Abstract
Carbapenemase-producing Klebsiella pneumoniae has been a major clinical threat worldwide because therapeutic options are limited. Although New Delhi metallo-β-lactamase (NDM) is an important carbapenemase responsible for carbapenem resistance, it is uncommon in carbapenemase-producing K. pneumoniae in China. In this study, we described strain HZW25, an NDM-7-producing K. pneumoniae strain belonging to sequence type 34 (ST34). HZW25 exhibited resistance to all β-lactams tested but was susceptible to aminoglycosides and fluoroquinolones. The whole genome of HZW25 was sequenced with Pacific Biosciences RSII SMRT technology. HZW25 was composed of one chromosomal DNA and four plasmids, and the resistance genes of HZW25 were all located on the chromosome, except blaNDM–7 was located on a conjugative plasmid belonging to type IncX3 designated P4. The results of conjugation and transformation experiments showed that blaNDM–7 could be horizontally transferred successfully from the donor strain, HZW25, to the recipient strains, E. coli J53 and E. coli DH5α. The NDM variant transposable elements of the blaNDM–7-harboring plasmid P4 were the ISL3 and IS3000 families. The upstream region of blaNDM–7 contained ΔISAba125, which was inserted near the IS5 or ΔIS5 sequence. Our study is the first report of metallo-β-lactamase NDM-7 in a carbapenemase-producing K. pneumoniae strain with ST34 in China. The emergence of NDM-producing K. pneumoniae would be troublesome during treatment using ceftazidime-avibactam. Therefore, the rapid and accurate identification of carbapenemase-producing K. pneumoniae is necessary.
Keywords: metallo-β-lactamase, NDM-7, IncX3, carbapenemase-producing Klebsiella pneumoniae, ceftazidime-avibactam
Introduction
Clinical treatment of carbapenem-resistant Enterobacteriaceae (CRE) is a critical challenge (Lee et al., 2016). Carbapenemase, which can be found as serine proteases and metalloproteinases, is responsible for carbapenem resistance. New Delhi metallo-β-lactamase (NDM) is a metallo-β-lactamase able to hydrolyze carbapenem (Khan et al., 2017). Although NDM-producing CRE has increased globally recently, the worldwide distribution and prevalence of NDM-positive strains appear to be variable between different countries and regions (Findlay et al., 2017; Khan et al., 2017; Lazaro-Perona et al., 2017; Wang et al., 2018; Perez-Vazquez et al., 2019; Wu et al., 2019). NDM-producing CRE strains have mainly spread in South Asia, the Baltans, North Africa and the Middle East (Dortet et al., 2014; Wu et al., 2019). Chinese national surveillance of carbapenem-resistant CRE in China has shown that NDM-producing CRE are less common than KPC-producing CRE, and NDM-positive strains are mainly E. coli (Zhang et al., 2017, 2018; Wang et al., 2018).
Since the first report of NDM in 2009, many variant NDMs have emerged (Yong et al., 2009; Dortet et al., 2014; Khan et al., 2017; Wu et al., 2019). Compared with NDM-1, NDM-7 has only two different amino acids, including Asp130Asn and Met154Leu, and NDM-7 has more enzymatic hydrolysis activity against carbapenem (Cuzon et al., 2013; Rahman et al., 2014). NDM-7 mainly exists in the IncX3-type plasmid and disseminates among different isolates (Chen et al., 2015). In this study, we analyzed the genomic sequence of the NDM-7-producing ST34 K. pneumoniae strain and the genetic surroundings and molecular characterization of NDM-7.
Materials and Methods
Patient and Bacterial Strain
Carbapenem-resistant K. pneumoniae was isolated from a bile sample collected from a young woman in Hangzhou First People’s Hospital in March 2017. The strain was identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS, Bruker MALDI-TOF Microflex LT/SH, Bruker Diagnostics, Germany) according to the manufacturer’s instructions. The patient had no history of traveling abroad. Informed consent was obtained for this study. The methods in this study were approved by the institutional ethics committee of Hangzhou First People’s Hospital and were carried out in accordance with the approval guidelines.
Antimicrobial Susceptibility Testing and Detection of Carbapenemase
Antimicrobial susceptibility was determined by the VITEK compact II automated microbiology system and interpreted according to the Clinical and Laboratory Standards Institute guidelines in 2018 (CLSI). The modified carbapenemase inactivation method (mCIM) and EDTA-modified mCIM (eCIM) were used to detect carbapenemase and metallo-β-lactamase as recommended by the CLSI in 2018 (CLSI, 2018).
Detection of Carbapenemase Genes
PCR was performed to screen carbapenem resistance genes, including blaKPC, blaIMP, blaVIM, blaNDM, and blaOXA–48. The amplicons were sequenced by Sanger sequencing.
Conjugation and Transformation Experiments
To evaluate the horizontal transferability of blaNDM, mixed broth mating was used as described in a previous study (Du et al., 2017). Sodium azide-resistant E. coli J53 was used as the recipient strain (donated by Professor YU, Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University). The transconjugants were selected on MacConkey agar containing 100 mg/liter sodium azide and 2 mg/liter meropenem for 24 h at 37°C. The electrotransformation assay was also performed to evaluate the dissemination of blaNDM using DH5α as the recipient strain as in a previous study (Findlay et al., 2017). The presumptive transconjugants were selected on MH agar plates supplemented with 2 mg/liter meropenem. All successful transformants were confirmed for the presence of blaNDM by PCR, and an antimicrobial susceptibility test was performed using the E-test method in parallel with the original strains and donor strains.
Whole Genome Sequencing
The DNA of HZW25 was extracted according to the manufacturer’s instructions and sequenced with Pacific Biosciences RSII SMRT technology SMRT technology (Menlo Park, CA, United States). Sequence reads were assembled using a hierarchical genome assembly process (HGAP) compiled specifically for quality trimming and de novo assembly. The graphical maps of the whole genome were converted by BLAST Ring Image Generator (BRIG). The whole-genome sequence was annotated using the Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) server available at NCBI.1 Multilocus sequence typing (MLST) analysis of HZW25 and cgMLST phylogenetic relationship analysis of all public sequences of ST34 were performed using the BacWGSTdb server with the entire genome sequence (Ruan and Feng, 2016). The antibiotic resistance genes were determined using ResFinder 3.0 with > 97% gene identity threshold (exception of β-lactamase variants with 100% identity) and 100% gene length and the comprehensive antibiotic resistance database (CARD) at https://card.mcmaster.ca/ with resistance gene identifier of > 90% identity. The virulence factors were detected using the virulence database of K. pneumoniae2 and a virulence factor database.3
Plasmid Analysis
All plasmid sequences were annotated using the PGAAP server. The plasmid replicon type was determined by PlasmidFinder4 with a 95% threshold for identity and 100% coverage. The blaNDM–7-carrying plasmid was compared to the publicly available plasmid references using BLAST at GenBank (www.ncbi.nlm.nih.gov/GenBank/). The plasmid comparisons were generated by Easyfig according to the online protocol,5 and presentations were generated by EdrawMax. The genetic environment of blaNDM–7 was analyzed and compared that of blaNDM variants.
Nucleotide Sequence Accession Number
The complete nucleotide sequence of HZW25 and four plasmids were deposited under the GenBank accession numbers CP025211–CP025215.
Results
Susceptibility Test
HZW25 was resistant to all β-lactams tested, while it was susceptible to aminoglycosides and fluoroquinolones. Carbapenemase activity was positive with mCIM and eCIM tests, suggesting the production of metallo-β-lactamase (MBL).
Antimicrobial Resistance Genes and Transfer Experiments
The acquired antibiotic resistance genes, including blaNDM–7, blaSHV–26, fosA, oqxA, and oqxB, were responsible for the resistance profile of HZW25. In addition, intrinsic antibiotic resistance genes with CARD resistance gene identifiers were also identified, including antibiotic efflux pumps of the major facilitator superfamily (MFS) (KpnE, KpnF, KpnG, KpnH, and ermR), an ATP-binding cassette (ABC) antibiotic efflux pump (msbA), regulators of the efflux pump (marA and marR), the porin membrane protein OmpK35 and a bleomycin resistance gene (BRP). There were no mutations in resistance genes on the chromosome. Only blaNDM–7 was located on the plasmid, while the other resistance genes were located on the chromosome. blaNDM–7 was successfully transferred to E. coli AzR J53 by conjugation and to E. coli DH5α by electroporation, and the transconjugants displayed resistance to broad-spectrum cephalosporins and carbapenems (Table 1). The presence of the blaNDM–7 gene in transconjugants was confirmed by PCR.
TABLE 1.
The minimum inhibitory concentration to antibiotics of transformed isolates with E-test methods (ug/mL).
| CRO | AMC | CTX | FEP | SCF | MEM | IPM | CIP | LEV | MH | ATM | TM | CN | |
| HZW25:J53 | >256 | >256 | >256 | >256 | >256 | >32 | >32 | 0.008 | 0.023 | 1 | 0.064 | 0.25 | 0.25 |
| J53 | <0.064 | 4 | <0.064 | <0.064 | <0.064 | <0.008 | 0.5 | <0.008 | 0.023 | 1 | 0.064 | 0.25 | 0.25 |
| DH5a:pHZW25 | >256 | >256 | >256 | >256 | 256 | 32 | >32 | 0.008 | 0.023 | 0.5 | 0.064 | 0.25 | 0.25 |
| DH5a | <0.064 | 2 | <0.064 | <0.064 | <0.064 | <0.008 | 0.008 | <0.008 | 0.023 | 0.5 | <0.064 | 0.25 | 0.25 |
| HZW25 | >256 | >256 | >256 | >256 | >256 | >32 | >32 | 0.25 | 0.75 | >256 | 0.5 | 0.25 | 0.25 |
CRO, Ceftriaxone; AMC, Amoxicillin-clavulanic acid; CTX, Cefotaxime; FEP, Cefepime; SCF, Sulbactam and Cefoperazone; MEM, Meropenem; IPM, Imipenem; CIP, Ciprofloxacin; LEV, Levofloxacin; MH, Minocycline; ATM, Aztreonam; TM, Tobramycin; CN, Gentamicin.
Molecular Grouping and Whole Genome Sequencing
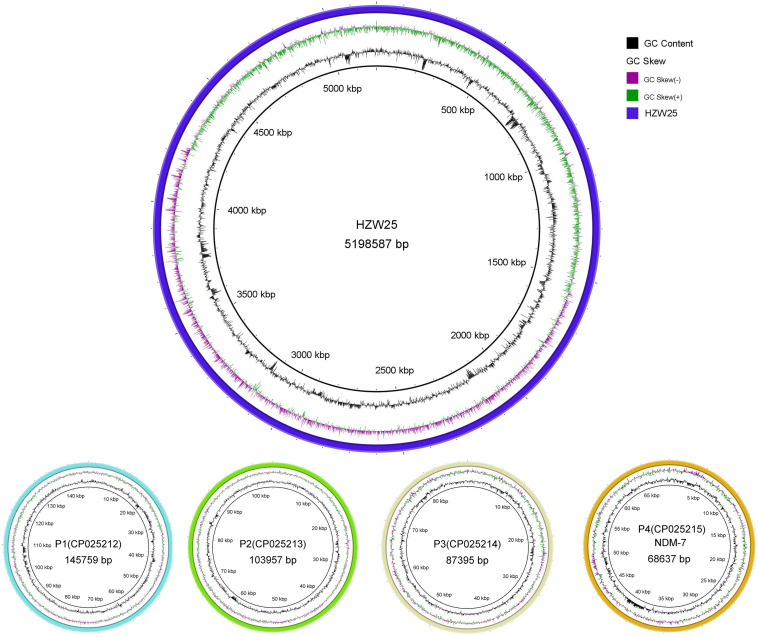
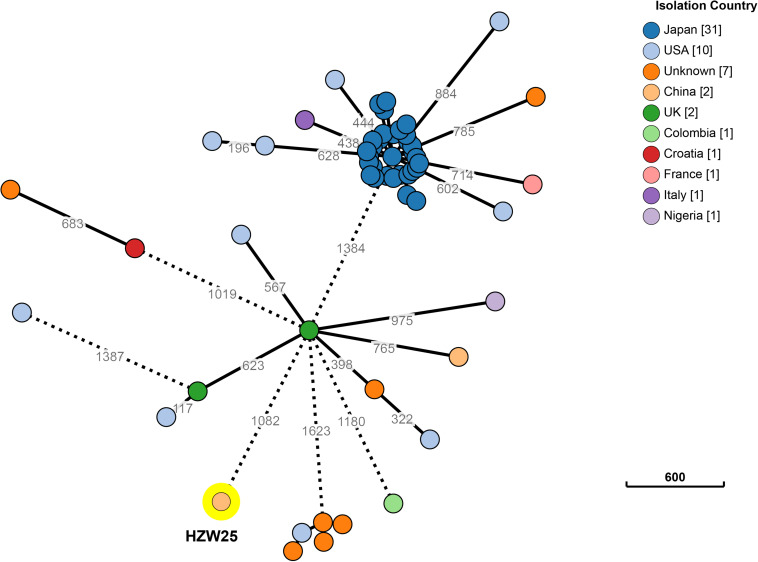
Approximately 1.2 Gb clean data was generated after whole genome sequencing with Pacific Biosciences RSII SMRT technology, providing a 221.0-fold average coverage of the genome. The data were assembled into five contigs, which contained one chromosome and four plasmids (Figure 1). HZW25 belonged to ST34 with the MLST sequence 2-3-6-1-9-7-4. The isolate was negative for the string test due to the deficiency of the rmpA and rmpA2 genes. HZW25 belonged to the K73 capsular type as determined by the wzi gene encoding the outer membrane protein of the cluster. Virulence genes were present in HZW25, including the mrkABCDFHIJ operon encoding type 3 fimbriae for biofilm formation, the fimABCDEFGHIK operon encoding type 1 fimbriae for adherence, the iutA gene encoding aerobactin, the entABCDEFS operon and the fepABCDG operon encoding enterobactin siderophore, and the iroE and iroN genes encoding salmochelin. HZW25 also had a secretion system including the clpV gene encoding T6SS-II, dotU, the impAFGHJ operon, ompA and sciN encoding T6SS-III. The four plasmids were designated P1, P2, P3 and P4, and they belonged to IncFIB(K), IncR, IncFII(pKPX1), and IncX3, respectively. The plasmids were characterized, and the results are shown in Table 2. Only P4 carried a resistance gene with blaNDM–7, and the other plasmids harbored no resistance genes. To date, there have been 57 K. pneumoniae strains belonging to ST34 worldwide, including Japan (n = 31), the United States (n = 10), China (n = 2), the United Kingdom (n = 2) and other countries (n = 12). Almost all strains were collected from humans and carried the blaSHV–26, fosA, oqxA, and oqxB resistance genes, but only HZW25 had blaNDM variants of blaNDM–7. Thirty-one strains from Japan carried blaIMP–1 and blaCTX–M–2 in addition to blaSHV–26 (Supplementary 1). All chromosomes of the ST34 K. pneumoniae strains harbored virulence clusters, such as the type I fimbriae cluster fim operon, type III fimbriae cluster mrk operon, and enterobactin siderophore cluster fep and ent operon (Supplementary 1). The phylogenetic trees of all ST34 K. pneumoniae strains in GenBank revealed that HZW25 was in a separate cluster (Figure 2), suggesting that the HZW25 strain had a long evolutionary distance from the others.
FIGURE 1.
The complete genome of K. pneumoniae strain HZW25.
TABLE 2.
The genomic information of HZW25 and four plasmids.
FIGURE 2.
The phylogenetic trees of all ST34 K. pneumoniae strains of released public sequences. The evolutionary distance showed that HZW25 was far from the other strains.
Characterization and Genetic Context of the blaNDM–7 Gene
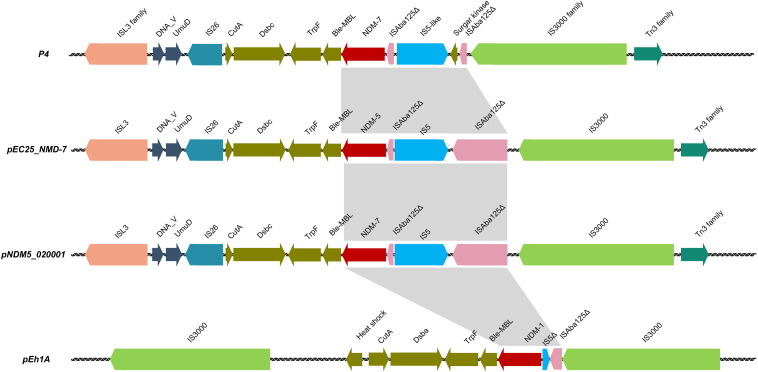
The blaNDM–7 gene was localized in plasmid P4 and could be horizontally transferred successfully from the donor strain HZW25 to the recipient strains E. coli J53 and E. coli DH5α. P4 was 68 637 bp in size with a G + C content of 45.69% and 93 open reading frames. Comparative DNA sequence analysis showed that P4 possessed an IncX3-type backbone. A comparison of the whole region between P4, pEC25_NMD-7, pNDM5_020001, and pEh1A showed that the genetic context of the regions flanking the NDM variants was similar, and the backbone of plasmids showed high degrees of conversation and similarity. The NDM variant transposable elements of P4, pEC25_NMD-7 and pNDM5_020001 were highly similar with the ISL3 family and IS3000 transposons. ΔISAba125 was upstream of the blaNDM variants and was inserted by the IS5 or ΔIS5 sequence (Figure 3).
FIGURE 3.
The comparative genetic context of blaNDM variants between P4, pEC25_NMD-7, pNDM5_020001, and pEH1A.
Discussion
Carbapenemase-resistant K. pneumoniae (CRKP) has been a significant clinical problem with very limited therapeutic options (Lee et al., 2016). Due to the high-level resistance of CRKP to almost all antibiotics and the high mortality rate of CRKP infections, carbapenemase-producing carbapenem resistance is a public health threat (Lee et al., 2016; Zhang et al., 2018).
In China, CRE strains are mainly composed of KPC-producing K. pneumoniae and NDM-producing E. coli (Zhang et al., 2017, 2018). NDM-7 has been reported in E. coli strains (Hao et al., 2018). Recently, NDM-7 carrying the IncA/C2 type plasmid harbored by K. pneumoniae strain ST147 was reported (Shankar et al., 2019). In our study, blaNDM–7 was first reported in ST34 K. pneumoniae. HZW25 was isolated from a young woman with acute cholangitis infection; based on multilocus sequence typing, HZW25 belongs to ST34 and contains several resistance genes responsible for beta-lactam resistance. The blaNDM–7 gene was located on an IncX3 plasmid, and the genetic environments and characteristics of the blaNDM–7-producing plasmid P4 were analyzed. The module of NDM-7 transposable elements was highly similar to the blaNDM variants harboring the IncX3-type plasmid, suggesting that this NDM variant module could disseminate among different clones.
In recent years, NDM variants, including NDM-7, have been increasingly reported in CRE isolates (Khan et al., 2017; Wu et al., 2019). In contrast to foreign countries, such as India, Spain and Canada, where NDM-7-producing strains are mainly K. pneumoniae strains (Chen et al., 2015; Seara et al., 2015; Lynch et al., 2016; Lazaro-Perona et al., 2017), in China, NDM variants are more common in carbapenemase-producing E. coli (Bi et al., 2018; Wang et al., 2018; Zhang et al., 2018). Wang L reported NDM-7-producing uropathogenic E. coli from a patient with bacteriuria in 2016 (Wang et al., 2016). Bi R reported a high prevalence of blaNDM variants in CRE E. coli strains, and blaNDM–7 ranked third among those variants (Bi et al., 2018). Hao Y analyzed the genotypic and phenotypic characterization of blaNDM–7 in E. coli from a patient with a urinary tract infection (Hao et al., 2018). Recently, Xu J and He F also reported NDM-7-producing E. coli from a urinary sample (Xu and He, 2019).
Although blaNDM variants are both located on bacterial chromosomes and plasmids, blaNDM variants positioned on plasmids play a vital role in the dissemination of resistance genes (Bonnin et al., 2012; Ho et al., 2012). blaNDM variants harboring plasmids are mainly of the IncFII, IncX3, and IncC(IncA/C2) types (Perez-Vazquez et al., 2019). IncX3 is the most common type of plasmid carrying blaNDM, and most IncX3 plasmids are present in E. coli (Paul et al., 2017; Bi et al., 2018). From the worldwide distribution of blaNDM-carrying plasmids in Enterobacteriaceae, IncX3 plasmids may serve as an important vehicle in the dissemination of NDM in East Asia, particularly in China (Ho et al., 2012). Hao Y compared the backbones of plasmids carrying NDM variants and collected from human and food animal origin, and the results showed that all plasmids were highly similar (> 99%) among patients (Hao et al., 2018). This suggested that IncX3 plasmids may serve as one of the major platforms on which blaNDM genes evolve with the generation of new NDM variants. Lee CS isolated NDM-7-producing K. pneumoniae and E. coli simultaneously from a patient, suggesting that blaNDM–7 might be transferred between K. pneumoniae and E. coli in vivo (Lee et al., 2014).
In our study, blaNDM–7 was in a 68 637 bp IncX3-type plasmid. blaNDM–7 was successfully transferred to E. coli AzR J53 by conjugation and to E. coli DH5a by electroporation. Compared to the common genetic contexts of blaNDM variants, blaNDM–7 had a similar surrounding environment, with ISAba125 (intact or truncated) upstream and bleMBL downstream; these elements are located in the transposon-like structure flanked by IS3000 and IS26, responsible for horizontal transfer of blaNDM–7 among Enterobacteriaceae.
The blaNDM–7 gene can be carried by different K. pneumoniae strains of different STs, including ST147, ST138, ST273, ST437, and ST278 (Lee et al., 2014; Chen et al., 2015; Seara et al., 2015; Chou et al., 2016; Lynch et al., 2016; Perez-Vazquez et al., 2019; Shankar et al., 2019). Although these strains were isolated in different countries and regions, these strains contain plasmids carrying blaNDM–7 with similar surroundings and are characterized by horizontal gene transmission (Chen et al., 2015). Chen described blaNDM-carrying plasmids from different Enterobacteriaceae isolates with identical structures, suggesting that very effective horizontal transfer events had occurred (Chen et al., 2015). Seara N reported the interhospital spread of NDM-7-producing K. pneumoniae in Spain (Seara et al., 2015). In our study, ST34 K. pneumoniae carrying blaNDM–7 was reported for the first time, and the evolutionary distance from known ST34 K. pneumoniae strains showed that the strain we isolated was far from the other isolated strains.
Ceftazidime-avibactam is an effective antibiotic for CRE. It has good antibacterial activity against KPC-producing K. pneumoniae but not NDM-producing K. pneumoniae or E. coli (van Duin and Bonomo, 2016; Shirley, 2018). The emergence of NDM-producing K. pneumoniae would be troublesome in CRE treatment using ceftazidime-avibactam. Therefore, it is important to screen carbapenemase types before ceftazidime-avibactam therapy, and it is necessary to detect serine proteases and metalloproteinases in epidemiological investigations.
Conclusion
This study describes blaNDM–7 in an ST34 K. pneumoniae strain for the first time. The blaNDM–7 gene is located on a conjugated and horizontally transmitted IncX3-type plasmid. The potential dissemination of blaNDM-like genes in IncX3-type plasmids should be considered. Before treatment with ceftazidime-avibactam, it is necessary to determine the types of carbapenemase in carbapenemase-resistant K. pneumoniae.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
MW, XW, and DY provided assistance and guidance in the research. QC and JZ wrote the manuscript. SW and YY assisted the manuscript checking. All authors checked the manuscript and submitted the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This research was supported by National Natural Science Foundation of China (81601799), Natural Science Foundation of Zhejiang Province (LQ20H200003), and Zhejiang Medical and Health Science and Technology Program (20200374292).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01885/full#supplementary-material
References
- Bi R., Kong Z., Qian H., Jiang F., Kang H., Gu B., et al. (2018). High prevalence of bla NDM variants among carbapenem-resistant Escherichia coli in northern jiangsu province. China. Front. Microbiol. 9:2704. 10.3389/fmicb.2018.02704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin R. A., Poirel L., Carattoli A., Nordmann P. (2012). Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS One 7:e34752. 10.1371/journal.pone.0034752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Peirano G., Lynch T., Chavda K. D., Gregson D. B., Church D. L., et al. (2015). Molecular characterization by using next-generation sequencing of plasmids containing blaNDM-7 in Enterobacteriaceae from Calgary, Canada. Antimicrob. Agents Chemother. 60 1258–1263. 10.1128/AAC.02661-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou A., Roa M., Evangelista M. A., Sulit A. K., Lagamayo E., Torres B. C., et al. (2016). Emergence of Klebsiella pneumoniae ST273 carrying blaNDM-7 and ST656 carrying blaNDM-1 in Manila, Philippines. Microb. Drug Resist. 22 585–588. 10.1089/mdr.2015.0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2018). Clinical and Laboratory Standards Institute:Performance Standards For Antimicrobial Susceptibility Testing:twenty-eighth Informational Supplement. Wayne, PA: CLSI. [Google Scholar]
- Cuzon G., Bonnin R. A., Nordmann P. (2013). First identification of novel NDM carbapenemase, NDM-7, in Escherichia coli in France. PLoS One 8:e61322. 10.1371/journal.pone.0061322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet L., Poirel L., Nordmann P. (2014). Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed. Res. Int. 2014:249856. 10.1155/2014/249856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Li B., Cao J., Wu Q., Chen H., Hou Y., et al. (2017). Molecular characterization and epidemiologic study of NDM-1-producing extensively drug-resistant Escherichia coli. Microb. Drug. Resist. 23 272–279. 10.1089/mdr.2015.0294 [DOI] [PubMed] [Google Scholar]
- Findlay J., Hopkins K. L., Alvarez-Buylla A., Meunier D., Mustafa N., Hill R., et al. (2017). Characterization of carbapenemase-producing Enterobacteriaceae in the west midlands region of England: 2007-14. J. Antimicrob. Chemother. 72 1054–1062. 10.1093/jac/dkw560 [DOI] [PubMed] [Google Scholar]
- Hao Y., Shao C., Bai Y., Jin Y. (2018). Genotypic and phenotypic characterization of incx3 plasmid carrying bla NDM-7 in Escherichia coli sequence type 167 isolated from a patient with urinary tract infection. Front. Microbiol. 9:2468. 10.3389/fmicb.2018.02468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P. L., Li Z., Lo W. U., Cheung Y. Y., Lin C. H., Sham P. C., et al. (2012). Identification and characterization of a novel incompatibility group X3 plasmid carrying bla NDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg. Microbes. Infect. 1:e39. 10.1038/emi.2012.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. U., Maryam L., Zarrilli R. (2017). Structure, genetics and worldwide spread of new delhi metallo-beta-lactamase (NDM): a threat to public health. BMC Microbiol. 17:101. 10.1186/s12866-017-1012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro-Perona F., Sarria-Visa A., Ruiz-Carrascoso G., Mingorance J., Garcia-Rodriguez J., Gomez-Gil R. (2017). Klebsiella pneumoniae co-producing NDM-7 and OXA-48 carbapenemases isolated from a patient with prolonged hospitalisation. Int. J. Antimicrob. Agents 49 112–113. 10.1016/j.ijantimicag.2016.09.013 [DOI] [PubMed] [Google Scholar]
- Lee C. R., Lee J. H., Park K. S., Kim Y. B., Jeong B. C., Lee S. H. (2016). global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. 10.3389/fmicb.2016.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. S., Vasoo S., Hu F., Patel R., Doi Y. (2014). Klebsiella pneumoniae ST147 coproducing NDM-7 carbapenemase and RmtF 16S rRNA methyltransferase in Minnesota. J. Clin. Microbiol. 52 4109–4110. 10.1128/JCM.01404-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T., Chen L., Peirano G., Gregson D. B., Church D. L., Conly J., et al. (2016). Molecular evolution of a Klebsiella pneumoniae ST278 isolate harboring blaNDM-7 and involved in nosocomial transmission. J. Infect. Dis. 214 798–806. 10.1093/infdis/jiw240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Bhattacharjee A., Ingti B., Choudhury N. A., Maurya A. P., Dhar D., et al. (2017). Occurrence of blaNDM-7 within IncX3-type plasmid of Escherichia coli from India. J. Infect. Chemother. 23 206–210. 10.1016/j.jiac.2016.12.009 [DOI] [PubMed] [Google Scholar]
- Perez-Vazquez M., Sola Campoy P. J., Ortega A., Bautista V., Monzon S., Ruiz-Carrascoso G., et al. (2019). Emergence of NDM-producing Klebsiella pneumoniae and Escherichia coli in Spain: phylogeny, resistome, virulence and plasmids encoding blaNDM-like genes as determined by WGS. J. Antimicrob. Chemother. 74 3489–3496. 10.1093/jac/dkz366 [DOI] [PubMed] [Google Scholar]
- Rahman M., Shukla S. K., Prasad K. N., Ovejero C. M., Pati B. K., Tripathi A., et al. (2014). Prevalence and molecular characterisation of New Delhi metallo-beta-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int. J. Antimicrob. Agents 44 30–37. 10.1016/j.ijantimicag.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Ruan Z., Feng Y. (2016). BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 44 D682–D687. 10.1093/nar/gkv1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seara N., Oteo J., Carrillo R., Perez-Blanco V., Mingorance J., Gomez-Gil R., et al. (2015). Interhospital spread of NDM-7-producing Klebsiella pneumoniae belonging to ST437 in Spain. Int. J. Antimicrob. Agents 46 169–173. 10.1016/j.ijantimicag.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Shankar C., Kumar S., Venkatesan M., Veeraraghavan B. (2019). Emergence of ST147 Klebsiella pneumoniae carrying blaNDM-7 on IncA/C2 with ompK35 and ompK36 mutations in India. J. Infect. Public Health 12 741–743. 10.1016/j.jiph.2019.03.023 [DOI] [PubMed] [Google Scholar]
- Shirley M. (2018). Ceftazidime-avibactam: a review in the treatment of serious gram-negative bacterial infections. Drugs 78 675–692. 10.1007/s40265-018-0902-x [DOI] [PubMed] [Google Scholar]
- van Duin D., Bonomo R. A. (2016). Ceftazidime/Avibactam and Ceftolozane/Tazobactam: second-generation beta-Lactam/beta-Lactamase inhibitor combinations. Clin. Infect. Dis. 63 234–241. 10.1093/cid/ciw243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Liu P. P., Wei D. D., Liu Y., Wan L. G., Xiang T. X., et al. (2016). Clinical isolates of uropathogenic Escherichia coli ST131 producing NDM-7 metallo-beta-lactamase in China. Int. J. Antimicrob. Agents 48 41–45. 10.1016/j.ijantimicag.2016.03.009 [DOI] [PubMed] [Google Scholar]
- Wang Q., Wang X., Wang J., Ouyang P., Jin C., Wang R., et al. (2018). Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in china (2012-2016). Clin. Infect. Dis. 67(Suppl._2), S196–S205. 10.1093/cid/ciy660 [DOI] [PubMed] [Google Scholar]
- Wu W., Feng Y., Tang G., Qiao F., McNally A., Zong Z. (2019). NDM metallo-beta-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32:e00115-18. 10.1128/CMR.00115-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., He F. (2019). Characterization of a NDM-7 carbapenemase-producing Escherichia coli ST410 clinical strain isolated from a urinary tract infection in China. Infect. Drug Resist. 12 1555–1564. 10.2147/IDR.S206211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D., Toleman M. A., Giske C. G., Cho H. S., Sundman K., Lee K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53 5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Liu L., Zhou H., Chan E. W., Li J., Fang Y., et al. (2017). Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19 98–106. 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang Q., Yin Y., Chen H., Jin L., Gu B., et al. (2018). Epidemiology of Carbapenem-Resistant Enterobacteriaceae Infections: report from the China CRE Network. Antimicrob. Agents Chemother. 62:e01882-17. 10.1128/AAC.01882-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.