETV6-RUNX1-like, 2 KMT2A (MLL)-rearranged, one DUX4 and 1 case without subtype data; Supplementary Table S1] were transplanted at limiting dilution from 250,000 cells to 10 cells into the femur of 8 to 12 week-old sublethally irradiated (225 cGy) female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. Engrafted tumor cells were harvested from bone marrow, spleen, and the central nervous system when mice displayed evidence of disease or at 30 weeks posttransplantation. Cells from the bone marrow and spleen were purified using the Miltenyi Mouse Cell Depletion Kit (Miltenyi Biotec; samples with >20% engraftment) or by cell sorting. All animal experiments were done in accordance with institutional guidelines approved by the University Health Network (Toronto, Canada) Animal Resource Centre (AUP#1117.37).
ddPCR
Seven relapse-associated mutations were genotyped by ddPCR technique (RainDance Technologies) using custom (NT5C2 p.P414A, CREBBP p.R1446C, NRAS p.G12R, and WHCS1 p.E1099K) or available (NT5C2 p.R39Q, KRAS p.G12S, and KRAS p. G12C) primers and probes (Supplementary Table S30). An average of 7 million droplets were generated by the RainDrop Source instrument, and emulsion PCR was performed using the C1000 Thermal Cycler (Bio-Rad). Droplet fluorescence of the amplified product was detected by the RainDrop Sense instrument, and data analysis was carried out using the RainDrop Analyst II Software. Detection limits were determined by testing serial dilutions of flow purified–mutant leukemia cells in wild-type REH cells. A frequency of >0.005% (>7 copies) could be consistently detected (Supplementary Fig. S9; Supplementary Table S24). ddPCR MAF correlated well with the MAFs called from WES (r = 0.971) or CapVal (r = 0.9964; Supplementary Fig. S9; Supplementary Table S23).
IGH and TCR Rearrangement Sequencing
IGH and TCRB loci were genotyped by ImmunoSeq (Adaptive Biotechnologies) to analyze clonal relationships of putative second tumors lacking shared genomic alteration (34 samples from 15 cases; Supplementary Table S19).
Germline Analyses
Germline copy number variants were filtered by the Database of Genomic Variants (62) and in-house databases. To prioritize germline SNV/indel variants, we filtered for rare variants (<0.01% population frequency in ExAC, dbSNP, GoNL, ESP, Wellderly, Kaviar, and Complete Genomics' 60 genomes databases) that were predicted to be deleterious (nonsense, frameshift, or canonical splice site variants, and missense variants with PhyloP>3) and were present in genes known to be associated with leukemia susceptibility and pediatric cancer syndromes (Supplementary Tables S31 and S32). Second, we used St. Jude's Medal Ceremony algorithm to identify Gold Medal variants (truncating mutations in tumor suppressors, matches to hotspots or truncating mutations in somatic mutation databases, and matches to locus-specific databases; ref. 63).
Mutational Signature Analysis
We defined hypermutation as samples containing >85 SNV/indels (∼1.3 mutations/Mb) based on the density histogram of the number of variants per sample in our cohort (Supplementary Fig. S4), which is more conserved than the cutoff determined by Segmented (ref. 24; >56 SNV/indels, 0.8 mutations/Mb).
For de novo extraction of somatic mutational signatures, we selected 22 hypermutated ALL samples from this cohort, and WES and WGS from patients with hypermutated B-ALL (n = 38) and WGS from B-cell lymphoma samples (N = 10, ICGC). Mutational signatures were extracted using NMF (64, 65) and MutationalPatterns (66). Similarity between two signatures, A and B, defined as nonnegative vectors with n mutation types, was calculated using cosine similarity. We defined two signatures to be the same if the cosine similarity is ≥0.95 (range 0–1). We calculated the cosine similarity between the mutational profiles of the samples and the mutational signatures and included the COSMIC mutational signatures and the de novo–extracted mutational signatures in this analysis.
Finally, we studied the strand asymmetry of signature A mutations in the context of transcription and replication. This analysis requires that individual mutations are assigned to a single signature. Using the number of trinucleotide changes and the signature probabilities per sample, we calculated the relative contributions of each signature for each trinucleotide context in a sample. Only trinucleotide changes with a relative contribution of ≥95% to one of the four signatures were assigned to that particular signature.
Replication timing in leukemia samples was determined using Repli-seq data obtained from five lymphoblastoid cell lines in the ENCODE project (ref. 67; GM06990, GM12801, GM12812, GM12813, GM12878), using median values per 1-kb bin (33). For intestinal organoids, we used Repli-seq data described previously (68). Predefined signature A mutations were assigned to early, intermediate, and late replicating regions, as described previously (33). Replication asymmetry analysis was done using the MutationalPatterns R package (66). On the basis of the distribution of rlog values from RNA-seq, genes were stratified into three groups for each sample: not expressed (genes with no or few supporting reads), genes with low expression (below the median level), and genes with high expression (median expression level or higher). We confirmed presence of SBS1 mutations by comparing their mutational profile with signature SBS1 for each sample (cosine similarity = 0.98; Supplementary Fig. S10). Gene expression stratification for healthy colon organoids was performed as described for the ALL samples. Testing of strand asymmetries within three groups was done using Poisson test for strand asymmetry significance testing.
Neoepitope Analyses
To characterize the antigenic potential of missense variants, we developed putative antigenicity scores that consider predicted patient-specific peptide: MHC binding variant-specific expression. WGS and RNA-seq data were used to infer class I HLA alleles to four-digit resolution for each patient using OptiType (35). For each missense SNV and patient HLA allele, we then used NetMHCcons 1.1 (36) to predict the binding affinities of all unique, mutated peptides of lengths 8–12 amino acids, excluding peptides that could be found elsewhere in the human proteome. Predicted binding affinities are often categorized as presumptive binders (≤ 500 nmol/L) and nonbinders, which can be useful for narrowing epitope targets (44), but to estimate total antigenic potential, we also conceptualized binding affinities as correlated with probabilities of peptide: MHC binding to consider all predicted binding affinities additively. The UPAS was calculated as the natural logarithm of the summation of the inverse of all putative binding affinities; for the subset of SNVs for which expression data were available, this value was then weighted by adding the natural logarithm of 0.01 + the fraction of expressed alternate-to-total bases to generate a WPAS specifically for those variants. Each of these scores is best considered in comparison across variants, with increasingly positive scores indicative of increasing putative antigenicity.
To investigate potential correlates of antigenic potential (e.g., disease, disease progression, hypermutator status, and interactions thereof) while controlling for the nonindependence of the data owed to multiple variants across patients, we used the lme4 R package (69) to fit linear mixed models with patient as a random effect. The car R package (70) was used to assess the statistical significance of the fixed effects, and residuals of models with significant effects were verified as unbiased and homoscedastic via visual inspection.
Data Availability
WES, WGS, transcriptome sequencing, and SNP array data are available at the European Genome-Phenome Archive (accession no. EGAS00001003975). The genomic landscape reported in this study can be explored at the St. Jude PeCan Data Portal https://stjuderesearch.org/site/data/relapsed-all.
Disclosure of Potential Conflicts of Interest
M.V. Relling reports receiving a commercial research grant from Servier Pharmaceuticals. P.G. Thomas has received speakers bureau honoraria from Illumina and PACT Pharma and has ownership interest (including patents) in PCT/US2016/064735. J.E. Dick is a Scientific Advisory Board member at Trillium Therapeutics, reports receiving a commercial research grant from Celgene, has ownership interest in a patent licensed to University Health Network. C.G. Mullighan reports receiving commercial research grants from Pfizer, AbbVie, and Loxo Oncology, has received speakers bureau honoraria from Amgen and Pfizer, and is a consultant/advisory board member for Illumina. No potential conflicts of interest were disclosed by the other authors.
Authors' Contributions
E. Waanders: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review, and editing. Z. Gu: Resources, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review, and editing. S.M. Dobson: Resources, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review, and editing. Ž. Antić: Software, formal analysis, investigation, methodology, writing–original draft, writing–review and, editing. J.C. Crawford: Resources, data curation, software, formal analysis, investigation, methodology, writing–original draft, writing–review, and editing. X. Ma: Resources, data curation, software, formal analysis, investigation, methodology, writing–original draft, writing–review, and editing. M.N. Edmonson: Data curation, software, formal analysis, and methodology. D. Payne-Turner: Data curation, software, formal analysis, investigation, and methodology. M. van de Vorst: Data curation and investigation. M.C.J. Jongmans: Data curation. I. McGuire: Resources, data curation, and visualization. X. Zhou: Resources and visualization. J. Wang: Resources and visualization. L. Shi: Resources, software, formal analysis, investigation, visualization, and methodology. S. Pounds: Software, formal analysis, investigation, and methodology. D. Pei: Software, formal analysis, investigation, and methodology. C. Cheng: Formal analysis. G. Song: Formal analysis. Y. Fan: Formal analysis. Y. Shao: Formal analysis, validation, and investigation. M. Rusch: Resources, validation, and investigation. K. McCastlain: Resources and investigation. J. Yu: Validation and investigation. R. van Boxtel: Formal analysis, validation, and investigation. F. Blokzijl: Formal analysis. I. Iacobucci: Resources, data curation, formal analysis, validation, investigation, and methodology. K.G. Roberts: Resources, data curation, validation, investigation, methodology, and project administration. J. Wen: Resources, formal analysis, investigation, and project administration. G. Wu: Formal analysis and investigation. J. Ma: Formal analysis and investigation. J. Easton: Formal analysis, validation, and investigation. G. Neale: Validation and investigation. S.R. Olsen: Investigation. K.E. Nichols: Resources and investigation. C.-H. Pui: Resources. J. Zhang: Resources, software, and formal analysis. W.E. Evans: Resources, software, and formal analysis. M.V. Relling: Resources. J.J. Yang: Resources. P.G. Thomas: Conceptualization, resources, data curation, software, formal analysis, and investigation. J.E. Dick: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review, and editing. R.P. Kuiper: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review, and editing. C.G. Mullighan: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review, and editing.
Supplementary Material
Supplementary results, figures, and table notes
Supplementary Tables
Acknowledgments
We thank the Biorepository, Genome Sequencing Facility of the St. Jude Hartwell Center for Bioinformatics and Biotechnology, and the Flow Cytometry and Cell Sorting Core Facility of St. Jude Children's Research Hospital. We acknowledge W.M. Nillesen and A.R. Mensenkamp for expert advice on germline mutation interpretation and N.B. Waanders for bioinformatic assistance. This work was supported in part by the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital, a St. Baldrick's Foundation Scholar Award, the Henry Schueler 41 & 9 Foundation, and a Robert J. Arceci Innovation Award (to C.G. Mullighan), the NCI grants P30 CA021765 (St. Jude Cancer Center Support Grant), P50 GM115279 (to C.G. Mullighan, M.V. Relling, W.E. Evans, G. Neale, J. Zhang, and J.J. Yang), Outstanding Investigator Award R35 CA197695 (to C.G. Mullighan), the Dutch Cancer Society (KUN2012-5366, to E.Waanders), the American Society of Hematology Scholar Award (to Z. Gu), the Leukemia & Lymphoma Society's Career Development Program Special Fellow (to Z. Gu), the NCI of the NIH under award number K99CA241297 (to Z. Gu), the KiKa Foundation (KiKa150 to R.P. Kuiper), the China Scholarship Council (CSC 201304910347, to J. Yu), and the Royal Netherlands Academy of Arts and Sciences Ter Meulen Grant to (J. Yu). Work to JED was supported by funds from: the Princess Margaret Cancer Centre Foundation, Ontario Institute for Cancer Research with funding from the Province of Ontario, Canadian Institutes for Health Research, Canadian Cancer Society Research Institute, Terry Fox Research Institute, Genome Canada through the Ontario Genomics Institute, and a Canada Research Chair.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
References
- 1. Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med 2015;373:1541–52. [DOI] [PubMed] [Google Scholar]
- 2. Nguyen K, Devidas M, Cheng SC, La M, Raetz EA, Carroll WL, et al. Factors influencing survival after relapse from acute lymphoblastic leukemia: a Children's Oncology Group study. Leukemia 2008;22:2142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iacobucci I, Mullighan CG. Genetic basis of acute lymphoblastic leukemia. J Clin Oncol 2017;35:975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun 2015;6:6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwartzman O, Savino AM, Gombert M, Palmi C, Cario G, Schrappe M, et al. Suppressors and activators of JAK-STAT signaling at diagnosis and relapse of acute lymphoblastic leukemia in Down syndrome. Proc Natl Acad Sci U S A 2017;114:E4030–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet 2013;45:290–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med 2013;19:368–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 2008;322:1377–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K, et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun 2014;5:3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 2016;113:11306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 2011;471:235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Irving J, Matheson E, Minto L, Blair H, Case M, Halsey C, et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood 2014;124:3420–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller CA, McMichael J, Dang HX, Maher CA, Ding L, Ley TJ, et al. Visualizing tumor evolution with the fishplot package for R. BMC Genomics 2016;17:880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miller CA, White BS, Dees ND, Griffith M, Welch JS, Griffith OL, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol 2014;10:e1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klco JM, Miller CA, Griffith M, Petti A, Spencer DH, Ketkar-Kulkarni S, et al. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA 2015;314:811–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet 2016;48:1481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pounds S, Cheng C, Li S, Liu Z, Zhang J, Mullighan C. A genomic random interval model for statistical analysis of genomic lesion data. Bioinformatics 2013;29:2088–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andries V, Vandepoele K, Staes K, Berx G, Bogaert P, Van Isterdael G, et al. NBPF1, a tumor suppressor candidate in neuroblastoma, exerts growth inhibitory effects by inducing a G1 cell cycle arrest. BMC Cancer 2015;15:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alexander TB, Gu Z, Iacobucci I, Dickerson K, Choi JK, Xu B, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature 2018;562:373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature 2011;469:362–7. [DOI] [PubMed] [Google Scholar]
- 21. Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 2011;469:356–61. [DOI] [PubMed] [Google Scholar]
- 22. Garcia Prat L, Dobson SM, Chan-Seng-Yue M, Vanner R, Murison A, Wintersinger J, et al. Relapse-initiating clones preexisting at diagnosis in B- cell acute lymphoblastic leukemia help predict molecular pathways of relapse. Blood 2018;132:915. [Google Scholar]
- 23. Dobson SM, García-Prat L, Vanner RJ, Wintersinger J, Waanders E, Gu Z, et al. Relapse-fated latent diagnosis subclones in acute B lineage leukemia are drug tolerant and possess distinct metabolic programs. Cancer Discov 2020;10:568–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja R, et al. Comprehensive analysis of hypermutation in human cancer. Cell 2017;171:1042–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The life history of 21 breast cancers. Cell 2012;149:994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012;149:979–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petljak M, Alexandrov LB, Brammeld JS, Price S, Wedge DC, Grossmann S, et al. Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell 2019;176:1282–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47:1402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Evensen NA, Madhusoodhan PP, Meyer J, Saliba J, Chowdhury A, Araten DJ, et al. MSH6 haploinsufficiency at relapse contributes to the development of thiopurine resistance in pediatric B-lymphoblastic leukemia. Haematologica 2018;103:830–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Ng AW, Boot A, et al. The repertoire of mutational signatures in human cancer. bioRxiv 2018:322859. [Google Scholar]
- 32. Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 2016;538:260–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 2018;556:457–62. [DOI] [PubMed] [Google Scholar]
- 34. Haradhvala NJ, Polak P, Stojanov P, Covington KR, Shinbrot E, Hess JM, et al. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell 2016;164:538–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 2014;30:3310–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karosiene E, Lundegaard C, Lund O, Nielsen M. NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics 2012;64:177–86. [DOI] [PubMed] [Google Scholar]
- 37. Eisen HN, Hou XH, Shen C, Wang K, Tanguturi VK, Smith C, et al. Promiscuous binding of extracellular peptides to cell surface class I MHC protein. Proc Natl Acad Sci U S A 2012;109:4580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tzoneva G, Dieck CL, Oshima K, Ambesi-Impiombato A, Sanchez-Martin M, Madubata CJ, et al. Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia. Nature 2018;553:511–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 2009;360:470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spinella JF, Richer C, Cassart P, Ouimet M, Healy J, Sinnett D. Mutational dynamics of early and late relapsed childhood ALL: rapid clonal expansion and long-term dormancy. Blood Adv 2018;2:177–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gawad C, Koh W, Quake SR. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc Natl Acad Sci U S A 2014;111:17947–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Swaminathan S, Klemm L, Park E, Papaemmanuil E, Ford A, Kweon SM, et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol 2015;16:766–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zamora AE, Crawford JC, Thomas PG. Hitting the target: how T cells detect and eliminate tumors. J Immunol 2018;200:392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zamora AE, Crawford JC, Allen EK, Guo XJ, Bakke J, Carter RA, et al. Pediatric patients with acute lymphoblastic leukemia generate abundant and functional neoantigen-specific CD8(+) T cell responses. Sci Transl Med 2019;11: pii:eaat8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pui CH, Pei D, Sandlund JT, Ribeiro RC, Rubnitz JE, Raimondi SC, et al. Long-term results of St Jude Total Therapy Studies 11, 12, 13A, 13B, and 14 for childhood acute lymphoblastic leukemia. Leukemia 2010;24:371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pui CH, Sandlund JT, Pei D, Campana D, Rivera GK, Ribeiro RC, et al. Improved outcome for children with acute lymphoblastic leukemia: results of Total Therapy Study XIIIB at St Jude Children's Research Hospital. Blood 2004;104:2690–6. [DOI] [PubMed] [Google Scholar]
- 47. Pui CH, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med 2009;360:2730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pounds S, Cheng C, Mullighan C, Raimondi SC, Shurtleff S, Downing JR. Reference alignment of SNP microarray signals for copy number analysis of tumors. Bioinformatics 2009;25:315–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 2007;23:657–63. [DOI] [PubMed] [Google Scholar]
- 50. Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 2013;45:242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012;481:157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Edmonson MN, Zhang J, Yan C, Finney RP, Meerzaman DM, Buetow KH. Bambino: a variant detector and alignment viewer for next-generation sequencing data in the SAM/BAM format. Bioinformatics 2011;27:865–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 2014;371:1005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nicorici D, Satalan M, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, et al. FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv 2014:11650. [Google Scholar]
- 55. Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K, et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol 2011;12:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gu Z, Churchman M, Roberts K, Li Y, Liu Y, Harvey RC, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun 2016;7:13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dang HX, White BS, Foltz SM, Miller CA, Luo J, Fields RC, et al. ClonEvol: clonal ordering and visualization in cancer sequencing. Ann Oncol 2017;28:3076–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 2014;42:D986–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med 2015;373:2336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 2010;11:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 2017;358:234–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bates D, Machler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J Stat Softw 2015;67:1–48. [Google Scholar]
- 70. Fox J, Weisberg S. An R companion to applied regression. Thousand Oaks: Sage; 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary results, figures, and table notes
Supplementary Tables
Data Availability Statement
WES, WGS, transcriptome sequencing, and SNP array data are available at the European Genome-Phenome Archive (accession no. EGAS00001003975). The genomic landscape reported in this study can be explored at the St. Jude PeCan Data Portal https://stjuderesearch.org/site/data/relapsed-all.