Abstract
Despite marked development in cancer therapies during recent decades, the prognosis for advanced cancer remains poor. The conventional tumor–cell‐centric view of cancer can only explain part of cancer progression, and thus a thorough understanding of the tumor microenvironment (TME) is crucial. Among cells within the TME, cancer‐associated fibroblasts (CAFs) are attracting attention as a target for cancer therapy. However, CAFs present a heterogeneous population of cells and more detailed classification of CAFs and investigation of functions of each subset is needed to develop novel CAF‐targeted therapies. In this context, application of newly developed approaches to single‐cell analysis has already made an impact on our understanding of the heterogeneity of CAFs. Here, we review the recent literature on CAF heterogeneity and function, and discuss the possibility of novel therapies targeting CAF subsets.
Keywords: cancer progression, cancer‐associated fibroblast, heterogeneity, single‐cell RNA‐sequencing, tumor microenvironment
The conventional tumor–cell‐centric view of cancer can only explain a part of cancer progression, thus understanding of the tumor microenvironment (TME) is crucial. Among cells within the TME, cancer‐associated fibroblasts (CAFs) are attracting attention as a target for cancer therapy. Here, we review the recent literature that has improved our understanding of heterogeneity in CAFs and function of each subset, and discuss the possibility of novel therapies targeting CAF subsets.

Abbreviations
- BM‐MSC
bone marrow mesenchymal stem cells
- CAF
cancer‐associated fibroblasts
- EMT
epithelial‐mesenchymal transition
- ER
estrogen receptor
- FACS
fluorescence‐activated cell sorting
- FAP
fibroblast activation protein
- FSP‐1
fibroblast specific protein 1
- GEMM
genetically engineered mouse model
- IPMN
intraductal papillary mucinous neoplasm
- MHC
major histocompatibility complex
- PDAC
pancreatic ductal adenocarcinoma
- PDGF
platelet‐derived growth factor
- PDGFR
platelet‐derived growth factor receptor
- scRNA‐seq
single‐cell RNA‐sequencing
- TME
tumor microenvironment
- α‐SMA
α‐smooth muscle actin
1. INTRODUCTION
Substantial progress has been made in our understanding of the molecular basis for cancer since the discovery of oncogenes. 1 To date, many oncogenes and tumor suppressor genes have been discovered, and this knowledge has contributed during the past decades to the development of molecularly targeted therapy. 2 Despite this marked improvement in cancer therapy, the prognosis for advanced cancer remains poor for many malignant diseases. Thus, there is an urgent need for the development of new and effective cancer therapies fueled by conceptually transformative basic science.
The conventional tumor–cell‐centric view of cancer can only partly explain the full process of cancer progression. The surrounding tumor microenvironment (TME) co‐evolves during malignant progression into an activated state through paracrine, juxtacrine, and autocrine communications, thus creating a dynamic signaling circuitry that contributes to cancer initiation, progression, and resistance to therapy. The significance of the TME is confirmed by several studies demonstrating association between gene expression patterns of microenvironmental cell types and prognosis in patients with breast, lung, and colorectal cancer, 3 , 4 , 5 among others. In this context, the TME is attracting attention as a target for cancer therapy and as a rich source of biomarkers that hold prognostic and/or predictive potential. The fundamental architecture of the tumor miniature organ consists of cancer cells, endothelial cells, pericytes, fibroblasts, various classes of leukocytes, and extracellular matrix. CAFs are among the most abundant cell types within a range of different tumor types, and the accumulating evidence points to a fundamental role for CAFs in influencing the malignant phenotype. 6
2. MARKERS AND ORIGINS OF CANCER‐ASSOCIATED FIBROBLASTS
The broad definition of CAFs is a fibroblast located within or in close proximity to the tumor mass. Pragmatically, CAFs are defined as spindle‐shaped cells, which are negative for epithelial, endothelial, and leukocyte markers derived from cancer tissue. It should be noted that, in this practical setting, cancer cells trans‐differentiated to fibroblast‐like or hybrid states through EMT could be included in this CAF population. CAFs have been isolated from a range of malignant tissues, including prostate cancer, lung cancer, breast cancer, gastric cancer, colorectal cancer, and pancreatic cancer. In contrast, CAFs are relatively rare in specimens from brain cancer, ovarian cancer, and kidney cancer. A selection of reported markers of CAFs is summarized in Table 1. Vimentin is considered to be a marker for quiescent CAFs, while α‐SMA, S100A4/fibroblast specific protein 1 (FSP‐1), FAP, Tenascin‐C, Periostin, Desmin, PDGFR‐α, PDGFR‐β, Thy‐1, Podoplanin, Integrin β1, Caveolin‐1 are considered as activated CAF markers. Each marker defines different cell populations that are partially overlapping but also show distinct expression profiles. In other words, no single marker can define the full CAF population, or distinguish CAFs from all other cell types. As an example, myofibroblasts are considered to be a subset of activated fibroblasts, characterized by de novo expression of α‐smooth muscle actin (α‐SMA), that have contractile and secretory profiles contributing to tissue repair during wound healing and cancer development. 7 Conceivably, the heterogeneity of CAFs may come from cells in different stages of differentiation from a common precursor, cells that have adopted different states depending on internal and external signaling cues, or alternatively from cells that have diverse origins. It has been reported that normal fibroblast can acquire a CAF phenotype through communication with cancer cells. 8 Conversely, it is also reported that CAFs can be reprogrammed to reduce the CAF phenotype, indicating that the transition is reversible. 9 Based on this, at least parts of the CAF phenotype are considered to be a cellular state of the fibroblast, not a fixed cell type. Indeed, CAFs may have diverse origins, including: (a) tissue‐resident fibroblasts, (b) cells trans‐differentiated from other cell types such as endothelial cell, epithelial cell, vascular smooth muscle cells, pericytes, adipocytes, and their progenitors, 10 , 11 , 12 (c) cancer cells trans‐differentiated to mesenchymal cells through EMT and (d) bone marrow‐derived precursors and bone marrow mesenchymal stem cells (BM‐MSC). 13 , 14
Table 1.
Markers for cancer‐associated fibroblasts
| CAF markers | Description of protein | Surface marker |
|---|---|---|
| Vimentin | Type III intermediate filament protein | No |
| α‐SMA | Actin isoform | No |
| FSP‐1/S100A4 | Calcium‐binding protein containing 2 EF‐hand calcium‐binding motifs | No |
| FAP | Membrane‐bound gelatinase | Yes |
| Tenascin‐C | Extracellular matrix glycoproteins | No |
| Periostin | Secreted extracellular matrix protein, a ligand for α‐V/β‐3 and α‐V/β‐5 integrins | No |
| Desmin | Type III intermediate filament protein | No |
| PDGFR‐α | Protein tyrosine kinase receptor | Yes |
| PDGFR‐β | Protein tyrosine kinase receptor | Yes |
| Thy‐1 | Glycophosphatidylinositol anchored protein | No |
| Podoplanin | Mucin‐type protein, heavily O‐glycosylated glycoprotein | Yes |
| Integrin β1 | Transmembrane receptor | Yes |
| Caveolin‐1 | Scaffolding protein within caveolar membranes | Yes |
Abbreviations: CAF, cancer‐associated fibroblasts; FAP, fibroblast activation protein; FSP‐1, fibroblast specific protein 1; PDGFR, platelet‐derived growth factor receptor; α‐SMA, α‐smooth muscle actin.
3. DEVELOPMENT OF METHODOLOGY TO STUDY CAFs
Several studies using cell culture of CAFs derived from multiple patients with cancer have revealed the heterogeneity of CAFs. Herrera et al 15 established primary CAF cultures from 15 primary human colon tumors and demonstrated differences among each CAF culture in fibroblast‐derived paracrine pro‐migratory effects on cancer cells. Moreover, the gene expression signature derived from the most pro‐migratory CAFs showed a marked prognostic value for the clinical outcome of patients with colon cancer. Hao et al 16 characterized 2 CAF subsets from 28 non–small cell lung cancers. They compared CAFs with high or low proliferation of fibroblasts and demonstrated that high desmoplastic CAFs showed an increased rate of collagen matrix remodeling, invasion, and tumor growth. These studies demonstrated a different phenotype of CAFs derived from different patients, and thus illustrate a high degree of inter‐patient variability in the CAF population. However, these studies did not directly address heterogeneity of CAFs within the same tumor.
Although it is known based on immunohistochemical studies that CAFs present a heterogeneous population of cells, experimental approaches to study gene expression or function of CAFs have been for a long time limited to bulk analysis until the development of single‐cell analysis. Because conventional bulk analysis can only provide an average of gene expression over a cell population, and mainly reflects the phenotype of the dominant cellular subset, information from minor populations can only be reflected in the results to a low degree. In this context, application of recent developments in genomics, transcriptomics, proteomics, epigenomics, and metabolomics to single‐cell analysis has quickly made an impact on understanding the heterogeneity of CAFs. In 2009, Tang et al 17 reported analysis of transcriptomes of single blastomeres from 4‐cell‐embryo‐stage mice by improving the amplification method for single‐cell transcriptomes. After that, Navin et al 18 sequenced 200 flow cytometry‐isolated cancer cells to study tumor evolution. Now, single‐cell RNA‐sequencing (scRNA‐seq) is applied in many fields in biology and contributes to a better understanding of various life phenomena. Comparison of bulk sequencing and scRNA‐seq analysis on CAFs from a solid tumor is shown in Figure 1.
Figure 1.
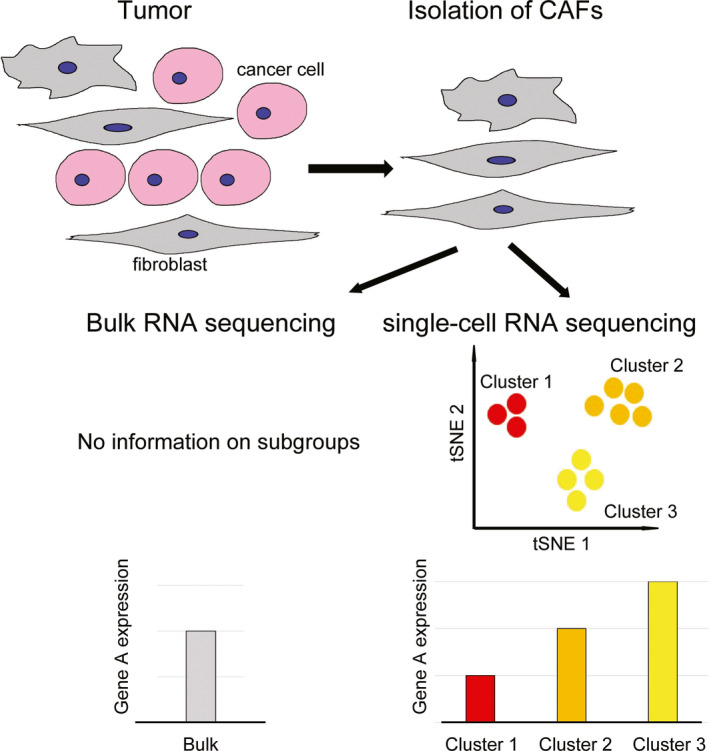
Comparison of bulk sequencing and single‐cell RNA‐sequencing analysis on cancer‐associated fibroblasts from a solid tumor. Cancer‐associated fibroblasts are isolated from cancer tissue. In bulk sequencing, information on subgroups cannot be obtained, and extracted RNA only provides expression data representing the average of a particular gene expression of the bulk population. In contrast, single‐cell RNA‐sequencing provides information on subgroups and cluster‐specific transcript information. CAF, cancer‐associated fibroblast; tSNE, t‐Distributed Stochastic Neighbor Embedding
4. STUDIES ON INTRA‐TUMORAL HETEROGENEITY OF CANCER‐ASSOCIATED FIBROBLASTS
To date, several studies detailing the heterogeneity of CAFs have been published. The classification of CAFs proposed in a selection of studies are summarized in Table 2.
Table 2.
Proposed classification of cancer‐associated fibroblasts
| Author | Year | Tumor type | Methodology for dividing CAFs | Species/experimental model | Name of subpopulation | Characteristics of population |
|---|---|---|---|---|---|---|
| Bartoschek 19 | 2018 | Breast cancer | Single‐cell RNA‐sequencing | Mouse/MMTV‐PyMT | vCAF (vascular CAF) | Enriched for vascular development and angiogenesis genes, representative marker: Desmin, enriched in tumor core |
| mCAF (matrix CAF) | Enriched for genes related to the extracellular matrix and EMT, representative marker: Fibulin‐1, PDGFR‐α, enriched in invasive front of tumors | |||||
| cCAF (cycling CAF) | Represent the proliferative segment of vCAFs | |||||
| dCAF (developmental CAF) | Distinguished by the expression of genes related to various kinds of stem cells, representative marker: Scrg1 | |||||
| Friedman 22 | 2020 | Breast cancer | Single‐cell RNA‐sequencing | Mouse/4T1 injection | pCAF (Pdpn) | Includes 6 subgroups (early immune regulatory, late immune regulatory, wound healing, extracellular fiber organization, inflammatory A, inflammatory B) |
| sCAF (S100a4) | Includes 2 subtypes (protein folding, antigen presentation) | |||||
| Costa 23 | 2018 | Breast cancer | FACS | Human resected sample | CAF‐S1 | Defined as CD29Med FAPHi FSP1Low‐Hi α‐SMAHi PDGFR‐βMed‐Hi CAV1Low, enriched in TNBC, observed in metastatic lymph nodes |
| CAF‐S2 | Defined as CD29Low FAPNeg FSP1Neg‐Low α‐SMANeg PDGFR‐βNeg CAV1Neg, enriched in luminal A tumor | |||||
| CAF‐S3 | Defined as CD29Med FAPNeg FSP1Med‐Hi α‐SMANeg‐Low PDGFR‐βMed CAV1Neg‐Low | |||||
| CAF‐S4 | Defined as CD29Hi FAPNeg FSP1Low‐Med α‐SMAHi PDGFR‐βLow‐Med CAV1Neg‐Low, enriched in TNBC, observed in metastatic lymph nodes | |||||
| Öhlund 26 | 2017 | Pancreatic cancer | Immunohistochemistry | Mouse/KPC, Human resected sample | myCAF (myofibroblastic CAF) | FAP + α‐SMA high expression, locate near tumor cell nests |
| iCAF (inflammatory CAF) | α‐SMA low, IL‐6 high expression, locate far from tumor cells in the desmoplastic area | |||||
| Elyada 27 | 2019 | Pancreatic cancer | Single‐cell RNA‐sequencing | Mouse/KPC, Human resected sample | apCAF (antigen‐presenting CAF) | Express MHC class II and CD74, activate CD4+ T cells in an antigen‐specific fashion |
| Lambrechts 33 | 2018 | Lung cancer | Single‐cell RNA‐sequencing | Human resected sample | Cluster 1 | Show a strong EMT and an extensive repertoire of extracellular matrix proteins and TGF‐β‐associated genes |
| Cluster 2 | Exhibit the highest expression of ACTA2 | |||||
| Cluster 4 | Enriched in the leading edge of the tumor | |||||
| Cluster 5 | Lower myogenesis and high mTOR signature expression, enriched in the tumor core | |||||
| Cluster 7 | Lower myogenesis and high mTOR signature expression, enriched in the tumor edge | |||||
| Li 34 | 2017 | Colorectal cancer | Single‐cell RNA‐sequencing | Human resected sample | CAF‐A | Express genes related to extracellular matrix remodeling, including the TGF‐β activator MMP2 |
| CAF‐B | Express markers of myofibroblasts such as ACTA2, TAGLN, and PDGFA | |||||
| Puram 35 | 2017 | HNSCC | Single‐cell RNA‐sequencing | Human resected sample | Myofibroblasts | Express ACTA2 and myosin light‐chain proteins (MYLK, MYL9) |
| CAF1 | Express FAP, PDPN, COL1A1, mesenchymal markers (eg, VIM, THY1) and ECM proteins (eg, MMP11, CAV1) | |||||
| CAF2 | Express FAP, PDPN, immediate early response genes (eg, JUN, FOS), ligands and receptors (eg, FGF7, TGFBR) | |||||
| Resting fibroblasts | Lacked expression of markers for myofibroblasts and CAFs |
Abbreviations: CAF, cancer‐associated fibroblasts; CAV1, Caveolin‐1; ECM, extracellular matrix; EMT, epithelial‐mesenchymal transition; FACS, fluorescence‐activated cell sorting; FAP, fibroblast activation protein; FSP‐1, fibroblast specific protein 1; HNSCC, head and neck squamous cell carcinoma; KPC, KrasLSL‐G12D/+; Trp53LSL‐R172H/+; Pdx‐1‐Cre; PDGFR, platelet‐derived growth factor receptor; TNBC, triple‐negative breast cancer.
4.1. Breast cancer
We have recently defined spatially and functionally distinct subpopulations of CAFs using scRNA‐seq of transcriptomes of mesenchymal cells from the MMTV‐PyMT GEMM of breast cancer (Figure 2A‐C). 19 MMTV‐PyMT mice, in which the long terminal repeat of mouse mammary tumor virus (MMTV‐LTR) is used as a promoter to drive the expression of mammary gland‐specific polyomavirus middle T‐antigen, develop spontaneous mammary tumors that closely resemble human breast cancers. 20 , 21 Analysis of scRNA‐seq of 768 transcriptomes of mesenchymal cells isolated from tumors of the MMTV‐PyMT mouse using a negative selection FACS strategy revealed 4 distinct subpopulations of CAFs. We named these vascular CAFs (vCAFs), matrix CAFs (mCAFs), cycling CAFs (cCAFs), and developmental CAFs (dCAFs), in accordance with the functional annotation of unique gene sets. Notably, Pdgfra was specifically expressed by cells in the mCAF cluster, whereas Pdgfrb was expressed by all cells, apart from dCAFs. The gene expression profile of vCAFs was found to be significantly enriched for genes functionally linked to vascular development and angiogenesis. Also, the expression of the vCAF marker Des (desmin) was distinctly higher in the tumor core, compared with the leading edge of the tumor. In contrast, the mCAF subset was enriched for transcripts related to the extracellular matrix and EMT. The mCAF markers Fbln1 and Pdgfra showed high prevalence of positive cells at the invasive front of tumors, in contrast with the relatively low abundance of mCAFs in the tumor core. The cluster of cCAFs contained cells that were in the G2, M, or S phase of the cell cycle, and were, upon closer inspection, found to represent the proliferative segment of vCAFs. Finally, dCAFs were distinguished by the expression of genes related to various kinds of stem cells (Scrg1, Sox9, and Sox10, among others). In dCAFs, expression of the transgenic PyMT oncogene was strongly detected, indicating a malignant cell‐origin for this subset of cells. In contrast, longitudinal studies suggested a peri‐vascular origin and an origin from resident fibroblasts for vCAFs and mCAFS, respectively. Taken together, our study delineated subclasses of breast CAFs derived from distinct origins, thus confirming the long‐standing notion that CAF heterogeneity is, in part, derived from recruitment of cells from different sources. Interestingly, based on our dataset, markers may now be developed to distinguish CAFs of different origins; as an example it may be noted that Thy1 distinguishes CAFs from non‐malignant (vCAF/mCAF) origin and malignant (dCAF) origins (Figure 2D). 19
Figure 2.
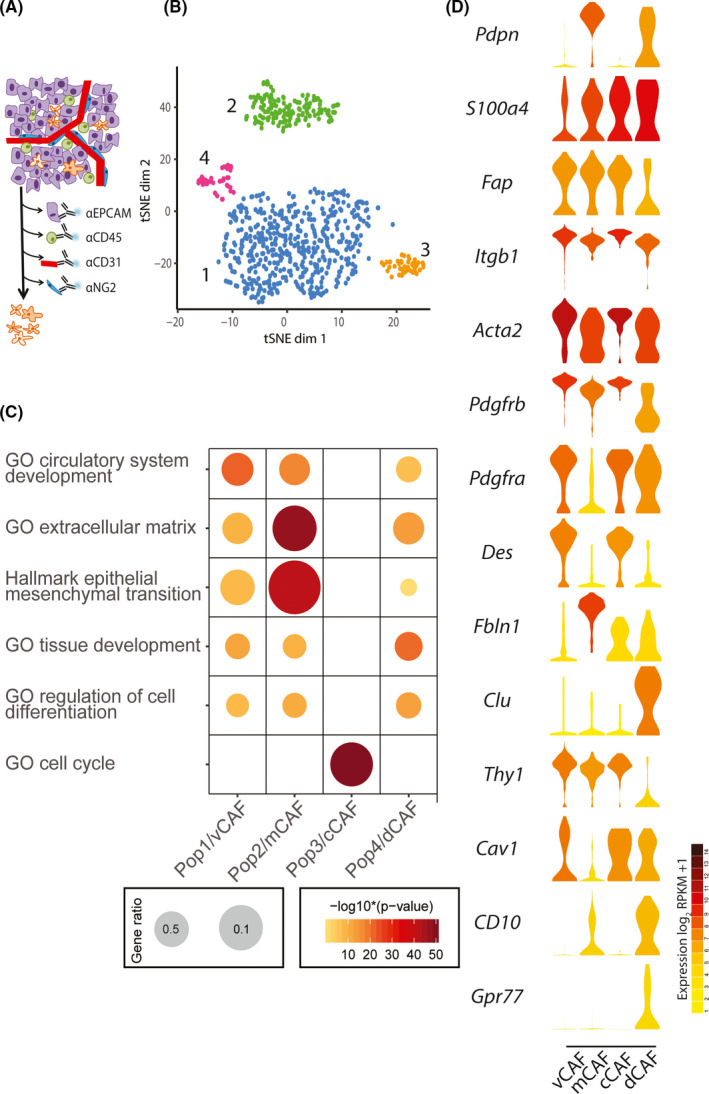
Analysis of single‐cell RNA‐sequencing of 768 transcriptomes of mesenchymal cells isolated from tumors of the MMTV‐PyMT mouse. A‐C, This figure are adapted from our previous report. 19 A, Schematic representation of negative selection strategy removing CD31+, CD45+, NG2+, and EPCAM+ cells to enrich for mesenchymal fibroblasts. B, t‐SNE layout of CAFs (n = 716) by RPKM‐normalized transcriptomic data. Colors represent clusters assigned by density‐based spatial clustering of applications with noise (DBSCAN). Populations 1‐4 are designated with discrete gene expression profiles. C, Enrichment of the 150 most significantly differentially expressed genes in gene ontology (GO) terms. Gene ratio is determined by the number of detected genes within a GO term compared with the total number of genes. Populations 1‐4 defined in (B) are defined as vCAF, mCAF, dCAF, and dCAF, respectively, and are shown as Pop1/vCAF, Pop2/mCAF, Pop3/cCAF, and Pop4/dCAF. D, Violin plots of genes in log2(RPKM + 1). Violin colors represent the mean expression of each population. Genes were selected based on classification of CAFs from Friedman et al, 22 Costa et al, 23 and Su et al. 25 CAF, cancer‐associated fibroblast; GO, gene ontology; tSNE, t‐Distributed Stochastic Neighbor Embedding
Friedman et al 22 analyzed subtypes of CAFs in the murine triple‐negative breast cancer model 4T1 and reported that CAF subtype compositions change with cancer progression (note that this study is currently only available as a pre‐print and thus has not yet gone through comprehensive peer review). They analyzed scRNA‐seq of 8987 transcriptomes of mesenchymal cells isolated using a negative FACS selection strategy. The analysis revealed 2 main groups of CAFs characterized by Pdpn expression (pCAF) and S100a4 expression (sCAF). Normal mammary fibroblasts expressed Pdpn, but were devoid of S100a4. More thorough analysis revealed that pCAFs included 6 subgroups (early immune regulatory, late immune regulatory, wound healing, extracellular fiber organization, inflammatory A, and inflammatory B), whereas sCAF included 2 subtypes (protein folding and antigen presentation). Interestingly, sCAFs were enriched for several classic BM‐MSC markers including Clu, which implies BM‐MSC origin. Notably, the proportion of the sCAF subsets were found to be dynamic, and the antigen‐presenting subpopulation expressing MHC class II takes dominance as tumors progress. For this review article, we overlaid the expression of 3 key genes from Friedman et al, 22 Pdpn, S100a4, and Clu, with the CAF subsets identified by our study (Figure 2D). 19 While Pdpn is predominantly expressed by mCAFs, and Clu is an exclusive marker for dCAFs, S100a4 is expressed by a fraction of cells in each of the CAF subsets of our study. While interesting similarities are noted, the discrepancies may illustrate heterogeneity between different types of mouse models (GEMM vs cell line), or be due to distinct starting materials related to differing strategies for the negative selection of cells.
Costa et al 23 performed a detailed characterization of CAFs of human breast cancer using FACS. They employed 6 markers: FAP, Integrin β1, α‐SMA, FSP‐1, PDGFR‐β, and Caveolin‐1 to divide CAFs into 4 different subpopulations: CAF‐S1, CAF‐S2, CAF‐S3, CAF‐S4. The authors further demonstrated that the CAF‐S1 subset was associated with an immunosuppressive microenvironment. Several of the CAF subsets were found to accumulate differentially in breast cancer molecular subtypes, enrichment of CAF‐S2 in luminal A tumors and accumulation of CAF‐S1 and CAF‐S4 in basal‐like breast cancer were observed. Pelon et al 24 from the same group reported that CAF‐S1 and CAF‐S4 accumulate in metastatic lymph nodes. Interestingly, they also demonstrated that patients with high levels of CAFs, particularly CAF‐S4, in lymph nodes at diagnosis were prone to develop late distant metastases. We also generated violin plots of genes of the 6 markers used in Costa et al 23 (Figure 2D) in our dataset. 19 Although direct comparison between our and their subtypes cannot be performed, no subtype of our classification seems identical with either of their subgroups simply based on these 6 markers.
Su et al 25 reported an important functional subset of CAFs, which was defined by cell surface molecules CD10 and GPR77 in breast cancer and lung cancer. This study demonstrated that CD10+GPR77+ CAFs promote tumor formation and chemoresistance by providing a survival niche for cancer stem cells. Intriguingly, both CD10 and GPR77 are prominently expressed by cells in the dCAF cluster revealed by our study (Figure 2D). 19 As stated above, dCAFs are suggested to originate from tumor cells that have undergone an EMT. Based on these findings, CD10+GPR77+ CAFs may, in fact, originate from the malignant cells themselves.
4.2. Pancreatic cancer
Heterogeneity of CAFs in pancreatic cancer is comprehensible according to the classification proposed by Tuveson's group. 26 , 27 At first, Öhlund et al 26 reported on 2 spatially separated, reversible, and mutually exclusive subtypes of CAFs using GEMMs; these were termed myofibroblastic CAFs (myCAFs) and inflammatory CAFs (iCAFs). Having a TGF‐β response gene profile, myCAFs were defined by FAP+ α‐SMAhigh expression, and locate near tumor cell nests. In contrast, iCAFs were defined by α‐SMAlow, IL‐6high expression, and locate far from tumor cells in the desmoplastic area. In addition, iCAFs have high expression of cytokine genes such as Il6, Il11, and Lif, and chemokines such as Cxcl1 and Cxcl2. Subsequently, Biffi et al 28 reported the mechanism through which these distinct fibroblast subtypes are established. They demonstrated using organoid and mouse models that IL1 induced leukemia inhibitory factor (LIF) expression and downstream JAK/STAT activation to generate iCAFs, and that TGF‐β antagonizes this process by downregulating IL1R1 expression to promote differentiation into myCAFs. Furthermore, Elyada et al 27 reported a third subtype of CAFs that expressed MHC class II and CD74, named antigen‐presenting CAFs (apCAFs). Intriguingly, apCAFs activate CD4+ T cells in an antigen‐specific fashion.
Two studies employing scRNA‐seq in pancreatic cancer support the classification described above. Hosein et al 29 conducted analysis of scRNA‐seq using GEMM and supports the classification of CAFs proposed by Öhlund et al 26 They sequenced 804 cells of late KIC (60‐d‐old KrasLSL−G12D/+Ink4a fl/flPtf1aCre/+ (KIC) mouse pancreas, a model for pancreatic ductal adenocarcinoma (PDAC)) and defined 2 CAF subgroups, FB1 and FB3. The FB1 group expressed insulin‐like growth factor signaling genes (Igfbp7, Igfbp4, and Igf1), Pdgfra, Cxcl12, Il6, and several other cytokines (Ccl11, Ccl7, Ccl2, and Csf1), and is considered to correspond to iCAFs. The FB3 population was positive for the myofibroblast markers Acta2 and Tagln and appears similar to the myCAF population. Notably, the FB3 group also expressed MHC II components and thus may also incorporate apCAFs. The classification of CAFs proposed by Öhlund et al 26 has been further validated by Bernard et al 30 using human samples, they performed scRNA‐seq on 5403 cells from 2 surgically resected low‐grade intraductal papillary mucinous neoplasms (IPMNs), 2 high‐grade IPMNs, and 2 PDACs. Interestingly, myCAFs were identified in all the 3 histologic types, although myCAFs were rare in low‐grade IPMNs and highly represented in high‐grade IPMNs. Conversely, iCAFs were identified exclusively in PDACs.
Neuzillet et al 31 employed a different strategy compared with the scRNA‐seq approach to study the heterogeneity of CAFs in PDAC by performing NanoString nCounter analysis of the expression of 770 genes by 16 primary cultures of CAFs. This study identified 4 subtypes of CAFs based on transcriptomic analysis, named subtypes A‐D. Also, it was confirmed by immunohistochemistry that multiple CAF subtypes co‐exist in individual patient samples. Comparing their classification, and that proposed by Öhlund et al, 26 subtypes B and D, which express ACTA2 and ECM components, resembles myCAFs whereas subtype C resembles iCAFs, and subtype A has characteristics of both iCAFs and myCAFs. Interestingly, the study also demonstrated that a prolonged exposure of non‐tumoral pancreatic stellate cells to conditioned medium from cancer cell lines induced a CAF‐like phenotype, as demonstrated by an increase in the expression of genes related to CAF subtypes B and C.
A recent report revealed a CAF subset that could be a potential target to boost responses of cancer patients to immune checkpoint blockade therapy. Dominguez et al 32 identified a population of CAFs that were programmed by TGF‐β and expressed the leucine‐rich repeat containing 15 (LRRC15) protein in PDAC. They further demonstrated that elevated levels of the LRRC15+ CAF signature correlated with poor response to anti–PD‐L1 therapy in clinical trials, demonstrating the potential of CAF subsets as predictive biomarkers for choice of treatment.
4.3. Lung cancer
Lambrechts et al 33 analyzed scRNA‐seq of 52 698 cells from resected samples of 5 patients with lung adenocarcinoma or lung squamous cell carcinoma. As a result, 52 stromal cell subtypes were identified. Out of them, 5 distinct types of fibroblasts, clusters 1, 2, 4, 5, and 7 were found. Clusters 1 and 4 were similar, cluster 1 showed a strong EMT and an extensive repertoire of extracellular matrix proteins and TGF‐β‐associated genes. Cluster 4 was enriched in the leading edge of the tumor, whereas cluster 2 exhibited the highest expression of ACTA2. Clusters 5 and 7 were highly similar, with lower myogenesis and high mTOR signature expression. The differences between clusters 5 and 7 were mainly related to the expression of glycolysis genes, indicating metabolic differences between various CAF subsets. Furthermore, cluster 5 was enriched in the tumor core, while cluster 7 was enriched in the tumor edge, further highlighting differences in spatial location as a key component in CAF heterogeneity.
4.4. Colorectal cancer
Li et al 34 analyzed scRNA‐seq of 969 cells from the resected specimen of 11 patients with colorectal cancer. They identified 2 distinct subtypes of CAFs, CAF‐A, and CAF‐B. CAF‐A cells expressed genes related to extracellular matrix remodeling, including the TGF‐β activator MMP2. CAF‐B cells expressed markers of myofibroblasts such as ACTA2, TAGLN, and PDGFA. However, it should be noted that this categorization was based on very few cells and without any spatial mapping in situ, and thus needs to be confirmed in further studies.
4.5. Head and neck squamous cell carcinoma
Puram et al 35 reported the analysis of scRNA‐seq of about 6000 cells from resected specimens from 18 patients with head and neck squamous cell carcinoma. Fibroblasts within the tumor were divided into 4 groups: myofibroblasts, CAFs (CAF1 and CAF2), and resting fibroblasts. The myofibroblast subset expressed ACTA2 and myosin light‐chain proteins (MYLK, MYL9). The CAF subset expressed receptors, ligands, and ECM genes, including FAP and PDPN. The resting fibroblasts lacked the expression of markers for myofibroblasts and CAFs. Further analysis partitioned CAFs into 2 types: CAF1 and CAF2. The CAF1 type expressed COL1A1, mesenchymal markers (eg, VIM, THY1) and ECM proteins (eg, MMP11, CAV1), while the CAF 2 type expressed immediate early response genes (eg, JUN, FOS), ligands and receptors (eg, FGF7, TGFBR). Interestingly, they also found that fibroblasts from regional lymph nodes were enriched for myofibroblasts and the CAF1 subset.
5. TARGETING OPPORTUNITIES FOR SUBSETS OF CAFs
Studies on the functional definition of CAF subsets implies the possibility that specific population of CAFs could be exploited as therapeutic targets. Although, to date, not clinically proven, several studies have reported preclinical studies of CAF subset‐targeted therapy. In our previous report, Pdgfra was expressed specifically by cells in the mCAF cluster. 19 Platelet‐derived growth factor (PDGF)‐CC is considered a selective ligand for PDGFR‐α based on in vitro studies and demonstrating that PDGF‐CC activates PDGFR‐α homodimers as well as PDGFR‐α/β heterodimers. 36 We demonstrated that paracrine signaling by PDGF‐CC between cancer cells and PDGFRα+ CAFs in the basal‐like breast tumor microenvironment controls breast cancer molecular subtype. Cancer cell‐derived PDGF‐CC activated mCAFs and induced them to secrete HGF, IGFBP3, and STC1, the action of which instigated an estrogen receptor (ER)α‐negative phenotype of breast cancer cells. Furthermore, genetic or pharmacologic blockade of PDGF‐CC prompted sensitization of previously resistant breast tumors to endocrine therapy through induction of a luminal phenotype. These findings suggested that administration of drugs that block signaling in the PDGF‐CC/PDGFRα axis could be a new mCAF‐targeted therapy in patients with ERα‐negative breast cancer.
As defined in the study by Friedman et al, 22 Podoplanin marks a specific subpopulation of CAFs, and is gathering attention as a target for therapy. Podoplanin‐expressing CAFs have been identified in various malignancies and have been reported to be a prognostic factor in breast cancer and lung cancer. 36 It has also been reported that Podoplanin expressed by CAFs is functionally responsible for the promotion of tumor formation in mouse subcutaneous tissue. 37 Antibodies, CAR‐T cells, biologics, and synthetic compounds that target Podoplanin are being developed and tested in preclinical models to date, and there is a possibility that these therapies may constitute targeted therapy for Podoplanin‐expressing CAFs. 38
In the study by Li et al 34 described previously, CAF‐A exclusively expressed FAP. Because FAP is overexpressed by CAFs in 85%‐90% of primary and metastatic colorectal cancers and not detectable in normal tissues, FAP has been considered as a suitable target with minimal toxicity for cancer therapy. Multiple clinical trials have explored targeting of cells expressing FAP; however to date they have all failed. 39
Immune checkpoint blockade has emerged as one of the most promising therapeutic options for patients. As described previously, Dominguez et al 32 demonstrated that LRRC15+ CAFs are associated with a poor response to anti–PD‐L1 therapy. This indicates that a specific CAF subset could be a potential target for improving immunotherapy. Future studies are needed to develop a new treatment targeting this specific CAF population.
6. LIMITATIONS OF STUDIES TO DATE AND FUTURE DIRECTIONS
Although a wealth of information has been brought by studies attempting to classify CAFs based on scRNA‐seq, there are some limitations in these studies. Firstly, it remains unclear whether each CAF population is preserved across cancer types. Secondly, it also remains unclear whether the classification of CAFs based on mouse models can be applied to human cancer. In breast cancer, studies using mouse models provide only information for one representative histologic type of breast cancer, while human breast cancer has multiple histologic types and thus a higher degree of complexity. Joint analysis of multiple scRNA‐seq datasets across cancer types and species will answer these questions in the future. 40 The third limitation is the lack of direct information on the origins of CAFs. Analysis of scRNA‐seq only provides information on the transcriptome at a specific time point, thus direct information that shows the origin of a specific population cannot be obtained. Because of the lack of highly specific Cre drivers for normal fibroblasts, lineage tracing studies in mice remain scarce, however based on the information gathered from scRNA‐seq studies novel tools can be developed and may help to reveal the origins of CAFs. Finally, spatially resolved analysis of CAF subsets using multiplexed visualization of RNA or protein are likely to be an increasingly valuable tool to comprehend the full complexity of heterogeneity among CAFs across different tumor types.
7. CONCLUDING REMARKS
Furthering our understanding of the TME is decisive to develop new cancer therapies and biomarkers. The function of CAFs, an important component of the TME, has to date not been satisfactorily perceived due to its heterogeneity. Development of single‐cell analysis, which enables us to further classify CAFs, is a significant breakthrough in cancer research and will further our understanding about subtypes and substates of CAFs. Detailed classification of CAFs and investigation of the functions of each subset provides us with crucial leads to develop novel CAF‐targeted precision therapies and biomarkers.
DISCLOSURE
KP is a minority shareholder in Paracrine Therapeutics, a company involved in the development of CAF‐targeted cancer therapies.
ACKNOWLEDGMENTS
KP is the Göran and Birgitta Grosskopf Professor of Molecular Medicine. This work was supported by grants to KP from the Swedish Cancer Society, the Swedish Research Council, the Biltema Foundation, the Cancera Foundation, the Region Skåne ALF funding, and the Sjöberg Foundation. RK is supported by postdoctoral fellowships from the Wenner‐Gren Foundation and the Swedish Cancer Society.
Kanzaki R, Pietras K. Heterogeneity of cancer-associated fibroblasts: Opportunities for precision medicine. Cancer Sci. 2020;111:2708–2717. 10.1111/cas.14537
REFERENCES
- 1. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57‐70. [DOI] [PubMed] [Google Scholar]
- 2. Biankin AV, Piantadosi S, Hollingsworth SJ. Patient‐centric trials for therapeutic development in precision oncology. Nature. 2015;526:361‐370. [DOI] [PubMed] [Google Scholar]
- 3. Navab R, Strumpf D, Bandarchi B, et al. Prognostic gene‐expression signature of carcinoma‐associated fibroblasts in non‐small cell lung cancer. Proc Natl Acad Sci USA. 2011;108:7160‐7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518‐527. [DOI] [PubMed] [Google Scholar]
- 5. Calon A, Lonardo E, Berenguer‐Llergo A, et al. Stromal gene expression defines poor‐prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320‐329. [DOI] [PubMed] [Google Scholar]
- 6. Pietras K, Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316:1324‐1331. [DOI] [PubMed] [Google Scholar]
- 7. Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton‐Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170:1807‐1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kojima Y, Acar A, Eaton EN, et al. Autocrine TGF‐beta and stromal cell‐derived factor‐1 (SDF‐1) signaling drives the evolution of tumor‐promoting mammary stromal myofibroblasts. Proc Natl Acad Sci USA. 2010;107:20009‐20014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ren YU, Zhou X, Liu X, et al. Reprogramming carcinoma associated fibroblasts by AC1MMYR2 impedes tumor metastasis and improves chemotherapy efficacy. Cancer Lett. 2016;374:96‐106. [DOI] [PubMed] [Google Scholar]
- 10. Zeisberg EM, Potenta S, Xie L, Zeisberg M, Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma‐associated fibroblasts. Cancer Res. 2007;67:10123‐10128. [DOI] [PubMed] [Google Scholar]
- 11. Zhang Y, Daquinag A, Traktuev DO, et al. White adipose tissue cells are recruited by experimental tumors and promote cancer progression in mouse models. Cancer Res. 2009;69:5259‐5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bochet L, Lehuede C, Dauvillier S, et al. Adipocyte‐derived fibroblasts promote tumor progression and contribute to the desmoplastic reaction in breast cancer. Cancer Res. 2013;73:5657‐5668. [DOI] [PubMed] [Google Scholar]
- 13. Ishii G, Sangai T, Oda T, et al. Bone‐marrow‐derived myofibroblasts contribute to the cancer‐induced stromal reaction. Biochem Biophys Res Commun. 2003;309:232‐240. [DOI] [PubMed] [Google Scholar]
- 14. Raz Y, Cohen N, Shani O, et al. Bone marrow‐derived fibroblasts are a functionally distinct stromal cell population in breast cancer. J Exp Med. 2018;215:3075‐3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herrera M, Islam A, Herrera A, et al. Functional heterogeneity of cancer‐associated fibroblasts from human colon tumors shows specific prognostic gene expression signature. Clin Cancer Res. 2013;19:5914‐5926. [DOI] [PubMed] [Google Scholar]
- 16. Hao J, Zeltz C, Pintilie M, et al. Characterization of distinct populations of carcinoma‐associated fibroblasts from non‐small cell lung carcinoma reveals a role for ST8SIA2 in cancer cell invasion. Neoplasia. 2019;21:482‐493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang F, Barbacioru C, Wang Y, et al. mRNA‐Seq whole‐transcriptome analysis of a single cell. Nat Methods. 2009;6:377‐382. [DOI] [PubMed] [Google Scholar]
- 18. Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single‐cell sequencing. Nature. 2011;472:90‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bartoschek M, Oskolkov N, Bocci M, et al. Spatially and functionally distinct subclasses of breast cancer‐associated fibroblasts revealed by single cell RNA sequencing. Nat Commun. 2018;9:5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578‐10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin EY, Jones JG, Li P, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113‐2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gil Friedman OL‐G, David E, Bornstein C, et al. Cancer‐associated fibroblast compositions change with breast‐cancer progression linking S100A4 and PDPN ratios with clinical outcome. bioRxiv. 2020. 10.1101/2020.01.12.903039 [DOI] [PubMed] [Google Scholar]
- 23. Costa A, Kieffer Y, Scholer‐Dahirel A, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463‐479 e410. [DOI] [PubMed] [Google Scholar]
- 24. Pelon F, Bourachot B, Kieffer Y, et al. Cancer‐associated fibroblast heterogeneity in axillary lymph nodes drives metastases in breast cancer through complementary mechanisms. Nat Commun. 2020;11:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Su S, Chen J, Yao H, et al. CD10(+)GPR77(+) cancer‐associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell. 2018;172:841‐856 e816. [DOI] [PubMed] [Google Scholar]
- 26. Öhlund D, Handly‐Santana A, Biffi G, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579‐596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elyada E, Bolisetty M, Laise P, et al. Cross‐species single‐cell analysis of pancreatic ductal adenocarcinoma reveals antigen‐presenting cancer‐associated fibroblasts. Cancer Discov. 2019;9:1102‐1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Biffi G, Oni TE, Spielman B, et al. IL1‐induced JAK/STAT signaling is antagonized by TGFbeta to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 2019;9:282‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hosein AN, Huang H, Wang Z, et al. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single‐cell resolution. JCI Insight. 2019;5:e129212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bernard V, Semaan A, Huang J, et al. Single‐cell transcriptomics of pancreatic cancer precursors demonstrates epithelial and microenvironmental heterogeneity as an early event in neoplastic progression. Clin Cancer Res. 2019;25:2194‐2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neuzillet C, Tijeras‐Raballand A, Ragulan C, et al. Inter‐ and intra‐tumoural heterogeneity in cancer‐associated fibroblasts of human pancreatic ductal adenocarcinoma. J Pathol. 2019;248:51‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dominguez CX, Muller S, Keerthivasan S, et al. Single‐cell RNA sequencing reveals stromal evolution into LRRC15(+) myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov. 2020;10:232‐253. [DOI] [PubMed] [Google Scholar]
- 33. Lambrechts D, Wauters E, Boeckx B, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24:1277‐1289. [DOI] [PubMed] [Google Scholar]
- 34. Li H, Courtois ET, Sengupta D, et al. Reference component analysis of single‐cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017;49:708‐718. [DOI] [PubMed] [Google Scholar]
- 35. Puram SV, Tirosh I, Parikh AS, et al. Single‐cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611‐1624 e1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cao R, Brakenhielm E, Li X, et al. Angiogenesis stimulated by PDGF‐CC, a novel member in the PDGF family, involves activation of PDGFR‐alphaalpha and ‐alphabeta receptors. FASEB J. 2002;16:1575‐1583. [DOI] [PubMed] [Google Scholar]
- 37. Ishii G, Ochiai A, Neri S. Phenotypic and functional heterogeneity of cancer‐associated fibroblast within the tumor microenvironment. Adv Drug Deliv Rev. 2016;99:186‐196. [DOI] [PubMed] [Google Scholar]
- 38. Krishnan H, Rayes J, Miyashita T, et al. Podoplanin: an emerging cancer biomarker and therapeutic target. Cancer Sci. 2018;109:1292‐1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jiang GM, Xu W, Du J, et al. The application of the fibroblast activation protein alpha‐targeted immunotherapy strategy. Oncotarget. 2016;7:33472‐33482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barkas N, Petukhov V, Nikolaeva D, et al. Joint analysis of heterogeneous single‐cell RNA‐seq dataset collections. Nat Methods. 2019;16:695‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]


