Abstract
The expression of classical human leukocyte antigen class I antigens (HLA‐I) on the surfaces of cancer cells allows cytotoxic T cells to recognize and eliminate these cells. Reduction or loss of HLA‐I is a mechanism of escape from antitumor immunity. The present study aimed to investigate the clinicopathological impacts of HLA‐I and non–classical HLA‐I antigens expressed on pancreatic ductal adenocarcinoma (PDAC) cells. We performed immunohistochemistry to detect expression of HLA‐I antigens in PDAC using 243 PDAC cases and examined their clinicopathological influences. We also investigated the expression of immune‐related genes to characterize PDAC tumor microenvironments. Lower expression of HLA‐I, found in 33% of PDAC cases, was significantly associated with longer overall survival. Higher expression of both HLA‐E and HLA‐G was significantly associated with shorter survival. Multivariate analyses revealed that higher expression of these three HLA‐I antigens was significantly correlated with shorter survival. Higher HLA‐I expression on PDAC cells was significantly correlated with higher expression of IFNG, which also correlated with PD1, PD‐L1 and PD‐L2 expression. In vitro assay revealed that interferon gamma (IFNγ) stimulation increased surface expression of HLA‐I in three PDAC cell lines. It also upregulated surface expression of HLA‐E, HLA‐G and immune checkpoint molecules, including PD‐L1 and PD‐L2. These results suggest that the higher expression of HLA‐I, HLA‐E and HLA‐G on PDAC cells is an unfavorable prognosticator. It is possible that IFNγ promotes a tolerant microenvironment by inducing immune checkpoint molecules in PDAC tissues with higher HLA‐I expression on PDAC cells.
Keywords: HLA class I antigens, HLA‐E, HLA‐G, IFNγ, pancreatic cancer
human leukocyte antigen class I antigens (HLA‐I) are needed for T cells to recognize target cells. Here, we showed that higher HLA‐I expression on pancreatic cancer cells is associated with poor prognosis, where formation of the tolerant microenvironment may be involved in IFNγ.
1. INTRODUCTION
Cytotoxic CD8+ T cells (CTL) eliminate cancer cells in responsive antitumor immune microenvironments. Classical human leukocyte antigen class I antigens (HLA‐A, HLA‐B and HLA‐C; HLA‐I) are constitutively expressed by nucleated cells. Surface expression of HLA‐I is necessary for cognate CTL to recognize cancer cells and is a prerequisite for CTL‐mediated immune therapy. Host immune surveillance against tumors functions in the early carcinogenic stage, although clinically established cancers usually progress even in the presence of a host anti–tumor immune response, as a result of tumor immune escape strategies, including evasion of immune surveillance and induction of immune tolerance. 1 , 2 , 3 , 4 One of the escape mechanisms is reduction or loss of HLA‐I on the surfaces of cancer cells. 5 This is an unfavorable prognosticator in many types of cancers, such as breast, 6 ovarian 7 and rectal cancers. 8
Non–classical HLA class I antigens, HLA‐E and HLA‐G, share many amino acid sequence similarities with classical HLA‐I. HLA‐E is thought to provide an important “self‐signal” to the immune system by accommodating and presenting peptide fragments from leader sequences of classical and nonclassical HLA‐I antigens. 9 , 10 HLA‐G is known to have a tolerogenic function in physiological and pathological conditions such as the maternal‐fetal interf ace and cancers. 1 , 11 Both HLA‐E and HLA‐G bind to inhibitory receptors on natural killer (NK) cells and inactivate their cytolytic function. 1 , 11 Low expression of HLA‐E is associated with longer survival in ovarian cancer 12 ; in contrast, cells positive for HLA‐G and negative for HLA‐E expression are associated with poorer outcomes compared to those negative for HLA‐G and positive for HLA‐E expression in patients with colorectal cancer. 13 Expression of HLA‐G is associated with poorer outcomes in several cancers such as breast cancer 14 and hepatocellular carcinoma, 15 although in high‐grade epithelial ovarian carcinoma, expression of HLA‐G is associated with longer progression‐free survival. 16 The prognostic significance of HLA‐G expression for pancreatic cancer patients is controversial. 17 , 18 , 19
Pancreatic ductal adenocarcinoma (PDAC) is one of the most devastating cancers found worldwide: the 5‐year survival rate is still <10%. 20 , 21 Apart from surgical resection, a curative treatment has not been developed. A thorough understanding of PDAC is required to develop new treatment modalities for patients with unresectable and recurrent PDAC. Recently, new cancer immunotherapies have been developed that re–stimulate host immune responses, resulting in sustained tumor elimination. Immune checkpoint inhibitors have led to marked and durable improvement in the outcome of patients with malignant tumors with poor prognoses, although they are not always effective and only patients with specific types of cancer or under limited conditions have benefitted. Reduction or loss of HLA‐I expression on cancer cells has been associated with reduced responses to immune checkpoint inhibitor therapy. 22 , 23 Responses to immunotherapy in PDAC are rare, 24 , 25 , 26 and, currently, combination therapies with immune checkpoint treatments are being examined.
Expression of HLA‐I on PDAC cells is important information for selecting cases that may be suitable for therapy. Two different groups reported HLA‐I expression on PDAC cells, 27 , 28 although these were not large‐scale studies and the association with patient outcomes was not determined. Ryschich et al examined 46 cases of PDAC, and reduced and lost expression of HLA‐I was found in 24% and 6% of cases, respectively. No significant difference in overall survival (OS) was found between patients with PDAC cells positive for HLA‐I and those with PDAC cells with reduced or no HLA‐I expression. 28 Imai et al reported that high and low expression of HLA‐I was found in 53% (19/36) and 47% (17/36) of PDAC patients, respectively, and high expression of HLA‐I was significantly associated with longer OS and recurrence‐free survival compared to that in the low expression group. 27
The aim of this study was to investigate the clinicopathological characteristics and prognoses associated with expression of classical and non–classical HLA‐I antigens on PDAC cells. We performed immunohistochemistry to detect expression of HLA‐I antigens in 243 PDAC cases and examined clinicopathological correlates. We also investigated the expression of immune‐related genes to characterize PDAC tumor microenvironments. Furthermore, we determined whether interferon gamma (IFNγ) affects expression of classical and non–classical HLA‐I antigens and immune checkpoint molecules using an in vitro assay with three PDAC cell lines.
2. MATERIALS AND METHODS
2.1. Ethics approval and consent to participate
This study was approved by the Institutional Review Board of the National Cancer Center, Japan (#2005‐077, #2011‐218). Informed consent was obtained from all participants involved in the study, and all clinical investigations were conducted in line with the principles of the Declaration of Helsinki.
2.2. Patients and samples
Clinical and pathological data and specimens used for immunohistochemical analysis were obtained through a detailed retrospective review of the medical records of 243 patients with PDAC: 146 consecutive patients who had undergone surgical resection between 1990 and 2000 and 98 patients who had undergone surgical resection between 2001 and 2005 at the National Cancer Center Hospital, Tokyo whose fresh frozen tissues were available from surgically resected specimens were included in the study. None of the patients had received any therapy before surgery. All patients included in this study underwent macroscopic curative resection, and all cases involved conventional ductal carcinomas. The clinicopathological characteristics of study participants are summarized in Table S1. The median follow‐up period after surgery for the patients as a whole and for living patients was 17.6 (2.6‐201) and 65.8 (2.6‐201) months, respectively. Patients were followed up every 1 to 2 months during the first year after surgery. Each follow up included a physical examination, blood chemistry test, and measurement of serum carbohydrate antigen 19‐9 (CA19‐9) and carcinoembrionic antigen. Ultrasonography and enhanced computed tomography were performed every 3 months. Recurrence was diagnosed when a new local or distant metastatic lesion was found on imaging studies or an increase in tumor marker levels with deterioration of patients’ condition was recognized. At the census date (September 2011), we checked whether the patients were dead or alive; 44 patients (18.1%) were alive, 170 (70.0%) had died of pancreatic cancer, and 29 (11.9%) had died of other causes. All M1 (TNM classification 29 ) patients had para‐aortic nodal metastasis, without any other form of metastasis.
2.3. Pathological examination
All the carcinomas were examined pathologically and classified according to the World Health Organization (WHO) classification, 30 the Union for International Cancer Control (UICC) TNM classification 29 and the Classification of Pancreatic Carcinoma of the Japan Pancreas Society. 31 Tertiary lymphoid organs and histological tumor necrosis were evaluated as previously described. 32 , 33 , 34
2.4. Immunohistochemistry
Immunohistochemistry was performed on 4‐µm‐thick formalin‐fixed, paraffin‐embedded tissue sections using the avidin–biotin complex method as described previously. 34 The CSA‐II System (Agilent) was used only for PD‐L2 staining. The list of antibodies used in this study is presented in Table S2. Immunohistochemistry without the primary antibody was used as a negative control. Tissue specimens were examined in this study using the maximum cut surfaces of tumors. We evaluated immunolabeled‐CD56 cells as previously mentioned. 35 We counted immunolabeled‐PD1 cell numbers using the same method with some modification. Briefly, we selected 10 areas in which the immunolabeled cells had infiltrated into the tumor representatively and PD1‐immunolabeled cells were counted at high magnification (×400). We considered PDAC tissue as PD1 positive when there were more than 20 PD1+ cells on average. When PD‐L1 or PD‐L2 labeled cells, both in cancer cells and stromal cells, were ≥5% of the entire cells in the PDAC tissue excluding lymphoid tissues, we considered them to be positive for PD‐L1 or PD‐L2.
2.5. Evaluation of HLA‐I, HLA‐E and HLA‐G expression
After immunohistochemistry, expression of HLA‐I (Figure 1) was evaluated and classified into four grades as follows: strongly positive (+++), with almost all cancer cells (≥90%) staining strongly positive for HLA‐I; moderately positive (++), with <90% and ≥50% of cancer cells staining strongly positive for HLA‐I; weakly positive (+), with <50% and >10% of cancer cells staining strongly positive or >10% of cancer cells staining weakly positive for HLA‐I; and negative (−), with ≤10% of cancer cells staining positive for HLA‐I. Strong positive staining was defined as staining intensity being equal to or stronger than that in lymphocytes or endothelial cells. Cells were considered positive for HLA‐I when expression was observed on plasma membranes. Cells with cytoplasmic HLA‐I staining were not considered positive. Expression of HLA‐E on cancer cells was evaluated using the same system as that of HLA‐I. When more than 5% of cancer cells in a PDAC tissue section expressed HLA‐G, the PDAC case was judged positive for HLA‐G; otherwise, it was considered negative. Three observers, having no access to the patient data, independently evaluated the expression grade of HLA‐I and HLA‐E, and the expression of HLA‐G. If more than one observer judged identical value, it became the final value. If there were three different judgments, the observers discussed the reasons for the difference and performed reevaluation until resolving three different judgments. To assess intraobserver reproducibility, several tissue sections were counted thrice by each observer. To assess interobserver reproducibility, 10 values counted by each observer for the same tumor were compared.
Figure 1.
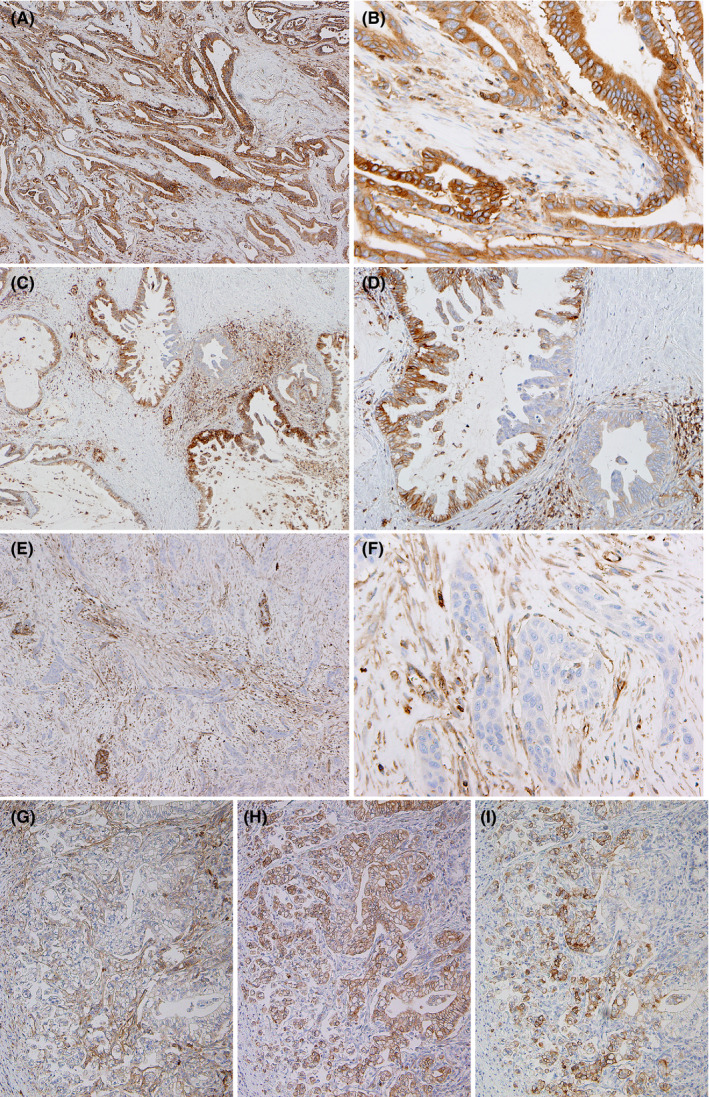
A‐D, Immunohistochemical detection of human leukocyte antigen class I antigens (HLA‐I) (A‐G), HLA‐E (H) and HLA‐G (I) in pancreatic ductal adenocarcinoma (PDAC). Low‐power view (A, C, E), medium‐power view (D, G‐I) and high‐power view (B, F). Strongly positive (A, B), moderately positive (C, D) and negative (E, F) for HLA‐I. Comparison of reactions to three antibodies in the same areas of PDAC tissue (G‐I)
2.6. Quantitative RT‐PCR
Total RNA was extracted from fresh frozen tissue, as described previously. 4 Quality of the extracted RNA was measured as described previously 33 ; the rRNA ratio [28s/18s] and RNA integrity number (RIN) were 1.21 ± 0.18 and 7.2 ± 0.9 (average ± SD), respectively. Quantitative RT‐PCR (RT‐qPCR) for target genes and non–target housekeeping control genes was performed on a 7500 Real‐Time PCR System (Applied Biosystems) using FastStart Universal Probe Master and probes from the Universal Probe Library (Roche Diagnostics). 4 The sequences of the primers and respective Universal Probe Library probes are given in Table S3. Expression levels were normalized to those of ACTB.
2.7. Cultivation of cancer cells and experimental stimulation
Human PDAC cells were obtained from the ATCC. AsPC‐1 cells were cultured in RPMI‐1640 medium with 10% FBS and 1% penicillin‐streptomycin‐glutamine (Invitrogen) at 37°C in a humidified incubator with 5% CO2. DMEM was used to cultivate Capan‐1 and Capan‐2 cells. Each of these cell lines was authenticated within 6 months of acquisition by short tandem repeat analysis (ATCC). The effect of IFNγ on cancer cells was examined after culture in medium with recombinant human IFNγ (100 ng/mL, R&D systems) for 48 hours. 36 , 37 Cells suspended in PBS with 5% FBS were stained. Before incubation with fluorescent dye‐labeled antibodies (Table S2), Fc receptors were blocked. Flow cytometry analyses were carried out using a FACSCalibur flow cytometer (BD Biosciences) and the data were analyzed using CellQuest Pro software (BD Biosciences). The experiments were repeated three times.
2.8. Microarray analysis
Total RNA was extracted from the three PDAC cell lines with or without IFNγ stimulation using RNeasy Mini Kits (Qiagen). The RIN was evaluated and the values were all confirmed to be >8.0. Microarray analysis was performed at the Chemical Evaluation Research Institute (CERI). Briefly, following a protocol of Agilent’s One‐Color Microarray‐Based Gene Expression Analysis Low Input Quick Amp Labeling ver.6.9, 100 ng of total RNA was used to generate Cy3‐labeled cRNA. Subsequently, samples were hybridized on a SurePrint_G3_Human_GE_8x60K_Microarray ver.3.0 (Agilent). Arrays were scanned with a DNA Microarray Scanner, and the acquired images were analyzed by Feature Extraction ver.10.7.1.1 (Agilent). The signals were normalized in Gene Spring GX 14.5 (Agilent). Hierarchical clustering of gene expression data was performed by a list on Gene Ontology. The gene list was made based on GO:0031294 (GO term: lymphocyte costimulation) and GO:0072676 (GO term: lymphocyte migration).
2.9. The Cancer Genome Atlas datasets
Normalized mRNA expression data and associated clinical annotations were obtained from The Cancer Genome Atlas (TCGA) study of PDAC. 38
2.10. Statistical analysis
Qualitative variables were compared using Fisher’s exact test. Pairwise comparisons of the subgroups were performed by Mann‐Whitney U test. Postoperative OS and disease‐free survival (DFS) rates were calculated using the Kaplan‐Meier method. A univariate analysis was performed for prognostic factors using the log‐rank test. The factors found to be significant by univariate analysis were subjected to multivariate analysis using the Cox proportional hazards model (backward elimination method). Differences at P < 0.05 were considered statistically significant. Statistical analyses were performed in StatView‐J 5.0 (Abacus Concepts).
3. RESULTS
3.1. Expression of human leukocyte antigen class I antigens on pancreatic ductal adenocarcinoma cells and prognostic associations with outcome
Expression of HLA‐I on PDAC cells was evaluated after immunohistochemistry (Figure 1) and classified into four grades. Strongly positive (+++) expression of HLA‐I was found in 21.0% of PDAC cases, moderately positive (++) expression was found in 46.1%, weakly positive (+) expression was found in 31.0%, and no (−) expression was in 2.1% (Figure 2A).
Figure 2.
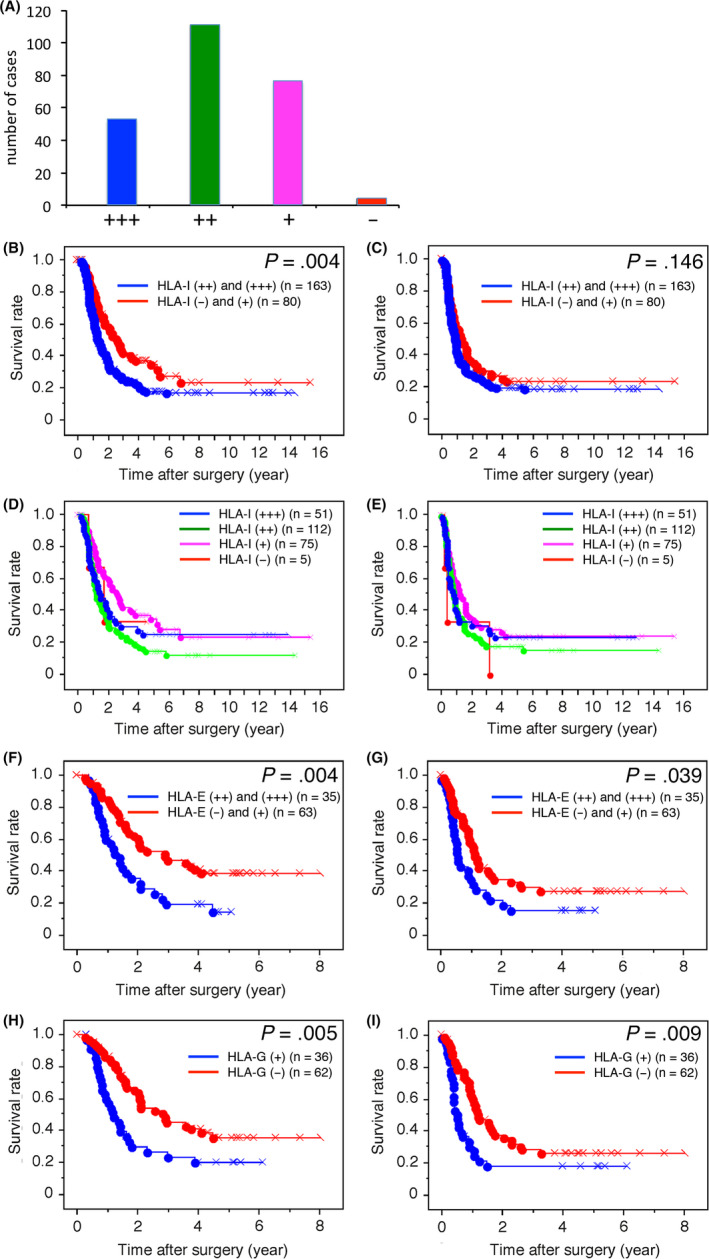
A, Bar graph showing the number of pancreatic ductal adenocarcinoma (PDAC) cases in each of the four human leukocyte antigen class I antigens (HLA‐I) expression level categories. B‐I, Kaplan‐Meier survival curves for overall survival (OS) in patients with PDAC according to HLA‐I (B, D), HLA‐E (F) HLA‐G (H) expression, and for disease‐free survival (DFS) in patients with PDAC according to HLA‐I (C, E), HLA‐E (G) and HLA‐G (I) expression
Kaplan‐Meier survival analysis revealed that higher expression of HLA‐I was significantly associated with shorter OS (Figure 2B). The weakly positive (+) and moderately positive (++) groups tended to have favorable and unfavorable OS, respectively (Figure 2D). Similar tendencies were observed for DFS, although this association was not statistically significant (Figure 2C and E). When the variables found to be significantly associated with outcomes by univariate COX analysis were subjected to multivariate COX analysis, higher HLA‐I expression was closely associated with shorter OS (Table 1).
Table 1.
Univariate and multivariate analyses of prognostic factors associated with overall survival in patients with pancreatic ductal adenocarcinoma (n = 243)
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P‐value | HR (95% CI) | P‐value | |
| Age (<65 y/≥65 y) | 1.082 (0.792‐1.489) | 0.615 | ||
| Gender (female/male) | 0.894 (0.654‐1.223) | 0.484 | ||
| Histological tumor necrosis (absence/presence) | 2.175 (1.576‐3.001) | <0.0001 | 1.895 (1.361‐2.637) | 0.0002 |
| Pathologic tumor status (T1 + T2/T3 + T4) | 1.467 (1.076‐2.001) | 0.015 | ||
| Pathologic node status (N0/N1 + N2) | 1.913 (1.257‐2.911) | 0.003 | ||
| Pathologic metastasis status (M0/M1) | 2.562 (1.637‐4.011) | <0.0001 | 2.097 (1.329‐3.308) | 0.002 |
| Histological grade (G1/G2 + G3) | 1.434 (1.005‐2.047) | 0.047 | ||
| Tumor margin status (negative/ positive) | 1.256 (0.904‐1.744) | 0.174 | ||
| Nerve plexus invasion (absence/presence) a | 1.374 (0.976‐1.934) | 0.068 | ||
| Lymphatic invasion (0, 1/2, 3) a | 2.086 (1.454‐2.991) | <0.0001 | 1.743 (1.183‐2.570) | 0.005 |
| Venous invasion (0, 1/2, 3) a | 1.714 (1.241‐2.369) | 0.001 | 1.484 (1.050‐2.097) | 0.025 |
| Intrapancreatic neural invasion (0, 1/ 2, 3) a | 1.589 (1.155‐2.185) | 0.004 | ||
| HLA Class I antigen expression (−, +/++, +++) | 1.621 (1.162‐2.261) | 0.005 | 1.527 (1.086‐2.146) | 0.015 |
Classified according to the classification of pancreatic carcinoma of Japan Pancreas Society. CI, confidence interval; HLA, human leukocyte antigen class I antigens; HR, hazard ratio. Bold letters indicate significant values.
3.2. Expression of HLA‐E and HLA‐G on pancreatic ductal adenocarcinoma cells and prognostic associations with outcome
Expression profiles of HLA‐E and HLA‐I were sometimes similar but other times quite different (Figure 1G and H). Profiles of HLA‐G and HLA‐I expression were usually different (Figure 1G and I). Higher expression of HLA‐E was found in 35.7% of PDAC cases, and expression of HLA‐G was found in 36.7% of PDAC cases. Kaplan‐Meier survival analysis revealed that higher expression of HLA‐E was significantly associated with both shorter OS and DFS (Figure 2F and G); expression of HLA‐G was also significantly associated with both shorter OS and DFS (Figure 2H and I). According to multivariate COX analysis, higher expression of HLA‐I and HLA‐E, and expression of HLA‐G were closely associated with shorter OS (Table S4), whereas higher expression of HLA‐E and expression of HLA‐G were closely associated with shorter DFS (Table S5).
3.3. Association between HLA‐I expression on pancreatic ductal adenocarcinoma cells and both clinicopathological variables and expression of immune‐related genes in pancreatic ductal adenocarcinoma tissues
We analyzed the relationship between HLA‐I expression on PDAC cells and various clinicopathological variables (Tables 2 and S1). HLA‐I expression showed a significant correlation with the presence of histological tumor necrosis. In addition, immunohistochemical detection of PD1, PD‐L1 and PD‐L2 in PDAC tissues was significantly correlated with HLA‐I expression but not with HLA‐E and HLA‐G. There was no correlation between HLA‐I expression and HLA‐E or HLA‐G expression on PDAC cells (Table 2).
Table 2.
Relationship between clinicopathological characteristics and HLA‐I, HLA‐E or HLA‐G expression in pancreatic ductal adenocarcinoma cells
| Characteristics | Total | HLA‐I expression | P | HLA‐E expression | P | HLA‐G expression | P | |||
|---|---|---|---|---|---|---|---|---|---|---|
| (−) or (+) | (++) or (+++) | (−) or (+) | (++) or (+++) | (−) or (+) | (++) or (+++) | |||||
| Age, years | ||||||||||
| <65 | 52 | 13 | 39 | 0.002 | 30 | 22 | 0.205 | 31 | 21 | 0.530 |
| ≥65 | 46 | 26 | 20 | 33 | 13 | 31 | 15 | |||
| Sex | ||||||||||
| Male | 62 | 25 | 37 | 1.00 | 40 | 22 | 1.00 | 43 | 19 | 0.129 |
| Female | 36 | 14 | 22 | 23 | 13 | 19 | 17 | |||
| Intratumoral tertiary lymphoid organs | ||||||||||
| Absence | 86 | 32 | 54 | 0.111 | 55 | 31 | 1.00 | 53 | 33 | 0.742 |
| Presence | 11 | 7 | 4 | 7 | 4 | 8 | 3 | |||
| Histological tumor necrosis | ||||||||||
| Absence | 30 | 18 | 12 | 0.008 | 24 | 11 | 0.659 | 25 | 10 | 0.274 |
| Presence | 68 | 21 | 47 | 58 | 23 | 36 | 25 | |||
| Pathologic tumor status | ||||||||||
| T1, T2 | 66 | 23 | 43 | 0.188 | 37 | 29 | 0.024 | 41 | 25 | 0.825 |
| T3, T4 | 32 | 16 | 16 | 26 | 6 | 21 | 11 | |||
| Pathologic node status | ||||||||||
| N0 | 21 | 8 | 13 | 1.00 | 14 | 7 | 1.00 | 15 | 6 | 0.451 |
| N1, N2 | 77 | 31 | 46 | 49 | 28 | 47 | 30 | |||
| Pathologic metastasis status | ||||||||||
| M0 | 88 | 35 | 53 | 1.00 | 58 | 30 | 0.323 | 57 | 31 | 0.491 |
| M1 | 10 | 4 | 6 | 5 | 5 | 5 | 5 | |||
| Tumor histological grade | ||||||||||
| G1 | 18 | 5 | 13 | 0.300 | 13 | 5 | 0.588 | 13 | 5 | 0.431 |
| G2, G3 | 80 | 34 | 46 | 50 | 30 | 49 | 31 | |||
| Tumor margin status | ||||||||||
| Negative | 75 | 27 | 48 | 0.224 | 50 | 25 | 0.457 | 46 | 29 | 0.622 |
| Positive | 23 | 12 | 11 | 13 | 10 | 16 | 7 | |||
| Nerve plexus invasion a | ||||||||||
| Absence | 38 | 15 | 23 | 1.00 | 20 | 18 | 0.083 | 25 | 13 | 0.830 |
| Presence | 60 | 24 | 36 | 43 | 17 | 37 | 23 | |||
| Lymphatic invasion a | ||||||||||
| 0, 1 | 27 | 11 | 16 | 1.00 | 19 | 8 | 0.488 | 18 | 9 | 0.815 |
| 2, 3 | 71 | 28 | 43 | 44 | 27 | 44 | 27 | |||
| Venous invasion a | ||||||||||
| 0, 1 | 28 | 9 | 19 | 0.369 | 21 | 7 | 0.243 | 21 | 7 | 0.166 |
| 2, 3 | 70 | 30 | 40 | 42 | 28 | 41 | 29 | |||
| Intrapancreatic neural invasion a | ||||||||||
| 0, 1 | 44 | 17 | 27 | 1.00 | 28 | 16 | 1.00 | 32 | 12 | 0.095 |
| 2, 3 | 54 | 22 | 32 | 35 | 19 | 30 | 24 | |||
| Tumor‐infiltrating CD56+ cells | ||||||||||
| Low | 48 | 29 | 19 | 1.00 | 27 | 21 | 0.085 | 30 | 18 | 1.00 |
| High | 47 | 28 | 19 | 35 | 12 | 29 | 18 | |||
| PD‐L1 expression | ||||||||||
| Negative | 39 | 21 | 18 | 0.034 | 26 | 13 | 0.830 | 23 | 16 | 0.525 |
| Positive | 59 | 18 | 41 | 37 | 22 | 39 | 20 | |||
| PD‐L2 expression | ||||||||||
| Negative | 68 | 32 | 36 | 0.043 | 43 | 25 | 0.822 | 44 | 24 | 0.657 |
| Positive | 30 | 7 | 23 | 20 | 10 | 18 | 12 | |||
| PD‐1 expression | ||||||||||
| Low | 67 | 32 | 35 | 0.026 | 45 | 22 | 0.500 | 44 | 23 | 0.505 |
| High | 31 | 7 | 24 | 18 | 13 | 18 | 13 | |||
| HLA Class I antigen expression | ||||||||||
| −, + | 39 | 28 | 11 | 0.282 | 27 | 12 | 0.394 | |||
| ++, +++ | 59 | 35 | 24 | 35 | 24 | |||||
| HLA‐E expression | ||||||||||
| −, + | 63 | 28 | 35 | 0.282 | 44 | 19 | 0.083 | |||
| ++, +++ | 35 | 11 | 24 | 18 | 17 | |||||
| HLA‐G expression | ||||||||||
| Negative | 62 | 27 | 35 | 0.394 | 44 | 18 | 0.083 | |||
| Positive | 36 | 12 | 24 | 19 | 17 | |||||
| Total | 39 | 59 | 63 | 35 | 62 | 36 | ||||
Classified according to the classification of pancreatic carcinoma of Japan Pancreas Society. Bold letters indicate significant values.
Next, we analyzed the relationship between HLA‐I expression on PDAC cells and immune‐related gene expression in PDAC tissue by RT‐qPCR. PDAC tissues with higher PDAC cell HLA‐I expression showed significantly lower expression of CD4 and RORC and higher expression of IFNG and ARG2 compared to those with lower expression of HLA‐I (Figure 3A). Expression of IFNG correlated with higher expression of PD1, PD‐L1, PD‐L2 and CD8 with high correlation coefficients (Figure 3B).
Figure 3.
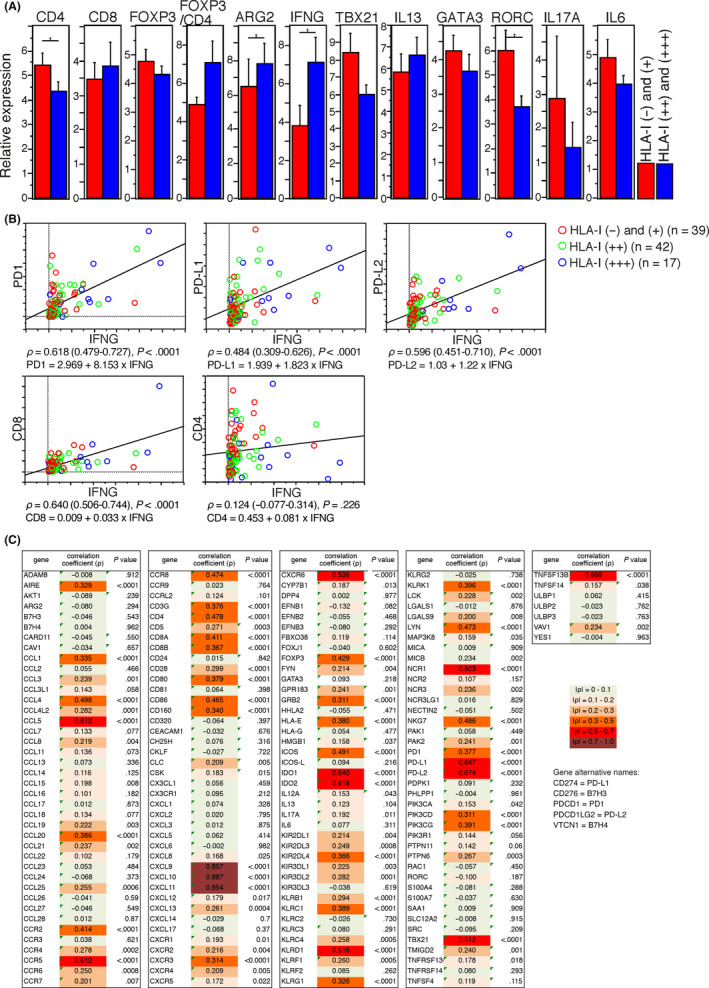
A, Expression of immune‐related genes in human leukocyte antigen class I antigens (HLA‐I) (+) and (–) (red) and HLA‐I (++) and (+++) (blue) pancreatic ductal adenocarcinoma (PDAC) tissues (n = 98) determined by by quantitative RT‐PCR. The y‐axis shows the relative expression of genes, and bars represent means ± standard errors. Differences are determined by Mann‐Whitney U test, with a significance value of P < 0.05 (*). B, Correlation between the expression of IFNG (x‐axis) and other genes (y‐axis) in PDAC tissues (n = 98) determined by quantitative RT‐PCR. Correlation coefficients (ρ) with 95% confidence intervals and the regression equation are shown under the graph. C, Correlation of immune‐related gene expression with IFNG expression in PDAC tissues in The Cancer Genome Atlas cohort (n = 176). Spearman’s correlation coefficients (ρ) with P‐values are shown
3.4. Gene expressions correlated with IFNG in The Cancer Genome Atlas cohort
To investigate the relationship of the tumor immune microenvironment of PDAC with IFNγ expression using another cohort, we analyzed the gene expression of immune‐related genes in the TCGA cohort. Expression of IFNG correlated with several genes, including PD1, PD‐L1, PD‐L2, CXCL9, CXCL10, CXCL11 and IDO1, with high correlation coefficients (Figures 3C and S1). Some of both NK cell activating and inhibitory receptors were correlated and had high correlation coefficients.
3.5. Expression of HLA‐I, HLA‐E, HLA‐G and immune checkpoint molecules was increased on the surfaces of pancreatic ductal adenocarcinoma cells by interferon gamma
To determine whether HLA‐I expression is induced by IFNγ stimulation in PDAC cells, we used an in vitro assay with three PDAC cell lines with low, moderate and high levels of HLA‐I expression in AsPC‐1, Capan‐2 and Capan‐1, respectively. As determined by flow cytometry, an IFNγ stimulus increased surface expression of HLA‐I in these three cell lines (Figure 4A). An IFNγ stimulus also increased surface expression of HLA‐E, HLA‐G and immune checkpoint molecules PD‐L1, PD‐L2, CD80, CD86, ICOS‐L and B7H4. Conversely, decreased B7H3 surface expression was found in AsPC‐1 and Capan‐2 (Figure 4A).
Figure 4.
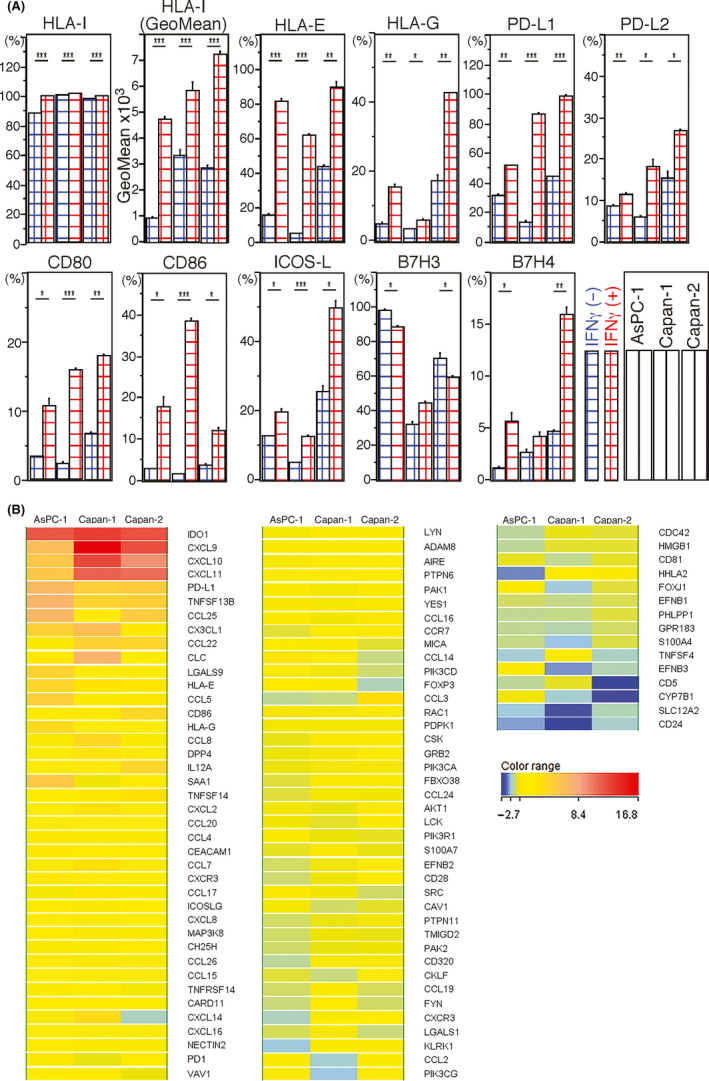
A, human leukocyte antigen class I antigens (HLA‐I) and other surface molecules induced by interferon gamma (IFNγ) stimulation in pancreatic ductal adenocarcinoma (PDAC) cells determined by flow cytometry. The y‐axis shows the ratio (%) of the positive cells for surface molecules. Red and blue meshed bars mean cells cultured with and without IFNγ, respectively. Bars represent means ± standard errors. In the graph box, the left, center and right two bars represent data for AsPC‐1, Capan‐1 and Capan‐2, respectively. Only the second bar graph shows mean fluorescent intensity (GeoMean) of PDAC cell surface HLA‐I expression. Differences are determined by Student’s t test, with significance values of P < 0.05 (*), <0.01 (**) and < 0.001(***). B, Heat map analyses delineating dynamic alteration of gene expression profiles by IFNγ stimulation of PDAC cells and immune checkpoint‐related and chemotaxis‐related molecules. Fold‐changes of gene expression in response to IFNγ stimulation in PDAC cells are shown and compared
Next, we obtained gene expression profiles of PDAC cells altered by IFNγ using a microarray technique. An IFNγ stimulus increased genes encoding classical and non–classical HLA‐I and HLA‐II antigens, as well as genes encoding immune checkpoint and co–stimulatory molecules (Figure 4B and Table S6). Furthermore, genes encoding several chemokines, most of which are chemoattractants for T cells (Figure 4B), were upregulated. Expression of CXCL9, CXCL10 and CXCL11, in particular, was markedly increased. In addition, expression of IDO1 and IDO2 was increased. These results suggest that IFNγ induces several chemokines to recruit T cells to PDAC cells. Several IFNγ‐affected gene, including those encoding immune‐suppressive molecules in PDAC cells, also showed strong correlations with IFNG expression in PDAC tissues (Figure 3D).
4. DISCUSSION
Host CTL can attack cancer cells by recognizing them via cancer antigens with HLA‐I expressed on their surfaces. Therefore, reduction or loss of HLA‐I expression on cancer cells allows cancer cells to disappear from CTL surveillance. Cancer cell escape from host immune surveillance can lead to poor patient outcomes in many types of cancer. 6 , 7 , 8 Furthermore, HLA‐I expression is a prerequisite for CTL‐based cancer immunotherapies. In this study, we investigated the clinicopathological significance of expression of classical HLA‐I, as well as non–classical HLA‐I, HLA‐E and HLA‐G, on PDAC cells. Unexpectedly, our results showed that lower expression of HLA‐I on PDAC cells was significantly associated with longer OS. According to two previous small‐scale studies using evaluation methods and antibodies different from ours, one found that higher expression of HLA‐I was significantly associated with longer patient survival, 27 but the other found no significant association between HLA‐I expression and patient outcome. 28 We used HLA‐I antibody clone EMR8‐5, which was employed in previous studies investigating the prognostic significance of HLA‐I expression in various types of cancer cells and cases of reduced or lost HLA‐I expression on cancer cells usually associated with unfavorable outcomes. 7
There are multiple molecular mechanisms underlying reduction or loss of HLA‐I expression on tumor cells, and the frequency of each of these mechanisms differs depending on cancer type. 39 , 40 There is few report on the causative molecular mechanisms and gene alterations related to downregulated HLA‐I in PDAC. IFNγ can induce expression of HLA‐I on cell surfaces. Indeed, surface expression of HLA‐I increased in all three PDAC cell lines stimulated with IFNγ (Figure 4A). IFNγ is usually a representative molecule in type I immune reactions that drive active antitumor adaptive immune responses. PDAC tissues with higher expression of HLA‐I on cells showed significantly higher expression of IFNG (Figure 3A), although other molecules representative of type I immune reactions such as TBX21 (Figure 3A) and IL12 (data not shown) were not upregulated. These findings suggested that higher expression of IFNG in PDAC tissues with higher expression of HLA‐I on PDAC cells is not related to an active‐phase type I immune reaction.
In 2019, Schreiber’s group reported that IFNγ becomes generally protumorigenic during the immune escape stage of “cancer immunoediting,” in which IFNγ induces inhibitory immune checkpoint molecules to create a tolerant immune microenvironment. 41 Similarly, IFNγ induces immune checkpoint molecules to limit T cell function during autoimmune diabetes. 42 In PDAC tumor microenvironments with higher PDAC cell HLA‐I expression in our cohort, IFNG expression correlated with expression of PD‐1, PD‐L1 and PD‐L2, with high correlation coefficients (Figure 3B). In the TCGA cohort, IFNG expression also highly correlated with expression of genes encoding inhibitory immune checkpoint molecules and the other immune suppressive molecules (Figures 3C and S1). In addition, our in vitro assay showed that IFNγ stimulation induced inhibitory immune checkpoint molecules PD‐L1, PD‐L2 and B7H4 on PDAC cell surfaces. Furthermore, Delitto et al reported that downstream mediators of an intratumoral IFNγ response suppress antitumor immunity and are associated with poor patient outcomes in PDAC. 36 It is possible that higher expression of IFNγ with the induction of immune checkpoint molecules in PDAC tissues with higher HLA‐I expression on PDAC cells leads to poor outcomes. Our in vitro assay showed that IFNγ stimulation also induced expression of co–stimulatory molecules CD86, CD80 and ICOS‐L on PDAC cell surfaces. Deviation to tolerance of the balance of various immune checkpoint factors is believed to be important to formation of an immune microenvironment.
In addition, based on a comparison of the gene expression profiles of PDAC cells with or without IFNγ expression, IFNγ strongly stimulates PDAC cells to recruit T cells through the expression of several chemokines, such as CXCL9, CXCL10 and CCL25 (Figure 4B). Some of these chemokines are also chemoattractants for NK cells. In addition, IFNγ stimulation induced a few chemokines that recruit monocytes and granulocytes. In this scenario, the recruited T cells can provide IFNγ, but their activity is suppressed and becomes exhausted in response to immune‐suppressive molecules such as IDO and contact with inhibitory immune checkpoint molecules expressed on PDAC cells. The cellular source of IFNγ has not been identified in the tolerant microenvironment, although these recruited NK and/or T cells, especially in CD8+ T cells, might be candidates. We observed the correlation between IFNG and CD8 in PDAC tissues with a high correlation coefficient (Figure 3B).
Tumor‐infiltrating CD8+ T cells are assumed to express PD‐1 and become exhausted in PDAC tumor microenvironments with higher PDAC cell HLA‐I expression. Such PDAC cases might seem to be good targets for immune checkpoint therapy, although the effect of immune checkpoint inhibitors in PDAC was shown to be very limited. 24 , 25 , 26 It is possible that an immune tolerant milieu in PDAC immune microenvironments with higher HLA‐I expression on PDAC cells is acquired during immune editing. A candidate mechanism is to strengthen inhibiting CTL reaction by CTL inactivation or exhaustion. Indeed, our in vitro study revealed that IFNγ induced expression of strong immune inhibitory molecules and several immune checkpoint molecules as well as increasing expression of HLA‐I on PDAC cell surfaces. Because IFNγ is an immune reactive agent, there might be a specific mechanism by which IFNγ becomes a mediator of immune tolerance in PDAC tissues. Understanding such tolerance mechanisms and ways to eliminate their function is important for recovering host tumor immune surveillance. These findings suggest that higher expression of HLA‐I on PDAC cells means more than maintaining the originally expressed HLA‐I without reduction or loss; rather, re–expression of HLA‐I expression altered by IFNγ may be needed. Thus, knowing HLA‐I expression profiles in PDAC cells is important and useful for defining tumor immune microenvironments in PDAC.
HLA‐E and HLA‐G bind to inhibitory receptors expressed on NK cells and inhibit the NK cell effector function. 11 We showed that the surface expression of both HLA‐E and HLA‐G on PDAC cells was associated with an unfavorable prognosis, in terms of both OS and DFS. To the best of our knowledge, this is the first report on the prognostic impact of HLA‐E in PDAC. However, two groups reported that HLA‐G was an unfavorable prognosticator, 17 , 18 and one group reported that it was a favorable prognosticator. 19 In addition to HLA‐E and HLA‐G inhibition of NK cell activity, HLA‐E can inhibit immune response through mechanisms that include inhibition of CD8+ T cell cytolytic function 12 and induction of NK cell apoptosis. 43 HLA‐G can also induce the expansion of myeloid‐derived suppressor cells, 44 inhibition of DC maturation and induction of tolerogenic DC, 45 and induction of NK cell apoptosis. 43 These immune‐suppressive effects could lead to the formation of immune‐tolerant tumor immune environments, which is assumed to be associated with unfavorable patient outcomes.
Pancreatic ductal adenocarcinoma tissues with higher PDAC cell expression of HLA‐I showed significantly lower expression of CD4 and RORC and higher expression of ARG2 compared to those with lower expression of HLA‐I (Figure 3A). CD4 and RORC are favorable prognosticators for patients with PDAC. 33 , 35 These findings are consistent with the idea that PDAC tumor microenvironment with higher expression of HLA‐I on PDAC cells might reflect immune tolerance. ARG2 molecules are expressed in limited cell types; that is, in cancer‐associated fibroblasts in hypoxic areas, in PDAC tissues. 34 Higher expression of ARG2 in PDAC tissues with higher PDAC cell HLA‐I expression was consistent with the significant correlation between higher expression of HLA‐I in PDAC cells and presence of histological tumor necrosis (Tables 2 and S1). Hypoxia upregulates inhibitory immune checkpoint molecules in nucleated cells 46 , 47 ; however, the relationship between hypoxia and HLA‐I expression is still controversial. 48 , 49 , 50 , 51 Although there is a discrepancy with observations from clinical samples, hypoxia is considered to augment HLA‐I expression in cancer cells. The hypoxic tumor microenvironment is linked to an unfavorable prognosis, 52 whereas higher HLA‐I expression in cancer cells is associated with favorable outcomes in many types of cancers. 6 , 7 , 8 Although it is not apparent how hypoxia‐induced HLA‐I expression is involved in altering characteristics of the tumor microenvironment in other types of cancers, it is likely that hypoxia induces higher HLA‐I expression in PDAC cells.
There are some limitations to this study. Data collection and analyses of our clinicopathological study were performed retrospectively and not much validated. We evaluated gene expression profiles of immune‐related genes by RT‐qPCR using the source of whole PDAC tissues (not single‐cell analysis), which could not determine detailed cellular information of expressing their genes and molecules. Based on the findings obtained from gene expression profiles in PDAC tissues and in vitro assays using PDAC cells, we speculated on the possible mechanisms to form an immune tolerant microenvironment of PDAC tissues with higher expression of HLA‐I on PDAC cells. Further studies are required to verify our findings and speculation.
In conclusion, higher expression of HLA‐I, HLA‐E and HLA‐G on PDAC cells were unfavorable independent prognosticators. It is possible that IFNγ was associated with higher expression of HLA‐I on PDAC cells and involved in the formation of a tolerant microenvironment through upregulation of immune checkpoint molecules.
DISCLOSURE
The authors declare no competing interests.
Supporting information
Fig S1
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
ACKNOWLEDGMENTS
This work was supported by a Grant‐in‐Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (NH), the Japan Agency for Medical Research and Development (NH) and the National Cancer Center Research and Development Fund (NH and YH). The authors thank Ms Keiko Gomisawa techniques of preparing tissue sections. We are grateful to the National Cancer Center Biobank for the tissue samples used in this study.
Hiraoka N, Ino Y, Hori S, et al. Expression of classical human leukocyte antigen class I antigens, HLA-E and HLA-G, is adversely prognostic in pancreatic cancer patients. Cancer Sci. 2020;111:3057–3070. 10.1111/cas.14514
REFERENCES
- 1. Leone P, De Re V, Vacca A, Dammacco F, Racanelli V. Cancer treatment and the KIR‐HLA system: an overview. Clin Exp Med. 2017;17:419‐429. [DOI] [PubMed] [Google Scholar]
- 2. Seliger B. Molecular mechanisms of HLA class I‐mediated immune evasion of human tumors and their role in resistance to immunotherapies. HLA. 2016;88:213‐220. [DOI] [PubMed] [Google Scholar]
- 3. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014‐1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hiraoka N, Yamazaki‐Itoh R, Ino Y, et al. CXCL17 and ICAM2 are associated with a potential anti–tumor immune response in early intraepithelial stages of human pancreatic carcinogenesis. Gastroenterology. 2011;140:310‐321. [DOI] [PubMed] [Google Scholar]
- 5. Torigoe T, Asanuma H, Nakazawa E, et al. Establishment of a monoclonal anti–pan HLA class I antibody suitable for immunostaining of formalin‐fixed tissue: unusually high frequency of down‐regulation in breast cancer tissues. Pathol Int. 2012;62:303‐308. [DOI] [PubMed] [Google Scholar]
- 6. de Kruijf EM, van Nes JG, Sajet A, et al. The predictive value of HLA class I tumor cell expression and presence of intratumoral Tregs for chemotherapy in patients with early breast cancer. Clin Cancer Res. 2010;16:1272‐1280. [DOI] [PubMed] [Google Scholar]
- 7. Mariya T, Hirohashi Y, Torigoe T, et al. Prognostic impact of human leukocyte antigen class I expression and association of platinum resistance with immunologic profiles in epithelial ovarian cancer. Cancer Immunol Res. 2014;2:1220‐1229. [DOI] [PubMed] [Google Scholar]
- 8. Speetjens FM, de Bruin EC, Morreau H, et al. Clinical impact of HLA class I expression in rectal cancer. Cancer Immunol Immunother. 2008;57:601‐609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Algarra I, Garcia‐Lora A, Cabrera T, Ruiz‐Cabello F, Garrido F. The selection of tumor variants with altered expression of classical and nonclassical MHC class I molecules: implications for tumor immune escape. Cancer Immunol Immunother. 2004;53:904‐910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei XH, Orr HT. Differential expression of HLA‐E, HLA‐F, and HLA‐G transcripts in human tissue. Hum Immunol. 1990;29:131‐142. [DOI] [PubMed] [Google Scholar]
- 11. Kochan G, Escors D, Breckpot K, Guerrero‐Setas D. Role of non–classical MHC class I molecules in cancer immunosuppression. Oncoimmunology. 2013;2:e26491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gooden M, Lampen M, Jordanova ES, et al. HLA‐E expression by gynecological cancers restrains tumor‐infiltrating CD8(+) T lymphocytes. Proc Natl Acad Sci USA. 2011;108:10656‐10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo ZY, Lv YG, Wang L, et al. Predictive value of HLA‐G and HLA‐E in the prognosis of colorectal cancer patients. Cell Immunol. 2015;293:10‐16. [DOI] [PubMed] [Google Scholar]
- 14. Kleinberg L, Florenes VA, Skrede M, et al. Expression of HLA‐G in malignant mesothelioma and clinically aggressive breast carcinoma. Virchows Arch. 2006;449:31‐39. [DOI] [PubMed] [Google Scholar]
- 15. Cai MY, Xu YF, Qiu SJ, et al. Human leukocyte antigen‐G protein expression is an unfavorable prognostic predictor of hepatocellular carcinoma following curative resection. Clin Cancer Res. 2009;15:4686‐4693. [DOI] [PubMed] [Google Scholar]
- 16. Rutten MJ, Dijk F, Savci‐Heijink CD, et al. HLA‐G expression is an independent predictor for improved survival in high grade ovarian carcinomas. J Immunol Res. 2014;2014:274584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu YF, Lu Y, Cheng H, et al. High expression of human leukocyte antigen‐G is associated with a poor prognosis in patients with PDAC. Curr Mol Med. 2015;15:360‐367. [DOI] [PubMed] [Google Scholar]
- 18. Zhou L, Niu ZY, Liang ZY, et al. HLA‐G impairs host immune response and predicts poor prognosis in pancreatic cancer. Am J Transl Res. 2015;7:2036‐2044. [PMC free article] [PubMed] [Google Scholar]
- 19. Sideras K, Biermann K, Yap K, et al. Tumor cell expression of immune inhibitory molecules and tumor‐infiltrating lymphocyte count predict cancer‐specific survival in pancreatic and ampullary cancer. Int J Cancer. 2017;141:572‐582. [DOI] [PubMed] [Google Scholar]
- 20. Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605‐1617. [DOI] [PubMed] [Google Scholar]
- 21. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7‐30. [DOI] [PubMed] [Google Scholar]
- 22. Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582‐587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19:133‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kabacaoglu D, Ciecielski KJ, Ruess DA, Algul H. Immune checkpoint inhibition for pancreatic ductal adenocarcinoma: current limitations and future options. Front Immunol. 2018;9:1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252‐264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Balachandran VP, Beatty GL, Dougan SK. Broadening the impact of immunotherapy to pancreatic cancer: challenges and opportunities. Gastroenterology. 2019;156:2056‐2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Imai D, Yoshizumi T, Okano S, et al. The prognostic impact of programmed cell death ligand 1 and human leukocyte antigen class I in pancreatic cancer. Cancer Med. 2017;6:1614‐1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ryschich E, Notzel T, Hinz U, et al. Control of T‐cell‐mediated immune response by HLA class I in human pancreatic carcinoma. Clin Cancer Res. 2005;11:498‐504. [PubMed] [Google Scholar]
- 29. Brierley JD, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumours, (8th edn). Hoboken, NJ: Wiley‐Blackwell; 2017. [Google Scholar]
- 30. Hruban RH, Boffetta P, Hiraoka N, et al. Ductal adenocarcinoma of the pancreas In: Bosman FT, Carneiro F, Hruban RH, Theise ND, eds. World Health Organization Classification of Tumours Pathology & Genetics Tumours of the Digestive System, (4th edn). Lyon, France: IARC Press; 2010:281‐291. [Google Scholar]
- 31. Japan‐Pancreas‐Society . Classification of Pancreatic Cancer, (3rd English edn). Tokyo, Japan: Kanehara; 2011. [Google Scholar]
- 32. Hiraoka N, Ino Y, Sekine S, et al. Tumour necrosis is a postoperative prognostic marker for pancreatic cancer patients with a high interobserver reproducibility in histological evaluation. Br J Cancer. 2010;103:1057‐1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hiraoka N, Ino Y, Yamazaki‐Itoh R, Kanai Y, Kosuge T, Shimada K. Intratumoral tertiary lymphoid organ is a favourable prognosticator in patients with pancreatic cancer. Br J Cancer. 2015;112:1782‐1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ino Y, Yamazaki‐Itoh R, Oguro S, et al. Arginase II expressed in cancer‐associated fibroblasts indicates tissue hypoxia and predicts poor outcome in patients with pancreatic cancer. PLoS One. 2013;8:e55146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ino Y, Yamazaki‐Itoh R, Shimada K, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer. 2013;108:914‐923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Delitto D, Perez C, Han S, et al. Downstream mediators of the intratumoral interferon response suppress antitumor immunity, induce gemcitabine resistance and associate with poor survival in human pancreatic cancer. Cancer Immunol Immunother. 2015;64:1553‐1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meng H, Lee Y, Ba Z, et al. In vitro production of IL‐6 and IFN‐gamma is influenced by dietary variables and predicts upper respiratory tract infection incidence and severity respectively in young adults. Front Immunol. 2015;6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cancer Genome Atlas Research Network . Electronic address aadhe, Cancer Genome Atlas Research N. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185‐203 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cai L, Michelakos T, Yamada T, et al. Defective HLA class I antigen processing machinery in cancer. Cancer Immunol Immunother. 2018;67:999‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aptsiauri N, Ruiz‐Cabello F, Garrido F. The transition from HLA‐I positive to HLA‐I negative primary tumors: the road to escape from T‐cell responses. Curr Opin Immunol. 2018;51:123‐132. [DOI] [PubMed] [Google Scholar]
- 41. Alspach E, Lussier DM, Schreiber RD. Interferon gamma and its important roles in promoting and inhibiting spontaneous and therapeutic cancer immunity. Cold Spring Harb Perspect Biol. 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Osum KC, Burrack AL, Martinov T, et al. Interferon‐gamma drives programmed death‐ligand 1 expression on islet beta cells to limit T cell function during autoimmune diabetes. Sci Rep. 2018;8:8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spaggiari GM, Contini P, Carosio R, et al. Soluble HLA class I molecules induce natural killer cell apoptosis through the engagement of CD8: evidence for a negative regulation exerted by members of the inhibitory receptor superfamily. Blood. 2002;99:1706‐1714. [DOI] [PubMed] [Google Scholar]
- 44. Agaugue S, Carosella ED, Rouas‐Freiss N. Role of HLA‐G in tumor escape through expansion of myeloid‐derived suppressor cells and cytokinic balance in favor of Th2 versus Th1/Th17. Blood. 2011;117:7021‐7031. [DOI] [PubMed] [Google Scholar]
- 45. Ristich V, Liang S, Zhang W, Wu J, Horuzsko A. Tolerization of dendritic cells by HLA‐G. Eur J Immunol. 2005;35:1133‐1142. [DOI] [PubMed] [Google Scholar]
- 46. Barsoum IB, Smallwood CA, Siemens DR, Graham CH. A mechanism of hypoxia‐mediated escape from adaptive immunity in cancer cells. Cancer Res. 2014;74:665‐674. [DOI] [PubMed] [Google Scholar]
- 47. Noman MZ, Desantis G, Janji B, et al. PD‐L1 is a novel direct target of HIF‐1alpha, and its blockade under hypoxia enhanced MDSC‐mediated T cell activation. J Exp Med. 2014;211:781‐790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kajiwara T, Tanaka T, Kukita K, et al. Hypoxia augments MHC class I antigen presentation via facilitation of ERO1‐alpha‐mediated oxidative folding in murine tumor cells. Eur J Immunol. 2016;46:2842‐2851. [DOI] [PubMed] [Google Scholar]
- 49. Kukita K, Tamura Y, Tanaka T, et al. Cancer‐associated oxidase ERO1‐alpha Regulates the expression of MHC class i molecule via oxidative folding. J Immunol. 2015;194:4988‐4996. [DOI] [PubMed] [Google Scholar]
- 50. Murthy A, Gerber SA, Koch CJ, Lord EM. Intratumoral hypoxia reduces IFN‐gamma‐mediated immunity and MHC Class I induction in a preclinical tumor model. Immunohorizons. 2019;3:149‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sethumadhavan S, Silva M, Philbrook P, et al. Hypoxia and hypoxia‐inducible factor (HIF) downregulate antigen‐presenting MHC class I molecules limiting tumor cell recognition by T cells. PLoS One. 2017;12:e0187314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Harris AL. Hypoxia–a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38‐47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6


