Abstract
Circular RNAs (circRNAs) were recently reported to be involved in the pathogenesis of ovarian cancer (OC); however, the molecular mechanisms of circRNAs in tumor progression and paclitaxel (PTX) resistance of OC remain largely undetermined. Here, we focused on circTNPO3 (hsa_circ_0001741), which is located on chromosome 7 (chr7): 128655032–128658211 and derived from TNPO3 gene, and thus we termed as circTNPO3. By microarray and qRT-PCR we identified circTNPO3 to be dramatically high expressed in OC samples and correlated with PTX resistance. Functionally, knockdown of circTNPO3 enhanced cell sensitivity to PTX via promoting PTX-induced apoptosis in vitro and in vivo. In mechanism, circTNPO3 acted as a sponge for microRNA-1299 (miR-1299), and NEK2 (NIMA-related kinase 2) was revealed to be target gene of miR-1299. Subsequently, functional assays illustrated that the oncogenic effects of circTNPO3 were attributed to the regulation of miR-1299/NEK2 axis. In conclusion, circTNPO3 contributed to PTX resistance of OC cells at least partly through upregulating NEK2 expression by sponging miR-1299. circTNPO3/miR-1299/NEK2 signaling pathway might play vital roles in the tumorigenesis and chemoresistance of OC.
Keywords: circular RNA, circTNPO3, microRNA, ceRNA, paclitaxel resistance, ovarian cancer
Graphical Abstract
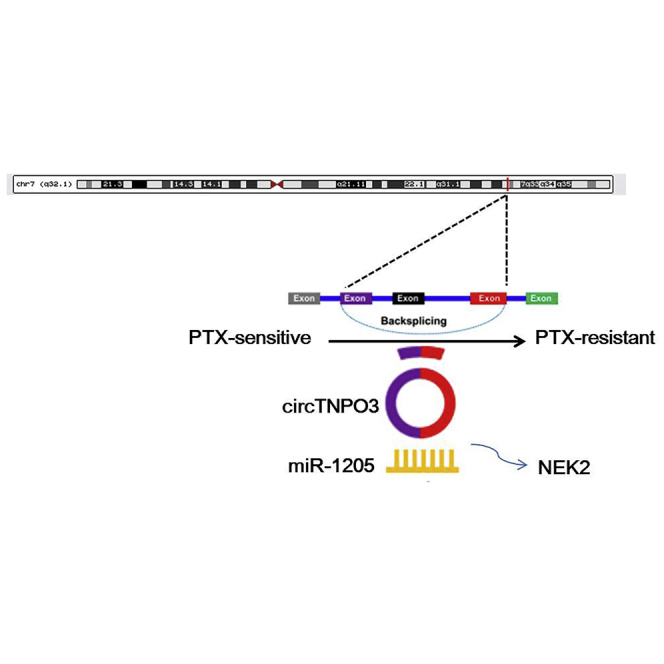
Bing et al. found that circTNPO3 contributed to PTX resistance of OC cells at least partly through upregulating NEK2 expression by sponging miR-1299. circTNPO3/miR-1299/NEK2 signaling pathway might play vital roles in the tumorigenesis and chemoresistance of OC.
Introduction
Ovarian cancer (OC) is the most lethal gynecological malignancy and the second leading cause of cancer-related death in women.1 About 75% of patients are diagnosed at the late stage of the disease.2 The standard therapeutic protocol, including paclitaxel (PTX)-based combination chemotherapy and cytoreductive surgery, leads to an overall 5-year survival rate of only 15%–30% for metastatic OC.3,4 However, drug resistance is a tough problem during cancer treatment. PTX resistance gradually develops during the course of chemotherapy, ultimately leads to the therapeutic failure.5 Thus, innovative therapeutic strategies for overcoming PTX resistance in OC are urgently needed.
The circular RNAs (circRNAs) group is a member of non-coding RNAs (ncRNAs) with a large variation in length ranging from hundreds to thousands of nucleotides.6 During the pre-mRNA splicing, the 5′ end of an upstream exon and the 3′ end of a downstream exon or the ends of an individual exon could join together to generate a circRNA.7 Accumulating studies proposed that circRNA is a pivotal regulator in cell functions and the development of various tumors, OC included.8, 9, 10 Moreover, abnormality of circRNA levels has been shown to be associated with the development of chemoresistance in various tumors.11, 12, 13 Nevertheless, further explorations are still needed to be performed for the illustration of the biological function and potential regulation mechanism of circRNA in PTX resistance of OC.
Here, we profiled the circRNA expression of PTX-resistant OC tissues in order to improve our understanding of the precise mechanisms of PTX resistance, as well as to identify potential circRNA biomarkers for OC patients. In the present study, we validated the significant differential expressions of circTNPO3 (hsa_circ_0001741) in OC tissues and cell lines. And also, circTNPO3 were found to be closely related to the PTX resistance of OC cells. Besides, based on in vitro models of PTX-resistant OC cells SKOV3/PTX and HeyA-8/PTX, we verified the underlying molecular mechanism of circTNPO3, acting as a sponge of miR-1299 to modulate NIMA-related kinase 2 (NEK2) expression and PTX resistance in OC cells, providing a new mechanism of chemoresistance of OC.
Results
circRNA Profiling in Human PTX-Resistant OC Tissues and circTNPO3 Characterization
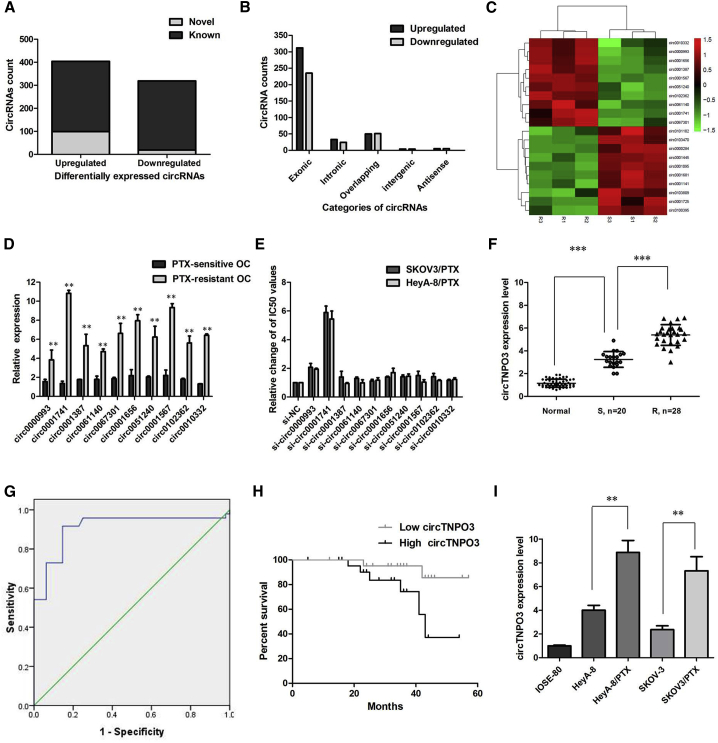
We performed microarrays to characterize the expression profiles of circRNAs in 3 pairs of PTX-sensitive OC tissues and PTX-resistant OC tissues. The differentially expressed circRNAs were identified by fold-change filtering (|fold change| > 2) and the Student’s t test (p < 0.01), which revealed 723 circRNAs that were significantly differentially expressed in the PTX-resistant tumor versus PTX-sensitive tumor set. Compared with PTX-sensitive OC tissues, there were 404 significantly upregulated and 319 significantly downregulated circRNAs in PTX-resistant OC tissues. Among the 723 differentially expressed circRNAs, 605 circRNAs (83.68%) have been identified in other studies in circBase (140,790 human circRNAs), and the other 118 (16.32%) are novel (Figure 1A). Furthermore, 624 circRNAs (86.31%) consisted of protein coding exons from 316 genes, and the length of most exonic circRNAs was less than 1,000 nucleotides (Figure 1B). Hierarchical clustering was then performed to demonstrate the most 10 up- and 10 downregulated circRNA expression patterns among the sets (Figure 1C). We then experimentally validated the 10 most upregulated circRNAs expression levels by qRT-PCR using PTX-resistant and PTX-sensitive tissue samples. The qRT-PCR results indicated hsa_circ_0001741 showed highest fold-change in the PTX-resistant OC tissues than in the PTX-sensitive OC tissues (Figure 1D). Furthermore, we found that inhibition of hsa_circ_0001741 reversed PTX resistance in both SKOV3/PTX and HeyA-8/PTX cell lines, while other 9 circRNAs showed little effect (Figure 1E). We therefore chose hsa_circ_0001741 for further analysis. According to the human reference genome, we further termed hsa_circ_0001741 (located at chromosome 7 [chr7]: 128655032–128658211, is derived from gene TNPO3, with a spliced mature sequence length of 432 bp) as “circTNPO3.”
Figure 1.
circRNA Profiling in Human PTX-Resistant OC Tissues and circTNPO3 Characterization
(A) Among the 723 differentially expressed circRNAs, 118 circRNAs were verified as novel circRNAs; 605 circRNAs were identified beforehand and listed in the circRNA database. (B) The 723 identified circRNAs were divided into five different categories on the basis of the way they were produced. (C) The heatmap showed the top 10 dysregulated circRNAs between PTX-resistant OC tissues and PTX-sensitive OC tissues. (D) Expression levels of top 10 dysregulated circRNAs were measured by qRT-PCR. (E) Inhibition of hsa_circ_0001741 reversed PTX resistance in both SKOV3/PTX and HeyA-8/PTX cell lines. (F) The level of circTNPO3 was significantly increased in PTX-resistant OC tissues compared to PTX-sensitive OC tissues. (G) Evaluation the diagnostic performance of circTNPO3 for OC diagnosis. (H) Kaplan-Meier curve revealed that high expression of circTNPO3 was relative to a poor overall survival in OC patients. (I) circTNPO3 expression was distinctively higher in PTX-resistant OC cells SKOV3/PTX and HeyA-8/PTX cells. All tests were performed at least three times. Data were expressed as mean ± SD. ∗∗∗p < 0.001; ∗∗p < 0.01.
circTNPO3 Was Highly Expressed in PTX-Resistant OC Tissues and Cells
Moreover, the expression level of circTNPO3 was examined by quantitative real-time PCR analysis in OC samples and paired adjacent normal tissues (ANTs) collected from 48 OC patients. The results suggested that circTNPO3 was found to be significantly upregulated in 48 OC tissues compared to ANT (p < 0.01, Figure 1F). There was an increasing trend in circTNPO3 levels from normal ovarian tissues to PTX-sensitive OC tissues (n = 20) and then to PTX-resistant OC tissues (n = 28), and the differences among the three groups were significant (p < 0.001). We used the ROC curve to examine the diagnostic value of circTNPO3 in OC tissues compared with ANT and found the area under the ROC curve (AUC) to be 0.910 (95% CI = 0.844–0.975, p < 0.0001; Figure 1G). We also investigated the clinical significance of circTNPO3 by analyzing the correlations between its expression level and clinicopathological characteristics. Using the median expression level of circTNPO3 as cutoff value, we divided the 48 OC patients into low and high expression groups. As listed in Table 1, circTNPO3 expression was significantly correlated with advanced FIGO stage and histological type of OC patients. Furthermore, OC patients with low expression of circTNPO3 displayed obviously longer overall survival times than those with high expression of circTNPO3 according to Kaplan-Meier survival curve analysis (p = 0.030; Figure 1H). Then, experiments were performed at the cellular level. circTNPO3 expression was distinctively higher in PTX-resistant OC cells SKOV3/PTX and HeyA-8/PTX cells, and obviously lower in normal ovarian surface epithelial cells IOSE-80 (Figure 1I; p < 0.01). In a word, these results suggested that circTNPO3 might be associated with PTX resistance in OC cells.
Table 1.
Association of circTNPO3 Expression with Clinicopathological Features of OC Patients
| Characteristics | circTNPO3 Low (n = 24) | Expression High (n = 24) | p |
|---|---|---|---|
| Age (years) <55 | 11 | 9 | 0.770 |
| ≥55 | 13 | 15 | |
| FIGO stage | |||
| I–II | 6 | 16 | 0.008 |
| III–IV | 18 | 8 | |
| Histological Type | |||
| I | 4 | 12 | 0.030 |
| II–III | 20 | 12 | |
| Positive lymph node | |||
| No | 5 | 8 | 0.517 |
| Yes | 19 | 16 | |
| Metastasis | |||
| No | 16 | 9 | 0.082 |
| Yes | 8 | 15 | |
| CA125 | |||
| <35 | 8 | 6 | 0.752 |
| ≥35 | 16 | 18 |
circTNPO3 Enhances the PTX Chemosensitivity of OC In Vitro
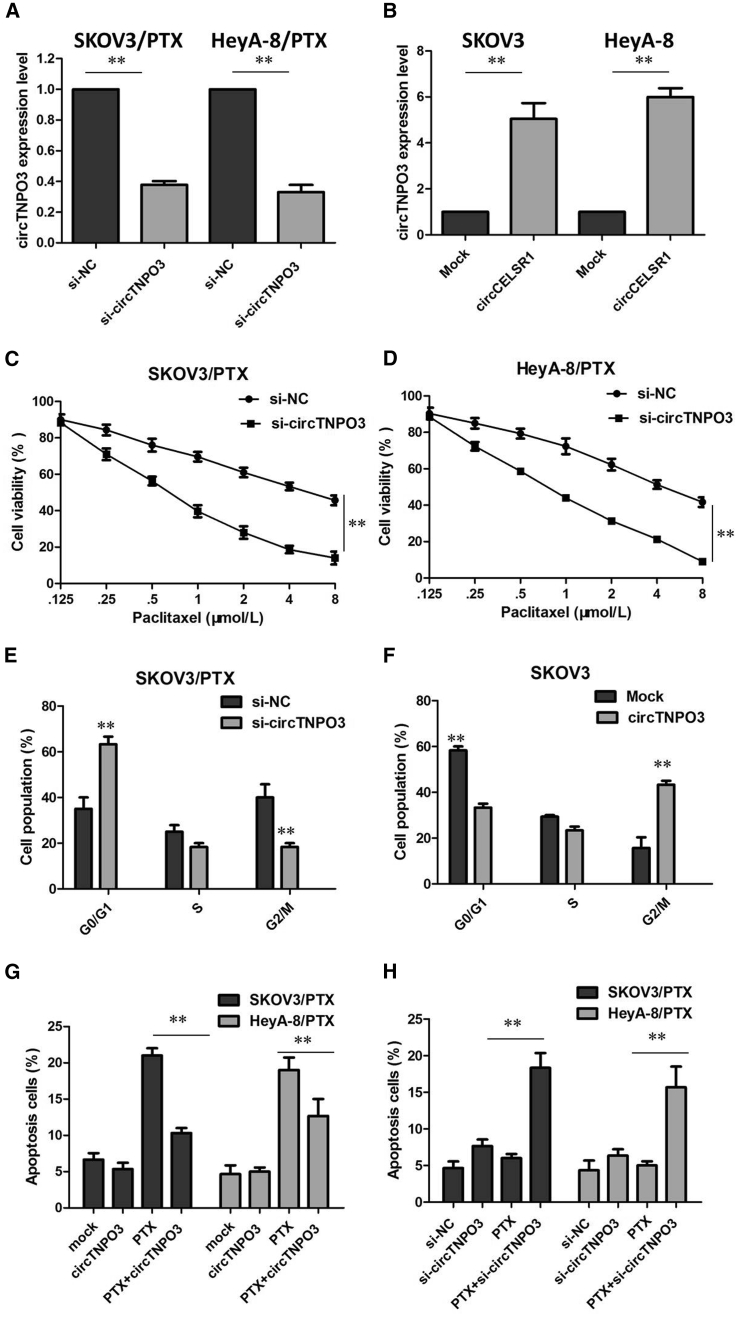
To further validate the expression level of circTNPO3 on PTX resistance, we performed loss- and gain-of-function studies by knockdown or overexpression of circTNPO3 in OC cells. First, small interfering RNA (siRNA) was designed to target the backsplice junction of circTNPO3, which did not affect the expression of linear species in SKOV3/PTX and HeyA-8/PTX cells (Figure 2A; p < 0.01). Meanwhile, we amplified the full-length cDNA of circTNPO3 and cloned it into the expression vector. circTNPO3 was significantly upregulated after transfecting the circTNPO3 expression vector in SKOV-3 and HeyA cells, whereas the transfection did not affect linear TNPO3 mRNA levels (Figure 2B; p < 0.01).
Figure 2.
circTNPO3 Enhances the PTX Chemosensitivity of OC In Vitro
(A) The expression of circTNPO3 was only downregulated by si-circTNPO3. (B) circTNPO3 was significantly upregulated after transfecting the circTNPO3 expression vector in SKOV-3 and HeyA cells. (C) Downregulation of circTNPO3 in SKOV3/PTX cells rendered both cell lines more sensitive to PTX compared with control group. (D) Downregulation of circTNPO3 in HeyA-8/PTX cells rendered both cell lines more sensitive to PTX compared with control group. (E) Downregulation of circTNPO3 significantly increased the percent of SKOV3/PTX cells in G0/G1 phase. (F) Overexpression of circTNPO3 significantly decreased the percent of SKOV3 cells in G0/G1 phase. (G) Downregulation of circTNPO3 significantly promoted the cell apoptosis of SKOV3/PTX and HeyA-8/PTX cells in the presence of PTX (100 nM). (H) Overexpression of circTNPO3 significantly inhibited the cell apoptosis of SKOV-3 and HeyA cells in the presence of PTX (100 nM). All tests were performed at least three times. Data were expressed as mean ± SD. ∗∗p < 0.01.
As a result, the downregulation of circTNPO3 in SKOV3/PTX and HeyA-8/PTX cells rendered both cell lines more sensitive to PTX compared with control group, as demonstrated by the decreased IC50 value of PTX following circTNPO3 downregulation (Figures 2C and 2D; p < 0.01). Further analysis revealed that the downregulation of circTNPO3 significantly increased the percent of cells in G0/G1 phase and promoted the cell apoptosis of SKOV3/PTX and HeyA-8/PTX cells in the presence of PTX (100 nM; Figures 2E and 2G; p < 0.01). However, overexpression of circTNPO3 did not promote cell proliferation of SKOV-3 and HeyA cells compared with the control group (data not show). The results of flow cytometry assays showed that overexpression of circTNPO3 significantly decreased the percent of cells in G0/G1 phase and inhibited the cell apoptosis of SKOV-3 and HeyA cells in the presence of PTX (100 nM; Figures 2F and 2H; p < 0.01). These assays collectively suggested that circTNPO3 silencing enhanced PTX-induced cytotoxicity in OC cells.
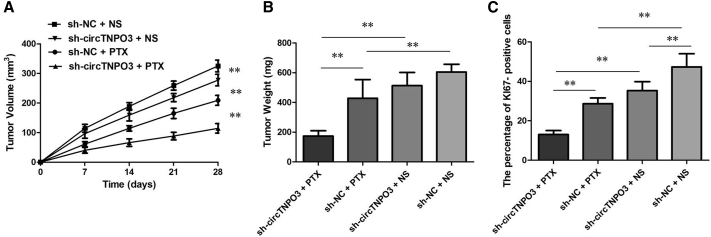
circTNPO3 Knockdown Enhanced the Anti-tumor Effect of PTX in OC In Vivo
To confirm the effects of circTNPO3 on the chemosensitivity of OC cells to PTX in vivo, we subcutaneously injected SKOV3/PTX cells stably infected with short hairpin (sh)-circTNPO3 or sh-control into each mouse, followed by administration with PTX. According to the treatment, the tumors on the mice were actually assigned to the following groups: sh-circTNPO3-transfected cells + PTX, sh-circTNPO3-transfected cells + NS, sh-NC-transfected cells + PTX, and sh-NC-transfected cells + NS. The results showed that PTX treatment significantly inhibited the growth of tumor cells when compared with control groups (p < 0.01). More importantly, with PTX treatment, tumor cells infected with sh-circTNPO3 grew lower than controls (p < 0.01), suggesting that circTNPO3 knockdown enhances the PTX chemosensitivity in vivo (Figure 3A). Tendencies in tumor weight were consistent with those in tumor volume (Figure 3B, p < 0.01). Moreover, immunohistochemistry assay showed that the tumors treated with sh-circTNPO3 plus PTX displayed an increased proliferation percentage of Ki-67 positive tumor cells compared with the control group (Figure 3C, p < 0.01). Collectively, these results implicated that circTNPO3 knockdown displayed a synergic effect with PTX in suppressing OC cell growth in vivo.
Figure 3.
circTNPO3 Knockdown Enhanced the Anti-tumor Effect of PTX in OC In Vivo
(A) Volume of tumors that developed in xenografts from different groups. (B) Weights of tumors that developed in xenografts from different groups. (C) The percentage of KI67-positive cells in xenografts from different groups. All tests were performed at least three times. Data were expressed as mean ± SD. ∗∗p < 0.01.
Confirmation of Subcellular Localization of circTNPO3
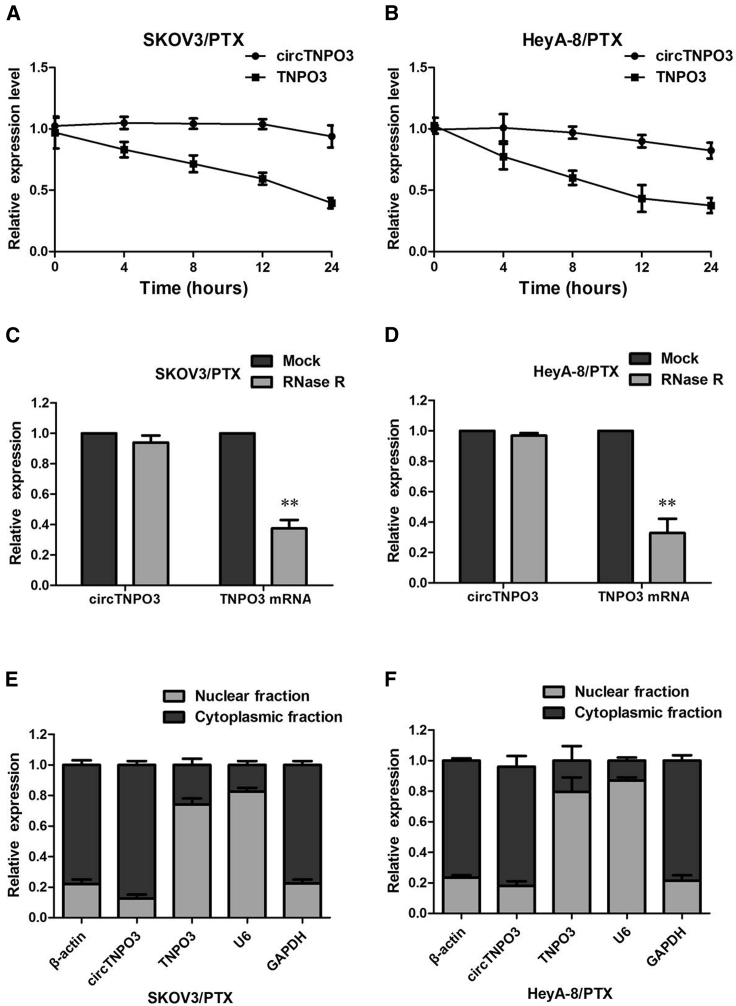
We investigated the stability and localization of circTNPO3 in SKOV3/PTX and HeyA-8/PTX cells. Total RNAs from SKOV3/PTX and HeyA-8/PTX cells were isolated at the indicated time points after treatment with Actinomycin D, an inhibitor of transcription. Analysis for stability of circTNPO3 and TNPO3 in SKOV3/PTX and HeyA-8/PTX cells treated with Actinomycin D, an inhibitor of transcription, revealed that the half-life of circTNPO3 transcript exceeded 24 h, with more stability than TNPO3 (Figures 4A and 4B). RNase R is an exoribonuclease that can degrade RNA from its 3′ to 5′ end but does not act on circRNA. In contrast to the linear TNPO3 mRNA, circTNPO3 was resistant to RNase R (Figures 4C and 4D). A nuclear and cytoplasmic protein extraction assay was performed to examine the subcellular localization of circTNPO3 in SKOV3/PTX and HeyA-8/PTX cells. It was found that circTNPO3 is abundant and stable in the cytoplasm of SKOV3/PTX and HeyA-8/PTX cells, while TNPO3 is abundant in the nucleus (Figures 4E and 4F). Our results implied that circTNPO3 harbored a loop structure and was predominantly localized in the cytoplasm.
Figure 4.
Confirmation of Subcellular Localization of circTNPO3
(A) qRT-PCR for the abundance of circTNPO3 and TNPO3 in SKOV3/PTX cells treated with Actinomycin D at the indicated time point. (B) qRT-PCR for the abundance of circTNPO3 and TNPO3 in HeyA-8/PTX cells treated with Actinomycin D at the indicated time point. (C) qRT-PCR for the expression of circTNPO3 and TNPO3 mRNA in SKOV3/PTX cells treated with or without RNase R. (D) qRT-PCR for the expression of circTNPO3 and TNPO3 mRNA in HeyA-8/PTX cells treated with or without RNase R. (E) Levels of circTNPO3 in the nuclear and cytoplasmic fractions of SKOV3/PTX cells. (F) Levels of circTNPO3 in the nuclear and cytoplasmic fractions of HeyA-8/PTX. All tests were performed at least three times. Data were expressed as mean ± SD. ∗∗p < 0.01.
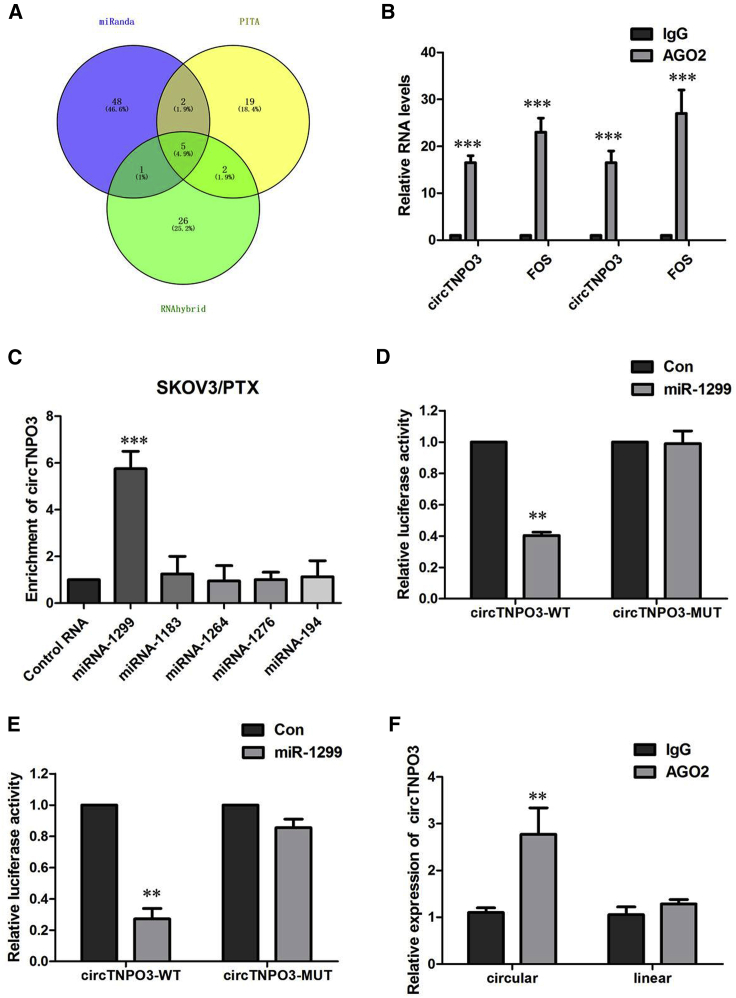
circTNPO3 Directly Binds to miR-1299 and Suppresses miR-1299 Activity
circRNAs have been shown to act as a miRNA sponge to regulate gene expression. We thus investigated the potential miRNAs associated with circTNPO3. We therefore analyzed the sequence of circTNPO3 using miRanda, PITA, and RNAhybrid and identified five candidate miRNAs by overlapping the prediction results of the miRNA recognition elements in the circTNPO3 sequence (miRNA-1183, miRNA-1264, miRNA-1276, miRNA-1299, and miRNA-194; Figure 5A). It is well known that miRNAs usually silence gene expression by combining with the AGO2 protein and form the RNA-induced silencing complex (RISC). In the context of competing endogenous RNA (ceRNA) mechanism, it might be a prevalent phenomenon that AGO2 could bind with both circRNAs and miRNAs. We therefore conducted an RNA immunoprecipitation (RIP) assay to pull down RNA transcripts that bind to AGO2 in SKOV3/PTX and HeyA-8/PTX cells. Indeed, endogenous circTNPO3 was efficiently pulled down by anti-Ago2 (Figure 5B). To further detect whether circTNPO3 could sponge miRNAs, we performed a miRNA pull-down assay using biotin-coupled miRNA mimics (miRNA-1183, miRNA-1264, miRNA-1276, miRNA-1299, and miRNA-194). Interestingly, circTNPO3 was only efficiently enriched by miR-1224, but not by the other four miRNAs (Figure 5C). To confirm the direct interaction between circTNPO3 and miR-1299, we carried out the dual luciferase reporter assay. As presented in Figures 5D and 5E, miR-1299 evidently decreased the luciferase activity of wild-type (WT)-circTNPO3 reporter plasmid, but it had little effect on the luciferase activity of mutant (MUT)-circTNPO3 reporter plasmid in SKOV3/PTX and HeyA-8/PTX cells. To further support this finding, we conducted an anti-Ago2 immunoprecipitation assay in SKOV3/PTX cells, which showed that, compared with immunoglobulin G (IgG), endogenous circTNPO3 but not TNPO3 was specifically enriched in the immunoprecipitation fraction pulled down by Ago2 (Figure 5F).
Figure 5.
circTNPO3 Directly Binds to miR-1299 and Suppresses miR-1299 Activity
(A) Schematic illustration showing the overlap of the target miRNAs of circTNPO3 predicted by miRanda, PITA, and RNAhybrid. (B) Endogenous circTNPO3 was efficiently pulled down by anti-Ago2. (C) miRNA pull-down assay showed that circTNPO3 was only efficiently enriched by miR-1299. (D) The luciferase reporter systems showed that miR-1299 mimic considerably reduced the luciferase activity of the WT-circTNPO3 luciferase reporter vector compared with negative control, while miR-1299 mimic did not pose any impact on the luciferase activity of MUT-circTNPO3-transfected SKOV3/PTX cells. (E) The luciferase reporter systems showed that miR-1299 mimic considerably reduced the luciferase activity of the WT-circTNPO3 luciferase reporter vector compared with negative control, while miR-1299 mimic did not pose any impact on the luciferase activity of MUT-circTNPO3-transfected HeyA-8/PTX cells. (F) circTNPO3 and miR-1299 simultaneously existed in the production precipitated by anti-AGO2. All tests were performed at least three times. Data were expressed as mean ± SD. ∗∗p < 0.01.
circTNPO3 Knockdown Inhibited PTX Resistance by Upregulating miR-1299 in PTX-Resistant OC Cells
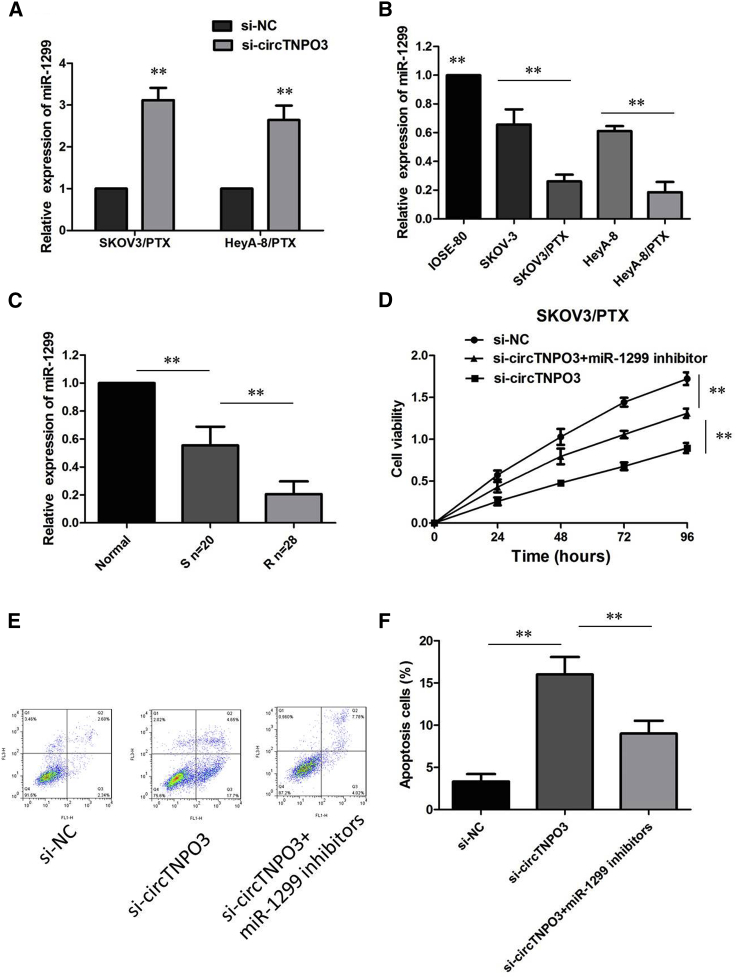
Meantime, qRT-PCR analysis showed that miR-1299 expression was obviously upregulated in si-circTNPO3-transfected SKOV3/PTX and HeyA-8/PTX cells (Figure 6A), suggesting that circTNPO3 interacted with miR-1299 to repress its expression. Not surprisingly, lower expression of miR-1299 was viewed in PTX-resistant OC cells versus their matched SKOV3 and HeyA-8 cells (Figure 6B). Also, miR-1299 was expressed at low level in PTX-resistant patients in contrast with that in PTX-sensitive patients (Figure 6C), indicating that miR-1299 was involved in PTX resistance in OC.
Figure 6.
circTNPO3 Knockdown Inhibited PTX Resistance by Upregulating miR-1299 in PTX-Resistant OC Cells
(A) qRT-PCR analysis showed that miR-1299 expression was obviously upregulated in si-circTNPO3-transfected SKOV3/PTX and HeyA-8/PTX cells. (B) Relative expression of miR-1299 in a panel of ovarian cancer cell lines. (C) Relative expression of miR-1299 in PTX-resistant ovarian cancer and PTX-sensitive ovarian cancer. (D) circTNPO3 knockdown-induced decrease of cell growth was partially restored by miR-1299 inhibition. (E) Silence of miR-1299 effectively attenuated the promoting effect of circTNPO3 knockdown on PTX-induced apoptosis. (F) Silence of miR-1299 effectively attenuated the promoting effect of circTNPO3 knockdown on PTX-induced apoptosis. All tests were performed at least three times. Data were expressed as mean ± SD. ∗∗p < 0.01.
To gain insight into whether circTNPO3 affected PTX resistance of OC cells via modulation of miR-1299, we further performed rescue assays to confirm how miR-1299 modulated PTX resistance. Cell counting kit-8 (CCK-8) assay showed that circTNPO3 knockdown-induced decrease of cell growth was partially restored by miR-1299 inhibition (Figure 6D). Simultaneously, flow cytometry analysis disclosed that deficiency of circTNPO3 expedited PTX-induced apoptosis, while silence of miR-1299 effectively attenuated the promoting effect of circTNPO3 knockdown on PTX-induced apoptosis (Figures 6E and 6F). In summary, deletion of miR-1299 partly abolished the promotion effect of circTNPO3 downregulation on PTX sensitivity in PTX-resistant OC cells.
circTNPO3 Positively Regulated NEK2 Expression by Interacting with miR-1299 in PTX-Resistant OC Cells
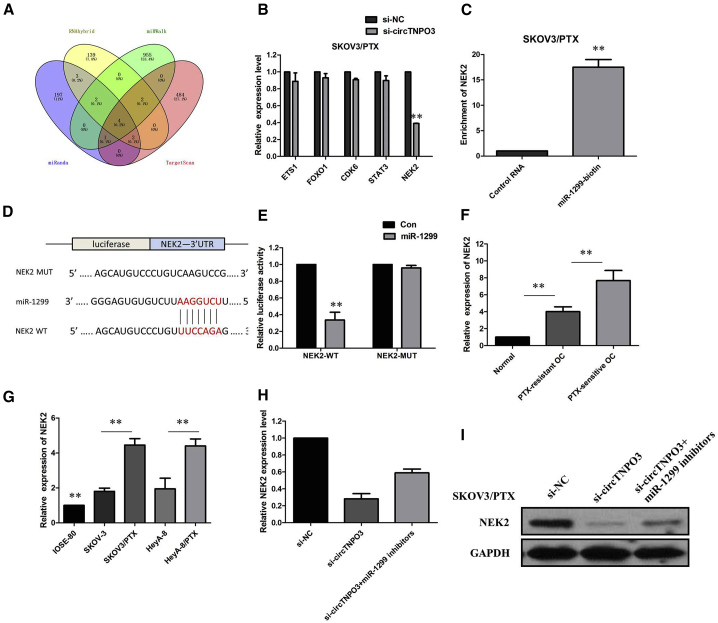
Given that miRNAs could play an important role in post-transcriptional gene regulation by partially base-paring with the 3′ untranslated region (3′ UTR) of target mRNAs, the target mRNAs of miR-1299 were predicted using four bioinformatic logarithms: miRWalk, miRanda, RNAhybrid, and Targetscan. We identified five target genes of miR-1299 by overlapping the prediction results of the four algorithms, and miR-1299 could target the 3′ UTRs of ETS1, FOXO1, CDK6, STAT3, and NEK2 (Figure 7A). To further verify the downstream targets of circTNPO3, we detected mRNA levels of five candidate target genes after silencing circTNPO3, and we found only NEK2 was downregulated (Figure 7B). To verify whether NEK2 was the direct target of miR-1299, we first performed the miRNA biotin pull-down assay. We found that miR-1299 could significantly enrich the 3′ UTR of NEK2 mRNA (Figure 7C), and NEK2 displayed the binding site for miR-1299 (Figure 7D). To further confirm the relationship between miR-1299 and NEK2, we performed a dual-luciferase reporter assay. Luciferase activity was significantly reduced when SKOV3/PTX cells were co-transfected with miR-1299 mimics and a luciferase reporter containing WT 3′ UTR of NEK2. However, the luciferase activity did not change significantly when SKOV3/PTX cells were co-transfected with miR-1299 mimics and a luciferase reporter containing a NEK2 3′ UTR in which the predicted miR-1299 binding site was mutated (Figure 7E). The results of real-time PCR analysis demonstrated that the NEK2 was higher in PTX-resistant OC tissues compared with those in PTX-sensitive OC tissues (Figure 7F; p < 0.01). The expression of NEK2 was obviously increased in PTX-resistant OC cell lines than that in their parental OC cell lines (Figure 7G; p < 0.01). Furthermore, we found that circTNPO3 knockdown or miR-1299 overexpression triggered a substantial decline of NEK2 mRNA and protein levels in SKOV3/PTX cells. Moreover, inhibition of circTNPO3 mediated decrease of NEK2 mRNA and protein levels were significantly recuperated following miR-1299 inhibitors (Figures 7H and 7I). All of these data made us draw a conclusion that circTNPO3 positively regulated NEK2 expression by interacting with miR-1299 in PTX-resistant OC cells.
Figure 7.
circTNPO3 Positively Regulated NEK2 by Interacting with miR-1299 in PTX-Resistant Ovarian Cancer Cells
(A) Venn diagram showing five genes that are putative miR-1299 targets computationally predicted by four algorithms (miRanda, RNAhybrid, miRWalk, and TargetScan). (B) mRNA levels of five candidate target genes were detected after silencing circTNPO3. (C) miR-1299 could significantly enrich the 3′ UTR of NEK2 mRNA. (D) The binding sequence between miR-1299 and NEK2. (E) Luciferase reporter assay demonstrated miR-1299 mimics significantly decreased the luciferase activity of NEK2-WT in OC cells. (F) The real-time PCR analysis demonstrated that the NEK2 was higher in PTX-resistant OC tissues. (G) The expression of NEK2 was obviously increased in PTX-resistant OC cell lines than that in their parental OC cell lines. (H) Inhibition of circTNPO3 mediated decrease of NEK2 mRNA expression was significantly recuperated following miR-1299 inhibitors. (I) Inhibition of circTNPO3-mediated decrease of NEK2 protein expression was significantly recuperated following miR-1299 inhibitors. All tests were performed at least three times. Data were expressed as mean ± SD. ∗∗p < 0.01.
Discussion
PTX treatment is one of the most predominant chemotherapeutic strategies for patients with OC.3,4 circRNAs have been identified to function as key regulators of diverse cellular processes including the initiation and progression of cancer.13, 14, 15 Convincing evidence has indicated the essential roles of circRNAs in drug resistance of diverse tumors.16, 17, 18 But the influence and related mechanisms of circRNAs on progression and drug resistance in OC are still unclear. Further analyses combining the circRNAs expression and drug sensitivity should be helpful to guide the OC clinical treatment. In this study, using microarray analysis, we determined that circRNA expression is associated with PTX resistance in OC. We found a novel circRNA termed circTNPO3 that was upregulated in tissue samples from patients with PTX-resistant OC tissues and in PTX-resistant cell lines and was correlated with OS. Then we focused on the function and mechanism of circTNPO3 in PTX resistance in OC. The data show that inhibition of circTNPO3 could increase the PTX sensitivity of OC cells. This presumption was subsequently validated in vivo. Functional and mechanistic studies manifested that circTNPO3 contributed to PTX resistance of OC cells possibly through upregulating NEK2 expression by sponging miR-1299. Taken together, our data demonstrate that circTNPO3 inhibits the sensitivity of PTX in OC partly via the circTNPO3/miR-1299/NEK2 axis.
In recent years, research on tumor drug sensitivity-related circRNA has been receiving increasing attention.19,20 In this study, we selected circTNPO3 to be dramatically upregulated in OC by microarray assay and then confirmed the high expression of circTNPO3 in PTX-resistant OC tissues and cell lines. Functional analysis further revealed that circTNPO3 knockdown improved PTX sensitivity, caused PTX-induced G0/G1 cell-cycle arrest and promoted PTX-induced apoptosis in PTX-resistant OC cells in vitro, suggesting that circTNPO3 knockdown may attenuate drug resistance of OC cells. Moreover, chemoresistance of circTNPO3 was validated on OC xenografts in nude mice. These results demonstrated that circTNPO3 conferred PTX resistance in PTX-resistant OC cells in vitro and in vivo.
Mechanistically, circRNAs have been demonstrated to be located in cytoplasm of cells and promoted tumorigenesis by regulating miRNA-mRNA axis.21,22 In this study, we found that circTNPO3 was located in cytoplasm, indicating that circTNPO3 might regulate gene expression at post-transcriptional level. Previous studies have demonstrated that circRNAs regulate tumor progression by sponging miRNAs to release mRNAs.23,24 Therefore, we investigated whether circTNPO3 exerted function in chemoresistance of OC via the miRNA-mRNA axis. In this study, we identified circTNPO3 as a new interactive molecule of miR-1299 and also confirmed that NEK2 was a new downstream target of miR-1299. Based on the bioinformatics analysis and mechanism experiments, we found that miR-1299 interacted with circTNPO3 in OC cells, and miR-1299 mimics reversed circTNPO3-mediated effects. We verified that circTNPO3 had an endogenous sponge-like effect on miR-1299 in OC. First, bioinformatics prediction and a luciferase reporter assay showed that circTNPO3 3ʹ UTR share identical miR-1299 response elements and might therefore bind competitively to miR-1299. Second, circTNPO3 and miR-1299 simultaneously existed in the production precipitated by anti-AGO2. Third, downregulation of circTNPO3 significantly promoted miR-1299 expression. Results of rescue assays indicated that downregulation of miR-1299 partially reversed the inhibitory effect of circTNPO3 silencing on OC cell proliferation, indicating that miR-1299 is involved in circTNPO3-mediated chemoresistance of OC. Furthermore, we found that the miR-1299 was significantly lower in PTX-resistant OC tissue and cell lines.
Furthermore, the potential targets of miR-1299 were predicted from four public bioinformatics websites. Among all these potential targets, NEK2 could be regulated by both circTNPO3 and miR-1299. Additionally, luciferase reporter assay and pull-down assay demonstrated the interaction between miR-1299 and NEK2. Therefore, we confirmed that circTNPO3 regulated chemoresistance of OC by miR-1299-NEK2 axis. As a member of the cell-cycle-regulating protein kinase family, NEK2 belongs to the centrosome-related protein kinase.25 An increasing number of studies have found that NEK2 was involved in the development and progression of many human tumors, including OC.26,27 Our findings were similar to the above results. NEK2 was higher in PTX-resistant OC tissues and cell lines. Furthermore, we found that circTNPO3 knockdown or miR-1299 overexpression triggered a substantial decline of NEK2 protein level in SKOV3/PTX cells. Moreover, inhibition of circTNPO3-mediated decrease of NEK2 protein expression was significantly recuperated following miR-1299 inhibitors. Taken together, the study revealed that a circTNPO3/miR-1299/NEK2 axis exists in chemoresistance of OC.
Conclusions
This study identified the involvement of circTNPO3 in PTX resistance of OC cells. circTNPO3 enhanced PTX resistance in ovarian cancer cells by working as a ceRNA to sponge miR-1299, thus reinforcing the protein level of NEK2. Therefore, our study contributes to better understanding of the molecular mechanism of drug resistance in OC, providing a promising circRNA-targeted therapy for OC.
Materials and Methods
Patient Specimens
48 paired resected OC tissues and adjacent normal tissues were collected from OC patients under the approval of the Institutional Review Boards of Hunan Cancer Hospital and the Affiliated Tumor Hospital of Xiang-Ya School of Medicine, Central South University. All patients in our study met the following criteria: all cases were confirmed by the postoperative pathologic examination, all patients before surgery only received PTX-based chemotherapy, and written informed consent from each patient was obtained prior to study initiation. A PTX-resistant case was distinguished when the primary tumor enlarged or relapsed within 12 months; otherwise, it can be defined as a PTX-sensitive case. Samples were promptly frozen in liquid nitrogen and maintained at −80°C until use.
RNA Isolation and Microarrays
The microarray experiments were performed by Shanghai Biotechnology Corporation (SBC) following the protocol of Agilent Technologies (CA, USA). Briefly, total RNA was isolated and purified using a mirVana miRNA Isolation Kit (Ambion, TX, USA), according to the manufacturer’s instructions. RNA samples from each group were then used to generate labeled complementary RNA (cRNA) targets using the Low Input Quick Amp WT Labeling Kit (Agilent Technologies, CA, USA) for the SBC human ceRNA microarray (4 × 180 K). The labeled cRNA targets were then hybridized with the slides and the slides were scanned on the Agilent Microarray Scanner (Agilent Technologies, CA, USA). Data were extracted with Feature Extraction software 10.7 (Agilent Technologies, CA, USA). circRNAs with high expression abundances, expression levels with more than 2-fold alteration, and a p value <0.01 were selected for further analysis.
Cell Culture
Human ovarian carcinoma cell lines (SKOV3 and HeyA-8) and normal ovarian epithelial cell line (IOSE-80) were purchased from ATCC (Manassas, VA, USA) and BNCC (Beijing, China), and maintained in RPMI-1640 medium (Solarbio, Beijing, China) plus 10% fetal bovine serum (FBS; Solarbio) and 1% penicillin/streptomycin (Solarbio) at 37°C with 5% CO2 and 95% air. PTX-resistant OC cell lines (SKOV3/PTX and MeyA-8/PTX) were established by progressively exposing their parent cells to increasing doses of PTX. Then, SKOV3/PTX and MeyA-8/PTX cells were incubated with 50 nM PTX to maintain their resistant phenotype.
Knockdown of circTNPO3
siRNAs that target the junction sequence of circTNPO3 were designed and synthesized by GenePharma (Shanghai, China). Cells were transfected with these siRNAs with GeneMute reagent (SignaGen Laboratories, Rockville, MD, USA). The circTNPO3 sequences were subcloned into pGLV3/H1/GFP/Puro vectors to construct sh-circTNPO3 for animal studies. All transfection assays were carried out in triplicate.
Overexpression of circTNPO3
Primers for constructing the pLO-ciR-circTNPO3 vector were designed. PLO-ciR vector was purchased from GENESEED (Guangzhou, China). Lentivirus particles were generated in the 293T packaging cells by transfection with the pMD2.G pseudotyping plasmid (Addgene, Teddington, UK, Cat# 12259), the psPAX2 packing plasmid (Addgene, Cat# 12260), and either the PLO-ciR-circTNPO3, or PLO-ciR viral vector plasmids. Transfections were performed with liptofectamine 3000 in a 6 cm dish.
RNA Extraction and qRT-PCR
The total RNA was isolated from tissues and cell lines using TRIzol reagent (Invitrogen, CA, USA), and exosomal RNA was extracted from plasma and culture medium using the exoRNeasy Midi Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s protocol. For circRNAs, RNase R was used to degrade linear RNA, which have poly(A), and amplified by divergent primer. Specific divergent primers spanning the back-splice junction site of circRNAs were designed. To quantify the amount of mRNA and circRNA, we synthesized cDNA using PrimeScript RT reagent Kit (TaKaRa, Dalian, China). The qRT-PCR analysis on circRNA and mRNA was performed using Prime Script RT reagent Kit (TaKaRa) and SYBR Premix Ex TaqII (TaKaRa). β-actin was used as an endogenous control. For miR-1299 analysis, miRNA was treated with DNase I to eliminate genomic DNA and cDNA was synthesized by Mir-X miR First-Strand Synthesis Kit (TaKaRa). SYBR Premix Ex TaqII (TaKaRa) was used for qRT-PCR. The expression was normalized to RNU6-2. The 2-ΔΔCT method was adopted to calculate relative expression.
Actinomycin D and RNase R Treatment
To block transcription, we added 2 mg/mL Actinomycin D or dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO, USA) as a negative control into the cell culture medium. For RNase R treatment, total RNA (2 μg) was incubated for 30 min at 37°C with or without 3 U/μg of RNase R (Epicenter Technologies, Madison, WI, USA). After treatment with Actinomycin D and RNase R, qRT-PCR was performed to determine the expression levels of circTNPO3 and TNPO3 mRNA.
Preparation of Nuclear and Cytoplasmic Extracts
The nuclear and cytoplasmic fraction of cells was isolated using the PARIS kit (Ambion, Austin, TX, USA, Cat# AM1921). About 107 cells were washed with PBS on ice followed by centrifugation at 500 × g for 5 min. Cell pellets were resuspended in 500 μL cell fraction buffer, incubated on ice for 10 min, and then centrifuged at 500 × g and 4°C for 5 min to separate the nuclear and cytoplasmic cell fractions. Nuclear pellets were homogenized with the cell disruption buffer.
CCK-8 Assay
Cell proliferation was assessed using the CCK-8 assay (Beyotime Biotechnology, Nantong, China). Cells (2 × 103) were seeded into each well of 96-well plates. 10 μL of CCK-8 solution was added to each well at six time points. After 1.5 h of incubation at 37°C, the absorbance at 450 nM was measured using Spectra Max 250 spectrophotometer (Molecular Devices, Sunnyvale, CA, USA). Experiments were independently performed in triplicate.
Cell-Cycle and Apoptosis Assay
SKOV3/PTX and HeyA-8/PTX cells were seeded into 6-well plates and treated with PTX for 48 h. To assess the cell cycle and apoptosis, we seeded 3 × 105 treated cells into 6-well plates and cultured them for 48 h at 37°C. The cells for cell-cycle analysis were digested using trypsin (Hyclone), washed twice with phosphate-buffered saline (PBS), and fixed in 70% ethanol overnight at 4°C. The cells were centrifuged at 500 × g for 5 min, washed twice with cold PBS, and centrifuged. After treatment with RNase A (0.1 mg/mL) and propidium iodide (PI, 0.05 mg/mL) purchased from 4A Biotech (Beijing, China) for 30 min at 37°C, cell-cycle analysis was performed through fluorescence-activated cell sorting flow cytometry (Beckman Coulter, Palo Alto, CA, USA). For the analysis of apoptosis, cells were trypsinized followed by two PBS washing steps. The cells were stained using the Annexin V/PI detection kit (4A Biotech, Beijing, China) for 5 min at room temperature. The apoptotic cells were measured using flow cytometry (Beckman Coulter). All experiments were repeated at least three times.
Luciferase Reporter Assay
The partial sequence of circTNPO3 containing the putative miR-1299-binding sites was fused into the psiCHECK-2 luciferase reporter vector from Promega (Madison, WI, USA) to generate circTNPO3-WT constructs. While mutations were introduced into putative miR-1299 binding sites for circTNPO3 sequence using the Site-Directed Mutagenesis Kit (TransGene, Beijing, China) and then fused into the psiCHECK-2 luciferase reporter vector to generate the circTNPO3-MUT constructs. SKOV3/PTX and MeyA-8/PTX cells were plated into 24-well plates and cultured for 24 h before transfection. The cells were transfected with circTNPO3-WT or circTNPO3-MUT constructs, together with miR-1299 or miR-con. Luciferase activity was evaluated 24 h post transfection using dual-luciferase assay system (Promega) referring to the manufacturer’s instruction.
RIP Assay
The interaction between circTNPO3 and miR-1299 was assessed by a Magna RIP RNA-Binding Protein Immunoprecipitation Kit from Millipore (Bedford, MA, USA) referring to manufacturer’s instruction. Briefly, SKOV3/PTX and MeyA-8/PTX cells were lysed in complete RIP lysis buffer on the ice for 5 min. Then the RIP lysate was incubated with magnetic beads conjugated with human anti-Argonaute 2 (AGO2; Millipore) or control anti-IgG (Millipore) antibody in RIP immunoprecipitation buffer for 6 h at 4°C. After incubating with proteinase K, purified RNA was extracted and analyzed for circTNPO3 and miR-1299 expression by qRT-PCR.
Western Blotting
Proteins were resolved on an SDS-denaturing polyacrylamide gel and transferred onto nitrocellulose membrane. Membranes were incubated with antibodies against NEK2 and GAPDH overnight at 4°C before blocking with 5% nonfat milk for 2 hr. Next, membranes were washed and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies at 4°C for 2 hr. Protein expression was assessed via enhanced chemiluminescence and band intensities quantified with LabWorks Image Acquisition and Analysis Software (UVP, Upland, CA, USA). All antibodies were purchased from Abcam (Cambridge, MA, USA).
Statistical Analysis
All data reported were verified in at least two different experiments and plotted as means ± standard deviations. The differences between control and experimental groups were analyzed by GraphPad Prism 7, using two-tailed unpaired t test. Pearson’s coefficient correlation was used for correlation assay. p values < 0.05 were considered as statistically significant.
Author Contributions
X.B. and Z.Z. performed primers’ design and experiments. Y. Wu and Y. Wang contributed cell and animal experiments. Y.Z. collected and classified the human tissue samples. J.W. analyzed the data. X.B. wrote the paper. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
We have received consents from individual patients who have participated in this study. The consent forms will be provided upon request. All authors have agreed to publish this manuscript. Funding: 13th Five-Year National Key R & D Program of the Ministry of Science and Technology, “focus on prevention and control of chronic diseases and non-communicable diseases” (2016YFC1303703).
Contributor Information
Yan Zhao, Email: yzzzh1618@yeah.net.
Jing Wang, Email: fupy1618@yeah.net.
References
- 1.Rebecca L. Siegel, Miller KD, Jemal A. cancer statistics. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Vaughan S., Coward J.I., Bast R.C., Jr., Berchuck A., Berek J.S., Brenton J.D., Coukos G., Crum C.C., Drapkin R., Etemadmoghadam D. Rethinking ovarian cancer: recommendations for improving outcomes. Nat. Rev. Cancer. 2011;11:719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mantia-Smaldone G.M., Edwards R.P., Vlad A.M. Targeted treatment of recurrent platinum-resistant ovarian cancer: current and emerging therapies. Cancer Manag. Res. 2011;3:25–38. doi: 10.2147/CMR.S8759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei W., Birrer M.J. Spleen tyrosine kinase confers paclitaxel resistance in ovarian cancer. Cancer Cell. 2015;28:7–9. doi: 10.1016/j.ccell.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Pinato D.J., Graham J., Gabra H., Sharma R. Evolving concepts in the management of drug resistant ovarian cancer: dose dense chemotherapy and the reversal of clinical platinum resistance. Cancer Treat. Rev. 2013;39:153–160. doi: 10.1016/j.ctrv.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 7.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 11.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 13.Ebbesen K.K., Kjems J., Hansen T.B. Circular RNAs: Identification, biogenesis and function. Biochim. Biophys. Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Beermann J., Piccoli M.T., Viereck J., Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 15.Han B., Chao J., Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol. Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Xiao G., Huang W., Zhan Y., Li J., Tong W. CircRNA_103762 promotes multidrug resistance in NSCLC by targeting DNA damage inducible transcript 3 (CHOP) J. Clin. Lab. Anal. 2020 doi: 10.1002/jcla.23252. Published online March 2, 2020. 10.1002/jcla.23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S., Cheng J., Quan C., Wen H., Feng Z., Hu Q., Zhu J., Huang Y., Wu X. circCELSR1 (hsa_circ_0063809) Contributes to Paclitaxel Resistance of Ovarian Cancer Cells by Regulating FOXR2 Expression via miR-1252. Mol. Ther. Nucleic Acids. 2020;19:718–730. doi: 10.1016/j.omtn.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao H.X., Miao C.F., Sang L.N., Huang Y.M., Zhang R., Sun L., Jiang Z.X. Circ_0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci. 2020;243:117255. doi: 10.1016/j.lfs.2020.117255. [DOI] [PubMed] [Google Scholar]
- 19.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Qiu L.P., Wu Y.H., Yu X.F., Tang Q., Chen L., Chen K.P. The emerging role of circular RNAs in hepatocellular carcinoma. J. Cancer. 2018;9:1548–1559. doi: 10.7150/jca.24566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai N., Peng E., Qiu X., Lyu N., Zhang Z., Tao Y., Li X., Wang Z. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J. Exp. Clin. Cancer Res. 2018;37:172. doi: 10.1186/s13046-018-0838-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He R., Liu P., Xie X., Zhou Y., Liao Q., Xiong W., Li X., Li G., Zeng Z., Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J. Exp. Clin. Cancer Res. 2017;36:145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song J., Wang H.L., Song K.H., Ding Z.W., Wang H.L., Ma X.S., Lu F.Z., Xia X.L., Wang Y.W., Fei-Zou, Jiang J.Y. CircularRNA_104670 plays a critical role in intervertebral disc degeneration by functioning as a ceRNA. Exp. Mol. Med. 2018;50:94. doi: 10.1038/s12276-018-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X.W., Zheng B.A., Hu Z.M., Qian Z.Y., Huang C.J., Liu X.Q., Wu W.D. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8:91674–91683. doi: 10.18632/oncotarget.21748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fry A.M. The Nek2 protein kinase: a novel regulator of centrosome structure. Oncogene. 2002;21:6184–6194. doi: 10.1038/sj.onc.1205711. [DOI] [PubMed] [Google Scholar]
- 26.Kokuryo T., Senga T., Yokoyama Y., Nagino M., Hamaguchi M. Abstract 1571: Nek2 as an effective molecular target for cancer treatment. Cancer Res. 2010;70:1571. [Google Scholar]
- 27.Liu X., Gao Y., Lu Y., Zhang J., Li L., Yin F. Upregulation of NEK2 is associated with drug resistance in ovarian cancer. Oncol. Rep. 2014;31:745–754. doi: 10.3892/or.2013.2910. [DOI] [PubMed] [Google Scholar]