Abstract
The immune system is a highly complex and dynamic biological system. It operates through intracellular molecular networks and intercellular (cell–cell) interaction networks. Systems immunology is an emerging discipline that applies systems biology approaches of integrating high-throughput multi-omics measurements with computational network modeling to better understand immunity at various scales. In this review, we summarize key omics technologies and computational approaches used for immunological studies at both population and single-cell levels. We highlight the hidden driver analysis based on data-driven networks and comment on the potential of translating systems immunology discoveries to immunotherapy of cancer and other human diseases.
Keywords: Systems immunology, scRNA-seq, gene regulatory network, cell–cell communication, hidden driver analysis
Graphical Abstract

Introduction
The immune system, one of the most complex and dynamic biological systems in mammals, is comprised of diverse cell types with varying functional states. Between the two major arms of the immune system, the innate immune system, comprised of macrophages, dendritic cells, neutrophils and other cells, serves as the first line of defense by mounting immediate and potent immune and inflammatory responses against invading pathogens and other immunological insults. Cells in the adaptive immune system, including T and B cells, have a more specialized role in immune reactions and are characterized by antigen specificity and long-term memory. These different elements interact as an integrative system to give rise to proper immune responses and regulation, and play crucial roles in protecting host health against viruses, bacteria, parasites and tumors. Dysfunctions in the immune network may lead to autoimmune, malignant, and inflammatory diseases. Characterizing these diverse cell types, their unique molecular features, and their interactions is the key to successfully manipulate the immune system for therapeutic applications. Advances in high-throughput profiling technologies, particularly the emerging single-cell omics platforms, enable comprehensive characterization of the immune components at multiple scales. However, immunity is not merely a sum of its components, and its behavior cannot be explained or predicted solely by examining individual components. Therefore, systems biology approaches are essential for decoding the cellular complexity, plasticity, and functional diversity of the immune system, leading to the emerging field of systems immunology to better understand how the immune system works as a whole in health and disease.
Gene regulatory networks function as the crucial molecular determinants of cell fate and state by governing gene expression programing and reprograming in immune development and homeostasis [1]. Signaling and epigenetic factors are also crucial drivers of immunological functions and are likely druggable, making them promising therapeutic targets. However, it is often difficult to identify many of these drivers (hence known as “hidden drivers”), because they may not be genetically altered or differentially expressed at the mRNA or protein levels but, rather, are altered by posttranslational modifications (PTMs; e.g., phosphorylation) or other mechanisms. Moreover, immune responses are mediated by both the intracellular gene networks and crosstalk between many types of immune cells in specific tissues and microenvironmental contexts; and their dysregulations can lead to diseases, including cancer and inflammatory disorders. Therefore, molecular and cellular networks, and their drivers and “hidden” drivers (cannot be easily detected by conventional approaches) must be systematically dissected to develop effective and curative immunotherapies for diseases such as cancer [2].
Omics technologies for immunology research
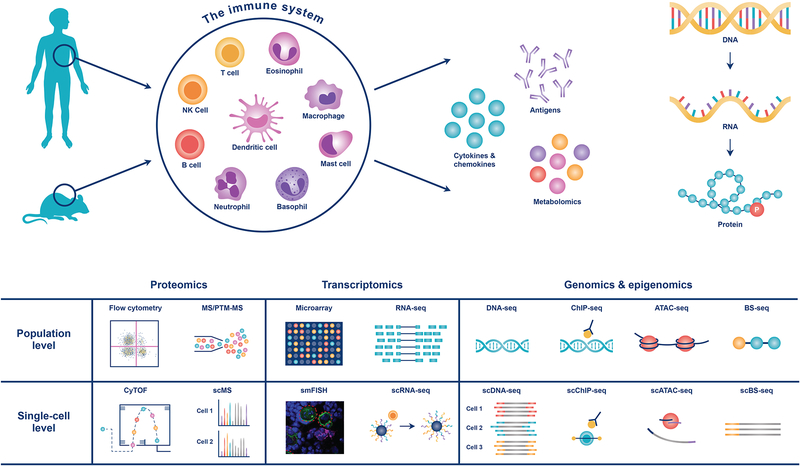
Technological advances in high-throughput and high-bandwidth profiling, phenotyping and perturbation assays have contributed to rapid advances of systems immunology. A variety of omics technologies at both population and single-cell levels have played important roles in improving our understanding of the immune system (Figure 1). Each technology has its advantages and limitations, and understanding these factors is essential to devise effective and reliable systems approaches that address immunological questions at the appropriate resolution. In this section, we discuss these technologies by summarizing the essential aspects of their proper use, with example applications and certain limitations.
Figure 1.
Overview of the omics profiling technologies to characterize the immune system of human and mouse at population and single-cell levels.
Population level
Transcriptome profiling by microarray or RNA-based next-generation sequencing (RNA-seq) is the most widely used omics method in immunology research. Transcriptome analysis has provided instrumental insights into the mechanisms of immune system development and homeostasis under steady state, and transcriptional dynamics during the immune response to antigens or pathogens, including the identification of diverse immune cell types and functional states [3]. As the cost of sequencing decreases, RNA-seq, particularly bulk RNA-seq, has become the more prevalent technology for gene expression profiling, with several advantages than microarray technology: high coverage and sensitivity (detecting low-abundance transcripts); detection of splicing events, gene fusions, and small RNAs; low background noise and batch effects; and the ability to handle low RNA input (down to 10 pg). The community-driven and publicly available databases of gene expression profiles, such as Gene Expression Omnibus (GEO), have enabled data mining across platforms, studies, and species. A few curated immune-specific databases with analysis and visualization tools have provided valuable resources for immunology researchers, including ImmGen [4], ImmPort [5], ImmuneSpace [6], and 10K Immunomes [7].
The expression levels of mRNA and protein can differ substantially for many genes [8], especially during the dynamic transitional state when there is a temporal delay between transcription and translation [9]. Moreover, posttranslational modifications, such as phosphorylation, are crucial regulators of protein functions and signaling, but are poorly correlated with mRNA or total protein expression. With the recent advances in mass spectrometry (MS) analytical technologies [10], in-depth proteomic profiling can now identify more than 10,000 proteins (whole proteomics) and 30,000 phosphopeptides (phosphoproteomics) across multiple samples simultaneously [11**,12,13]. The tandem mass tagging (TMT) [14] and the label-free quantitation (LFQ) are two common proteomic methods to quantify the differential abundance of expressed proteins, with the TMT method recently shown to have higher precision and coverage than the LFQ method [15,16]. Despite the challenge to cover the entire proteome and PTM landscape, current MS-based proteomic technologies are capable of providing comprehensive characterizations of proteome dynamics and biological insights into gene regulation and signaling circuits in immunology, such as T-cell activation [11**,17] and host-pathogen interaction [18]. Proteomics by affinity purification-mass spectrometry (AP-MS) is also commonly used to identify protein-protein interactions (PPIs) [19**] that help dissect the molecular mechanisms of crucial immunological modulators, for example, Mst1 signaling in regulatory T cells [20]. Recently, advanced MS-based platforms have been developed to profile and explore the metabolome that may shape the functions of immune cells (e.g., metabolomics [21] and lipidomics [22,23]).
DNA-based next-generation sequencing (NGS) has revolutionized the study of many fields in biology, including immunology [24**]. Whole-genome or -exome sequencing and targeted DNA sequencing are now routinely used to identify somatic genetic alterations associated with cancer and other diseases, spurring the advent of precision medicine [25]. In basic immunology research, NGS is commonly used to dissect protein-DNA interactions (ChIP-seq) [26], protein-RNA interactions (CLIP-seq) [27], DNA methylation (Bisulfite-seq) [28], chromosomal interactions (Hi-C) [29], and chromatin accessibility (ATAC-seq) [30].
The revolutionary CRISPR/Cas-based genome engineering technologies enable the use of genome-wide functional perturbation screening [31] to systematically interrogate novel players and circuits that regulate or modulate immune development, homeostasis and response [32,33**]. CRISPR and conventional RNAi screens perform comparably for identifying essential genes [34]. Novel CRISPRi/a technologies provide a complementary but superior approach to RNAi by repressing or activating gene expression at the transcriptional level, while RNAi represses gene expression at the mRNA level [35].
Single-cell level
The immune system encompasses various cell types and functional states. Population- or bulk-based profiling performed by averaging results from thousands of cells of distinct types presents an inherent heterogeneity problem for data analysis and interpretation. However, the advent of single-cell technologies to profile the transcriptome (scRNA-seq) [36], proteome (mass cytometry or CyTOF, NanoLC-MS) [37,38], genome [39], and chromatin accessibility or epigenome (scATAC-seq, scChIP-seq, scBS-seq, scHi-C) [40,41,42,43,44] has provided an unprecedented opportunity to overcome this challenge by simultaneously quantifying molecular features at the single-cell resolution. Indeed, single-cell technology was recognized as the breakthrough of the year for 2018. In immunology research, scRNA-seq [45**] and mass cytometry [37] are widely adapted.
In the last few years, advances in technologies of cell suspension, automation, microfluidics and implementation of unique molecular identifiers have boosted the scRNA-seq field by improving the throughput (the number of cells), sensitivity (the number of uniquely-detected genes), precision (level of noise), and reproducibility [46**]. The scRNA-seq technology has been widely used in immunology to reveal immune cell heterogeneity and dynamics in healthy and malignant conditions [47*,48*,49*]. Significant efforts have been invested to profile the entire human and mouse cell atlas [50,51,52]. Because of their high-throughput of cells, droplet-based scRNA-seq platforms, including 10X Genomics Chromium [53], inDrop [54], and Drop-seq [55], are becoming more popular than FACS- or plate-based protocols for immunology studies [56]. However, plate-based methods have no sequencing bias on the 5’ or 3’ end of transcript tags and capture more molecules than droplet-based platforms [57]. The combined use of both platforms can provide more comprehensive and in-depth information [58]. Imaging-based, single-molecule fluorescence in situ hybridization (smFISH) [59,60] is another powerful, emerging technology for high-throughput single-cell transcriptomics with additional spatial information integrated, but is yet to be applied to the immune system.
Flow cytometry uses fluorescent antibodies to simultaneously profile multiple proteins per cell and has been the mainstay for immune-phenotyping. Mass cytometry overcomes the limitation associated with the spectral overlap of fluorophores in flow cytometry by using metal-conjugated antibodies that increase the dimension [37]. It has enabled the identification and characterization of a variety of immune cell types and states in the mammalian immune system with emerging applications in the clinic [61]. However, this technology is limited to a small number of pre-defined parameters (e.g., surface markers), and the profiling of these parameters depends on the availability of protein-specific antibodies. More recently, a multiplexed immunofluorescence method has been developed to obtain 40 protein readouts of thousands of cells in situ [62], which may also be adopted in immunology.
To understand cellular behaviors in-depth, strategies to integrate multiple single-cell omics technologies or combine them with population-based profiling to simultaneously profile various dimensions of biological information from the same cell have emerged [63]. For instance, recent studies have combined profiles of single-cell and bulk transcriptomes [64]; transcriptomes and chromatin states [65,66*]; transcriptomes and protein epitopes [67,68,69]; transcriptomes obtained by scRNA-seq and those obtained by smFISH [70*]; epitomes and protein epitopes [71]; transcriptomes and functional genomes [72,73,74]; and genome, transcriptome, and methylome data [75]. Application of these cutting-edge integrative technologies to immunological questions will likely provide new insight in our understanding of the immune system.
Computational approaches for systems immunology
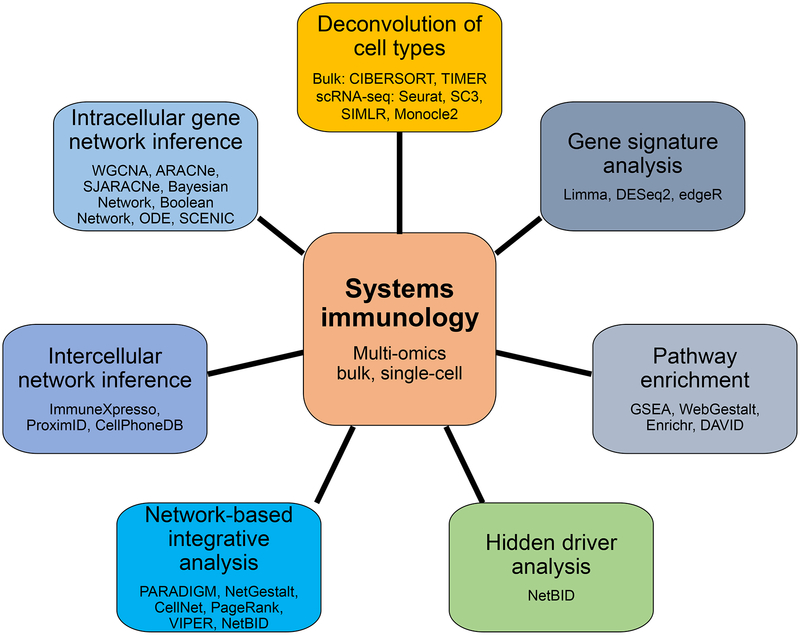
Multi-omics technologies providing population– and single-cell–level information give rise to remarkably rich and complex datasets with which to tackle immunological questions. However, interpretation and integration of such “big” data remain a challenge and a barrier to broad implementation of systems approaches in immunological studies. In this section, we review common computational algorithms and strategies for in-depth analysis and integration of multi-omics data in systems immunology (Figure 2). We start with the immune cell deconvolution to identify proportions of cells within heterogenous populations, and then discuss various systems biology strategies to dissect the molecular pathways or features associated with immune cell identity, function and response, with an emphasis on hidden driver analysis based on data-driven networks to decode regulatory mechanisms of the immune system.
Figure 2.
Overview of common computational analyses and algorithms in systems immunology.
Deconvolution of the immune cellular heterogeneity
One of the most frequent analyses in immunology is immune-cell phenotyping because extensive cellular heterogeneity underpins the functional diversity of the immune system. For bulk microarray or RNA-seq gene expression profiles, linear regression-based deconvolution algorithms [76,77,78] have been developed to predict the frequency of diverse cell subsets based on predefined signatures. However, these approaches rely on prior knowledge on existing immune cell types. Instead, the widespread adoption of single-cell profiling enables unbiased identification of known and unknown subsets of immune cells. Several algorithms for clustering analysis, cell-type identification, and visualization from single-cell transcriptomics data have emerged. For instance, SC3 employs consensus k-means clustering method with a combination of various distance metrics and initial conditions that improves the accuracy and robustness of clustering in comparison with previous approaches [79]. For more detailed discussion of cluster algorithms for scRNA-seq, we refer readers to other comprehensive reviews [80**,81]. However, more advanced and efficient algorithms of scRNA-seq analysis remain needed to capture the nonlinear cell–cell correlations, to reduce noise from the “dropout” effects, and to handle datasets with millions of cells.
Gene signature and pathway enrichment analysis
Genome-wide transcriptomic and proteomic profiles of immune cells following treatments, stimulations or genetic perturbations provide valuable insights into molecular signatures and pathways that define cell identity, gene regulation, and immune responses. Differential gene expression analysis is the mainstream strategy to define a gene signature, followed by functional or pathway enrichment by hypergeometric test or gene set enrichment analysis (GSEA)-type approaches [82,83,84,85]. However, the signature analysis may be limited by poor correlation between different studies, as signature genes derived from independent experiments may not be entirely consistent. Additionally, the pathway databases may lack context-specific information and are limited by incomplete or inaccurate prior knowledge. Immune cell deconvolution gives the proportion of heterogeneous cell types while functional enrichment analysis defines the molecular features in each cell type. A combination of these two approaches facilitates downstream analysis of intracellular and intercellular interactions.
Intracellular gene network inference
The availability of large-scale profiling platforms enables the study of relationships among the molecular elements (i.e., intracellular gene networks) in the immune system. Most of the network reconstruction methods are based on gene expression profiles of perturbation experiments (e.g., gene silencing, deletion or overexpression), as previously reviewed [86,87]. Here, we highlight two common network inference strategies that use baseline transcriptomic data. One is co-expression network analysis by WGCNA [88] based on Pearson or Spearman correlations. However, co-expression networks usually contain a large number of redundant interactions that lack biological relationships. To overcome this problem, ARACNe [89] uses mutual information to capture nonlinear gene–gene relationships and applies data processing inequality to remove redundant edges. It has been widely used to infer transcription factor (TF) regulatory networks from gene expression data. Recently, SJARACNe [90] was developed to scale up and extend ARACNe to infer both TF regulatory and signaling networks from large-input datasets, including scRNA-seq data. For example, SJARACNe was used to reverse-engineer the signaling interactome of dendritic cells (DCs), leading to novel molecular insights into the functions of DC subsets [91**]. For scRNA-seq data, SCENIC utilizes TF motif databases to reconstruct regulatory networks that improves clustering and reduces batch effects [92]. Other modeling approaches including Bayesian network, Boolean network, and diffusion or differential equation–based network approaches, are used for inference of small-sized networks [93]. For example, Bayesian network was used to identify causal correlations of molecular and clinical features of Alzheimer’s disease [94]. However, it remains challenging to scale up these approaches for genome-wide networks, because of the high complexity of parameters and limited samples [95]. To complement and improve networks predicted in silico, experimental approaches are also used to directly infer subnetworks of proteins of interest (e.g., TF regulatory network by ChIP-seq [96], post-transcriptional networks by CLIP-seq [27], PPI by AP-MS [19], enzyme-substrate network by PTM-enriched proteomics [97], and metabolic networks by metabolomics [21]). However, these networks are limited to selected proteins and lack generalizability.
Network-based integrative analysis
Integration of multi-tier omics data increases the sensitivity and reliability of discoveries in the immune and other complex biological systems by aggregating information at multi-layers to increase the signal-to-noise ratio [93,98]. This approach is particularly important in understanding immune system function, given the high complexity of cellular components and molecular circuits in the immune system. However, different omics platforms have distinct features and dimensions, making the meta-analysis challenging. The most popular strategy is to superimpose co-expression or regulatory networks, constructed from transcriptomes and/or knowledge-based network databases (e.g., MSigDB, PPI, TF-target, kinase-substrate) on various omics data to identify network modules that control immune cell development and response [99]. For example, this strategy has been applied to integrate temporal transcriptome, proteome, and phosphoproteome data, leading to the identification of novel signaling circuits and bioenergetics pathways that mediate T-cell quiescence exit [11]. Additionally, PARADIGM [100] integrates genomic and transcriptomic alterations to identify dysregulated pathways. NetGestalt [101] defines the hierarchical architecture in the network of omics data clustering. CellNet [102] utilizes co-expression networks to determine the cell identity and master regulators of cell types/states. PageRank combines ATAC-seq and transcriptomic datasets to identify master regulators of T-cell residency in non-lymphoid tissues and tumors [103**]. Both VIPER [104] and NetBID [91**] use ARACNe/SJARACNe-derived regulatory networks to infer protein activities in individual samples and master regulators associated with phenotypes. While VIPER is focused primarily on gene expression data, NetBID uses a distinct activity inference algorithm and a Bayesian framework to integrate multiple omics data.
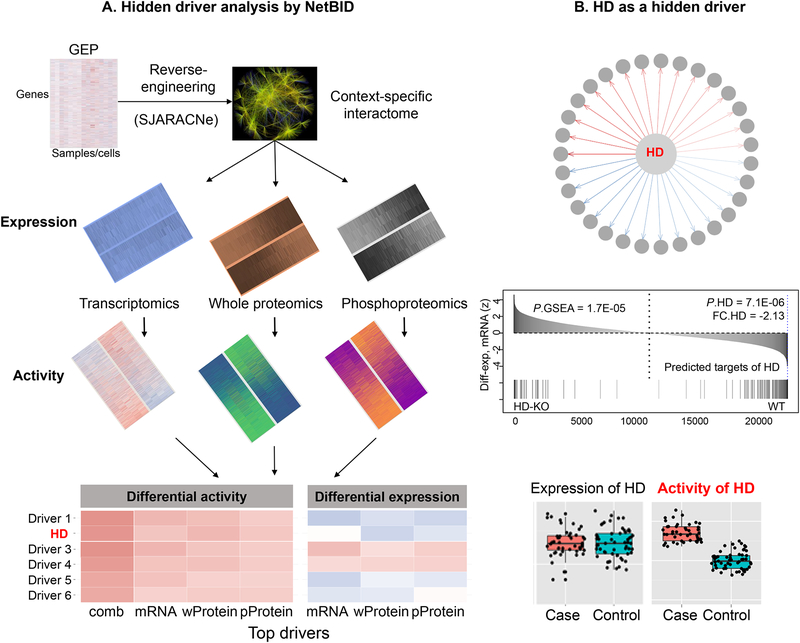
Hidden driver analysis based on data-driven and context-specific networks
In addition to transcription factors that are the focus of most network-based algorithms, signaling and epigenetic factors are also crucial drivers of immunological functions. However, many of these factors are hidden drivers, because their activities are associated with PTM but not with genetic alterations or expression abundance. PTM proteomics-based direct measurements of protein activities are technically challenging. Here, we highlight NetBID [91] (Figure 3), a recently developed algorithm to identify hidden drivers from multi-omics data by using data-driven networks and Bayesian inference. In our study of DC subset functions [91], NetBID superimposed a DC-specific signaling interactome, which was computationally reconstructed from a set of transcriptomic profiles of total DCs, onto multi-layer omics datasets (transcriptome, proteome, phosphoproteome) to infer activities of signaling proteins in CD8α+ and CD8α− DCs, followed by a Bayesian approach to integrate information at all levels, leading to the identification of putative hidden drivers that selectively modulate functions of DC subsets. In particular, NetBID has identified the Hippo kinase Mst1/Stk4 as a hidden driver, selectively active in CD8α+ DCs, which was further validated by genetic and functional experiments. Of note, there is no differential expression of Mst1 at mRNA levels, while Mst1 protein expression is even lower in CD8α+ than CD8α− DCs. One advantage of NetBID for successfully capturing Mst1 is that the Mst1 subnetwork inferred in silico is enriched in its putative downstream targets as defined by perturbation experiments, enabling inference of its true functional activity. NetBID currently relies on bulk omics data. An improved version that handles single-cell omics data to infer cell-type–specific hidden drivers remains to be developed.
Figure 3.
Hidden driver analysis by NetBID. (A) The overview flowchart of NetBID analysis to identify hidden drivers of phenotype case vs. control. (B) An illustration of an example hidden driver (HD) that has no differential expression but has network enrichment and activity. Diff-exp, differential expression.
Intercellular network inference
Cell–cell communication fundamentally regulates how the immune system operates as a network to effectively respond to infection and other insults. Systematically decoding intercellular networks that modulate immunity has been a longstanding challenge. Recently, an algorithm developed by text mining the literature has predicted previously unappreciated cell–cytokine interactions [105*], but the attempt is limited by the inherent bias of existing knowledge. Single-cell technologies have provided a unique opportunity to tackle this challenge in a more unbiased manner. Systematic inference of intercellular communications is still in early development, with a few limited examples based on scRNA-seq to date [106,107*,108,109**,110].
Closing remarks: towards translational systems immunology
Technological advancement is driving fundamental discoveries in immunology. Recent advances in single-cell technologies enable the study of immunological diversity and complexity at an unprecedented resolution. Next-generation, single-cell omics methods are able to simultaneously capture additional information, such as spatial organization [111], dynamic clonality via lineage barcoding [112], and immune receptor repertoire [113*,114,115]. An equally important and complementary effort is to develop sophisticated computational algorithms to analyze and integrate high-throughput multi-omics and multi-sourced data [116].
The importance of translational immunology is illustrated by the remarkable success of cancer immunotherapies that demonstrate durable responses in the clinic, including CAR-T-cell therapies [117] and checkpoint-blockade therapies [118], which were recently recognized by the Nobel prize [119]. For example, tumor cells escape immune surveillance by up-regulating PD-L1 that interacts with PD-1 receptor on T cells to elicit the immune checkpoint response. Therefore, blocking the crosstalk between PD-L1 on tumor cells and PD-1 on T cells will reactivate the cytotoxic T cells to kill tumor cells. However, immunotherapies are efficacious for only a fraction of patients, and existing biomarkers based on tumor mutation burden and single protein expression (e.g., PD-L1) have limited prediction power. The emerging systems immunology approaches could be translated to tackle pressing issues in the clinic [98] by dissecting the heterogeneity and interactions of tumor and immune microenvironment [116]. For instance, integrative systems biology analysis of bulk omics data from over 10,000 patient samples of 33 cancer types has provided instrumental insights into the immune landscape of cancer [120]. More recently, scRNA-seq and high-dimensional flow cytometry analyses of human tumors have revealed a unique CD8 T cell subset that infiltrates tumors and responds to checkpoint blockade immunotherapy to mediate effective tumor immunity [121,122], and this control mechanism is also observed and validated in murine tumor models [123,124]. The state-of-the-art technologies are enabling comprehensive molecular characterization of tumor cells and their microenvironment from large cohorts of patient samples at the single-cell resolution. The development of immune-competent and humanized mouse models is facilitating immune-related functional and mechanistic studies. We envision that network-based systems immunology analysis of multi-omics data, from both the human and mouse model systems, will enable identification of hidden drivers of resistance to existing cancer immunotherapies, novel predictive biomarkers to better stratify patients, and novel therapeutic targets and combination strategies to overcome drug resistance and develop more precise immunotherapy. These strategies may also manifest legitimate therapeutic opportunities for other immune-related disorders, including autoimmune, inflammatory and neurodegenerative diseases.
Highlights.
Systems immunology is emerging with omics tools at population & single-cell levels
Integrative analysis of multi-omics data has revealed novel insights in immunology
Single-cell sequencing technology is driving immunology research
Data-driven and context-specific networks enable capture of hidden drivers
Acknowledgements
The authors apologize to all colleagues whose key contributions could not be individually cited due to space limitations. We thank Chenxi Qian, Koon-Kiu Yan, and Xinran Dong for help with the artwork, and Nicole Chapman, Yanyan Wang, Yogesh Dhungana and Hao Shi for editing and scientific inputs.
Funding: This work was supported by ALSAC, the St. Jude Comprehensive Cancer Center Developmental Fund [to J.Y.], by the National Institutes of Health [AG047928, AG053987, GM114260 to J.P.; AI105887, AI131703, AI140761, CA176624, CA221290, NS064599 to H.C.].
Abbreviation
- ChIP-seq
chromatin immunoprecipitation sequencing
- CLIP-seq
crosslinking immunoprecipitation sequencing
- BS-seq
bisulfite sequencing
- ATAC-seq
assay for transposase-accessible chromatin using sequencing
- Hi-C
all-vs-all chromosome conformation capture by sequencing
- CyTOF
cytometry by time of flight
- LC-MS
liquid chromatography-mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Yui MA, Rothenberg EV: Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol 2014, 14:529–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P, Allison JP: Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 2015, 161:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burel JG, Apte SH, Doolan DL: Systems Approaches towards Molecular Profiling of Human Immunity. Trends Immunol 2016, 37:53–67. [DOI] [PubMed] [Google Scholar]
- 4.Shay T, Kang J: Immunological Genome Project and systems immunology. Trends Immunol 2013, 34:602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J, et al. : ImmPort: disseminating data to the public for the future of immunology. Immunol Res 2014, 58:234–239. [DOI] [PubMed] [Google Scholar]
- 6.Sauteraud R, Dashevskiy L, Finak G, Gottardo R: ImmuneSpace: Enabling integrative modeling of human immunological data. The Journal of Immunology 2016, 196:124.165–124.165.26573834 [Google Scholar]
- 7.Zalocusky KA, Kan MJ, Hu Z, Dunn P, Thomson E, Wiser J, Bhattacharya S, Butte AJ: The 10,000 Immunomes Project: Building a Resource for Human Immunology. Cell Rep 2018, 25:1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walley JW, Sartor RC, Shen Z, Schmitz RJ, Wu KJ, Urich MA, Nery JR, Smith LG, Schnable JC, Ecker JR, et al. : Integration of omic networks in a developmental atlas of maize. Science 2016, 353:814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Beyer A, Aebersold R: On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165:535–550. [DOI] [PubMed] [Google Scholar]
- 10.Aebersold R, Mann M: Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537:347–355. [DOI] [PubMed] [Google Scholar]
- 11.**.Tan H, Yang K, Li Y, Shaw TI, Wang Y, Blanco DB, Wang X, Cho JH, Wang H, Rankin S, et al. : Integrative Proteomics and Phosphoproteomics Profiling Reveals Dynamic Signaling Networks and Bioenergetics Pathways Underlying T Cell Activation. Immunity 2017, 46:488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper uses co-expression network-based approach to integrate time-series proteomics, phosphoproteomics and transcriptomics data to study T cell activation.
- 12.Stewart E, McEvoy J, Wang H, Chen X, Honnell V, Ocarz M, Gordon B, Dapper J, Blankenship K, Yang Y, et al. : Identification of Therapeutic Targets in Rhabdomyosarcoma through Integrated Genomic, Epigenomic, and Proteomic Analyses. Cancer Cell 2018, 34:411–426 e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, et al. : Proteogenomic characterization of human colon and rectal cancer. Nature 2014, 513:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rauniyar N, Yates JR, 3rd: Isobaric labeling-based relative quantification in shotgun proteomics. J Proteome Res 2014, 13:5293–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Connell JD, Paulo JA, O’Brien JJ, Gygi SP: Proteome-Wide Evaluation of Two Common Protein Quantification Methods. J Proteome Res 2018, 17:1934–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogrebe A, von Stechow L, Bekker-Jensen DB, Weinert BT, Kelstrup CD, Olsen JV: Benchmarking common quantification strategies for large-scale phosphoproteomics. Nat Commun 2018, 9:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ron-Harel N, Santos D, Ghergurovich JM, Sage PT, Reddy A, Lovitch SB, Dephoure N, Satterstrom FK, Sheffer M, Spinelli JB, et al. : Mitochondrial Biogenesis and Proteome Remodeling Promote One-Carbon Metabolism for T Cell Activation. Cell Metab 2016, 24:104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weekes MP, Tomasec P, Huttlin EL, Fielding CA, Nusinow D, Stanton RJ, Wang EC, Aicheler R, Murrell I, Wilkinson GW, et al. : Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell 2014, 157:1460–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.**.Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et al. : Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper defines the largest PPI network (BioPlex) to date using AP-MS approaches.
- 20.Shi H, Liu C, Tan H, Li Y, Nguyen TM, Dhungana Y, Guy C, Vogel P, Neale G, Rankin S, et al. : Hippo Kinases Mst1 and Mst2 Sense and Amplify IL-2R-STAT5 Signaling in Regulatory T Cells to Establish Stable Regulatory Activity. Immunity 2018, 49:899–914 e896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai Z, Tsugawa H, Wohlgemuth G, Mehta S, Mueller M, Zheng Y, Ogiwara A, Meissen J, Showalter M, Takeuchi K, et al. : Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat Methods 2018, 15:53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang K, Han X: Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem Sci 2016, 41:954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, Kogadeeva M, Picotti P, Meissner F, Mann M, et al. : L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 2016, 167:829–842 e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.**.Goodwin S, McPherson JD, McCombie WR: Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016, 17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review summarizes the technology development of NGS.
- 25.Ashley EA: Towards precision medicine. Nat Rev Genet 2016, 17:507–522. [DOI] [PubMed] [Google Scholar]
- 26.Northrup DL, Zhao K: Application of ChIP-Seq and related techniques to the study of immune function. Immunity 2011, 34:830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee FCY, Ule J: Advances in CLIP Technologies for Studies of Protein-RNA Interactions. Mol Cell 2018, 69:354–369. [DOI] [PubMed] [Google Scholar]
- 28.Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, Carter R, Awad W, Neale G, Thomas PG, et al. : De Novo Epigenetic Programs Inhibit PD-1 Blockade-Mediated T Cell Rejuvenation. Cell 2017, 170:142–157 e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burren OS, Rubio Garcia A, Javierre BM, Rainbow DB, Cairns J, Cooper NJ, Lambourne JJ, Schofield E, Castro Dopico X, Ferreira RC, et al. : Chromosome contacts in activated T cells identify autoimmune disease candidate genes. Genome Biol 2017, 18:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ: Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 2013, 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shalem O, Sanjana NE, Zhang F: High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 2015, 16:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. : A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell 2015, 162:675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.**.Shifrut E, Carnevale J, Tobin V, Roth TL, Woo JM, Bui CT, Li PJ, Diolaiti ME, Ashworth A, Marson A: Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper combines CRISPR screens with scRNA-seq to identify regulators of T cell stimulation and suppression.
- 34.Morgens DW, Deans RM, Li A, Bassik MC: Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol 2016, 34:634–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. : Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159:647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA: RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc 2010, 5:516–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spitzer MH, Nolan GP: Mass Cytometry: Single Cells, Many Features. Cell 2016, 165:780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, Clair G, Chrisler WB, Shen Y, Zhao R, Shukla AK, Moore RJ, Misra RS, Pryhuber GS, Smith RD, et al. : Proteomic Analysis of Single Mammalian Cells Enabled by Microfluidic Nanodroplet Sample Preparation and Ultrasensitive NanoLC-MS. Angew Chem Int Ed Engl 2018, 57:12370–12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gawad C, Koh W, Quake SR: Single-cell genome sequencing: current state of the science. Nat Rev Genet 2016, 17:175–188. [DOI] [PubMed] [Google Scholar]
- 40.Mezger A, Klemm S, Mann I, Brower K, Mir A, Bostick M, Farmer A, Fordyce P, Linnarsson S, Greenleaf W: High-throughput chromatin accessibility profiling at single-cell resolution. Nat Commun 2018, 9:3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. : A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 2018, 174:1309–1324 e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark SJ, Lee HJ, Smallwood SA, Kelsey G, Reik W: Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol 2016, 17:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O’Shaughnessy-Kirwan A, et al. : 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 2017, 544:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE: Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol 2015, 33:1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.**.Giladi A, Amit I: Single-Cell Genomics: A Stepping Stone for Future Immunology Discoveries. Cell 2018, 172:14–21. [DOI] [PubMed] [Google Scholar]; This review discusses current applications and future perspectives of single-cell genomics in immunology research.
- 46.**.Tanay A, Regev A: Scaling single-cell genomics from phenomenology to mechanism. Nature 2017, 541:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reivews recent advances in single-cell genomics and applications to transform observational studies to mechanistic insights.
- 47.*.Papalexi E, Satija R: Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 2018, 18:35–45. [DOI] [PubMed] [Google Scholar]; This paper reviews recent stuides using scRNA-seq to dissect the immune system heterogeneity.
- 48.*.Ren X, Kang B, Zhang Z: Understanding tumor ecosystems by single-cell sequencing: promises and limitations. Genome Biol 2018, 19:211. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review discusses recent studies of single-cell omics in understanding tumor microenvironment.
- 49.*.Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, Gao R, Kang B, Zhang Q, Huang JY, et al. : Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018, 564:268–272. [DOI] [PubMed] [Google Scholar]; This study uses scRNA-seq to dissect the dynamics of tumor infiltrating T cells in colorectal cancer patients.
- 50.Adlung L, Amit I: From the Human Cell Atlas to dynamic immune maps in human disease. Nat Rev Immunol 2018, 18:597–598. [DOI] [PubMed] [Google Scholar]
- 51.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. : The Human Cell Atlas. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, et al. : Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 2018, 173:1307. [DOI] [PubMed] [Google Scholar]
- 53.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. : Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017, 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW: Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161:1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. : Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X, Li T, Liu F, Chen Y, Yao J, Li Z, Huang Y, Wang J: Comparative Analysis of Droplet-Based Ultra-High-Throughput Single-Cell RNA-Seq Systems. Mol Cell 2018. [DOI] [PubMed] [Google Scholar]
- 57.Tabula Muris Consortium: Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 2018, 562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wallrapp A, Riesenfeld SJ, Burkett PR, Abdulnour RE, Nyman J, Dionne D, Hofree M, Cuoco MS, Rodman C, Farouq D, et al. : The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 2017, 549:351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eng CL, Shah S, Thomassie J, Cai L: Profiling the transcriptome with RNA SPOTs. Nat Methods 2017, 14:1153–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, et al. : Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 2018, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hartmann FJ, Babdor J, Gherardini PF, Amir E-AD, Jones K, Sahaf B, Marquez DM, Krutzik P, O’Donnell E, Sigal N, et al. : Comprehensive Immune Monitoring of Clinical Trials to Advance Human Immunotherapy. Biorxiv 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gut G, Herrmann MD, Pelkmans L: Multiplexed protein maps link subcellular organization to cellular states. Science 2018, 361. [DOI] [PubMed] [Google Scholar]
- 63.Macaulay IC, Ponting CP, Voet T: Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends Genet 2017, 33:155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hagai T, Chen X, Miragaia RJ, Rostom R, Gomes T, Kunowska N, Henriksson J, Park JE, Proserpio V, Donati G, et al. : Gene expression variability across cells and species shapes innate immunity. Nature 2018, 563:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee MJ, Majeti R, Chang HY, Greenleaf WJ: Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 2018, 173:1535–1548 e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.*.Karmaus PWF, Chen X, Lim SA, Herrada AA, Nguyen TM, Xu B, Dhungana Y, Rankin S, Chen W, Rosencrance C, et al. : Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 2019, 565:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper integrates scRNA-seq with ATAC-seq to delineate novel mechanisms and regualtors that underlie TH17 cell stemness and plasticity.
- 67.Genshaft AS, Li S, Gallant CJ, Darmanis S, Prakadan SM, Ziegler CG, Lundberg M, Fredriksson S, Hong J, Regev A, et al. : Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Genome Biol 2016, 17:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frei AP, Bava FA, Zunder ER, Hsieh EW, Chen SY, Nolan GP, Gherardini PF: Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Methods 2016, 13:269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P: Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017, 14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.*.Zhu Q, Shah S, Dries R, Cai L, Yuan GC: Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data. Nat Biotechnol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper integrates scRNA-seq and seqFISH to identify spatially unique subpopulations.
- 71.Chen X, Litzenburger UM, Wei Y, Schep AN, LaGory EL, Choudhry H, Giaccia AJ, Greenleaf WJ, Chang HY: Joint single-cell DNA accessibility and protein epitope profiling reveals environmental regulation of epigenomic heterogeneity. Nat Commun 2018, 9:4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. : Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 2016, 167:1853–1866 e1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C: Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods 2017, 14:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, Amit I: Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 2016, 167:1883–1896 e1815. [DOI] [PubMed] [Google Scholar]
- 75.Bian S, Hou Y, Zhou X, Li X, Yong J, Wang Y, Wang W, Yan J, Hu B, Guo H, et al. : Single-cell multiomics sequencing and analyses of human colorectal cancer. Science 2018, 362:1060–1063. [DOI] [PubMed] [Google Scholar]
- 76.Li B, Liu JS, Liu XS: Revisit linear regression-based deconvolution methods for tumor gene expression data. Genome Biol 2017, 18:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al. : Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016, 17:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA: Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015, 12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, et al. : SC3: consensus clustering of single-cell RNA-seq data. Nat Methods 2017, 14:483–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.**.Kiselev VY, Andrews TS, Hemberg M: Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet 2019. [DOI] [PubMed] [Google Scholar]; This review summarizes existing algorithms and challenges for clustering analysis of scRNA-seq data.
- 81.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA: Single-cell transcriptomics to explore the immune system in health and disease. Science 2017, 358:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Godec J, Tan Y, Liberzon A, Tamayo P, Bhattacharya S, Butte AJ, Mesirov JP, Haining WN: Compendium of Immune Signatures Identifies Conserved and Species-Specific Biology in Response to Inflammation. Immunity 2016, 44:194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chapman NM, Zeng H, Nguyen TM, Wang Y, Vogel P, Dhungana Y, Liu X, Neale G, Locasale JW, Chi H: mTOR coordinates transcriptional programs and mitochondrial metabolism of activated Treg subsets to protect tissue homeostasis. Nat Commun 2018, 9:2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang K, Blanco DB, Neale G, Vogel P, Avila J, Clish CB, Wu C, Shrestha S, Rankin S, Long L, et al. : Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature 2017, 548:602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karmaus PWF, Herrada AA, Guy C, Neale G, Dhungana Y, Long L, Vogel P, Avila J, Clish CB, Chi H: Critical roles of mTORC1 signaling and metabolic reprogramming for M-CSF-mediated myelopoiesis. J Exp Med 2017, 214:2629–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Amit I, Regev A, Hacohen N: Strategies to discover regulatory circuits of the mammalian immune system. Nat Rev Immunol 2011, 11:873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marbach D, Costello JC, Kuffner R, Vega NM, Prill RJ, Camacho DM, Allison KR, Consortium D, Kellis M, Collins JJ, et al. : Wisdom of crowds for robust gene network inference. Nat Methods 2012, 9:796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Langfelder P, Horvath S: WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008, 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A: ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 2006, 7 Suppl 1:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Khatamian A, Paull EO, Califano A, Yu J: SJARACNe: a scalable software tool for gene network reverse engineering from big data. Bioinformatics 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.**.Du X, Wen J, Wang Y, Karmaus PWF, Khatamian A, Tan H, Li Y, Guy C, Nguyen TM, Dhungana Y, et al. : Hippo/Mst signalling couples metabolic state and immune function of CD8alpha(+) dendritic cells. Nature 2018, 558:141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper is the first for hidden driver inference by integrating multi-omics data, using NetBID algorithm based on data-driven networks and Bayesian inference.
- 92.Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, et al. : SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017, 14:1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kidd BA, Peters LA, Schadt EE, Dudley JT: Unifying immunology with informatics and multiscale biology. Nat Immunol 2014, 15:118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mostafavi S, Gaiteri C, Sullivan SE, White CC, Tasaki S, Xu J, Taga M, Klein HU, Patrick E, Komashko V, et al. : A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat Neurosci 2018, 21:811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang S, Chaudhary K, Garmire LX: More Is Better: Recent Progress in Multi-Omics Data Integration Methods. Front Genet 2017, 8:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. : Architecture of the human regulatory network derived from ENCODE data. Nature 2012, 489:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mischnik M, Sacco F, Cox J, Schneider HC, Schafer M, Hendlich M, Crowther D, Mann M, Klabunde T: IKAP: A heuristic framework for inference of kinase activities from Phosphoproteomics data. Bioinformatics 2016, 32:424–431. [DOI] [PubMed] [Google Scholar]
- 98.Davis MM, Tato CM, Furman D: Systems immunology: just getting started. Nat Immunol 2017, 18:725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chaussabel D, Baldwin N: Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol 2014, 14:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D, Stuart JM: Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 2010, 26:i237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shi Z, Wang J, Zhang B: NetGestalt: integrating multidimensional omics data over biological networks. Nat Methods 2013, 10:597–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morris SA, Cahan P, Li H, Zhao AM, San Roman AK, Shivdasani RA, Collins JJ, Daley GQ: Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell 2014, 158:889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.**.Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, Wang D, Getzler AJ, Nguyen T, Crotty S, et al. : Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumours. Nature 2017, 552:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper combines RNAi screening, ATAC-seq with RNA-seq to reveal novel immune regulators.
- 104.Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Ye BH, Califano A: Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet 2016, 48:838–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.*.Kveler K, Starosvetsky E, Ziv-Kenet A, Kalugny Y, Gorelik Y, Shalev-Malul G, Aizenbud-Reshef N, Dubovik T, Briller M, Campbell J, et al. : Immune-centric network of cytokines and cells in disease context identified by computational mining of PubMed. Nat Biotechnol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper develops a literature-mining algorithm to identify intercellular networks.
- 106.Boisset JC, Vivie J, Grun D, Muraro MJ, Lyubimova A, van Oudenaarden A: Mapping the physical network of cellular interactions. Nat Methods 2018, 15:547–553. [DOI] [PubMed] [Google Scholar]
- 107.*.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polanski K, Goncalves A, et al. : Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018, 563:347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper develops a database of ligand-receptor complexes and a tool to predict cell–cell communication from scRNA-seq data.
- 108.Cohen M, Giladi A, Gorki AD, Solodkin DG, Zada M, Hladik A, Miklosi A, Salame TM, Halpern KB, David E, et al. : Lung Single-Cell Signaling Interaction Map Reveals Basophil Role in Macrophage Imprinting. Cell 2018, 175:1031–1044 e1018. [DOI] [PubMed] [Google Scholar]
- 109.**.Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, Ashenberg O, Su CW, Smillie C, Shekhar K, et al. : T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell 2018, 175:1307–1320 e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses scRNA-seq to identify novel interactions between intestinal stem cells and T helper subsets that module distinct intestinal stem cell fates.
- 110.Camp JG, Sekine K, Gerber T, Loeffler-Wirth H, Binder H, Gac M, Kanton S, Kageyama J, Damm G, Seehofer D, et al. : Multilineage communication regulates human liver bud development from pluripotency. Nature 2017, 546:533–538. [DOI] [PubMed] [Google Scholar]
- 111.Moor AE, Itzkovitz S: Spatial transcriptomics: paving the way for tissue-level systems biology. Curr Opin Biotechnol 2017, 46:126–133. [DOI] [PubMed] [Google Scholar]
- 112.Biddy BA, Kong W, Kamimoto K, Guo C, Waye SE, Sun T, Morris SA: Single-cell mapping of lineage and identity in direct reprogramming. Nature 2018, 564:219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.*.Villani AC, Sarkizova S, Hacohen N: Systems Immunology: Learning the Rules of the Immune System. Annu Rev Immunol 2018, 36:813–842. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review summarizes recent progresses in transcriptome and TCR/BCR reportire analyses.
- 114.Wendel BS, Del Alcazar D, He C, Del Rio-Estrada PM, Aiamkitsumrit B, Ablanedo-Terrazas Y, Hernandez SM, Ma KY, Betts MR, Pulido L, et al. : The receptor repertoire and functional profile of follicular T cells in HIV-infected lymph nodes. Sci Immunol 2018, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ma KY, He C, Wendel BS, Williams CM, Xiao J, Yang H, Jiang N: Immune Repertoire Sequencing Using Molecular Identifiers Enables Accurate Clonality Discovery and Clone Size Quantification. Front Immunol 2018, 9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hackl H, Charoentong P, Finotello F, Trajanoski Z: Computational genomics tools for dissecting tumour-immune cell interactions. Nat Rev Genet 2016, 17:441–458. [DOI] [PubMed] [Google Scholar]
- 117.June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC: CAR T cell immunotherapy for human cancer. Science 2018, 359:1361–1365. [DOI] [PubMed] [Google Scholar]
- 118.Ribas A, Wolchok JD: Cancer immunotherapy using checkpoint blockade. Science 2018, 359:1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wolchok J: Putting the Immunologic Brakes on Cancer. Cell 2018, 175:1452–1454. [DOI] [PubMed] [Google Scholar]
- 120.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, et al. : The Immune Landscape of Cancer. Immunity 2018, 48:812–830 e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. : Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 2018, 175:998–1013 e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brummelman J, Mazza EMC, Alvisi G, Colombo FS, Grilli A, Mikulak J, Mavilio D, Alloisio M, Ferrari F, Lopci E, et al. : High-dimensional single cell analysis identifies stem-like cytotoxic CD8(+) T cells infiltrating human tumors. J Exp Med 2018, 215:2520–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, et al. : Intratumoral Tcf1(+)PD-1(+)CD8(+) T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 2019, 50:195–211 e110. [DOI] [PubMed] [Google Scholar]
- 124.Kurtulus S, Madi A, Escobar G, Klapholz M, Nyman J, Christian E, Pawlak M, Dionne D, Xia J, Rozenblatt-Rosen O, et al. : Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1(−)CD8(+) Tumor-Infiltrating T Cells. Immunity 2019, 50:181–194 e186. [DOI] [PMC free article] [PubMed] [Google Scholar]