Abstract
A growing number of studies have focused on the involvement of non-coding RNAs (ncRNAs) in the internal ribosome entry site (IRES)-mediated translation in tumorigenesis; however, the underlying mechanisms in colorectal cancer (CRC) remain elusive. In this study, we show that LINC00473 (LNC473) exerted its functions as a tumor suppressor in promoting apoptotic protease-activating factor 1 (APAF1) IRES activity through competitively sponging miR574-5p and miR15b-5p in CRC initiation and pathogenesis. Specifically, LNC473 and its downstream target APAF1 were significantly downregulated accompanied by upregulated miR574-5p and miR15b-5p in CRC cells and tissues, which had a significant prognostic impact on clinical outcomes in our CRC cohort (n = 157). Furthermore, ectopic LNC473 significantly sponged endogenous miR574-5p or miR15b-5p and thereby inhibited cell proliferation and colony formation capacity, and it accelerated cell apoptosis through activating the APAF1-CASP9-CASP3 pathway. Notably, LNC473 overexpression resulted in dramatic promotion of APAF1 IRES activity and translation, whereas rescue experiments confirmed the recovery by the existence of LNC473 and miR574/15b-5p. Mechanistically, LNC473 overexpression promoted IRES binding domain exposure and removed the constraints controlling from miR574-5p and miR15b-5p, and subsequently enhanced IRES-mediated APAF1 expression in vitro and in vivo. Therefore, our results uncover a novel LNC473-miR574/miR15b-APAF1 signaling axis, which provides new targets and crosstalk regulation mechanism for CRC prevention and treatment.
Keywords: LNC473, IRES-mediated translation, miR574-5p, miR15b-5p, ceRNA, APAF1, colorectal cancer
Graphical Abstract
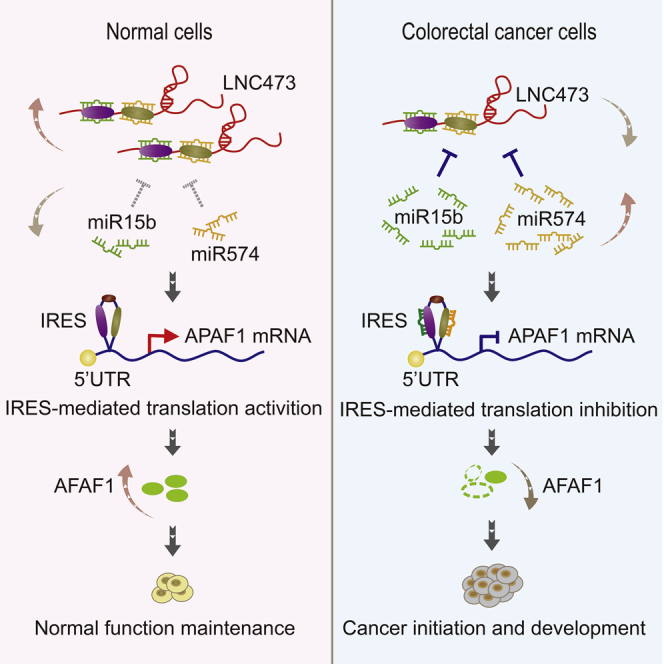
The authors show that LNC473 is critically involved in the tumorigenesis, progression, and prognosis in CRC by competitively sponging hsa-miR574/miR15b-5p, yet it regulates APAF1 IRES-mediated translation initiation. The novel signaling axis will provide strong support for finding efficient molecular biomarkers and pathways for CRC diagnosis and treatment.
Introduction
Colorectal cancer (CRC) is one of the most common digestive system malignancies in the world, causing more than 53,200 cancer-related deaths with approximately 147,950 new cases diagnosed each year.1 Deficiencies in screening biomarkers hinder the effective evaluation of the severity and prognosis of CRCs. Approximately 40% of the CRC patients were diagnosed at the early clinical stage, which, however, resulted in large variations of clinical outcomes, such as the overall 5-year survival times.2 Mounting evidence exists on the important participation of non-coding RNAs (ncRNAs), especially long ncRNAs (lncRNAs) and microRNAs (miRNAs), in the initiation and progression in the variety of tumors.3, 4, 5, 6 Efficient biomarkers among them have been identified for cancer diagnosis and prognosis evaluation. Thus, insight into molecular biomarkers for CRC could considerably facilitate clinical diagnosis and treatment and promote the survival time of cancer patients.
LINC00473 (LNC473), located at the 6q27 locus, encodes an intergenic 1,832-bp ncRNA, which consists of three exons and three introns, forming two annotated transcript isoforms. Recent studies found that dysregulation of LNC473 occurred in several types of cancers, including hepatic cellular cancer (HCC), non-small-cell lung cancer (NSCLC), cervical cancer, and Wilms’ tumor.7, 8, 9, 10. In HCC, LNC473 acts as an essential modulator by recruiting deubiquitinase USP9X to prompt cell proliferation and invasion and induce the epithelial-mesenchymal transition (EMT) process.7 In NSCLC, LNC473 expression was significantly increased, and such overexpression predicted a poor prognosis. Furthermore, LNC473 overexpression was necessary for the growth and survival of LKB1-inactivated NSCLC cells by direct regulation of the NONO protein and CREB-mediated transcription. Recently, the classical “competing endogenous RNA (ceRNA)” mechanism suggested the regulatory crosstalk dialogs between lncRNAs and miRNAs.11 As expected, LNC473 also exerted its functions by sponging endogenous miRNAs and regulated its target genes by the ceRNA hypothesis.8 In Wilms’ tumor, LNC473 knockdown effectively promoted the regression of miR195/IKKα-mediated cell growth inhibition in vitro and in vivo.9 In cervical cancer, miR34a reduced the stability of LNC473 by specifically recognizing miRNA response elements (MREs), leading to the degradation of oncogenic ILF2 and subsequent tumor inhibition.10 However, note that the involvement of LNC473 in CRC tumorigenesis concerning its underlying molecular mechanism and the crosstalk regulation pattern has not yet been reported and needs further investigation.
Previous studies supported that degradation of genes was aroused by miRNAs based on the complementary base pairing of the 3′ untranslated region (UTR) in the target mRNAs and subsequently accelerated the degradation of the downstream genes at the post-transcriptional regulation level.12 In recent years, the internal ribosome entry site (IRES)-mediated translation mechanism aroused increasing attention due to its emerging roles under pathological stress conditions in cells (e.g., cellular hypoxic stress and DNA damage), especially during tumorigenesis.13, 14, 15, 16, 17 The miRNAs exert the functions by an alternative novel regulation pattern, which acted on the IRES region within the 5′ UTR of target genes (not the classical 3′ UTR) and then mediated IRES-dependent translation and expression.18 An earlier study revealed the IRES involved in the translation of several critical tumor suppressor mRNAs (e.g., cat-1, N-myc, apoptotic protease-activating factor 1 [APAF1], and s-Src).19 Interestingly, the 5′ UTR of APAF1 was found to contain a stable hairpin, to be rich in G-C, and to form an active IRES, and the translation of IRES-dependent initiation was dominant through its enhanced activity caused by the alteration of secondary structure, which differed from findings that identified c-myc mRNAs.20 Thus, a pivotal role was suggested in IRES-mediated translation and thereby in the control of cell-fate decisions, especially in tumorigenesis. A recent study established that in human neuroblastoma, MYCN mRNA is suppressed by miR375 through the MYCN IRES-dependent translation regulation pattern as investigated in vitro and in vivo.21 Such findings prompted us to explore whether the APAF1 IRES-mediated translation is involved in tumorigenesis by specifically binding with the domain of these existing miRNAs, which might provide novel targets or function mechanisms for altering the development and progression of CRC.
In the present study, we discovered a novel mechanism regulated by the LNC473-miR574/miR15b-APAF1 signaling axis in CRC tumorigenesis and pathogenesis. In human CRC, LNC473 exerts its functions as a tumor suppressor by sponging miR574-5p and miR15b-5p based on the ceRNA mechanism and promotes the expression of its critical target APAF1 through IRES-mediated post-transcriptional regulation. Our findings disclose a novel regulation manner and shed new light on the understanding of the relationship between lncRNAs and IRES-mediated translation in CRC diagnosis and prognosis evaluation and treatment.
Results
Correlations of LNC473 and miR574 and miR15b Expression in CRC Cells and Tissues
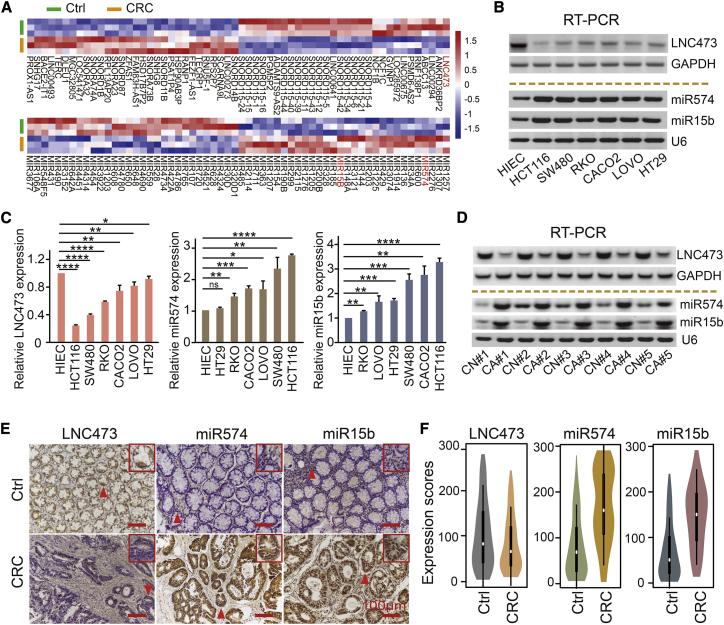
To explore the dysregulated lncRNAs-miRNAs and their potential functions in CRC tumorigenesis, we analyzed the differential expression profiling between CRC tissues and matched tumor-adjacent controls based on an Affymetrix GeneChip microarray dataset obtained from Gene Expression Omnibus (GEO): GSE137511 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137511). The statistical differences were set at p < 0.05, as illustrated in Figure 1A and Tables S1 and S2. Specifically, we established significantly downregulated LNC473 expression (log2 fold change [FC] = 3.48) accompanied by the upregulation of miR574-5p and miR15b-5p (log2 FC = 2.22 and 1.96, respectively). Consistently, a dramatically decreased expression of LNC473 was observed in CRC samples based on The Cancer Genome Atlas (TCGA) dataset (Figure S1A). More importantly, our further examination identified that LNC473 expression was significantly decreased, whereas the levels of miR574-5p and miR15b-5p were remarkably increased in the six CRC cells lines, including HT29, RKO, LOVO, Caco-2, HCT116, and SW480, compared with the normal control HIEC cells as detected by RT-PCR and qPCR assays (Figures 1B and 1C). Similar results were demonstrated based on the paired CRC tissues and matched controls (n = 20), which showed a reduced LNC473 expression accompanied with elevated expression of miR574-5p and miR15b-5p in CRC tissues (Figure 1D; Figures S1B and S1C), indicating the potential negative relationship of LNC473 with miR574-5p or miR15b-5p in CRC cells and tissues.
Figure 1.
Screening of the Differentially Expressed lncRNAs and miRNAs in CRC Tissues and Cells
(A) The hierarchical cluster heatmap illustrating the most differentially expressed lncRNAs and miRNAs in CRC tissues and matched tumor-adjacent controls (n = 3, p < 0.05). Red in the heatmap denotes upregulation; blue denotes downregulation. (B and C) The levels of LNC473, miR574-5p, and miR15b-5p in normal intestinal epithelial HIEC and CRC cells detected by RT-PCR (B) and qPCR (C) assays. n = 3 independent experiments. Data are expressed as mean ± SD. ns (not significant), p > 0.05; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. (D) Representative data of LNC473, miR574-5p, and miR15b-5p expression in CRC tissues and matched tumor-adjacent controls detected by RT-PCR assay (5 representative data are shown). (E) ISH assay determining the cellular localization of LNC473, miR574-5p, and miR15b-5p in CRC tissues and matched adjacent-tumor controls (n = 157). Scale bars, 100 μm. (F) Violin charts displaying the differences in expression scores of the indicators in our included cohort (n = 157). Median (interquartile range) is shown.
Thereafter, we detected the expression of LNC473, miR574-5p, and miR15b-5p in a relatively large cohort including 157 CRC tissues and matched tumor-adjacent controls from CRC patients using tissue microarray (TMA) and in situ hybridization (ISH) assays with some modification.22 Based on the statistics of the expression scores, we first determined the cutoff score of low/high expression for each indicator calculated by the receiver operating characteristic (ROC) curve method (Figure S1D). Moreover, these indicators mainly expressed in the cytoplasm of the CRC tissues (Figure 1E), and the dysregulation of LNC473, miR574-5p, and miR15b-5p expression reappeared (Figure 1F). Of note, we confirmed a negative linear correlation pattern of expression between LNC473 and miR574-5p or miR15b-5p in this cohort (Figure S1E). Collectively, our results suggest that LNC473 is downregulated and negatively correlated with miR574-5p or miR15b-5p expression in human CRC cells and tissues, indicating the possible interaction and negative regulation manner between LNC473 and the miRNAs in CRC tumorigenesis and progression.
Impact of LNC473, miR574, and miR15b Expression on Prognosis of CRC Patients
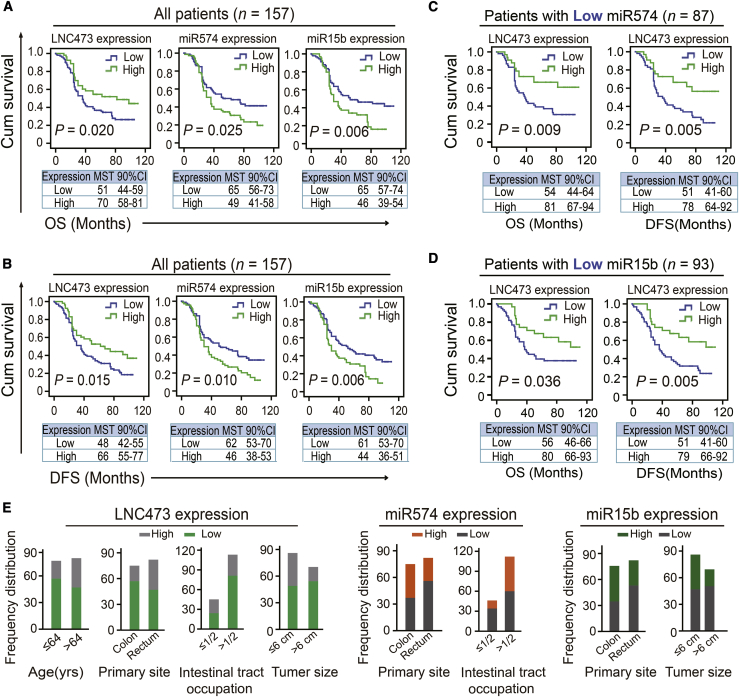
To explore the independent prognostic values of these indicators, we investigated the association of LNC473, miR574-5p, and miR15b-5p expression with clinical outcomes and clinicopathological variables in our cohort. First, Kaplan-Meier curves and log-rank tests revealed that LNC473 high expression had a better overall survival (OS, p = 0.020; high/low expression: mean survival time [MST] = 70/51 months) and disease-free survival (DFS, p = 0.015, high/low expression: MST = 66/48 months), as outlined in Figures 2A and 2B and Table S4. In contrast, miR574-5p high expression was significantly correlated with shortened OS (p = 0.025; high/low expression: MST = 49/65 months) and DFS (p = 0.010; high/low expression: MST = 46/62 months). Similarly, miR15b-5p high expression had a poor impact on OS (p = 0.006; high/low expression: MST = 46/65 months) and DFS (p = 0.006; high/low expression: MST = 44/61 months), as shown in Figures 2A and 2B and Table S4. Further adjusted multivariate Cox regression analysis also confirmed the prognostic values of these indicators in our included CRC cohort, illustrated in Table S5. More importantly, stratification analysis confirmed the synergistic prognostic values of LNC473 low/high expression based on the subgroup of low expression of miR574-5p (n = 87) or miR15b-5p (n = 93). Specifically, in the subgroup of miR574-5p low expression (n = 87), the LNC473 high expression cohort showed a strengthened better clinical prognosis, including OS (p = 0.009; high/low expression: MST = 81/54 months) and DFS (p = 0.005; high/low expression: MST = 78/51 months) (Figure 2C). Consistent results of the miR15b-5p low expression cohort (n = 93) manifested that LNC473 high expression carriers had prolonged OS (p = 0.036; high/low expression: MST = 80/56 months) and DFS (p = 0.005; high/low expression: MST = 79/51 months) (Figure 2D). Moreover, stratified analysis of adjusted multivariate COX regression also proved the identical results (Table S5).
Figure 2.
Correlations of LNC473, miR574, and miR15b Expression with the Clinical Variables in CRC patients
(A and B) Kaplan-Meier curves and log-rank tests showing the association of LNC473, miR574-5p, and miR15b-5p with the OS (A) and DFS (B) in the included CRC patients. (C and D) Stratified Kaplan-Meier curves and log-rank tests illustrating the impact of LNC473 on prognosis (OS and DFS) in the subgroup of patients with miR574-5p low expression (C) or miR15b-5p low expression (D). (E) Correlation between high/low expression of the indicators and clinicopathological parameters analyzed by chi-square or Fisher’s exact tests and unconditioned logistic regression.
In addition, we performed the Chi-square tests and unconditional logistic regression analysis adjusted by diagnosis ages and pathological pattern to determine the interaction of these indicators with the clinical variables, as outlined in Figure 2E and Table S6. As a result, we found that LNC473 low expression was distributed frequently in the tissues of patients with age ≤64 years, colon cancer, intestinal tract occupation (>1/2), or tumor size (>6 cm). Nevertheless, miR574-5p high expression was associated with the samples with intestinal tract occupation (>1/2) and colon cancer; meanwhile, miR15b-5p high expression was easier to be found in samples with colon cancer and tumor size (>6 cm) (Figure 2E; Table S6). These data indicate the independent prognostic values of LNC473, miR574-5p, and miR15b-5p in clinical outcome evaluation of CRC patients.
Effect of LNC473 on Biological Characters by Regulating miR574 and miR15b in CRC Cells
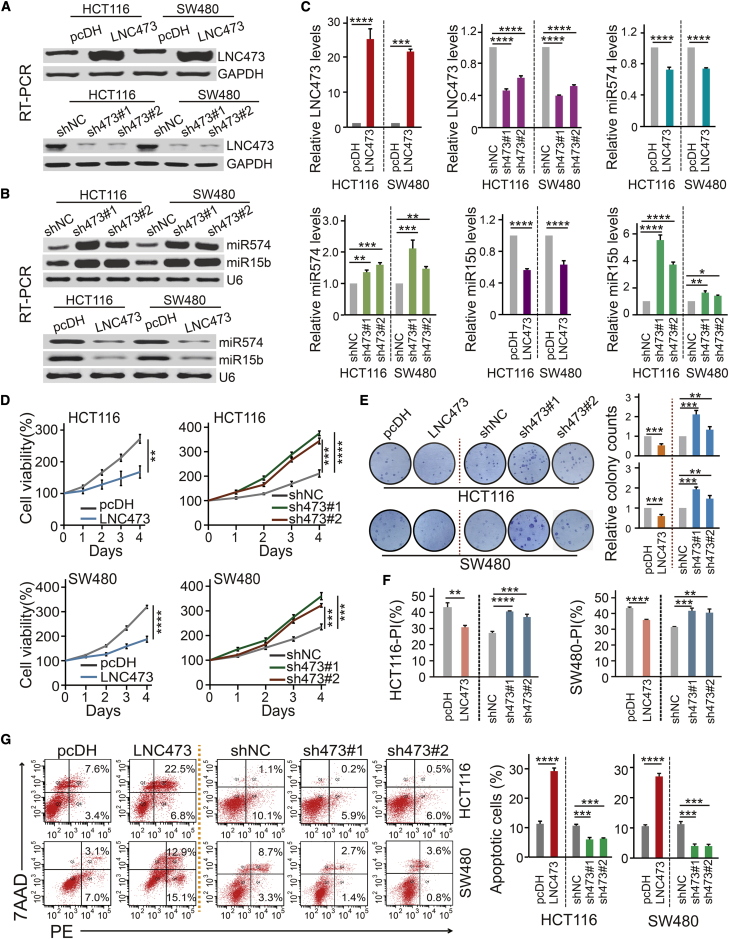
Next, we demonstrated the biological activities and regulation pattern of LNC473 interacting with miR574-5p and miR15b-5p in CRC cells identified by loss- and gain-of-function experiments. RT-PCR and qPCR assays identified that LNC473 overexpression caused substantial inhibition of miR574-5p and miR15b-5p expression in both HCT116 and SW480 CRC cells (Figures 3A–3C). Conversely, LNC473 knockdown enhanced the expression of miR574-5p and miR15b-5p in the two types of CRC cells (Figures 3A–3C). Furthermore, Cell Counting Kit-8 (CCK8) assays and colony formation assays (CFAs) revealed that LNC473 depletion significantly promoted cell viability and colony formation capacity, whereas LNC473 overexpression suppressed the cell proliferative and colonic abilities in HCT116 and SW480 cells (p < 0.05, Figures 3D and 3E). Moreover, the rescue experiments, where miR574-5p and miR15b-5p were upregulated after LNC473 overexpression in CRC cells, confirmed the recovery of cell proliferation and colony formation capacity in comparison to the negative control (NC) transfection, as shown in Figures S2A and S2B. Thereafter, the cell cycle and apoptosis analysis indicated that ectopic LNC473 expression led to phase G0/G1 of the cell cycle blockade, as well as induction of cellular apoptosis in the CRC cells. In contrast, LNC473 knockdown substantially promoted the proliferation index (PI) and inhibited the cellular apoptosis in both HCT116 and SW480 cells as detected by flow cytometry, outlined in Figures 3F, 3G, and S2C. Similarly, the rescue experiments demonstrated that the cell-cycle profiles and cell apoptosis rates were recovered in HCT116 and SW480 cells co-transfected with the pcDH-LNC473 vector and miR574-5p or miR15b-5p mimic (Figures S2D and S2E).
Figure 3.
Effect of LNC473 on Biological Characters by Regulating miR574 and miR15b in HCT116 and SW480 Cells
(A–C) The levels of LNC473 (A), miR574-5p, and miR15b-5p (B) after LNC473 overexpression or knockdown in HCT116 and SW480 cells detected by RT-PCR assay and qPCR assay (C). (D and E) Cell proliferation (D) and colony formation ability (E) were detected after interfering LNC473 expression by a CCK8 assay and colony formation assay. Representative images of cells in six-well plates (E, left) and the histograms (E, right) are shown. (F and G) The proliferation index (PI) of cell cycle (F) and percentage (%) of cell apoptosis (G) were detected upon interfering LNC473 expression by flow cytometry. All tests were performed at least three times. Data are expressed as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
APAF1 Is a Novel Target of LNC473 Interacting with miR574 and miR15b
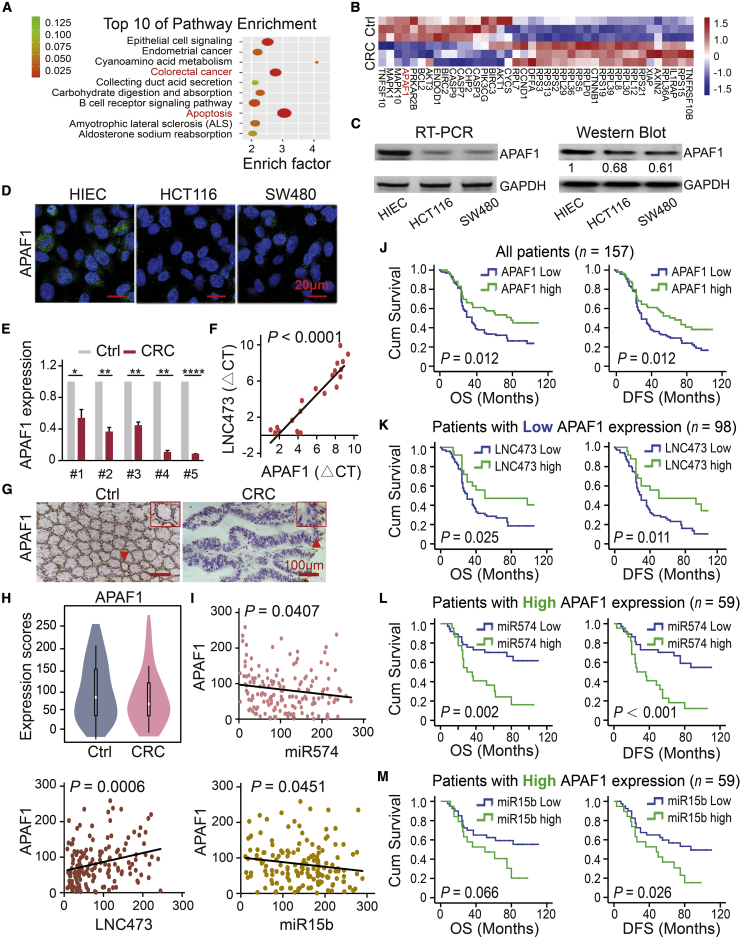
To identify the downstream targets of LNC473 sponging miR574-5p and miR15b-5p in CRC tumorigenesis, we conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis to enrich co-expressed mRNAs in the top 10 cellular functional components and signaling pathways including “apoptosis” and “CRC” based on Affymetrix GeneChip microarray (Figure 4A). The heatmap cluster identified the significantly downregulated APAF1 among the mRNAs related to the two pivotal pathways (Figure 4B; Table S3). Consistently, the analysis based on datasets confirmed the reduced APAF1 expression in CRC samples compared with controls from TCGA database upon Oncomine https://www.oncomine.org/index.jsp), outlined in Figures S3A and S3B. Importantly, our examination results demonstrated that APAF1 was significantly decreased in both HCT116 and SW480 cells compared with HIEC control cells as detected by RT-PCR, qPCR, western blot (WB), and immunofluorescence (IF) assays, as illustrated in Figures 4C, 4D, and S3C. Similarly, decreased APAF1 expression was confirmed in CRC tissues compared with that of the matched tumor-adjacent controls (n = 20) using a qPCR assay (representative data shown in Figure 4E), and subsequently resulting in a positive linear correlation between LNC473 and APAF1 expression (R2 = 0.863, p < 0.0001), as shown in Figure 4F.
Figure 4.
Identify Downstream Targets of LNC473 Interacting with miR574 and miR15b
(A) KEGG analysis enriched the co-expressed target mRNAs in the top 10 functions and signaling pathways affected by LNC473, miR574-5p, and miR574-5p. (B) The hierarchical clustering heatmap shows the related target mRNAs in CRC patient tissues and the matched adjacent-tumor controls (n = 3). (C and D) APAF1 mRNA levels detected by RT-PCR (C, left), APAF1 protein levels detected by western blot (C, right), and IF (D) among HIEC, HCT116, and SW480 cells. Scale bars, 20 μm. n = 3 independent experiments. (E) APAF1 mRNA expression in CRC tissues and matched controls assayed by RT-PCR. n = 3 independent experiments. Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001). (F) Linear regression analysis of LNC473 and APAF1 relative copy numbers. (G and H) IHC assay detecting cellular localization (G) and violin charts comparing differences in expression scores (H) of APAF1 protein in 157 pairs of tissues. Scale bars, 100 μm. (I) Linear correlation pattern showing the relationship of APAF1 expression with included ncRNAs in CRC samples. (J) Kaplan-Meier curves showing the association of APAF1 high/low expression with the prognosis of patients. (K–M) Stratified Kaplan-Meier curves illustrating the impact of LNC473 (K), miR574-5p (L), and miR574-5p (M) on prognosis in the subgroup of patients with APAF1 low expression.
Subsequently, we detected the APAF1 expression by TMA and immunohistochemistry (IHC) assays and assessed the low/high expression of APAF1 based on the cutoff score using the ROC curve method in our included CRC cohort (n = 157) (Figures 4G and 4H; Figure S3D). As expected, obviously decreased APAF1 expression was determined in the CRC patient tissues, which exhibited the characteristics that a tumor suppressor possessed (Figures 4G and 4H). Furthermore, we confirmed the positive linear correlation between APAF1 and LNC473 expression, whereas a negative relationship was identified between APAF1 and miR15b-5p or miR574-5p expression in the cohort, as shown in Figure 4I. Notably, Kaplan-Meier analysis and log-rank tests showed that APAF1 low expression was significantly associated with shortened OS (p = 0.012; high/low expression: MST = 69/50 months) and DFS (p = 0.012; high/low expression: MST = 65/48 months), as shown in Figure 4J and Table S4. Consistently, the adjusted multivariate Cox regression analysis also established APAF1 as the independent prognostic indicator in this cohort (Table S5). In the stratified subgroup of APAF1 low expression patients (n = 98), synergistic prognosis results indicated that carrying LNC473 low expression showed worse clinical outcomes, including OS (p = 0.025; MST of high/low expression: 65/45 months) and DFS (p = 0.011; MST of high/low expression: 63/42 months), as shown in Figures 4K and S3E. Nevertheless, in the subgroups of APAF1 high expression (n = 59), miR574-5p low expression showed a more significant improvement in OS (p = 0.002; MST of high/low expression: 47/81 months) and DFS (p < 0.001; MST of high/low expression: 43/79 months) (Figure 4L). Meanwhile, patients with miR15b-5p low expression showed a prolonged DFS (p = 0.026; MST of high/low expression: 49/73 months) (Figure 4M). Additionally, inverted stratification analysis revealed a more obvious correlation between APAF1 expression and prognosis of CRC patients in the subgroups with low LNC473 (n = 104), miR574-5p (n = 87), and miR-15b expression (n = 93) (Figure S3F). Similarly, based on the stratification above, adjusted multivariate Cox regression analysis was used to perform an in-depth validation (Table S5). Thus, our results indicate that APAF1 might be an independent prognostic biomarker and a potential target of LNC473 sponging miR15b-5p and miR574-5p. However, its underlying molecular mechanism is still unclear in CRC tumorigenesis and pathogenesis.
LNC473 Acts as a Sponge of miR574/15b Regulating APAF1 IRES Activity in CRC Cells
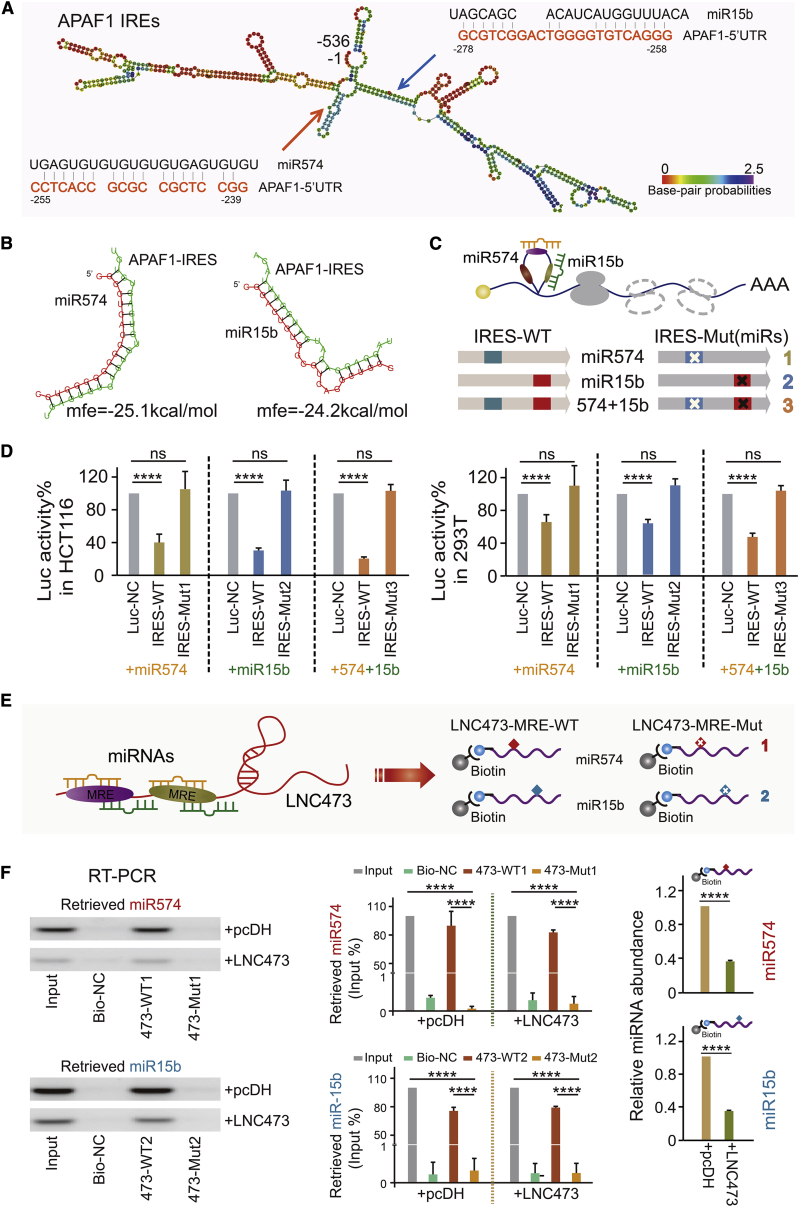
APAF1 IRES, within the 5′ UTR of APAF1 mRNA, displayed an enhanced activity and mediated APAF1 translation in tumorigenesis.20 Thus, based on the confirmation that the 3′ UTR region of APAF1 was not a target of miR574-5p and miR15b-5p, we hypothesized that the levels of APAF1 mediated by IRES-dependent translation were regulated by LNC473 via sponging miR574-5p and miR15b-5p. As shown in Figure 5A, the IRESite database provided an experimentally verified sequence of APAF1 IRES (http://www.iresite.org/). Of note, the complementary domain and binding sites presented in Figure 5A were between APAF1 IRES and miR574-5p (+239 to +255)/miR15b-5p (+258 to +278); meanwhile, the minimum free energy (MFE) between them was evaluated by RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) (Figure 5B). Since a relatively higher affinity was predicted among the indicators, we subsequently evaluated the APAF1 IRES activities after co-transfecting with IRES-wild-type (WT) or IRES-mutant (Mut)1/2/3 luciferase reporter and miR574-5p and/or miR15b-5p mimics in HCT116 and HEK293T cells (Figure 5C). As expected, the dual-luciferase reporter assay showed the suppression of APAF1 IRES activities after co-transfecting miR574-5p and/or miR15b-5p mimics with an IRES-WT luciferase vector. However, no significant alteration was observed with IRES-Mut1/2/3 luciferase vectors, suggesting that miR574-5p and miR15b-5p could directly bind to APAF1 IRES by identifying specific sequences (Figure 5D).
Figure 5.
The Molecular Mechanism of LNC473 Regulating APAF1 IRES by Sponging miR574 and miR15b
(A and B) Predicting the secondary structure of the APAF1 IRES (A) and evaluating the location and MFE of miR574-5p/miR15b-5p binding to APAF1 IRES (B) by RNAhybrid. (C) Pattern diagram of luciferase vectors of APAF1 IRES with wild-type or mutated sequences. (D) Dual-luciferase reporter assay identifying the specific binding activities of APAF1 IRES with miR574-5p/miR15b-5p in HCT116 cells. (E) Pattern diagram of the biotin-labeled probes of LNC473 harboring wild-type or mutant sequences of MREs. (F) RIP assay detecting the specific binding activities of LNC473 with miR574-5p/miR15b-5p in HCT116 cells. All tests were performed at least three times. Data are expressed as mean ± SD. ns, p > 0.05; ∗∗∗∗p < 0.0001.
Next, to demonstrate LNC473 acting as an endogenous miRNA sponge, we predicted that the MREs, existing in LNC473, stably bound to miR574 or miR15b based on the RNAhybrid database (Figure S4A). After that, we designed and synthesized the biotin-labeled LNC473 probes harboring sequences of the MREs (LNC473-MRE-WT1/2) or corresponding mutant MREs (LNC473-MRE-Mut1/2), as presented in Figure 5E. As expected, an RNA immunoprecipitation (RIP) assay confirmed that LNC473-MRE-WT probes dramatically pulled down the endogenous miR574-5p or miR15b-5p, but not in the presence of LNC473-MRE-Mut1/2 probes detected in both HCT116 (Figure 5F) and HEK293T cells (Figure S4B). Additionally, LNC473 overexpression resulted in a significant depletion of the dissociative miR574-5p and miR15b-5p (Figure 5F; Figures S4B and S4C). Conversely, we further altered the expression of the two miRNAs, and the results of RT-PCR and qPCR assays identified that miR574-5p and miR15b-5p mimics inhibited LNC473 expression in both HCT116 and SW480 cells. In contrast, miR574-5p and miR15b-5p inhibitors improved the levels of LNC473 in the two CRC cells (Figures S4D and S4E). These findings indicate a direct ceRNA regulation mechanism between LNC473 and miR574-5p or miR15b-5p in CRC cells.
LNC473-miR574/miR15b-APAF1 Signaling Axis in CRC Cells
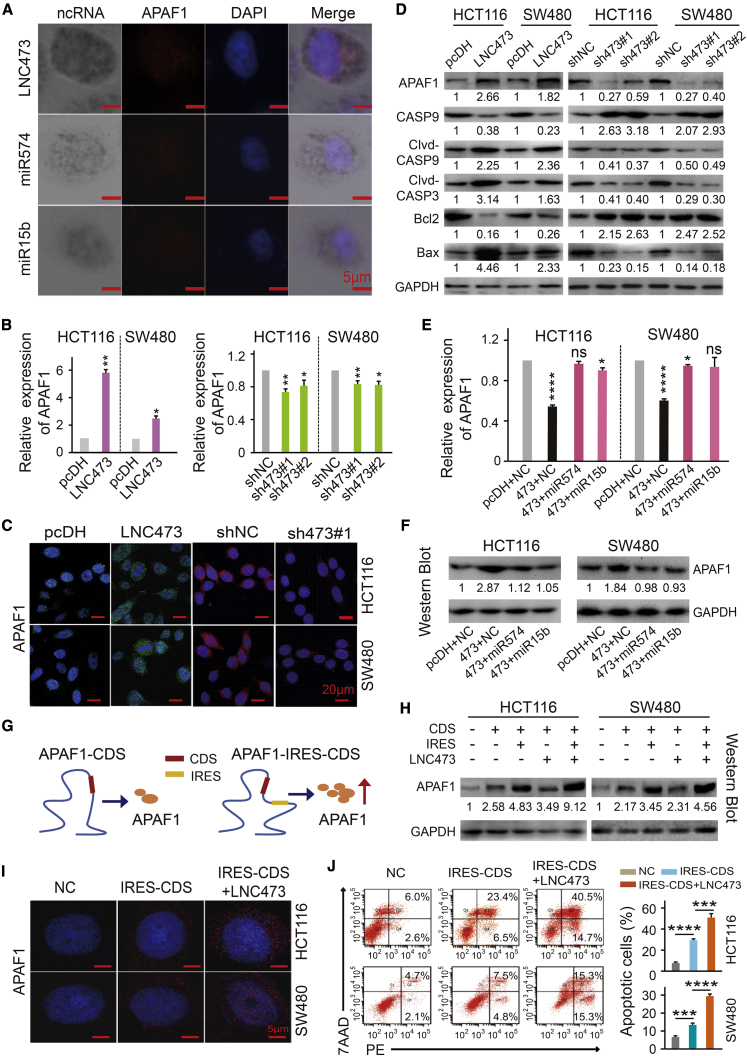
To elucidate the functions of the critical regulating axis in the context of LNC473 involved in CRC tumorigenesis, we for the first time conducted the co-localization of these indicators using the ISH and IF combination assay. Of note, cellular co-localization results indicated that APAF1 distributed consistently with LNC473, miR574-5p, and miR15b-5p each in the cytoplasm of HCT116 cells (Figure 6A). Furthermore, we identified the positive regulation pattern between LNC473 and APAF1 expression. Specifically, LNC473 overexpression significantly enhanced APAF1 mRNA levels in HCT116 and SW480 cells. Nevertheless, LNC473 knockdown resulted in the inhibition of APAF1 levels established by qPCR and RT-PCR assays (Figure 6B; Figure S5A). Consistently, IF and WB assays confirmed the same manner of APAF1 protein expression in HCT116 and SW480 cells, as shown in Figures 6C, 6D, and S5B. Meanwhile, the loss- and gain-of-function experiments demonstrated that mRNA and protein levels of APAF1 substantially recovered in HCT116 and SW480 cells treated with pcDH-LNC473 vector and miR574-5p or miR15b-5p mimic by qPCR, WB, and RT-PCR assays, as illustrated in Figures 6E, 6F, and S5C. Then, we explored the expression of APAF1 downstream modulators and revealed that LNC473 overexpression facilitated the cleaved active form of CASP9 and CASP3, subsequently led to an increased expression ratio of Bax/Bcl-2 and thereby inducing apoptosis of CRC cells. Conversely, LNC473 depletion resulted in suppressing the protein cleavage of CASP9 and CASP3 and reducing the expression ratio of Bax/Bcl-2 in both HCT116 and SW480 cells, as shown in Figure 6D. Thus, our results suggested that the APAF1 protein, involved in CASP9-CASP3/Bax/Bcl-2 apoptotic signaling pathway, was regulated by LNC473 via competitive sponging of miR574/15b-5p in CRC cells.
Figure 6.
LNC473-miR574/miR15b-APAF1 Signaling Axis in HCT116 and SW480 Cells
(A) IF and ISH combination assay revealing the co-localization of APAF1 protein with included ncRNAs in HCT116 cells. Scale bars, 5 μm. (B and C) The levels of APAF1 mRNA (B) and protein (C) were determined after interfering LNC473 expression in HCT116 and SW480 cells by qPCR and IF assays. Scale bars, 20 μm. (D) The expression of apoptosis-related proteins including APAF1 was detected after interfering LNC473 expression in HCT116 and SW480 cells by western blot assay. (E and F) Rescue experiments showing the APAF1 levels in HCT116 and SW480 cells with exposure to the co-transfection of pcDH-LNC473 vector and miR574-5p or miR15b-5p mimic by qPCR (E) and western blot (F) assays. (G) Pattern diagram of APAF1-CDS and APAF1-IRES-CDS vectors. (H and I) APAF1 protein expression was determined in HCT116 and SW480 cells treated with APAF1-IRES-CDS vector, or pcDH-LNC473 and APAF1-IRES-CD co-transfection by western blot (H) and IF assays (I). Scale bars, 5 μm. (J) The percentage (%) of cell apoptosis in cells upon co-overexpressing APAF1-CDS or APAF1-IRE-CDS and LNC473 as assayed by flow cytometry. All tests were performed at least three times. Data were expressed as mean ± SD. ns (nonsignificant), p > 0.05; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
To further investigate the roles of LNC473 in APAF1 IRES-mediated translation in CRC cells, we first successfully constructed APAF1-IRES-CDS (with IRES sequence) and APAF1-CDS (without IRES sequence) vectors for subsequent transfection into HCT116 and SW480 cells, as shown in Figure 6G. As expected, the WB assay verified that the presence of the IRES element significantly increased APAF1 protein expression by comparing cells transfected with the two vectors, respectively, exactly suggesting the role of IRES in enhancing APAF1 translation activities. Deeply, pcDH-LNC473, co-transfected with APAF1-IRES-CDS, led to a more dramatically enhanced APAF1 expression than did only APAF1-IRES-CDS; instead, no significant alteration with APAF1-CDS was detected by WB (Figure 6H) and IF assays (Figure 6I; Figure S5D). Similarly, the cell apoptosis rates appeared to be significantly promoted relying on the LNC473-overexpressing and APAF1-IRES exposure (Figure 6J), suggesting a novel APAF1 IRES-dependent function regulated by LNC473 in CRC cells.
LNC473 Inhibits Cell Proliferation by Exposing APAF1 IRES In Vivo
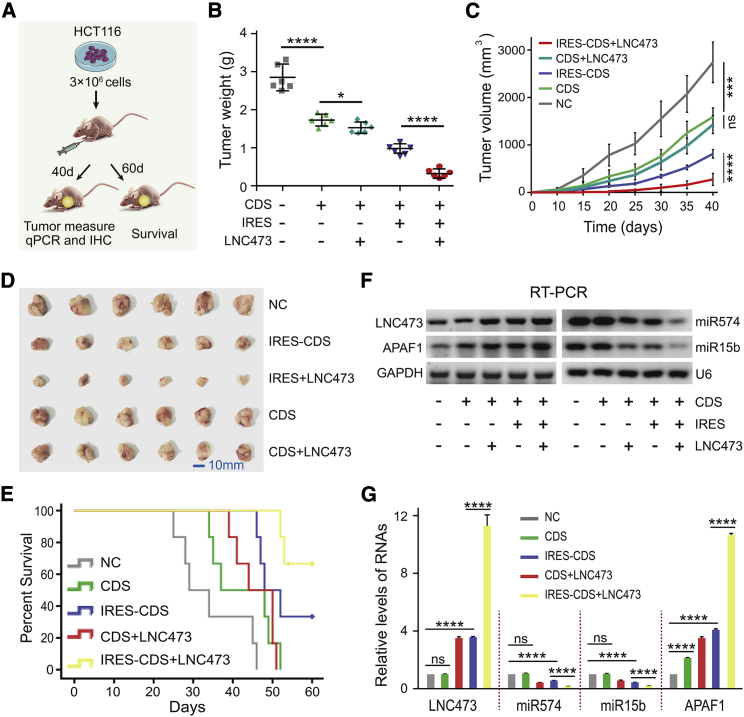
We further evaluated the biological effects of LNC473 on the CRC cells in vivo. Xenografts in BALB/c nude mice were established by inoculating HCT116 cells treated with empty vector (NC), APAF1-CDS vector, APAF1-IRES-CDS vector, pcDH-LNC473 + APAF1-CDS vectors, and pcDH-LNC473 + APAF1-IRES-CDS vectors at the right armpits, respectively (Figure 7A). We sacrificed six mice of each group and removed the xenograft tumors at day 40 post-implantation, and meanwhile followed up the other six mice until death or at day 60 for each group. As a result, the weight and volume of xenograft tumors developed from the inoculated HCT116 cells treated with APAF1-IRES-CDS appeared to have more noticeable decrease than that with APAF1-CDS (without IRES element) or NC. Furthermore, the tumor appeared to have the most remarkable growth inhibition in the group where the HCT116 cells were co-transfected with the pcDH-LNC473 and APAF1-IRES-CDS, compared with the group treated only by APAF1-IRES-CDS and other intervention groups (Figures 7B–7D). The findings above indicate that the impaired tumorigenic potential of CRC cells is due to the specific targeting of APAF1 IRES by LNC473 overexpression in the xenograft mice model. To further confirm the view, we compared the survival times of xenograft mice and found a remarkably prolonged survival time in the group treated by pcDH-LNC473 and APAF1-IRES-CDS co-transfection (Figure 7E), suggesting that the interaction of LNC473 with APAF1 IRES played a synergistic role on improving the prognosis in CRC. Afterward, to verify whether LNC473 promoted IRES-mediated APAF1 expression by sponging miR574-5p and miR15b-5p, RT-PCR and qPCR assays were performed to detect miR574-5p and miR15b-5p levels in vivo. The results confirmed the significant suppression of miR574-5p and miR15b-5p expression in the presence of pcDH-LNC473 and APAF1-IRES-CDS (Figures 7F and 7G), suggesting an IRES-independent function regulated by LNC473 restraining miR574-5p and miR15b-5p. Collectively, the results demonstrate that LNC473, as an endogenous sponge of miR574-5p and miR15b-5p, inhibits the cell proliferation and shortens the survival time in vivo by targeting the APAF1 IRES-mediated translation in CRC tumorigenesis.
Figure 7.
LNC473-miR574/miR15b-APAF1 Signaling Axis in Vivo
(A) Schematic diagram of xenografts in BALB/c nude mice by inoculating HCT116 cells. (B and C) Weights (B) and volumes (C) of tumors in xenografts from different groups. (D) Representative images of excised tumors are shown. Scale bar, 10 mm. (E) Kaplan-Meier curves showing the overall survival of nude mice from different groups. (F and G) RT-PCR (F) and qPCR (G) assays detecting miR574-5p and miR15b-5p levels upon improving LNC473 or APAF1 IRES expression in vivo. All tests were performed at least three times. Data are shown as mean ± SD. n = 6 for each group. ns, p > 0.05; ∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Discussion
Herein, we demonstrated that LNC473 is critically involved in the tumorigenesis, progression, and prognosis in CRC by competitively sponging miR574-5p and miR15b-5p, yet regulates APAF1 IRES-mediated translation initiation. APAF1, a critical apoptotic regulator of cell cycle arrest and apoptosis in normal enterocytes and CRC cells,23,24 is essential for determining the CRC tumor formation and cell fate decisions.25, 26, 27 In contrast, miR574-5p, which is upregulated in a variety of cancers28, 29, 30, 31 during carcinogenesis, was considered to be a master oncogene and even act as a serum biomarker for early-stage NSCLC.32 Similar to miR574-5p, the oncogenic functions of miR15b-5p in tumor development and progression were increasingly investigated.33, 34, 35, 36 Thus, our findings placed LNC473 and its direct target APAF1 (IRES-dependent translation) in the center of the master MRE networks that govern cell fates. Briefly, in normal enterocytes, LNC473, as well as APAF1 (IRES-dependent translation), is highly expressed, whereas the abundance of miR574-5p and miR15b-5p (interacting with MREs of LNC473) is at a lower level. In CRC cells and tissues, the abnormal downregulation of LNC473 attenuates its competing endogenous sponge effect on miR574-5p and miR15b-5p, causing them to be released and upregulated, which in turn leads to the inactivation of APAF1 IRES and blockage of APAF1 protein translation. Such blockage could be recovered and even reversed by exposure on synergistic overexpression of LNC473 and APAF1-IRES-CDS (with an IRES sequence) in vitro and in vivo, subsequently reversing the expression patterns of the above genes.
Although IRES-mediated translation was investigated in a variety of viral models, it has not been extensively studied in human cellular transcripts until recently.37 Cellular IRES-mediated translation can be substantially increased by multiple diverse physiological, pathophysiological, and stress conditions (e.g., endoplasmic reticulum stress, hypoxia, nutrient limitation, mitosis), with an inhibition of the cap-dependent translation.37 Note that IRES elements can be found in a vast majority of cellular mRNAs (e.g., c-myc, APAF1, FGF, XIAP, p53, VEGF), whose encoded proteins are master regulators of cell survival, proliferation, or apoptosis, and they tend to protect cells from stress, alternatively, and induce programmed cell death.37 APAF1 interacts with procaspase-9, leading to cleavage and activation of this initiator caspase, which exerts a central role in the induction of cell apoptosis. Importantly, the 5′ UTR of APAF1 mRNA is relatively long, G-C rich, and has the potential to fold into IRES, which is effectively active in a range of human cancer cells, including HEK293T, HeLa, human hepatic carcinoma cell HepG2, and breast cancer MCF7 cells.38 The IRES-mediated translation was maintained at a constant cellular level of APAF1 protein, even under conditions where cap-independent translation was compromised.20
Currently, in contrast to an improved perception of the mechanism of viral IRES element action, exceedingly little is known about the underlying mechanism and the importance of cellular IRES-mediated translation.21 In this study, we focused on a novel mechanism regarding the structure and function of cellular APAF1 IRES and revealed a regulation pattern negatively controlled by miR574-5p or miR15b-5p. The relatively higher affinity between the APAF1 5′ UTR and miR574-5p and/or miR15b-5p was evaluated by calculating MFE. Furthermore, remarkably decreased APAF1 IRES luciferase activities were observed after co-transfection of the APAF1 IRES-WT vector and miR574-5p and/or miR15b-5p mimics, which were abrogated when the IRES was mutated, demonstrating an IRES-dependent pattern of miR574-5p and miR15b-5p specifically modulating APAF1. It is striking that in CRC cells, a dramatically enhanced APAF1 expression was observed in the co-existence of pcDH-LNC473 and APAF1-IRES-CDS vectors. However, rescue experiments confirmed the substantial recovery of APAF1 mRNA and protein levels after exposure to the pcDH-LNC473 vector and miR574-5p or miR15b-5p mimic, which revealed the critical role of LNC473 in directly restraining miR574-5p and miR15b-5p, causing the targeted facilitation of APAF1 IRES-dependent translation.
All previous studies on miR574-5p or miR15b-5p modulation of gene expression investigated binding to the 3′ UTR of target mRNA and promotion of mRNA degradation, followed by inhibition of the process of translation.31,39, 40, 41 Interestingly, our results established that the IRES activity of human APAF1 can be suppressed by existing miRNAs (miR574-5p, miR15b-5p), and the process could be inhibited by LNC473, an endogenous miRNA sponge. This novel mechanism might provide a useful strategy for intervention seeking to alter the development and progression of CRC. Increasing numbers of studies have provided evidence that lncRNAs act as crucial molecules in the initiation and progression of the tumor by their involvement in transcriptional and post-transcriptional regulation processing.42 Several lncRNAs (e.g., LNC473, HOTAIR, and MALAT1) were reported to dysregulate and exert their functions as a tumor promoter or suppressor to regulate cell proliferation, apoptosis, migration, and invasion.10,43,44. Notably, in contrast to the results of recent studies showing the oncogenic effects of LNC473 in NSCLC and HCC,7,8 our findings demonstrated that LNC473 functions as a suppressor in CRC cells and tissues, whose expression and function are critical for both the tumor progression and the prognosis of patients. It is noteworthy that LNC473 overexpression in our study inhibited cell proliferation and colony formation capacity. Furthermore, it blocked the G1/G0 phase of the cell cycle and promoted cell apoptosis through the CASP9-CASP3/Bax/Bcl-2 pathway accompanied by the suppression of miR574-5p or miR15b-5p expression, and vice versa in CRC cells. Meanwhile, the expression patterns explicitly showed a significant impact on the prognostic survival times in CRC patients. The elevated LNC473 expression was associated with better OS and DFS in our cohort. However, the higher expression levels of miR574-5p and miR15b-5p were significantly associated with poor clinical outcomes. Even stratified prognostic analysis confirmed the synergistic impact on survival times of these indicators in CRC patients, suggesting a negative correlation or specifically direct binding among them. Zhu et al.9 reported that LNC473 served as a sponge of miR195, and, through regulating the miR195/IKKα axis, it promoted the initiation and progression of Hodgkin’s lymphoma. Our results further revealed that the LNC473 probes with MREs specifically sponged the endogenous miR574-5p and miR15b-5p through particular sequences. Nevertheless, it is noteworthy that such a pattern was entirely abrogated by mutations of the binding sequences in CRC cells, showing competitive behaviors to endogenous RNAs (ceRNAs) through direct binding regulation.
In summary, our research provides insights into a novel molecular mechanism for the involvement of LNC473 in regulating the initiation and pathogenesis in CRC. By sponging miR574-5p and miR15b-5p in CRC cells and tissues, LNC473 functions as a tumor suppressor mediating CRC cell proliferation inhibition and apoptosis promotion by modifying the translation initiation activity of APAF1 IRES in vitro and in vivo. Our study is the first report on the involvement of the signaling axis LNC473-miR574/miR15b-APAF1 IRES in CRC development or progression. Therefore, our work is beneficial to better understanding the potential applications of lncRNAs, miRNAs, and cellular IRE-dependent translation networks in CRC prevention and clinical therapy.
Materials and Methods
Collection of CRC Patient Specimens
In our study, we obtained 157 paired CRC tissues and matched adjacent-tumor control tissues from the First Hospital of China Medical University and Cancer Hospital of China Medical University from September 2014 to September 2015. All enrolled patients were diagnosed histopathologically with clear CRC. We excluded those patients with a history of other malignant tumors, or without receiving chemotherapy or radiation before surgery. All patients signed a written informed consent form before being enrolled in this study. The Medical Ethics Committee of China Medical University approved this study. The collected specimens were stored at −80°C before using. The clinicopathological characteristics of included CRC patients are presented in Table S7.
lncRNA Microarray Assays
The GeneChip Human Transcriptome Array 2.0 (HTA 2.0, Affymetrix, USA) contains probes covering coding and non-coding transcripts, with more than 6 million distinct human genomes detected by Shanghai OE Biotech Technology (Shanghai, China). We obtained the accession no. GEO: GSE137511 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137511) from the GEO website for raw data deposition. We selected databases, including Ensembl, UCSC, NONCODE, RefSeq, and lncRNAdb, to annotate the detected transcripts.
Gene Expression Analysis
The raw data were normalized based on a robust multichip analysis (RMA) algorithm as explored with GeneSpring software (version 13.1; Agilent Technologies). Differentially expressed genes (DEGs) were screened through log2 fold change as well as a p value obtained from a t test. The threshold of upregulated and downregulated DEGs was set at p ≤0.05. KEGG analysis was applied to determine the critical roles of these DEGs played in relative pathways. Hierarchical clustering was performed to reveal the relationships of these differentially expressed lncRNAs, miRNAs, and mRNA in these included samples.
Cell Culture
The human normal intestinal epithelial cell line HIEC and CRC cells, including HCT116, SW480, RKO, Caco-2, LOVO, and HT29 cells, were purchased from Peking Union Medical College Cell Resource Center (PUMCCRC, Beijing, China). HCT116 and SW480 cells were cultured in RPMI 1640 medium (BI, Jerusalem, Israel), and HT29 cells were grown in L15 medium (HyClone, Logan, UT, USA) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco, NY, USA). RKO, LOVO, HIEC, and HEK293T cells (from PUMCCRC) were maintained in DMEM (Invitrogen, NY, USA) containing 10% (v/v) FBS. Caco-2 cells were grown in DMEM containing 20% (v/v) FBS. All of the cells were maintained at 37°C with 100 U/mL penicillin/streptomycin (Invitrogen, CA, USA) in a humidified atmosphere with 5% CO2. In this study, an absence of mycoplasma or bacterial contamination of the cells was detected.
Cell Transfection
The LNC473 (NR_026860.1) knockdown vector (short hairpin RNA [shRNA]473#1, shRNA473#2), the LNC473 overexpression vector (pcDH-LNC473), and their corresponding NC vectors were purchased from GenePharma (Shanghai, China). The mimics of hsa-miR574-5p (miRBase: MIMAT0004795) and hsa-miR15b-5p (miRBase: MIMAT0000417) were obtained from Ribobio (Guangzhou, China). The APAF1-IRES-WT or APAF1-IRES-Mut1/2/3 luciferase reporters and the APAF1-CDS (without IRES sequence) or APAF1-IRES-CDS vector (with IRES sequence) were constructed by Genewiz (GenBank: NM_181861.2, Suzhou, China). The shRNA sequences of LNC473 and luciferase reporter sequence for IRES activity detection are summarized in Tables S8 and S9. Cells were cultured on six-well plates with a density of 60%–70%, and then transiently transfected using Lipofectamine 2000 (Invitrogen, USA) or stably transfected by lentiviral transduction particles (MOI of 10:1) with 5 μg/mL Polybrene. Cells were collected after transfection at 24 or 48 h for further experiments.
RT-PCR and qPCR Assays
Briefly, the total RNAs and miRNAs were extracted with TRIzol reagent (Invitrogen, CA, USA) and a miRNA extraction kit (ComWin Biotech, Beijing, China), respectively. The reaction of RT-PCR of total RNAs (Toyobo, Osaka, Japan) and miRNAs (Takara, Dalian, China) was examined using reverse transcription kits, and the detailed reaction conditions for each indicator are illustrated in Table S10. For quantitative real-time PCR assay we conducted the reverse transcription with an RT kit QuantStudio 3 (Thermo Fisher Scientific, MA, USA). Thereafter, qPCR was determined (in triplicate) using SYBR Green mix (Toyobo, Japan). The normalized relative expression of the indicator was analyzed with the 2−ΔΔCt method with the reference genes GAPDH and/or U6. The primers used for RT-PCR and qPCR amplification are listed in Table S11.
WB Assay
In brief, an equal amount of protein was separated with 10%–12% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to 0.22-μm polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA). Thereafter, the membranes were blocked with 5% milk and immunoblotted with primary antibodies. The antibodies included APAF1 (1:500; Proteintech, Chicago, IL, USA), GAPDH (1:2,000; Zsbio, Beijing, China), and apoptotic-associated proteins such as pro- or cleaved-CASP9, cleaved-CASP3, Bcl2, and Bax (1:1,000; Affinity, OH, USA).
Cell Co-localization and IF Assay
Briefly, cells were cultured in 24-well plates with glass coverslips, fixed, and stained with indicated antibodies as previously described.45 An RNA ISH assay was performed according to the kit instructions, except for the counterstaining with 0.1% hematoxylin. Thereafter, the cells were stained with the indicated APAF1 antibody, Alexa Fluor anti-rabbit immunoglobulin G (IgG), and DAPI. The antibodies used for IF detection included APAF1 (#21710-1-AP, 1:100, Proteintech, USA) and Alexa Fluor anti-rabbit IgG (#SA00007-2, 1:200, Proteintech, USA). The cell nuclei were counterstained with DAPI (Beyotime, Shanghai, China). Finally, a Nikon C2 plus confocal microscope was used to obtain images under ×40 magnification (Nikon, Tokyo, Japan).
Cell Proliferation and Colony Formation Assays
For the cell viability assay, transfected HCT116 and SW480 cells were seeded in 96-well plates (100 μL/well) at a density of 5 × 103 cells/well for 24 h. Then, CCK8 assay (BestBio, Shanghai, China) results were determined at 24, 48, 72, and 96 h. The optical density 450 (OD450) absorbance values were obtained in triplicate and replicated six times. For the colony formation assay, transfected cells were seeded into six-well plates (2 × 103 cells/well) for 2 weeks of culturing. Visible colonies were washed with 1× PBS, fixed with 4% paraformaldehyde for 30 min, and stained with 0.1% hematoxylin for 10 min. Finally, the number of colonies with more than 50 cells was calculated in this study.
Flow Cytometry Assays
The transfected cells (1 × 106) were harvested and washed twice with cold 1× PBS. For the cell cycle assay, collected cells were fixed with 70% ethanol and treated with 20 μL of RNase A (2 μg/mL) and incubated at 37°C for 30 min. Then cells were stained with propidium iodide (PI; 50 μg/mL, BD Biosciences, San Jose, CA, USA) for 1 h at 4°C. For cell apoptosis analysis, cells were resuspended with 400 μL of annexin V binding buffer and incubated with 5 μL of annexin V-phycoerythrin (PE) for 10 min and 5 μL of 7-aminoactinomycin D (7-AAD) for 1 min in the darkroom at 4°C. Finally, cells were detected using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
IHC and ISH Assays
TMAs and IHC assays were performed according to a previous publication with some modification.46 Briefly, each 4-μm TMA section was deparaffinized and rehydrated in a graded ethanol series. After washing with PBS, the section was incubated with a monoclonal antibody against APAF1 (dilution 1:200, Proteintech, Chicago, IL, USA) overnight at 4°C. Thereafter, the section was incubated in the secondary antibody for 30 min at 37°C, and then horseradish peroxidase-coupled streptavidin (Dako, Glostrup, Denmark) was added and the section was stained with DAB (3,3′-diaminobenzidine). For the RNA ISH assay, we determined the indicators according to the manufacturer’s protocol (Boster, Wuhan, China). Briefly, a TMA section was incubated with digoxin-labeled probes solutions overnight at 37°C. Then, it was exposed in the streptavidin-peroxidase reaction system and stained with DAB. Finally, the expression of these indicators, including LNC473, miR574-5p, miR15b-5p, and APAF1, were counted under a microscope (Nikon, Tokyo, Japan) for subsequent analysis. The specific sequences of the probe of these indicators are listed in Table S12.
Luciferase Reporter Assay
HEK293T and HCT116 cells were seeded into 96-well plates overnight before transfection. Then, cells were co-transfected with luciferase reporter vectors (APAF1-IRES-WT, APAF1-IRES-Mut1 [miR574-5p], APAF1-IRES-Mut2 [miR15b-5p], APAF1-IRES-Mut3 [miR574-5p and miR15b-5p]) (100 ng/well) and hsa-miR-574-5p and/or hsa-miR-15b-5p mimics (50 nM/well) by Lipofectamine 2000 (Invitrogen, CA, USA). At 48 h post-transfection, the cells were collected and lysed in passive lysis buffer (Promega, WI, USA). The luciferase activities of the cell lysates were determined in a Nano-Glo reporter assay system.
RIP Assay
3,000 ng of each RNA extract was incubated with 200 μM of the biotin-labeled probe (Sangon, Shanghai, China) of LNC473-MRE (containing the MRE sequence) with miR-574 (473-WT1 and 473-Mut1) or miR-15b (473-WT2 and 473-Mut2). Then, the total volume was adjusted to 1 mL with binding buffer and the mixture was incubated at 30°C for 60 min. 20 μL of agarose beads was added and rotated slowly overnight at 4°C. After washing five times, the mixture was heated with 95°C for 15 min. Purified RNA was obtained for further RT-PCR and qPCR assays.47,48 Additionally, the specific sequence of probes used in this study is shown in Table S13.
Tumor Formation Assay In Vivo
Sixty male BALB/c-nu mice (5 weeks old) obtained from Hua Fukang Biological Technologies (Beijing, China) were acclimatized to the new environment for 1 week. HCT116 cells (3 × 106) were implanted in the right armpit region. The mice’s weight and tumor growth were examined every 5 days. The tumor volume was calculated by the equation V = 0.5 × D × d 2, where V is equal to the tumor volume, and D and d represent the longitudinal diameter and latitudinal diameter, respectively. Forty days post-implantation, six mice of each group were sacrificed and the xenograft tumors were removed and measured. The rest of the mice in each group were followed up until death or at the 60th day of observation. The protocols were approved by the Committee on the Ethics of Animal Experiments of China Medical University. This study complied with the Regulations of Experimental Animal Administration issued by the Ministry of Science and Technology of China.
Statistical Analysis
In the present study, all of the data analyses were performed using SPSS 19.0 software package (SPSS, Chicago, IL, USA). The Student’s t test (two-tailed) and Wilcoxon t test were used to compare the significant differences of the paired and unpaired continuous variables between groups. A Pearson’s χ2 or Fisher’s exact test was conducted to analyze the expression or distribution frequency differences of the variables. The Kaplan-Meier method and multivariate Cox proportional hazard regression analysis were selected to estimate the potential prognosis of associated indicators. The data are presented as mean ± standard deviation (SD) or median (quartile). Statistical significance was established when p < 0.05 in all tests.
Author Contributions
M.W., R.Z., and H.W. conceived and designed the project. X.H., Y.L., B.F., and Q.C. designed and supervised experiments conducted in the laboratories. X.H., Z.W., T.S., Y.Q., and W.Q. performed experiments and/or data analyses; M.W., H.W., and H.Z. contributed reagents/analytic tools and/or grant support; M.W., H.W., X.H., and B.F. wrote the paper. All authors discussed the results and commented on the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (nos. 31828005, 81872905, 81673475); the National Natural Science Foundation of China and Liaoning Joint Fund Key Program (no. U1608281); and by the Liaoning Revitalization Talents Program (XLYC1807155), China Postdoctoral Science Foundation (2019T120225) and Shenyang S&T Projects (19-109-4-09).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.07.009.
Contributor Information
Rui Zhang, Email: zhangrui@cancerhosp-ln-cmu.com.
Minjie Wei, Email: mjwei@cmu.edu.cn.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Burke C., Kaul V., Pohl H. Polyp resection and removal procedures: insights from the 2017 Digestive Disease Week. Gastroenterol. Hepatol. (N. Y.) 2017;13(19, Suppl 2):1–24. [PMC free article] [PubMed] [Google Scholar]
- 3.Shen X., Guo H., Xu J., Wang J. Inhibition of lncRNA HULC improves hepatic fibrosis and hepatocyte apoptosis by inhibiting the MAPK signaling pathway in rats with nonalcoholic fatty liver disease. J. Cell. Physiol. 2019;234:18169–18179. doi: 10.1002/jcp.28450. [DOI] [PubMed] [Google Scholar]
- 4.Xiong T., Li J., Chen F., Zhang F. PCAT-1: a novel oncogenic long non-coding RNA in human cancers. Int. J. Biol. Sci. 2019;15:847–856. doi: 10.7150/ijbs.30970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu T., Zhang K., Shi J., Huang B., Wang X., Qian K., Ma T., Qian T., Song Z., Li L. MicroRNA-940 inhibits glioma progression by blocking mitochondrial folate metabolism through targeting of MTHFD2. Am. J. Cancer Res. 2019;9:250–269. [PMC free article] [PubMed] [Google Scholar]
- 6.Sekar D., Mani P., Biruntha M., Sivagurunathan P., Karthigeyan M. Dissecting the functional role of microRNA 21 in osteosarcoma. Cancer Gene Ther. 2019;26:179–182. doi: 10.1038/s41417-019-0092-z. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Yang F., Li X., Gong Z.J., Wang L.W. Long noncoding RNA LNC473 inhibits the ubiquitination of survivin via association with USP9X and enhances cell proliferation and invasion in hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2018;499:702–710. doi: 10.1016/j.bbrc.2018.03.215. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z., Li J.L., Lin S., Cao C., Gimbrone N.T., Yang R., Fu D.A., Carper M.B., Haura E.B., Schabath M.B. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J. Clin. Invest. 2016;126:2267–2279. doi: 10.1172/JCI85250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu S.B., Fu W., Zhang L.Y., Fu K., Hu J.H., Jia W., Liu G. LINC00473 antagonizes the tumour suppressor miR-195 to mediate the pathogenesis of Wilms tumour via IKKα. Cell Proliferat. 2018;51:e12416. doi: 10.1111/cpr.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi C., Yang Y., Yu J., Meng F., Zhang T., Gao Y. The long noncoding RNA LINC00473, a target of microRNA 34a, promotes tumorigenesis by inhibiting ILF2 degradation in cervical cancer. Am. J. Cancer Res. 2017;7:2157–2168. [PMC free article] [PubMed] [Google Scholar]
- 11.Abdollahzadeh R., Daraei A., Mansoori Y., Sepahvand M., Amoli M.M., Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J. Cell. Physiol. 2019;234:10080–10100. doi: 10.1002/jcp.27941. [DOI] [PubMed] [Google Scholar]
- 12.Li H., Mao S., Wang H., Zen K., Zhang C., Li L. MicroRNA-29a modulates axon branching by targeting doublecortin in primary neurons. Protein Cell. 2014;5:160–169. doi: 10.1007/s13238-014-0022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long J.M., Maloney B., Rogers J.T., Lahiri D.K. Novel upregulation of amyloid-β precursor protein (APP) by microRNA-346 via targeting of APP mRNA 5′-untranslated region: implications in Alzheimer’s disease. Mol. Psychiatry. 2019;24:345–363. doi: 10.1038/s41380-018-0266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James C.C., Smyth J.W. Alternative mechanisms of translation initiation: an emerging dynamic regulator of the proteome in health and disease. Life Sci. 2018;212:138–144. doi: 10.1016/j.lfs.2018.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao G., Dhar S., Bedford M.T. PRMT5 regulates IRES-dependent translation via methylation of hnRNP A1. Nucleic Acids Res. 2017;45:4359–4369. doi: 10.1093/nar/gkw1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji B., Harris B.R.E., Liu Y., Deng Y., Gradilone S.A., Cleary M.P., Liu J., Yang D.Q. Targeting IRES-mediated p53 synthesis for cancer diagnosis and therapeutics. Int. J. Mol. Sci. 2017;18:E93. doi: 10.3390/ijms18010093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaunay S., Rapino F., Tharun L., Zhou Z., Heukamp L., Termathe M., Shostak K., Klevernic I., Florin A., Desmecht H. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J. Exp. Med. 2016;213:2503–2523. doi: 10.1084/jem.20160397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreucci E., Bianchini F., Biagioni A., Del Rosso M., Papucci L., Schiavone N., Magnelli L. Roles of different IRES-dependent FGF2 isoforms in the acquisition of the major aggressive features of human metastatic melanoma. J. Mol. Med. (Berl.) 2017;95:97–108. doi: 10.1007/s00109-016-1463-7. [DOI] [PubMed] [Google Scholar]
- 19.Cobbold L.C., Spriggs K.A., Haines S.J., Dobbyn H.C., Hayes C., de Moor C.H., Lilley K.S., Bushell M., Willis A.E. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell. Biol. 2008;28:40–49. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coldwell M.J., Mitchell S.A., Stoneley M., MacFarlane M., Willis A.E. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H., Liu T., Yi S., Gu L., Zhou M. Targeting MYCN IRES in MYCN-amplified neuroblastoma with miR-375 inhibits tumor growth and sensitizes tumor cells to radiation. Mol. Oncol. 2015;9:1301–1311. doi: 10.1016/j.molonc.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Xin S., Wu H., Xing C., Duan L., Sun W., Hu X., Lin R., Zhang F. High expression of microRNA-31 and its host gene LOC554202 predict favorable outcomes in patients with colorectal cancer treated with oxaliplatin. Oncol. Rep. 2018;40:1706–1724. doi: 10.3892/or.2018.6571. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., Guo Y., Wang M., Dong H., Zhang J., Zhang L. Quercetrin from Toona sinensis leaves induces cell cycle arrest and apoptosis via enhancement of oxidative stress in human colorectal cancer SW620 cells. Oncol. Rep. 2017;38:3319–3326. doi: 10.3892/or.2017.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jagadish N., Parashar D., Gupta N., Agarwal S., Purohit S., Kumar V., Sharma A., Fatima R., Topno A.P., Shaha C., Suri A. A-kinase anchor protein 4 (AKAP4) a promising therapeutic target of colorectal cancer. J. Exp. Clin. Cancer Res. 2015;34:142. doi: 10.1186/s13046-015-0258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang J., Yang F., Wang Y., Wang Y., Xue G., Mei Q., Wang F., Sun S. MicroRNA-23a antisense enhances 5-fluorouracil chemosensitivity through APAF-1/caspase-9 apoptotic pathway in colorectal cancer cells. J. Cell. Biochem. 2014;115:772–784. doi: 10.1002/jcb.24721. [DOI] [PubMed] [Google Scholar]
- 26.Zlobec I., Minoo P., Baker K., Haegert D., Khetani K., Tornillo L., Terracciano L., Jass J.R., Lugli A. Loss of APAF-1 expression is associated with tumour progression and adverse prognosis in colorectal cancer. Eur. J. Cancer. 2007;43:1101–1107. doi: 10.1016/j.ejca.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 27.Umetani N., Fujimoto A., Takeuchi H., Shinozaki M., Bilchik A.J., Hoon D.S.B. Allelic imbalance of APAF-1 locus at 12q23 is related to progression of colorectal carcinoma. Oncogene. 2004;23:8292–8300. doi: 10.1038/sj.onc.1208022. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z., Li X., Xiao Q., Wang Z. miR-574-5p mediates the cell cycle and apoptosis in thyroid cancer cells via Wnt/β-catenin signaling by repressing the expression of Quaking proteins. Oncol. Lett. 2018;15:5841–5848. doi: 10.3892/ol.2018.8067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji S., Ye G., Zhang J., Wang L., Wang T., Wang Z., Zhang T., Wang G., Guo Z., Luo Y. miR-574-5p negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and the development of colorectal cancer. Gut. 2013;62:716–726. doi: 10.1136/gutjnl-2011-301083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q., Li X., Guo Z., Xu F., Xia J., Liu Z., Ren T. MicroRNA-574-5p was pivotal for TLR9 signaling enhanced tumor progression via down-regulating checkpoint suppressor 1 in human lung cancer. PLoS ONE. 2012;7:e48278. doi: 10.1371/journal.pone.0048278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyers-Needham M., Ponnusamy S., Gencer S., Jiang W., Thomas R.J., Senkal C.E., Ogretmen B. Concerted functions of HDAC1 and microRNA-574-5p repress alternatively spliced ceramide synthase 1 expression in human cancer cells. EMBO Mol. Med. 2012;4:78–92. doi: 10.1002/emmm.201100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foss K.M., Sima C., Ugolini D., Neri M., Allen K.E., Weiss G.J. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J. Thorac. Oncol. 2011;6:482–488. doi: 10.1097/JTO.0b013e318208c785. [DOI] [PubMed] [Google Scholar]
- 33.Sun L.N., Zhi Z., Chen L.Y., Zhou Q., Li X.M., Gan W.J., Chen S., Yang M., Liu Y., Shen T. SIRT1 suppresses colorectal cancer metastasis by transcriptional repression of miR-15b-5p. Cancer Lett. 2017;409:104–115. doi: 10.1016/j.canlet.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Pan W.Y., Zeng J.H., Wen D.Y., Wang J.Y., Wang P.P., Chen G., Feng Z.B. Oncogenic value of microRNA-15b-5p in hepatocellular carcinoma and a bioinformatics investigation. Oncol. Lett. 2019;17:1695–1713. doi: 10.3892/ol.2018.9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen R., Sheng L., Zhang H.J., Ji M., Qian W.Q. miR-15b-5p facilitates the tumorigenicity by targeting RECK and predicts tumour recurrence in prostate cancer. J. Cell. Mol. Med. 2018;22:1855–1863. doi: 10.1111/jcmm.13469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao C., Li Y., Chen G., Wang F., Shen Z., Zhou R. Overexpression of miR-15b-5p promotes gastric cancer metastasis by regulating PAQR3. Oncol. Rep. 2017;38:352–358. doi: 10.3892/or.2017.5673. [DOI] [PubMed] [Google Scholar]
- 37.Komar A.A., Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen F.C. Alternative RNA structure-coupled gene regulations in tumorigenesis. Int. J. Mol. Sci. 2014;16:452–475. doi: 10.3390/ijms16010452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ku T., Li B., Gao R., Zhang Y., Yan W., Ji X., Li G., Sang N. NF-κB-regulated microRNA-574-5p underlies synaptic and cognitive impairment in response to atmospheric PM2.5 aspiration. Part. Fibre Toxicol. 2017;14:34. doi: 10.1186/s12989-017-0215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weng Y., Shen Y., He Y., Pan X., Xu J., Jiang Y., Zhang Q., Wang S., Kong F., Zhao S. The miR-15b-5p/PDK4 axis regulates osteosarcoma proliferation through modulation of the Warburg effect. Biochem. Biophys. Res. Commun. 2018;503:2749–2757. doi: 10.1016/j.bbrc.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 41.Luo H., Li Y., Liu B., Yang Y., Xu Z.D. MicroRNA-15b-5p targets ERK1 to regulate proliferation and apoptosis in rat PC12 cells. Biomed. Pharmacother. 2017;92:1023–1029. doi: 10.1016/j.biopha.2017.05.140. [DOI] [PubMed] [Google Scholar]
- 42.Tano K., Akimitsu N. Long non-coding RNAs in cancer progression. Front. Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tung M.C., Wen Y.C., Wang S.S., Lin Y.W., Chow J.M., Yang S.F., Chien M.H. Impact of long non-coding RNA HOTAIR genetic variants on the susceptibility and clinicopathologic characteristics of patients with urothelial cell carcinoma. J. Clin. Med. 2019;8:E282. doi: 10.3390/jcm8030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu D., Sun W.W., Li L., Ma L., Sun L., Jin X., Li T., Hou W., Wang J.H. Long noncoding RNA MALAT1 releases epigenetic silencing of HIV-1 replication by displacing the polycomb repressive complex 2 from binding to the LTR promoter. Nucleic Acids Res. 2019;47:3013–3027. doi: 10.1093/nar/gkz117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin X., Liu M.Y., Zhang D.F., Zhong X., Du K., Qian P., Yao W.F., Gao H., Wei M.J. Baicalin mitigates cognitive impairment and protects neurons from microglia-mediated neuroinflammation via suppressing NLRP3 inflammasomes and TLR4/NF-κB signaling pathway. CNS Neurosci. Ther. 2019;25:575–590. doi: 10.1111/cns.13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H., Guan S., Sun M., Yu Z., Zhao L., He M., Zhao H., Yao W., Wang E., Jin F. Ano1/TMEM16A overexpression is associated with good prognosis in PR-positive or HER2-negative breast cancer patients following tamoxifen treatment. PLoS ONE. 2015;10:e0126128. doi: 10.1371/journal.pone.0126128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He M., Wu H., Jiang Q., Liu Y., Han L., Yan Y., Wei B., Liu F., Deng X., Chen H. Hypoxia-inducible factor-2α directly promotes BCRP expression and mediates the resistance of ovarian cancer stem cells to adriamycin. Mol. Oncol. 2019;13:403–421. doi: 10.1002/1878-0261.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma M.T., He M., Wang Y., Jiao X.Y., Zhao L., Bai X.F., Yu Z.J., Wu H.Z., Sun M.L., Song Z.G., Wei M.J. miR-487a resensitizes mitoxantrone (MX)-resistant breast cancer cells (MCF-7/MX) to MX by targeting breast cancer resistance protein (BCRP/ABCG2) Cancer Lett. 2013;339:107–115. doi: 10.1016/j.canlet.2013.07.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.