Abstract
Resistant disease is still a main obstacle in acute myeloid leukemia (AML) treatment. Therefore, individual genetic variations affecting therapy response are gaining increasing importance. Both SNPs and ABC transporter genes could already be associated with drug resistance. Here, we report allelic variants of MRP1 (ABCC1) SNPs rs129081, rs212090, and rs212091 with significant influences on survival in AML patients. DNA was extracted from bone marrow samples (n = 160) at diagnosis. Genotyping 48 SNPs within seven different ABC transporter genes using real-time PCR revealed rs129081 GG variant with a significant higher OS (p = 0.035) and DFS (p = 0.01). Comparing TT and AA rs212090 variants showed significant influences on DFS (p = 0.021). SNP rs212091 GG expression was associated with worse OS (p = 0.006) and a significant difference in DFS between alleles GG and AA (p = 0.018). The multivariable models confirmed a significant influence on OS for rs212091 (AA HR = 0.296, 95% CI 0.113–0.774, p = 0.013 and GG p = 0.044). Rs129081 variant CG, TT of rs212090, AA, and AG of rs212091 demonstrated significant impact on DFS (p = 0.024, p = 0.029, p = 0.017, and p = 0.042, respectively). This analysis demonstrates a significant influence of MRP1 SNPs on survival in AML. As they were not associated to prognostic characteristics, we suggest these SNPs to be independent prognostic markers for AML.
Electronic supplementary material
The online version of this article (10.1007/s00277-020-04163-7) contains supplementary material, which is available to authorized users.
Keywords: Acute myeloid leukemia, Multidrug resistance, MRP1, SNPs, Prognosis, Survival
Background
Acute myeloid leukemia (AML) is characterized by uncontrolled proliferation of undifferentiated myeloid blasts requiring rapid induction chemotherapy to induce a remission. Although approximately 75% of patients achieve complete remission (CR) after induction treatment consisting of cytarabine (100 mg/m2, days 1–7) and daunorubicin (60 mg/m2, on three consecutive days), two-thirds of AML patients under the age of 60 relapse after successful induction chemotherapy, with an even higher rate in older patients [1–3]. Drug resistance affecting standard chemotherapeutic compounds in AML results in induction failure or relapse. Very often relapse presents with a highly aggressive AML clone insensitive to salvage chemotherapy with the urgent need of intensified treatment and more invasive procedures like allogeneic stem cell transplantation, thus far the only available curative option for these patients.
Molecular mechanisms causing therapy failure leading to inferior prognosis of AML patients are still incompletely understood and one of the most difficult obstacles in AML therapy. Within the context of clinically relevant inter-individual differences in treatment response or susceptibility to cytostatic agents and other drugs alike, single nucleotide polymorphisms (SNPs) were identified as individual genetic variations accountable for treatment failure. Several of these polymorphisms have already been described with different therapy outcomes, individual alterations of pharmaceutical responses like an elevated risk of chemoresistance, and risk to develop a malignancy in general and AML alike [4–8]. For example, SNPs located within cytarabine metabolizing enzyme genes were associated with therapeutic effects concerning AML patients’ outcome [9]. Illmer et al. could further demonstrate a significant impact of ABCB1 (MDR1, P-glycoprotein) SNPs on therapy outcome in AML patients [4].
Transmembrane ATP-binding cassette transporters (ABC transporters) are important for substrate efflux and therefore providing a physiological function in multiple tissues, protecting cells against toxic metabolites. Some ABC transporters have certain substrate specificities for anticancer drugs, especially for cytarabine and daunorubicin. For cytarabine experimental analyses revealed that certain ABC transporter expressions lead to lower drug levels in leukemic blasts and thereby causing poor therapy response or even drug resistance [10].
ABC transporters have been extensively associated with the underlying principle of a so-called multidrug resistance (MDR), causing resistance to both multiple physiologic substrates and therapeutic drugs alike, leading to therapy failure, disease progression, and resistant disease [11–13].
ABC transporters ABCB1, ABCC1 (MRP1), and ABCG2 (BCRP) could be identified as the main MDR-generating transporters within the ABC family characterized by a very broad range of substrates and substrate specifity [11, 12, 14]. Furthermore, ABCC11 (MRP8) mRNA expression in blast progenitor cells of AML patients was demonstrated to have a significant influence on treatment response and long-term survival by conferring resistance to cytarabine [15].
Hence, the goal of this study was to further investigate whether so far non-investigated SNPs located within ABC-transporter genes responsible for daunorubicin efflux have an impact on treatment outcome in intensively treated younger AML patients.
Patients and methods
This analysis included AML patients (non-APL AML) within the prospective AML2003 trial (NCT00180102) for patients under the age of 60 years with a normal karyotype (NK) from whom bone marrow aspirate with sufficient DNA extraction was available. Bone marrow aspirates obtained at diagnosis from a total of 160 Caucasian AML patients according to WHO criteria were investigated [16]. Written informed consent was obtained from all patients and the study was approved by the local ethics committee of the University of Dresden (EK153092003). The patients’ characteristics are summarized in Table 1. The median age at diagnosis was 45 years (IQR 38–54 years). Only patients with a normal karyotype were selected for this study, thereby minimizing heterogeneity of the underlying data set.
Table 1.
Patients’ characteristics
| Characteristics n = 160 |
N (%) | Median (range) |
|---|---|---|
| Sex | ||
| Female | 82 (51) | |
| Male | 78 (49) | |
| Age at diagnosis (years) | 46 (18–60) | |
| White blood cell count (×109/l) | 31.2 (0.3–353) | |
| Bone marrow blasts (%) | 71 (8–95.5) | |
| CD34 expression positive (%) | 6 (0–93) | |
| Lactat dehydrogenase (IU/l) | 569 (167–5184) | |
| Peroxidase expression positive | 65 (1–100) | |
| ECOG | ||
| 0/1 | 122 (76.3) | |
| 2/3 | 29 (18.1) | |
| 4/5 | 1 (0.6) | |
| Missing | 8 (5) | |
| FAB subtype | ||
| M0 | 2 (1.3) | |
| M1 | 50 (31.3) | |
| M2 | 46 (28.7) | |
| M4 | 25 (15.6) | |
| M5 | 22 (13.7) | |
| M6 | 2 (1.3) | |
| M7 | 1 (0.6) | |
| Missing | 12 (7.5) | |
| Disease status | ||
| De novo AML | 149 (93.1) | |
| Therapy-related AML | 3 (1.9) | |
| Secondary AML (preceeding MDS) | 8 (5) | |
| FLT3-ITD mutation status | ||
| Mutated | 68 (42.5) | |
| Wildtype | 92 (57.5) | |
| NPM1 mutation status | ||
| Mutated | 89 (55.6) | |
| Wildtype | 71 (44.4) | |
| CR | 125 (78.1) | |
| No CR | 22 (13.8) | |
| Missing | 13 (8.1) | |
ECOG, Eastern Co-operative Oncology Group performance index; FAB, French American British classification of acute leukemia; AML, acute myeloid leukemia; MDS, myelodysplastic syndrome; FLT3-ITD, FMS-like tyrosine kinase 3 internal tandem duplication; NPM1, Nucleophosmin 1; CR, complete remission
All patients received double induction chemotherapy with daunorubicin (60 mg/m2, on three consecutive days) and cytarabine (100 mg/m2, days 1–7). Complete remission (CR) was evaluated according to standard criteria [17]. A total of 78% of the patients achieved CR after standard induction treatment (Table 1). Adverse side effects and organ toxicity were graded according to the Common Terminology Criteria for Adverse Events of the National Cancer Institute (CTCAE) [18]. Post-remission therapy was in accordance with previously published data [19].
DNA extraction from AML cells was performed using the QIAamp DNA blood mini kit (Qiagen, Hilden, Germany). Real-time PCR was performed with the Universal Master Mix and SNP assays supplied by Applied Biosystems (Applied Biosystems, Foster City, CA, USA). A total of 48 SNPs, located within the genes of 7 different ABC transporters (ABCA2, ABCA3, ABCB1, ABCB2, ABCB5, ABCB7, and ABCC1) were investigated (Supplementary Table 1). Each SNP was analyzed with a single-tube assay. The identification of the corresponding SNPs was performed as in silico analysis using NIH dbSNP database and HapMap, respectively. Genotyping of the observed SNP was performed using the 7500 Sequence Detection Software (Version 1.3.1) by Applied Biosystems (Applied Biosystems, Foster City, CA, USA). Hereby the detection of each SNP variant was based on the fluorescence of associated markers (VIC or FAM as reporter markers). Statistical analyses on the impact of different SNP genotypes in ABC transporter genes were performed using SPSS (Version 19.0, Chicago, IL, USA).
Continuous variables were analyzed using the U test according to Mann-Whitney while categorical variables were analyzed with the χ2 test, respectively. Calculation of survival probabilities was performed according to the method of Kaplan-Meier. Overall survival (OS) and disease-free survival (DFS) were defined according to standard criteria [17]. The differences in OS and DFS for the respective genotypes of the SNPs were analyzed with the log rank test. Cox regression was applied to identify independent prognostic variables for survival in univariate and multivariable analyses. The significance level was terminated at 0.05. The 95% confidence interval (CI) of hazard ratios (HR) was computed to provide quantitative information on the relevance of results. The deviation from the Hardy-Weinberg equilibrium was analyzed using the χ2 test. For pairwise linkage disequilibrium between the genetic markers, the three estimators D, D′, and r were calculated. These analyses were carried out using the free statistical computing environment R (Version 2.3.1) and its library genetics for genetic analyses.
Results
A significant impact on major outcomes (OS and DFS) was detected for three ABCC1 (MRP1) transporter SNPs rs129081 (CACCCC[C/G]ACTCCA), rs212090 (TTACTG[A/T]TCCCAC), and rs212091 (ACCTTA[A/G]AGAACA).
Patients carrying the homozygous rs129081 GG-SNP (n = 65) had a significant higher 5-year-OS and 5-year-DFS, compared to the homozygous wild type CC (n = 27) and heterozygous CG (n = 68) patients (OS: GG 68% [95% CI 55–80%] vs. CC 40% [95% CI 18–61%] vs. CG 64% [95% CI 52–75%], p = 0.035; DFS: GG 64% [95% CI 50–77%] vs. CC 35% [95% CI 16–53%] vs. CG 50% [95% CI 38–62%], p = 0.01) in univariate analysis, Figs. 1 and 2. Although these results did not have a significant impact on OS in multivariable analysis (Table 2), heterozygous CG demonstrated a significant impact on DFS in multivariable analysis (p = 0.024), while the other allele variants of rs129081 had no independent influence on DFS (Table 3).
Figure 1.
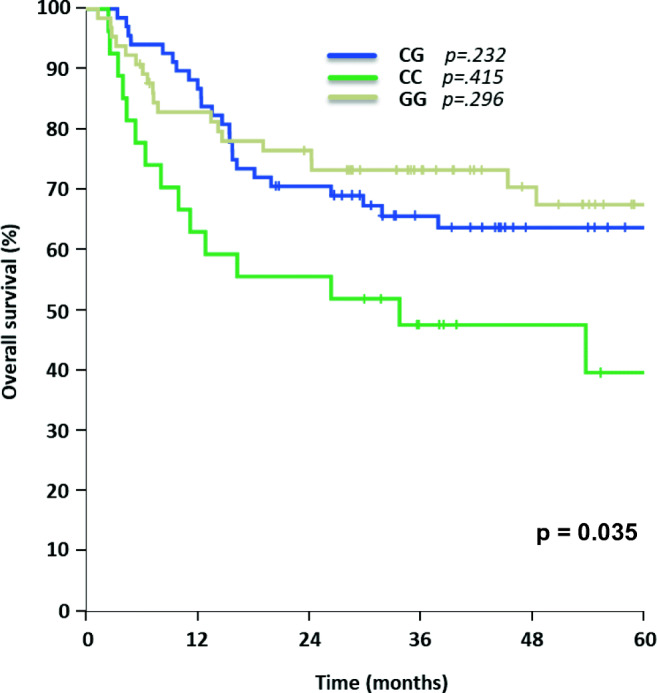
Overall survival for AML patients with different allele variants of rs129081
Fig. 2.
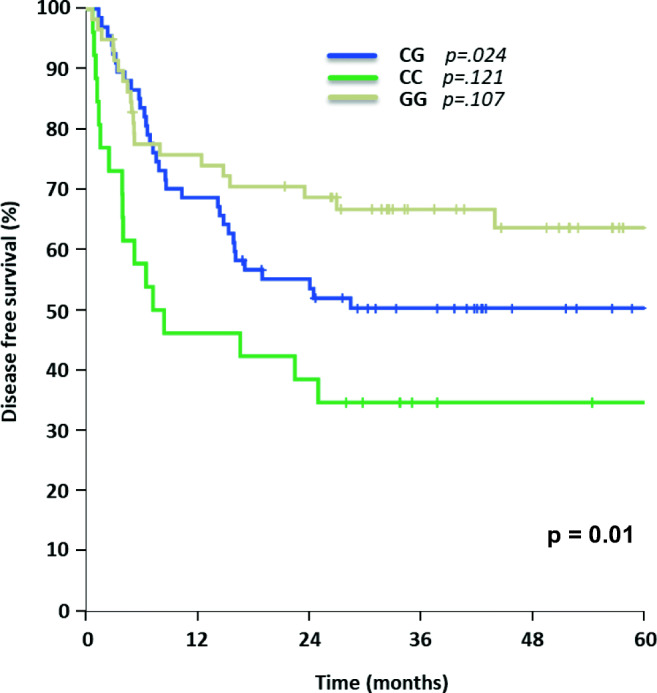
Disease-free survival for AML patients carrying allele variants of rs129081
Table 2.
Multivariate analysis—overall survival
| SNP | Hazard ratio (HR) | 95% CI | p value |
|---|---|---|---|
| rs129081 | |||
| CG | Baseline | 0.232 | |
| CC | 1.346 | 0.659–2.750 | 0.415 |
| GG | 0.716 | 0.383–1.340 | 0.296 |
| rs212090 | |||
| AA | Baseline | 0.590 | |
| AT | 1.165 | 0.569–2.387 | 0.676 |
| TT | 1.469 | 0.678–3.180 | 0.329 |
| rs212091 | |||
| GG | Baseline | 0.044 | |
| AG | 0.363 | 0.127–1.040 | 0.059 |
| AA | 0.296 | 0.113–0.774 | 0.013 |
Results of multivariate testing for overall survival of rs129081, rs212090, and rs212091 including hazard ratios, 95% CIs, and p values. CI, confidence interval
Table 3.
Multivariate analysis—disease-free survival
| SNP | Hazard ratio (HR) | 95% CI | p value |
|---|---|---|---|
| rs129081 | |||
| CG | Baseline | 0.024 | |
| CC | 1.647 | 0.876–3.095 | 0.121 |
| GG | 0.626 | 0.354–1.107 | 0.107 |
| rs212090 | |||
| AA | Baseline | 0.089 | |
| AT | 1.712 | 0.841–3.481 | 0.138 |
| TT | 2.321 | 1.092–4.932 | 0.029 |
| rs212091 | |||
| GG | Baseline | 0.058 | |
| AG | 0.312 | 0.101–0.958 | 0.042 |
| AA | 0.280 | 0.098–0.797 | 0.017 |
Results of multivariate testing for disease free survival of rs129081, rs212090, and rs212091 including hazard ratios, 95% CIs, and p values. CI, confidence interval
For allele variants of SNP rs212090 (AA n = 38, AT n = 80, TT n = 42), no statistically significant impact on OS both in univariate and multivariate testing was observed (AA 66% [95% CI 50–82%] vs. AT 64% [95% CI 53–75%] vs. TT 50% [95% CI 33–68%], p = 0.289, Fig. 3 and Table 2), but a trend towards worse OS for genotype TT could be identified. Although rs212090 showed no significant influence on DFS with genotype TT leading to shortest DFS (AA 68% [95% CI 52–83%] vs. AT 52% [95% CI 41–64%] vs. TT 40% [95% CI 25–55%], p = 0.064, Fig. 4), it revealed a significant difference in DFS when comparing homozygous alleles AA and TT (p = 0.021) in univariate testing. The significant impact of homozygous variant TT of rs212090 on DFS was confirmed in multivariate analysis (p = 0.029, HR 2.321, 95% CI 1.092–4.932, Table 3), respectively.
Fig. 3.
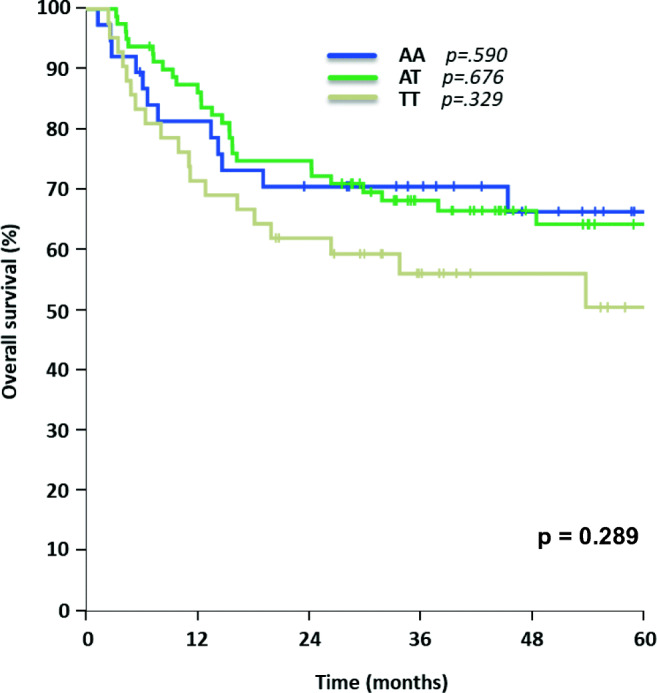
Overall survival for AML patients concerning different rs212090 allele variants
Fig. 4.
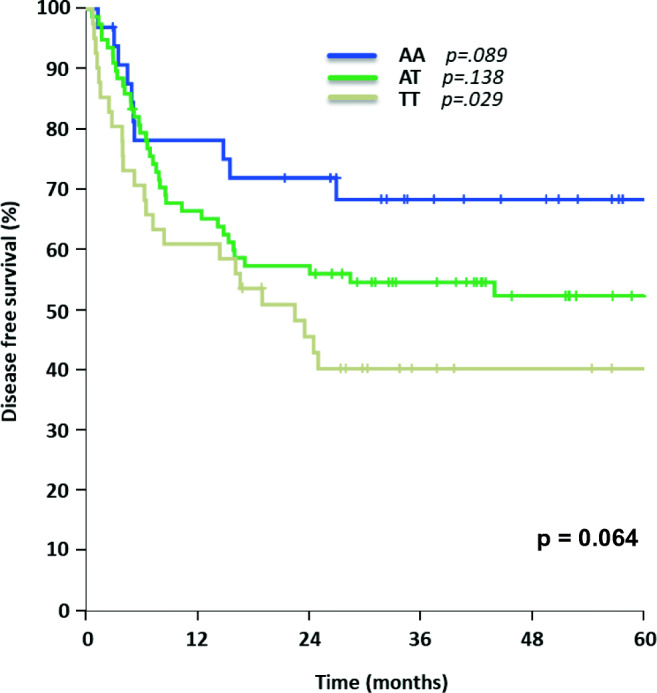
Disease-free survival for AML patients with different rs212090 allele expressions
SNP rs212091 showed a significant difference for OS with the homozygous allele GG (n = 6) leading to inferior OS (GG 0% vs. AA 64% [95% CI 55–74%] vs. AG 59% [95% CI 44–75%], p = 0.006, Fig. 5). In multivariable testing, an independent influence on OS for AA (n = 114, p = 0.044) and GG (HR = 0.296, 95% CI 0.113–0.774, p = 0.013) was demonstrated (Table 2). Regarding DFS for rs212091, no significant influence could be demonstrated in a univariate model (AA 55% [95% CI 46–65%] vs. 49% AG [95% CI 33–64%] vs. GG 0%, p = 0.058, Fig. 6). Comparing the homozygous genotypes of rs212091, GG was correlated with a significant reduction of DFS in contrast to homozygous counterpart AA (GG 0% vs. AA 55%, p = 0.018). These findings could be confirmed in multivariable analysis with independent significant impact on DFS for GG (HR = 0.28, 95% CI 0.098–0.797, p = 0.017) and AG (HR = 0.312, 95% CI 0.101–0.958, p = 0.042), Table 3.
Fig. 5.
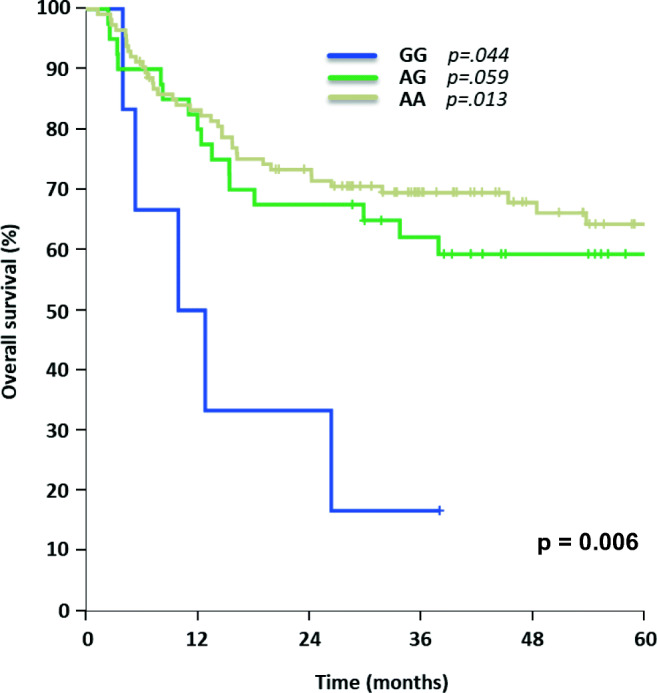
Overall survival for AML patients carrying different allele variants of rs212091
Fig. 6.
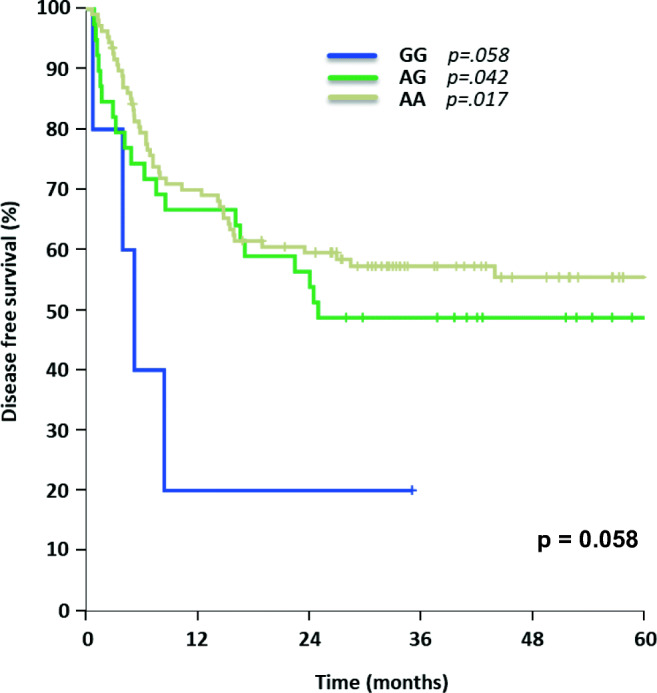
Disease-free survival for AML patients with different rs212091 allele expressions
A total of 78% (n = 125) of the here analyzed study cohort achieved CR. CR rate was not affected by these MRP1 polymorphisms (rs129081 p = 0.078, rs212090 p = 0.148, rs212091 p = 0.420). A trend towards a higher incidence of relapse of the investigated SNP variants was in accordance with worse OS and DFS, respectively, but did not reach statistical significance (p = 0.152, p = 0.426, and p = 0.231).
Furthermore, our data also showed that there were no significant differences of standard clinical baseline characteristics, FLT3-ITD, or NPM1 mutation status in the above described SNP groups. Regarding the combination of a co-occurring FLT3-ITD and NPM1 mutation, rs129081, rs212090, and rs212091 allele variants were not significantly associated with a FLT3-ITD/NPM1 co-expression (Table 4). Though SNP rs212090 alleles were characterized with significant differences in FLT3-ITD ratio, differences in FLT3-ITD ratios of rs129081 and rs212091 did not reach statistical significance (Table 4).
Table 4.
AML risk factors
| rs129081 | rs212090 | rs212091 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CG | GG | AA | AT | TT | AA | AG | GG | ||||
| FLT3-ITD mut (%) | 59.3 | 38.2 | 40 | p = 0.15 | 39.5 | 41.3 | 47.6 | p = 0.73 | 42.1 | 40 | 66.7 | p = 0.46 |
| NPM1 mut (%) | 44.4 | 55.9 | 60 | p = 0.39 | 63.2 | 56.3 | 47.6 | p = 0.37 | 59.6 | 45 | 50 | p = 0.27 |
| FLT3-ITD/NPM1 (%) | 33.3 | 27.9 | 27.7 | p = 0.28 | 28.9 | 28.7 | 28.6 | p = 0.77 | 27.2 | 30 | 50 | p = 0.11 |
| FLT3-ITD ratio > 0.5 (%) | 88 | 54 | 65 | p = 0.08 | 73 | 48 | 90 | p = 0.01 | 69 | 50 | 100 | p = 0.13 |
| LDH (IU/l) | 1126 | 788 | 688.5 | p = 0.06 | 662.8 | 769.1 | 1001.2 | p = 0.36 | 761.9 | 916.1 | 909.3 | p = 0.97 |
| WBC (×109/l) | 86.3 | 49.6 | 58.9 | p = 0.39 | 57.1 | 53.5 | 73.4 | p = 0.47 | 60.7 | 58.2 | 48.5 | p = 0.97 |
Characteristic risk factors for worse (FLT3-ITD mutation status, FLT3-ITD ratio > 0.5, high LDH count at diagnosis, and high WBC at diagnosis) and good prognosis (NPM1 mutation status) in AML, and their frequency among the analyzed allele variants of SNP rs129081, rs212090, and rs212091 are listed above. FLT3-ITD, FMS-like tyrosine kinase 3 internal tandem duplication; NPM1, Nucleophosmin 1; LDH, lactat dehydrogenase; WBC, white blood count
The patients’ age had no influence on the investigated MRP1 polymorphisms (rs129081: p = 0.962, rs212090: p = 0.585, rs212091: p = 0.706). The quality and intensity of toxicities according to CTCAE after chemotherapy were not significantly different between allele variants. Genotype frequencies of rs129081, rs212090, and rs212091 were in accordance with the Hardy-Weinberg equation. Calculation of the linkage disequilibrium identified linked SNPs (rs129081/rs212090 p = 0, D′ = 1; rs129081/rs212091 p = 0, D′ = 1; and rs212090/rs212091 p = 1.62^–14, D′ = 1, respectively).
Discussion
As AML presents clinically as a very heterogeneous hematopoietic malignancy with considerable variations in treatment response and patient outcome, prognostic markers need to be identified to offer individualized and risk-stratified therapy options to improve survival. Therapy resistance with progressive disease and limited therapeutic options still remains a major obstacle in the treatment of AML patients. Especially younger AML patients which represent a population with curative potential and long-term remission after intensive treatment, still need further adjusted treatment, as a considerable number of younger patients also suffer from progressive disease and relapse, but are uniformly treated with daunorubicin and cytarabine [20]. The clinical dilemma and pressure is about identifying mechanisms of these chemotherapy-resistances. During the last decade, several molecular prognostic factors for AML have been detected and included in standard routine diagnostics leading to improved risk stratification and prognosis evaluation and therefore adjusted therapeutic approaches [21–23]. With this analysis, we identified promising predictors for OS and DFS with SNP rs129081, rs212090, and rs212091 in NK-AML patients and indicate these SNPs to be prognostic markers for OS and DFS, respectively.
MRP1 SNPs rs129081, rs212090, and rs212091 are located within the 3′UTR region of the MRP1 transporter gene and therefore cause no primary change in amino acid sequence. As the 3′UTR region is a binding site for regulatory miRNAs, the influence of these SNPs by altering DNA sequence could unfold on a post-transcriptional level with different miRNAs leading to modification of the synthesized DNA strand and hereby potentially altering protein structures of ABC transporters after DNA transcription. In solid tumors as well as in AML, miRNAs have been identified as critical multiple key elements in tumorigenesis [24]. Whether these differences in survival are due to differential sensitivity towards anthracyclines, altered substrate-susceptibility or substrate binding-sites, or whether they can be applied to other functional and structural properties still remains unclear and needs further exploration. If chemotherapeutic agents themselves induce increased expression of transporter genes or increased transcription activity is also still a matter of investigation.
Interestingly, these polymorphisms did not confer any differences with regard to other AML specific characteristics and known risk factors for worse survival like secondary AML, white blood count, serum LDH, FLT3-ITD, or NPM1-mutation status at diagnosis. There were no differences concerning CR rates in these three MRP1 SNP patient cohorts. Though CR was not significantly affected, genotypes CC for rs129081, TT for rs212090, and GG for rs212091 predicted inferior OS and DFS. These findings are in concordance with data by Schaich et al., who demonstrated an influence of MRP1 gene expression on DFS, but not on CR [25]. Post-remission therapy had also no significant impact on outcome. Though a trend towards worse outcome and a higher incidence of relapse could be associated with MRP1 polymorphisms, validation of our data in an independent AML cohort is necessary to confirm these observations for further research.
Moreover, no significant effect of these SNPs on chemotherapy-related toxicities was seen. In contrast, Cao et al. demonstrated an association of MRP1 SNPs with gastrointestinal toxicities after chemotherapy in a Chinese population, which we could not confirm in our study of Caucasian patients [26]. No correlation was observed regarding age, gender, ECOG performance status, FAB subtypes, bone marrow blasts at diagnosis, or baseline blood count parameters (data not shown). As these three MRP1 SNPs were in allele frequency accordance to healthy Caucasian individuals (data from dbSNP), we suggest a genetic predisposition for altered chemotherapy response, therapy resistance, and worse survival. The identification of linked polymorphisms indicates the influence of certain SNP haplotypes on therapy sensitivity and outcome in AML therapy.
On a molecular level, we hypothesize these SNPs as possible causal factors for differential anthracycline susceptibility. Therapy resistance conferred by MRP1 SNPs might be an effect dominant in relapsed or progressive disease, when sensitive AML subclones are eliminated by cytotoxic chemotherapy and resistant blasts expressing MRP1 SNPs proliferate in contrast to their chemosensitive counterparts. ABC transporter-associated MDR via drug efflux could be addressed in several studies [27, 28]. Therefore, AML patients carrying these genotypes associated with a poor prognosis may benefit from alternative therapeutic consolidation strategies like early allogeneic stem cell transplantation, higher dosages of daunorubicin, or alternative formulations of daunorubicin (e.g., CPX-351) during induction treatment [29]. Recently, higher dosages of daunorubicin during induction chemotherapy have also been demonstrated to be beneficial in certain subgroups of AML patients [30]. Therapeutic strategies trying to modulate the efflux of these transporters have failed in most clinical AML trials so far [31].
In conclusion, we demonstrate the influence of MRP1 transporter polymorphisms CG of rs129081 on DFS, rs212090 TT allelic variant on DFS, and homozygous allele expressions GG and AA of rs212091 on OS, as well as AA and AG allelic rs212091 variants on DFS in AML patients with a normal karyotype for the first time to the best of our knowledge. Hence, we suggest these SNPs to be independent prognostic markers for AML concerning survival. As SNPs in miRNA binding sites may have an effect on gene transcription and protein expression, certain SNPs influencing prognosis in AML should be evaluated in addition to established mutational changes in AML to improve prognostic algorithms and individual treatment intensity and survival of AML patients.
Electronic supplementary material
(DOCX 34 kb)
Acknowledgments
The participation of nurses, staff, and doctors of centers of the Study Alliance Leukemia (SAL) is highly acknowledged.
Availability of data and material
The datasets generated and analyzed during the study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
Authors’ contributions
Desiree Kunadt and Friedrich Stölzel performed the research. Friedrich Stölzel, Markus Schaich, and Christian Dransfeld designed the research study. Desiree Kunadt, Claudia Dill, and Maria Schmiedgen performed the experiments. All authors analyzed and interpreted the data. Desiree Kunadt and Friedrich Stölzel wrote the paper. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding Information
Open Access funding provided by Projekt DEAL.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008.
Consent to participate
Informed consent was obtained from all patients for being included in the study.
Consent for publication
All authors approved the manuscript and gave their consent for publication.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mayer RJ, Davis RB, Schiffer CA, Berg DT, Powell BL, Schulman P, Omura GA, Moore JO, McIntyre OR, Frei E. Intensive postremission chemotherapy in adults with acute myeloid leukemia. Cancer and leukemia group B. N Engl J Med. 1994;331(14):896–903. doi: 10.1056/NEJM199410063311402. [DOI] [PubMed] [Google Scholar]
- 2.Büchner T, Berdel WE, Wörmann B, Schoch C, Haferlach T, Schnittger S, Kern W, Aul C, Lengfelder E, Schumacher A, Reichle A, Staib P, Balleisen L, Eimermacher H, Grüneisen A, Rasche H, Sauerland MC, Heinecke A, Mesters RM, Serve HL, Kienast J, Hiddemann W. Treatment of older patients with AML. Crit Rev Oncol Hematol. 2005;56(2):247–259. doi: 10.1016/j.critrevonc.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Fernandez HF, Sun Z, Yao X, Litzow MR, Luger SM, Paietta EM, Racevskis J, Dewald GW, Ketterling RP, Bennett JM, Rowe JM, Lazarus HM, Tallman MS. Anthracycline dose intensification in acute myeloid leukemia. NEJM. 2009;361(13):1249–1259. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Illmer T, Schuler US, Thiede C, Schwarz UI, Kim RB, Gotthard S, Freund D, Schäkel U, Ehninger G, Schaich M. MDR1 gene polymorphisms affect therapy outcome in acute myeloid leukemia patients. Cancer Res. 2002;62(17):4955–4962. [PubMed] [Google Scholar]
- 5.Kukreti R, Bhatnagar P, B-Rao C, Gupta S, Madan B, Das C, Guleria R, Athavale AU, Brahmachari SK, Ghosh B. Beta(2)-adrenergic receptor polymorphisms and response to salbutamol among Indian asthmatics*. Pharmacogenomics. 2005;6(4):399–410. doi: 10.1517/14622416.6.4.399. [DOI] [PubMed] [Google Scholar]
- 6.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, Kinsey SE, Lightfoot T, Roman E, Irving JAE, Allan JM, Tomlinson IP, Taylor M, Greaves M, Houlston RS. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rashed R, Shafik RE, Shafik NF, et al. Associations of interleukin-10 gene polymorphisms with acute myeloid leukemia in human (Egypt) J Cancer Res Ther. 2018;14(5):1083–1086. doi: 10.4103/0973-1482.187367. [DOI] [PubMed] [Google Scholar]
- 8.Zhu KW, Chen P, Zhang DY, Yan H, Liu H, Cen LN, Liu YL, Cao S, Zhou G, Zeng H, Chen SP, Zhao XL, Chen XP. Association of genetic polymorphisms in genes involved in Ara-C and dNTP metabolism pathway with chemosensitivity and prognosis of adult acute myeloid leukemia (AML) J Transl Med. 2018;16(1):90. doi: 10.1186/s12967-018-1463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahlknecht U, Dransfeld CL, Bulut N, Kramer M, Thiede C, Ehninger G, Schaich M. SNP analyses in cytarabine metabolizing enzymes in AML patients and their impact on treatment response and patient survival: identification of CDA SNP C-451T as an independent prognostic parameter for survival. Leukemia. 2009;23(10):1929–1932. doi: 10.1038/leu.2009.113. [DOI] [PubMed] [Google Scholar]
- 10.Michieli M, Damiani D, Ermacora A, Masolini P, Raspadori D, Visani G, Scheper RJ, Baccarani M. P-glycoprotein, lung resistance-related protein and multidrug resistance associated protein in de novo acute non-lymphocytic leukaemias: biological and clinical implications. Br J Haematol. 1999;104(2):328–335. doi: 10.1046/j.1365-2141.1999.01172.x. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 12.Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5(3):219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 13.Fletcher JI, Haber M, Henderson MJ, Norris MD. ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer. 2010;10(2):147–156. doi: 10.1038/nrc2789. [DOI] [PubMed] [Google Scholar]
- 14.Lage H. An overview of cancer multidrug resistance: a still unsolved problem. Cell Mol Life Sci. 2008;65(20):3145–3167. doi: 10.1007/s00018-008-8111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, Köck K, Ritter CA, Chen ZS, Grube M, Jedlitschky G, Illmer T, Ayres M, Beck JF, Siegmund W, Ehninger G, Gandhi V, Kroemer HK, Kruh GD, Schaich M. Expression of ABCC-type nucleotide exporters in blasts of adult acute myeloid leukemia: relation to long-term survival. Clin Cancer Res. 2009;15(5):1762–1769. doi: 10.1158/1078-0432.CCR-08-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 17.Cheson BD, Bennett JM, Kopecky KJ, Büchner T, Willman CL, Estey EH, Schiffer CA, Doehner H, Tallman MS, Lister TA, Lo-Coco F, Willemze R, Biondi A, Hiddemann W, Larson RA, Löwenberg B, Sanz MA, Head DR, Ohno R, Bloomfield CD. Revised recommendations of the International Working Group for Diagnosis, standardization of response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol. 2003;21(24):4642–4649. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 18.National Institute of Cancer. Common terminology criteria for adverse events (CTCAE). NIH Publication. 1999. p.0–35
- 19.Schaich M, Parmentier S, Kramer M, Illmer T, Stölzel F, Röllig C, Thiede C, Hänel M, Schäfer-Eckart K, Aulitzky W, Einsele H, Ho AD, Serve H, Berdel WE, Mayer J, Schmitz N, Krause SW, Neubauer A, Baldus CD, Schetelig J, Bornhäuser M, Ehninger G. High-dose cytarabine consolidation with or without additional amsacrine and mitoxantrone in acute myeloid leukemia: results of the prospective randomized AML2003 trial. J Clin Oncol. 2013;31(17):2094–2102. doi: 10.1200/JCO.2012.46.4743. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez HF, Sun Z, Yao X, Litzow MR, Luger SM, Paietta EM, Racevskis J, Dewald GW, Ketterling RP, Bennett JM, Rowe JM, Lazarus HM, Tallman MS. Anthracycline dose intensification in acute myeloid leukemia. N Engl J Med. 2009;361:1249–1259. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, Wermke M, Bornhäuser M, Ritter M, Neubauer A, Ehninger G, Illmer T. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99(12):4326–4335. doi: 10.1182/blood.V99.12.4326. [DOI] [PubMed] [Google Scholar]
- 22.Verhaak RG, Goudswaard CS, van Putten W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106(12):3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 23.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, Levine RL, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz M, Sierra J, Tallman MS, Tien HF, Wei AH, Löwenberg B, Bloomfield CD. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabra MM, Salmena L (2017) MicroRNAs and acute myeloid leukemia chemoresistance: a mechanistic overview. Front Oncol 7:255. 10.3389/fonc.2017.00255.eCollection2017 [DOI] [PMC free article] [PubMed]
- 25.Schaich M, Soucek S, Thiede C, Ehninger G, Illmer T, the SHG AML96 study group MDR1 and MRP1 gene expression are independent predictors for treatment outcome in adult acute myeloid leukaemia. Br J Haematol. 2005;128(3):324–332. doi: 10.1111/j.1365-2141.2004.05319.x. [DOI] [PubMed] [Google Scholar]
- 26.Cao HX, Miao CF, Yan L, Tang P, Zhang LR, Sun L. Polymorphisms at microRNA binding sites of Ara-C and anthracyclines-metabolic pathway genes are associated with outcome of acute myeloid leukemia patients. J Transl Med. 2017;15(1):235. doi: 10.1186/s12967-017-1339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu Rev Pharmacol Toxicol. 1999;39:361–398. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 28.Broxterman HJ, Sonneveld P, Pieters R, Lankelma J, Eekman CA, Loonen AH, Schoester M, Ossenkoppele GJ, Löwenberg B, Pinedo HM, Schuurhuis GJ. Do P-glycoprotein and major vault protein (MVP/LRP) expression correlate with in vitro daunorubicin resistance in acute myeloid leukemia? Leukemia. 1999;13(2):258–265. doi: 10.1038/sj.leu.2401331. [DOI] [PubMed] [Google Scholar]
- 29.Lancet JE, Uy GL, Cortes JE, Newell LF, Lin TL, Ritchie EK, Stuart RK, Strickland SA, Hogge D, Solomon SR, Stone RM, Bixby DL, Kolitz JE, Schiller GJ, Wieduwilt MJ, Ryan DH, Hoering A, Banerjee K, Chiarella M, Louie AC, Medeiros BC. CPX-351 (cytarabine and daunorubicin) liposome for injection versus conventional cytarabine plus daunorubicin in older patients with newly diagnosed secondary acute myeloid leukemia. J Clin Oncol. 2018;36(26):2684–2692. doi: 10.1200/JCO.2017.77.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luskin MR, Lee JW, Fernandez HF, Abdel-Wahab O, Bennett JM, Ketterling RP, Lazarus HM, Levine RL, Litzow MR, Paietta EM, Patel JP, Racevskis J, Rowe JM, Tallman MS, Sun Z, Luger SM. Benefit of high-dose daunorubicin in AML induction extends across cytogenetic and molecular groups. Blood. 2016;127(12):1551–1558. doi: 10.1182/blood-2015-07-657403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daenen S, van der Holt B, Verhoef GE, Löwenberg B, Wijermans PW, Huijgens PC, van Marwijk Kooy R, Schouten HC, Kramer MH, Ferrant A, van den Berg E, Steijaert MM, Verdonck LF, Sonneveld P. Addition of cyclosporin A to the combination of mitoxantrone and etoposide to overcome resistance to chemotherapy in refractory or relapsing acute myeloid leukaemia: a randomised phase II trial from HOVON, the Dutch-Belgian Haemato-oncology working group for adults. Leuk Res. 2004;28(10):1057–1067. doi: 10.1016/j.leukres.2004.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 34 kb)
Data Availability Statement
The datasets generated and analyzed during the study are available from the corresponding author on reasonable request.


