Summary
Histone H3 lysine 9 (H3K9) methylation is dynamically regulated by methyltransferases and demethylases. In spermatogenesis, prospermatogonia differentiate into differentiating or undifferentiated spermatogonia after birth. However, the epigenetic regulation of prospermatogonia to spermatogonia transition is largely unknown. We found that perinatal prospermatogonia have extremely low levels of di-methylated H3K9 (H3K9me2) and that H3K9 demethylases, JMJD1A and JMJD1B, catalyze H3K9me2 demethylation in perinatal prospermatogonia. Depletion of JMJD1A and JMJD1B in the embryonic germline resulted in complete loss of male germ cells after puberty, indicating that H3K9me2 demethylation is essential for male germline maintenance. JMJD1A/JMJD1B-depleted germ cells were unable to differentiate into functional spermatogonia. JMJD1 isozymes contributed to activation of several spermatogonial stem cell maintenance genes through H3K9 demethylation during the prospermatogonia to spermatogonia transition, which we propose is key for spermatogonia development. In summary, JMJD1A/JMJD1B-mediated H3K9me2 demethylation promotes prospermatogonia to differentiate into functional spermatogonia by establishing proper gene expression profiles.
Keywords: histone demethylation, transcription, spermatogonia, prospermatogonia
Highlights
-
•
JMJD1A/JMJD1B catalyze H3K9 demethylation in prospermatogonia
-
•
JMJD1A/JMJD1B are required for prospermatogonia to spermatogonia transition
-
•
JMJD1A/JMJD1B activate genes required for spermatogonial stem cell maintenance
In this article, Kuroki et al. show that the H3K9 demethylases, JMJD1A and JMJD1B, are essential for differentiation and maintenance of male germ cells in mice. JMJD1A/JMJD1B-mediated H3K9 demethylation regulates prospermatogonia to spermatogonia transition by ensuring proper gene expression.
Introduction
Developmental gene expression is tuned by transcription factors and the levels of post-translational modifications. Covalent modifications of the core histone tail are important epigenetic marks linked to a wide variety of nuclear functions, including transcriptional regulation. The discovery of enzymes that add/remove methyl groups to/from histones suggests that histone methylation levels are not static but are dynamically regulated by a variety of processes (Kooistra and Helin, 2012). Histone H3 lysine 9 (H3K9) methylation, a representative epigenetic mark of transcriptionally silenced heterochromatin, is dynamically regulated by H3K9 methyltransferases and demethylases.
Spermatogenesis is a highly coordinated differentiation process in male germ cells. Dynamic regulation of H3K9 methylation plays a crucial role in spermatogenesis. Mice lacking H3K9 tri-methyltransferases, Suv39h1 and Suv39h2, displayed male germ cell arrest at the mid-to-late pachytene stage of meiosis (Peters et al., 2001). The germline-specific depletion of G9a, which catalyzes H3K9 di-methylation, resulted in early pachytene stage arrest in the male and female germline (Tachibana et al., 2007). Germline-specific depletion of H3K9 tri-methyltransferase, Eset, caused postnatal hypogonadism in both sexes (Liu et al., 2014). Finally, loss of H3K9 demethylase, JMJD1A, impeded spermatid elongation (Okada et al., 2007) (Liu et al., 2010).
Prospermatogonia are male germ cells located in the center of testis cords in the pre- and perinatal period. After birth, prospermatogonia differentiate into undifferentiated- or differentiating spermatogonia (de Rooij, 1998; Yoshida et al., 2006). How epigenetic regulation contributes to prospermatogonia to spermatogonia transition is largely unknown. We previously revealed that H3K9me2 in embryonic prospermatogonia is maintained at extremely low levels compared with somatic cells (Deguchi et al., 2013). We proposed that a low abundance of GLP/G9a heteromeric complex, a primary enzyme for H3K9 di-methylation (Tachibana et al., 2002, 2005), might partly account for H3K9me2 hypomethylation in prospermatogonia (Deguchi et al., 2013). However, contribution of H3K9 demethylase(s) to H3K9me2 hypomethylation was not determined.
Here, we show that H3K9me2 hypomethylation is maintained in postnatal prospermatogonia. We also show that H3K9me2 hypomethylation is achieved enzymatically via JMJD1A and JMJD1B, the enzymes responsible for H3K9me2 demethylation. We established a mouse line in which JMJD1A and/or JMJD1B were depleted in a germline-specific manner. Depletion of JMJD1A and JMJD1B resulted in a complete loss of male germ cells after puberty. Cytological analysis revealed that JMJD1A/JMJD1B-depleted germ cells could not differentiate into functional spermatogonia. Furthermore, transcriptome analysis indicated that JMJD1A/JMJD1B-depleted germ cells did not undergo a change in gene expression from prospermatogonia to spermatogonia. Finally, we demonstrated that activation of serial spermatogonial stem cell (SSC) maintenance genes was dependent upon JMJD1-mediated H3K9 demethylation. In summary, we demonstrate the pivotal role of H3K9 demethylation in spermatogenesis, where JMJD1 isozymes promote the differentiation of prospermatogonia into functional spermatogonia.
Results
Expression Profiles of JMJD1A and JMJD1B in Prospermatogonia
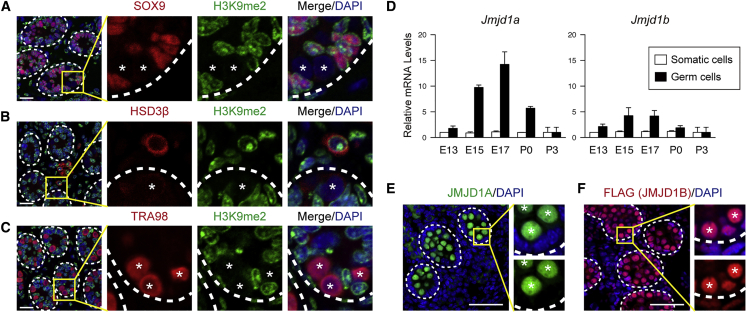
We previously demonstrated that H3K9me2 levels were significantly lower in prenatal prospermatogonia than in somatic cells during mouse embryogenesis (Deguchi et al., 2013). To examine whether H3K9me2 hypomethylation was maintained in postnatal prospermatogonia, we performed immunostaining for H3K9me2 on testis sections at postnatal day 3 (P3). Sections were stained with antibodies against SOX9 and HSD3b, markers for Sertoli cells and Leydig cells, respectively. Male germ cells were stained with a TRA98 antibody, which recognizes a mouse testicular germ cell-specific antigen (Tanaka et al., 1997). H3K9me2 signals were abundantly detected in the nuclei of Sertoli cells and Leydig cells in postnatal testes (Figures 1A and 1B, respectively). In contrast, H3K9me2 signals of prospermatogonia were extremely low, indicating that H3K9me2 hypomethylation was maintained from prenatal to postnatal prospermatogonia (Figure 1C).
Figure 1.
Expression Profiles of JMJD1A and JMJD1B in Prospermatogonia
(A–C) P3 wild-type mouse testes sections stained with an anti-H3K9me2 antibody. Sertoli cells and Leydig cells were marked with antibodies against SOX9 (A) and HSD3β (B), respectively. A TRA98 antibody was used to detect germ cells (C). Nuclei were stained with DAPI. Dashed lines represent basement membranes. Asterisks represent germ cells. Scale bars, 20 μm.
(D) Quantitative mRNA analysis for Jmjd1a (left) and Jmjd1b (right) in germ cells and somatic cells separated from Oct4-EGFP Tg testes at the indicated time points. Relative mRNA levels in E13.5 somatic cells were arbitrarily defined as 1. Data are presented as the mean ± SD (n = 3 mice for each time point).
(E and F) Immunofluorescence analysis of JMJD1A and JMJD1B in embryonic testes sections. Dashed lines represent basement membranes. Asterisks represent germ cells. Scale bars, 50 μm. (E) E17.5 wild-type testis sections were stained with an anti-JMJD1A antibody and DAPI. Enlarged boxes represent double staining for JMJD1A and DAPI (top) and single staining for JMJD1A (bottom). (F) E17.5 Jmjd1b+/FLAG−KI testes were stained with an anti-FLAG antibody and DAPI. Enlarged boxes represent double staining for FLAG and DAPI (top) and single staining for FLAG (bottom).
We previously demonstrated that H3K9me2 hypomethylation in embryonic prospermatogonia was associated with reduced expression of GLP, a regulatory subunit of the GLP/G9a H3K9 methyltransferase complex (Deguchi et al., 2013; Tachibana et al., 2005; Ueda et al., 2006). Introduction of a GLP transgene increased H3K9me2 in prospermatogonia. However, the H3K9me2 levels were still lower than in somatic cells in this transgenic line (Deguchi et al., 2013). One explanation for this observation is that H3K9me2 demethylation machinery also functions in prospermatogonia. The Jmjd1 family of proteins, JMJD1A and JMJD1B, possess intrinsic H3K9 demethylating activity and are involved in transcriptional activation (Yamane et al., 2006). We recently demonstrated the redundant but essential function of JMJD1A and JMJD1B in the peri-implantation stage of embryogenesis (Kuroki et al., 2018). To address the potential contribution of JMJD1A and JMJD1B to H3K9me2 hypomethylation in prospermatogonia, male germ cells were isolated from prenatal and postnatal testes for mRNA analysis. qRT-PCR analysis revealed that Jmjd1a mRNA was expressed more highly in germ cells than in somatic cells from embryonic day 15.5 (E15.5) to P3 (Figure 1D, left). Of note, Jmjd1a expression in prospermatogonia increased during late embryogenesis, reaching a peak at E17.5. Furthermore, prospermatogonia expressed higher levels of Jmjd1b mRNA compared with somatic cells, with peak expression at E15.5 (Figure 1D, right). Immunofluorescence analysis of E17.5 sections indicated that JMJD1A signals were abundant in germ cells but not in somatic cells (Figure 1E) consistent with the mRNA expression analyses (Figure 1D, left). To detect the endogenous JMJD1B protein, we established a knockin mouse carrying the Jmjd1bFLAG−KI allele, which produced a JMJD1B protein with a C-terminal FLAG tag (Kuroki et al., 2018). JMJD1B-FLAG was detected both in germ and somatic cells, but the levels of JMJD1B-FLAG were substantially higher in germ cells compared with somatic cells (Figure 1F) consistent with the mRNA analyses (Figure 1D, right). In summary, expression profiles of JMJD1A and JMJD1B do not contradict our hypothesis that JMJD1 isozymes might contribute to active H3K9me2 demethylation in prospermatogonia.
JMJD1A and JMJD1B Catalyze H3K9 Demethylation in Prospermatogonia
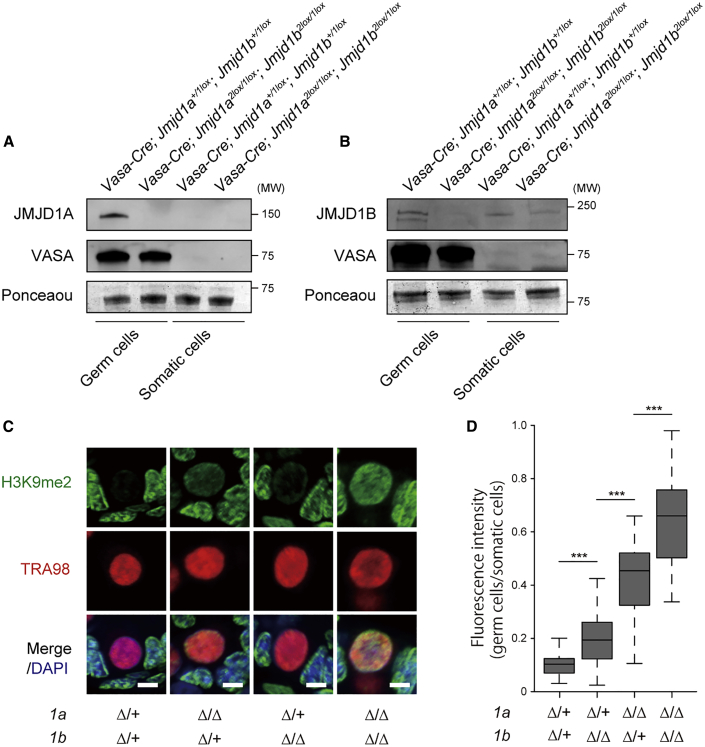
To address the role of JMJD1 isozymes in H3K9me2 hypomethylation in the male germline, we performed loss-of-function analyses of JMJD1A and/or JMJD1B. The combined loss of JMJD1A and JMJD1B led to lethality in the early post-implantation period (Kuroki et al., 2018); therefore, we generated mice in which Jmjd1a and Jmjd1b were conditionally deleted by Cre-loxP-mediated recombination. The loxP-flanked (2lox) conditional alleles of Jmjd1a and Jmjd1b are shown in Figure S1A and are described in our previous report (Kuroki et al., 2018). Mice carrying 2lox alleles of Jmjd1a and Jmjd1b were crossed with Vasa-Cre mice, to produce progeny in which Cre/loxP-mediated recombination occurs in germ cells from E15 (Gallardo et al., 2007). To address the accuracy of cell-type-specific depletion of JMJD1A and JMJD1B, germ cells were isolated from neonatal testes for immunoblot analysis. JMJD1A and JMJD1B were not detected in Vasa-Cre; Jmjd1a2lox/1lox; Jmjd1b2lox/1lox germ cells, but were abundant in control Vasa-Cre; Jmjd1a+/1lox; Jmjd1b+/1lox germ cells (Figures 2A and 2B). Importantly, in somatic cells, JMJD1B was still detected in Vasa-Cre; Jmjd1a2lox/1lox; Jmjd1b2lox/1lox mice at a similar level to that in control mice (Figure 2B). These data indicate that JMJD1 isozymes were successfully depleted before birth in a germ cell-specific manner.
Figure 2.
JMJD1A and JMJD1B Catalyze H3K9me2 Demethylation in Prospermatogonia
(A and B) Immunoblot analysis of JMJD1A (A) and JMJD1B (B) in neonatal testicular cells. Germ cells and somatic sells were separated from P0 testes of the indicated genotypes for immunoblot analysis. Enrichment of germ cells was confirmed by immunoblotting with an anti-VASA antibody. Ponceau staining was used as a loading control.
(C) Immunofluorescence analysis of H3K9me2 in P3 testes sections from the indicated genotypes. A TRA98 antibody was used to detect germ cells. Scale bars, 5 μm.
(D) Relative fluorescence intensities of H3K9me2 in P3 germ cells of the indicated genotypes. Fluorescence intensities of germ cells and neighboring Sertoli cells were measured using ImageJ software. Relative fluorescence intensity values were calculated by dividing the fluorescence intensities of germ cells by those of Sertoli cells. We examined >50 germ cells per genotype. ∗∗∗p < 0.001 (one-way ANOVA with Tukey's test).
To address the potential contribution of JMJD1A and JMJD1B to H3K9me2 hypomethylation in prospermatogonia, we examined H3K9me2 in Jmjd1a and Jmjd1b mutant testes at P3. A schematic illustration of the generation of serial mutant mice is shown in Figure S1B. We hereafter describe the genotype of germ cells in Vasa-Cre; Jmjd1a2lox/+; Jmjd1b2lox/+ mice as 1aΔ/+; 1bΔ/+. Anti-H3K9me2 immunofluorescence analysis revealed that H3K9me2 in 1aΔ/Δ; 1bΔ/+ cells and 1aΔ/+; 1bΔ/Δ cells was significantly increased compared with that in 1aΔ/+; 1bΔ/+ cells (Figure 2C and summarized in Figure 2D). Notably, H3K9me2 was profoundly increased by the 1aΔ/Δ mutation and moderately increased by the 1bΔ/Δ mutation, indicating the predominant role of JMJD1A in H3K9me2 demethylation in prospermatogonia. Remarkably, the 1aΔ/Δ; 1bΔ/Δ mutation induced a >6-fold increase in H3K9me2 compared with that in the 1aΔ/+; 1bΔ/+ control (Figures 2C and 2D). H3K9me3 but not H3K9me1 was also increased in 1aΔ/Δ; 1bΔ/Δ male germ cells (Figures S2A and S2B, respectively). These data indicate that JMJD1A and JMJD1B preferentially demethylate H3K9me2/3 but not me1 in prospermatogonia. An in vitro enzymatic study demonstrated that recombinant JMJD1A demethylates H3K9me1/2, but not me3 (Yamane et al., 2006). It is reasonable that specific partner protein(s) may alter the substrate specificity of JMJD1 enzymes in vivo.
JMJD1A and JMJD1B Are Required for Maintenance of the Male Germline
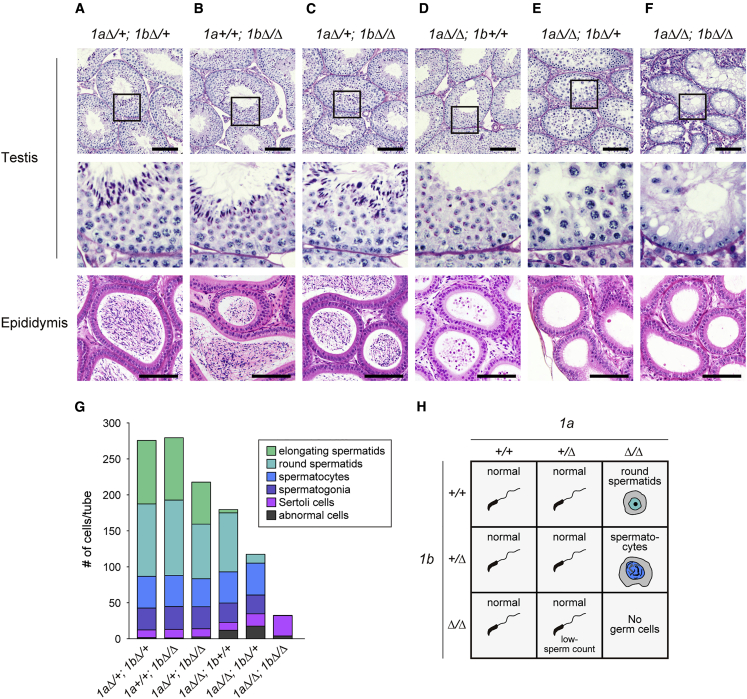
To address the impact of JMJD1A/JMJD1B depletion on spermatogenesis, we performed histological analysis of reproductive organs in 3-month-old male mice (Figures 3A–3F) and summarized the number of germ cells per seminiferous tubule section (Figure 3G). Spermatogenesis in 1aΔ/+; 1bΔ/+ and 1a+/+; 1bΔ/Δ germ cells proceeded normally (Figures 3A and 3B). 1aΔ/+; 1bΔ/Δ germ cells could also differentiate into mature sperm, but populations of spermatids were moderately decreased compared with those in 1aΔ/+; 1bΔ/+ and 1a+/+; 1bΔ/Δ cells (Figures 3C and 3G). Male mice of these three mutants were fertile in natural mating. In contrast, a single homozygous mutation in Jmjd1a (1aΔ/Δ; 1b+/+) resulted in failure of the elongation step of spermiogenesis and the corresponding male mice were infertile (Figure 3D), indicating that JMJD1A is required for spermiogenesis. Previous reports demonstrated that the zygotic loss of JMJD1A results in failure of the elongation step of spermiogenesis (Liu et al., 2010; Okada et al., 2007). Taken together, our conditional knockout studies confirmed that the spermiogenesis defect in JMJD1A-deficient mice was caused by a malfunction in the germ cell itself. Importantly, an additional 1b mutation on the 1aΔ/Δ background had a profound effect on spermatogenesis (Figures 3E and 3G). Haploid spermatids were barely detected in testes carrying 1aΔ/Δ; 1bΔ/+ germ cells, suggesting that meiosis was perturbed in this mutant. Consistent with this, haploid germ cells were markedly reduced in adult testis carrying 1aΔ/Δ; 1bΔ/+ germ cells (Figure S3). These results indicate that JMJD1 isozymes may play a crucial role in meiosis progression. Most importantly, loss of all Jmjd1a and Jmjd1b alleles resulted in the absence of germ cells at any developmental stage; only Sertoli cells were present in the corresponding seminiferous tubules of adult mice (Figures 3F and 3G). The final developmental stages of spermatogenesis in each mutant are summarized in Figure 3H. We found redundant but partially different roles of Jmjd1 alleles in spermatogenesis. First, at least one allele of either Jmjd1a or Jmjd1b is required for the maintenance of germ cells. Second, at least one allele of Jmjd1a or two alleles of Jmjd1b is required for the completion of meiosis. Third, at least one allele of Jmjd1a is required for the completion of the elongation step. In the latter two cases, the Jmjd1a allele is more important than the Jmjd1b allele.
Figure 3.
JMJD1A and JMJD1B Are Required for Maintenance of the Male Germline
(A–F) Histological analysis of testes and epididymis of 3-month-old male mice of the genotypes indicated above each figure (A) to (F). Paraffin-embedded testis and epididymis sections were stained with PAS-hematoxylin and hematoxylin-eosin, respectively. Enlarged views of testes sections (boxed areas in the top panel) are shown in the middle panel. Scale bars, 100 μm.
(G) Population analysis of testicular cells in 3-month-old testes of the indicated genotypes. The number of cells per PAS-hematoxylin-stained cross-tubular section is presented. Testicular cells were classified into six types according to their PAS-hematoxylin-staining profile, morphology, and intratubular localization. Abnormal cells represent those that could not be classified. More than 10 tubular sections were analyzed per sample.
(H) Summary of the developmental phenotypes of male germ cells lacking Jmjd1a and/or Jmjd1b alleles. Terminal developmental stages of male germ cells of the indicated genotypes are illustrated.
Germline-Specific Depletion of JMJD1A and JMJD1B Impairs Spermatogonia Development
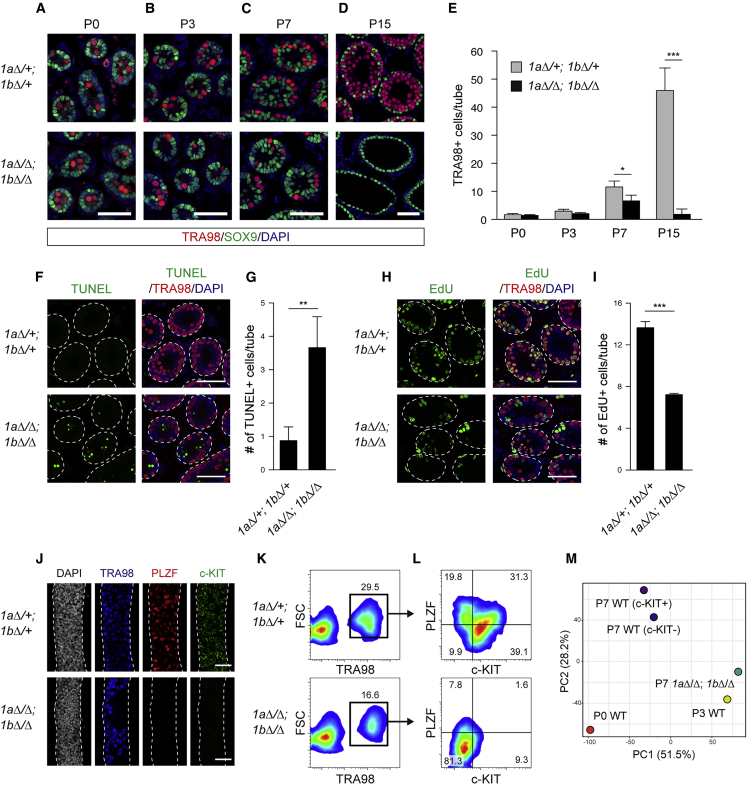
To identify the critical step of germ cell development influenced by JMJD1A/JMJD1B depletion, we examined when germ cells are exhausted in the mutant testes (Figures 4A–4D). Immunohistochemical analysis revealed that germ cells were barely detected in mutant testes at P15, in which only SOX9+ Sertoli cells were detected (Figure 4D). This indicated that 1aΔ/Δ; 1bΔ/Δ germ cells do not enter first-round spermatogenesis. In contrast, germ cells were detected in mutant testes until P7 (Figures 4A–4C). We counted germ cells per tubule of the mutant testes, as summarized in Figure 4E. No significant reduction was observed in the number of germ cells in mutant testes from P0 to P3. However, the number of germ cells was reduced in mutant testis at P7 (Figures 4C and 4E). Because spermatogonia are the dominant population in normal testes at P7, spermatogonia development may be influenced by JMJD1A/JMJD1B depletion. To address the cause of the reduction in germ cell number, we performed cell death and proliferation analyses of mutant testes at P7 (Figures 4F–4I). The number of apoptotic cells was markedly increased in testes carrying 1aΔ/Δ; 1bΔ/Δ germ cells (Figures 4F and 4G). 5-Ethynyl-2′-deoxyuridine (EdU) incorporation analysis indicated that proliferation was also affected in 1aΔ/Δ; 1bΔ/Δ germ cells (Figures 4H and 4I). We therefore concluded that an increase in cell death and a decrease in cell proliferation both account for the reduced number of 1aΔ/Δ; 1bΔ/Δ germ cells.
Figure 4.
Germline-Specific Depletion of JMJD1A and JMJD1B Impairs Spermatogonia Development
(A–D) Immunofluorescence analysis of control (1aΔ/+; 1bΔ/+, top) and mutant (1aΔ/Δ; 1bΔ/Δ, bottom) testes sections at the developmental times indicated above each figure (A) to (D). Germ cells and Sertoli cells were marked with TRA98 and anti-SOX9 antibodies, respectively. Scale bars, 50 μm.
(E) Summarized number of TRA98+ germ cells per cross-tubular section at the indicated times. Error bars represent ± SEM for >100 tubules from three mice. ∗∗∗p < 0.001 (t test).
(F–I) Cell death and proliferation analyses of JMJD1A/JMJD1B-depleted germ cells at P7. (F) TUNEL-stained sections of P7 testes of the indicated genotypes were counterstained with a TRA98 antibody and DAPI. Scale bars, 50 μm. (G) Average numbers of TUNEL+ cells in cross-tubular sections are summarized. Error bars represent ± SEM (n = 3 mice each). ∗∗p < 0.05 (t test). (H) P7 testicular cells incorporating EdU were counterstained with a TRA98 antibody and DAPI. Scale bars, 50 μm. (I) Average numbers of EdU+ cells per cross-tubular section are summarized. Error bars represent ± SEM (n = 3 mice each). ∗∗∗p < 0.001 (t test).
(J) Whole-mount immunofluorescence analysis of seminiferous tubules from P7 control (1aΔ/+; 1bΔ/+, top) and P7 mutant (1aΔ/Δ; 1bΔ/Δ, bottom) testes. A TRA98 antibody was used to mark germ cells. Antibodies against PLZF and c-KIT were used to detect undifferentiated spermatogonia and differentiating spermatogonia, respectively. Nuclei were counterstained with DAPI.
(K and L) Population analysis of P7 germ cells by fluorescence-activated cell sorting (FACS). Germ cells were dissociated from control (top) and mutant testes (bottom), stained with a TRA98 antibody and antibodies against PLZF and c-KIT, and then used for FACS analysis. (K) P7 testicular cells were fractionated to examine cell profiles for forward scatter (FSC) and TRA98 staining. Boxes represent the germ cell population. (L) Population analysis of P7 male germ cells. TRA98-sorted germ cells (boxes shown in K) were used for the classification of PLZF+ undifferentiated spermatogonia and c-KIT+ differentiating spermatogonia.
(M) PCA plot visualizing the degree of dissimilarity of gene expression in male germ cells between the indicated genotypes and developmental times.
Spermatogonia are classified as two cell types, differentiating and undifferentiated spermatogonia. During the prospermatogonia to spermatogonia transition, prospermatogonia give rise to differentiating or undifferentiated spermatogonia. Thus, we examined whether 1aΔ/Δ; 1bΔ/Δ male germ cells can differentiate into spermatogonia. Seminiferous tubules of P7 testes were stained with an antibody against PLZF (Buaas et al., 2004; Costoya et al., 2004), a marker for undifferentiated spermatogonia, and with an antibody against c-KIT, a marker of differentiating spermatogonia (Schrans-Stassen et al., 1999), in combination with a TRA98 antibody (Figure 4J). TRA98+ cells were present in control and mutant tubules at P7. PLZF+ and c-KIT+ cells were abundant in control tubules but were not detected in mutant tubules (Figure 4J). To quantitatively evaluate PLZF+ and c-KIT+ cell populations, we performed fluorescence-activated cell sorting (Figures 4K and 4L). TRA98+ germ cells were sorted from P7 testicular cells (Figure 4K), and were then analyzed using antibodies against PLZF and c-KIT (Figure 4L). JMJD1A/JMJD1B depletion reduced germ cell numbers to about half that in control testes (Figure 4K), consistent with Figures 4C and 4E. Population analysis indicated that the c-KIT+/PLZF− cell population, representing differentiating spermatogonia, was predominant in P7 control germ cells, constituting approximately 40% of all cells (Figure 4L, top). The c-KIT-/PLZF+ population, representing undifferentiated spermatogonia, constituted approximately 20% of all cells in the control testes (Figure 4L, top). In addition, we found another population (∼30%) composed of c-KIT+/PLZF+ cells. This population might represent an intermediate type of cell between undifferentiated spermatogonia and differentiating spermatogonia (Figure 4L, top). Strikingly, c-KIT+/PLZF−, c-KIT-/PLZF+, and c-KIT+/PLZF+ populations were almost absent in 1aΔ/Δ; 1bΔ/Δ germ cells at P7 (Figure 4L, bottom). Instead, the c-KIT−/PLZF− population was predominant in 1aΔ/Δ; 1bΔ/Δ germ cells, constituting approximately 80% of all cells. This c-KIT−/PLZF− population was barely detected in control testes (Figure 4L, top).
To determine the cellular characteristics of JMJD1A/JMJD1B-depleted germ cells, we performed mRNA sequencing (mRNA-seq) analysis using germ cells purified from P7 testes. We compared the gene expression of 1aΔ/Δ; 1bΔ/Δ germ cells with that of wild-type germ cells at several developmental stages by principal-component analysis (PCA). For this analysis, we utilized previously published mRNA-seq data of wild-type neonatal prospermatogonia (P0 WT), P7 wild-type c-KIT− spermatogonia (P7 WT [c-KIT−]), and P7 wild-type c-KIT+ spermatogonia (P7 WT [c-KIT+]) (Kubo et al., 2015). In addition to these data, we prepared mRNA-seq data of wild-type germ cells purified from P3 testis of this study (P3 WT) (Figure 4M). Genes that respond to environmental and biological stimuli were significantly enriched both in PC1 and PC2 (Figure S4). We noticed similar distances between P0 WT, P3 WT, and P7 WT. Interestingly, the distance from P7 1aΔ/Δ; 1bΔ/Δ to P3 WT was much closer than that from P7 1aΔ/Δ; 1bΔ/Δ to P7 WT (c-KIT+ or c-KIT−) (Figure 4M), indicating that the prospermatogonia to spermatogonia transition was perturbed by JMJD1A/JMJD1B depletion. Considering that 1aΔ/Δ; 1bΔ/Δ germ cells did not enter the first round of spermatogenesis (Figure 4D) and did not express typical spermatogonia markers (Figure 4L), JMJD1A/JMJD1B depletion might inhibit the generation of functional spermatogonia.
JMJD1A/JMJD1B Depletion Perturbs the Prospermatogonia to Spermatogonia Transition
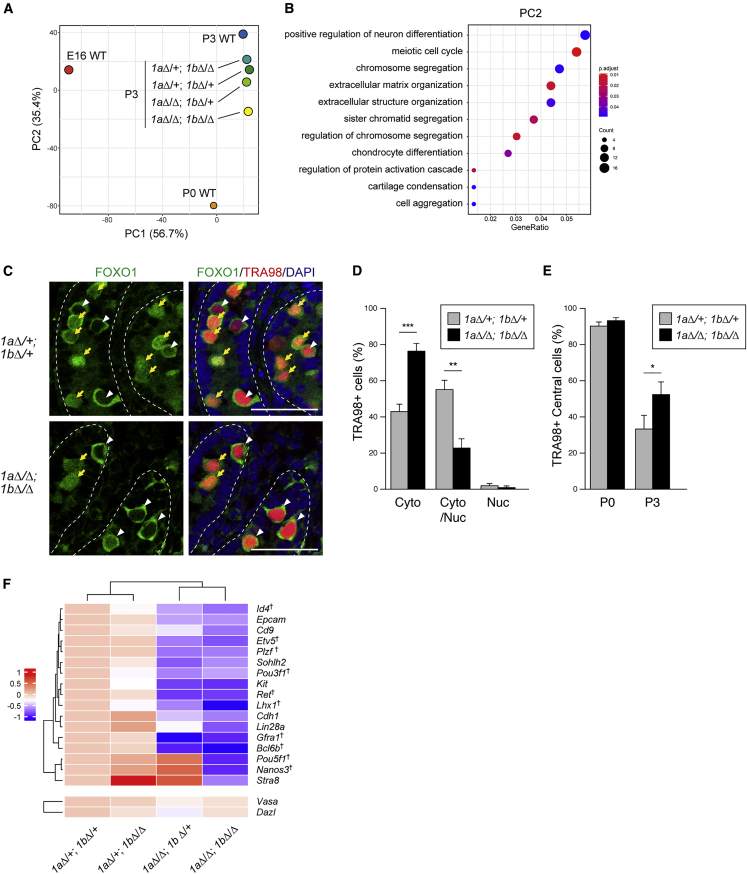
To address how JMJD1 isozymes control the prospermatogonia to spermatogonia transition, we performed transcriptomic analysis of postnatal germ cells lacking JMJD1A and/or JMJD1B. In brief, 1aΔ/+; 1bΔ/+, 1aΔ/+; 1bΔ/Δ, 1aΔ/Δ; 1bΔ/+, and 1aΔ/Δ; 1bΔ/Δ germ cells were isolated from the corresponding P3 testes by immunoaffinity purification, and were then subjected to mRNA-seq analysis. For PCA, we used previously published datasets for E16 prospermatogonia (E16 WT) and P0 WT (Kubo et al., 2015), and a P3 WT dataset prepared in this study. We extracted differentially expressed genes between P0 WT and P3 WT, and then merged our transcriptome datasets for P3 germ cells (Figure 5A). As shown in Figure 5A, serial mutants of P3 germ cells were located near the line connecting P0 to P3. Of these, 1aΔ/Δ; 1bΔ/+ was further away from P3 WT than 1aΔ/+; 1bΔ/Δ, indicating that JMJD1A is more crucial than JMJD1B in this process. Importantly, among the mutants, 1aΔ/Δ; 1bΔ/Δ was farthest from P3 WT and nearest to P0 WT (Figure 5A), indicating that the loss of all four Jmjd1 alleles induced the most severe developmental delay from P0 to P3. Notably, the PCA plot showed that PC2 (y axis) mainly contributed to the P0 to P3 transition (Figure 5A). Genes related to neuron differentiation, meiosis, and chromosome segregation were significantly enriched in PC2 (Figure 5B); therefore, perturbed expression of these genes might account for the developmental delay of germ cells lacking JMJD1A and/or JMJD1B.
Figure 5.
JMJD1A/JMJD1B Depletion Perturbs the Prospermatogonia to Spermatogonia Transition
(A) A PCA plot was used to visualize the degree of gene expression dissimilarity in male germ cells between the indicated genotypes and developmental times. We extracted differentially expressed genes between P0 WT and P3 WT and used them for PCA.
(B) Gene ontology analysis of genes contributing to PC2 shown in Figure 5A. We calculated the loading of PC2 and used the top 5% of genes for gene ontology analysis.
(C) Immunofluorescence analysis of FOXO1 subcellular localization in control and JMJD1A/JMJD1B-depleted germ cells at P3. We classified localization patterns of FOXO1 as cytoplasmic (indicated by white arrowheads), nuclear, or both (indicated by yellow arrows). Scale bars, 50 μm.
(D) Comparative evaluation of the subcellular localization of FOXO1 between control and JMJD1A/JMJD1B-depleted germ cells. Error bars represent ± SEM (n = 3 mice per genotype).
(E) Intratubular localization of prenatal and postnatal germ cells in the seminiferous tubule. Control and mutant testes sections at the indicated developmental times were stained with antibodies to TRA98 and DAPI. “Peri” and “Central” represent TRA98+ germ cells in contact with the basement membrane and not in contact with the basement membrane, respectively. Error bars represent ± SEM (n = 3 mice per indicated genotype and developmental time).
(F) Expression of spermatogonial stem cell marker genes in postnatal male germ cells was visualized with a heatmap. mRNA-seq data of P3 germ cells of the indicated genotypes was utilized. Vasa and Dazl were used as pan-germ cell marker genes. Daggers represent genes required for the maintenance of male germ cells. The heatmap represents log2-transformed FPKM values of each sample normalized to those of the control.
To further confirm the requirement of JMJD1 isozymes for prospermatogonia to spermatogonia transition, we examined the subcellular localization of FOXO1, a marker of prospermatogonia/spermatogonia (Figures 5C and 5D). FOXO1 translocates from the cytoplasm to the nucleus during the prospermatogonia to c-KIT− spermatogonia transition (Goertz et al., 2011). In control P3 germ cells, a population in which FOXO1 was present in the cytoplasm and nucleus reached over 50%. In contrast, in 1aΔ/Δ; 1bΔ/Δ germ cells, this population was minor (∼20%) but another population in which FOXO1 was detected only in the cytoplasm was predominant (∼80%) (Figure 5D). This cytological analysis confirmed perturbation of the prospermatogonia to spermatogonia transition in 1aΔ/Δ; 1bΔ/Δ germ cells. Prospermatogonia migrate into the basement membrane to form an SSC pool in seminiferous tubules during the prospermatogonia to spermatogonia transition. To address whether this process was inhibited by JMJD1A/JMJD1B depletion, we examined the intratubular localization of germ cells in P0 and P3 testes. No significant difference in intratubular localization was observed between mutant and control germ cells at P0 (Figure 5E). In contrast, ∼70% of control germ cells had migrated into the basement membrane, whereas only ∼50% of mutant cells had migrated at P3, further confirming that the prospermatogonia to spermatogonia transition was perturbed by JMJD1A/JMJD1B depletion.
To understand how JMJD1A/JMJD1B depletion affects the expression of SSC marker genes, we compared mRNA levels of P3 germ cells by heatmap analysis (Figure 5F). The expression levels of pan-germ cell marker genes, such as Vasa and Dazl, were not affected by JMJD1A/JMJD1B depletion, whereas the expression levels of SSC marker genes were significantly reduced in 1aΔ/Δ; 1bΔ/Δ cells (Figure 5F), indicating that JMJD1A/JMJD1B selectively and positively regulates SSC marker genes during the prospermatogonia to spermatogonia transition. Notably, reduced expression of SSC maker genes was more prominent in 1aΔ/Δ; 1bΔ/+ cells than in 1aΔ/+; 1bΔ/Δ cells, indicating a predominant role of JMJD1A in the activation of SSC marker genes. Among these genes, several are required for SSC maintenance (as discussed later). We speculate that the inactivation of multiple SSC maintenance genes may account for the failure of functional spermatogonia development in 1aΔ/Δ; 1bΔ/Δ germ cells.
To investigate the cause of the disturbed prospermatogonia to spermatogonia transition, we performed proliferation and cell death analyses in mutant testes at P3 (Figures S5A and S5B). EdU incorporation analysis indicated decreased proliferation of 1aΔ/Δ; 1bΔ/Δ germ cells in P3 testes (Figures S5A). In contrast, the frequency of apoptotic cells was not changed by JMJD1A/JMJD1B depletion (Figure S5B). These data indicate that perturbed prospermatogonia to spermatogonia transition in 1aΔ/Δ; 1bΔ/Δ germ cells might occur by inhibition of cell-cycle progression. A mitotic wave is initiated in male germ cells after birth; therefore, dysregulation of the genes related to chromosomal segregation (Figure 5B) might inhibit this mitotic wave in 1aΔ/Δ; 1bΔ/Δ germ cells.
JMJD1 Isozymes Preferentially Activate Transcription in Gene-Dense Regions of Chromosomes in Prospermatogonia
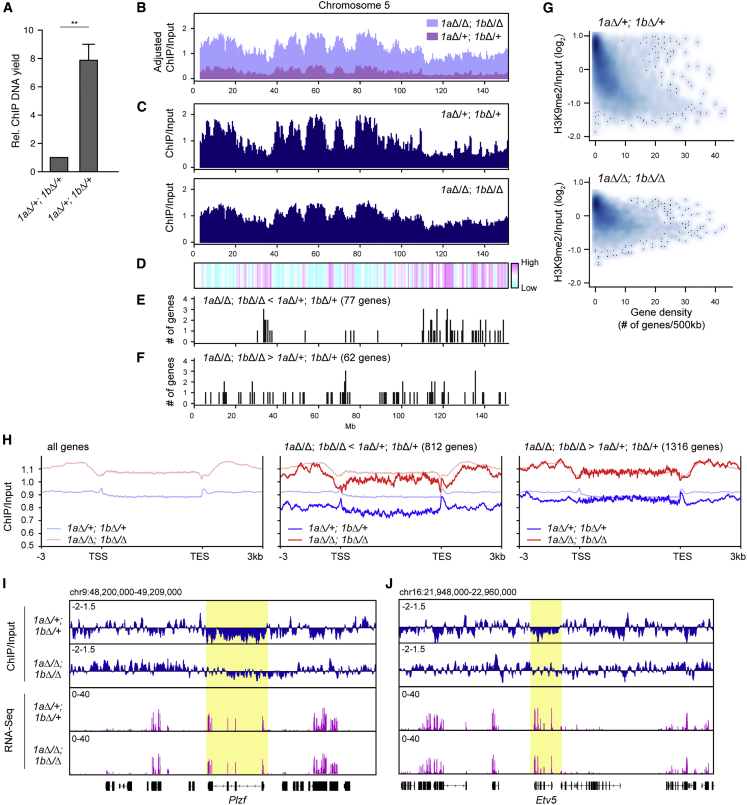
To examine the contribution of JMJD1-mediated H3K9me2 demethylation to transcription at the single-gene level, we performed H3K9me2 chromatin immunoprecipitation sequencing (ChIP-seq) analysis using purified P3 male germ cells. H3K9me2 ChIP analysis demonstrated that an 8- to 10-fold greater amount of DNA was immunoprecipitated from 1aΔ/Δ; 1bΔ/Δ cells compared with control cells (Figure 6A). This is in accord with the immunofluorescence analysis of H3K9me2 (Figure 2D). We present two patterns of H3K9me2 distribution. First is the absolute distribution of H3K9me2, in which data were normalized to the immunoprecipitated amounts of DNA (Figure 6B, chromosome 5 was used as a representative chromosome). Second is the relative distribution of H3K9me2, in which similar numbers of reads from control and mutant cells are displayed (approximately 40 million reads) (Figure 6C). As shown in Figure 6B, an increase in H3K9me2 was prominent throughout chromosome 5, indicating that JMJD1 isozymes target global regions of chromosomes. Relative H3K9me2 distribution profiles showed that H3K9me2 was unevenly distributed in P3 control germ cells, with H3K9me2-high and -low regions (Figure 6C). Next, we calculated the fold increase of H3K9me2 induced by JMJD1A/JMJD1B depletion by dividing the H3K9me2 value of 1aΔ/Δ; 1bΔ/Δ cells by that of control cells (Figure 6D). We found that chromosomal regions preferentially demethylated by JMJD1A/JMJD1B (shown in red in Figure 6D) overlap with originally H3K9me2-low regions in control cells (Figure 6C).
Figure 6.
JMJD1 Isozymes Preferentially Activate Transcription in Gene-Dense Chromosomal Regions in Prospermatogonia
(A) P3 germ cells of the indicated genotypes were subjected to native ChIP analysis with an anti-H3K9me2 antibody and the yield of ChIP DNA was quantified. DNA collected from control cells was assumed as 1 for data normalization. Data are presented as the mean ± SEM (n = 3 independent experiments.) ∗∗p < 0.01 (t test).
(B–F) Correlation between H3K9me2 distribution and JMJD1A/JMJD1B-regulated genes in prospermatogonia. P3 germ cells were isolated from control (1aΔ/+; 1bΔ/+) and JMJD1A/JMJD1B-depleted (1aΔ/Δ; 1bΔ/Δ) germ cells at P3 for ChIP-seq analysis of H3K9me2. We used a genomic view of chromosome 5 as representative data. Data were normalized (B) and not normalized (C) according to the amount of immunoprecipitated DNA. (D) Heatmap showing the log2-transformed ratio of ChIP/input of JMJD1A/JMJD1B-depleted germ cells divided by that of control germ cells. (E and F) Chromosomal distribution of genes that were downregulated (E) and upregulated (F) in 1aΔ/Δ; 1bΔ/Δ P3 germ cells compared with those in 1aΔ/+; 1bΔ/+ P3 germ cells.
(G) Correlation between gene density and H3K9me2 levels in control germ cells (top) and JMJD1A/JMJD1B-depleted germ cells (bottom). The x and y axes indicate the number of mm10 RefSeq genes and the ChIP/input ratio of H3K9me2, respectively.
(H) Averaged profile of H3K9me2 around the gene bodies of control (1aΔ/+; 1bΔ/+: blue) and JMJD1A/JMJD1B-depleted (1aΔ/Δ; 1bΔ/Δ: red) germ cells. H3K9me2 profiles of all genes (left), downregulated (middle) and upregulated (right) in 1aΔ/Δ; 1bΔ/Δ P3 germ cells compared with that in 1aΔ/+; 1bΔ/+ P3 germ cells are presented.
(I and J) Distribution profile of H3K9me2 and expression profile for the Plzf (I) and Etv5 (J) loci. Log2-transformed ratios of ChIP/Input (top) and FPKM counts from RNA-seq analysis (bottom) are shown.
Gene expression analysis demonstrated that 812 and 1,316 genes were down- and upregulated in 1aΔ/Δ; 1bΔ/Δ P3 germ cells compared with 1aΔ/+; 1bΔ/+ P3 germ cells, respectively (listed in Table S1). The former and latter genes are hereafter referred to as dKO < Ctrl and dKO > Ctrl, respectively. Mapping analysis of JMJD1A/JMJD1B-regulated genes on chromosome 5 revealed that dKO < Ctrl genes were enriched in the regions where H3K9me2 was preferentially demethylated by JMJD1A/JMJD1B (Figures 6D and 6E), but this feature was not found in dKO > Ctrl genes (Figures 6D and 6F).
We previously demonstrated that JMJD1 isozymes preferentially targeted gene-dense regions of chromosomes in embryonic stem cells (Kuroki et al., 2018). To determine whether this feature was conserved in prospermatogonia, we examined the correlation between gene density and H3K9me2 in P3 germ cells (Figure 6G). There was an inverse relationship between gene density and H3K9me2 levels in control germ cells (Figure 6G, top). Importantly, this feature was dampened by JMJD1A/JMJD1B depletion (Figure 6G, bottom), indicating that JMJD1 isozymes preferentially target gene-dense regions of chromosomes in prospermatogonia.
We next examined the correlation between altered gene expression and altered H3K9me2 levels in JMJD1A/JMJD1B-depleted male germ cells at P3 (Figure 6H). As H3K9me2 levels in dKO > Ctrl genes were similar to those in all genes, upregulation of dKO > Ctrl genes might occur independently of H3K9 demethylation. In contrast, H3K9me2 levels were markedly lower in dKO < Ctrl genes compared with all genes, indicating that H3K9me2 in dKO < Ctrl genes was more actively targeted by JMJD1 isozymes compared with H3K9me2 in other loci. Finally, we examined the impact of JMJD1A/JMJD1B-mediated H3K9 demethylation on gene expression at the single-gene level. For this purpose, genomic loci around Plzf and Etv5 were chosen as representative regions (Figures 6I and 6J, respectively) because the expression levels of these SSC marker genes were downregulated by JMJD1A/JMJD1B depletion (Figure 5H). In control cells, H3K9me2 levels of Plzf and Etv5 gene bodies were much lower than those of adjacent genes. In 1aΔ/Δ; 1bΔ/Δ cells, the increase of H3K9me2 was more remarkable in Plzf and Etv5 gene bodies compared with adjacent genes (Figures 6I and 6J). Importantly, the expression levels of Plzf and Etv5 were reduced but those of adjacent genes were not affected by JMJD1A/JMJD1B depletion. As shown in Figure S6, these characteristics were also found in SSC marker genes whose expression was regulated by JMJD1A/JMJD1B. Collectively, the transcriptional activation of SSC marker genes in prospermatogonia is highly dependent on JMJD1A/JMJD1B-mediated H3K9 demethylation.
Discussion
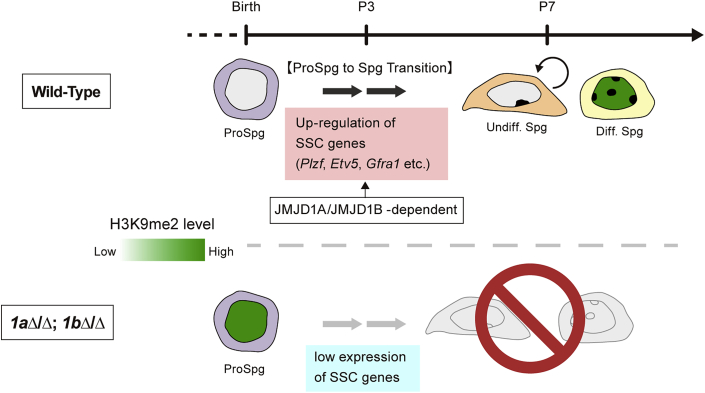
Our present studies are summarized in Figure 7. We propose that activation of SSC maintenance genes in prospermatogonia is critical for the subsequent differentiation into spermatogonia. JMJD1A and JMJD1B are redundant but substantially contribute to activation of these genes by removing repressive H3K9 methylation marks. Depletion of JMJD1A/JMJD1B results in increased H3K9 methylation and decreased transcription of SSC maintenance genes, thereby blocking the generation of functional (i.e., undifferentiated and differentiating) spermatogonia. Below, we discuss our results with reference to recent findings related to epigenetic regulation and germ cell development.
Figure 7.
Role of JMJD1A and JMJD1B in the Prospermatogonia to Spermatogonia Transition
In wild-type spermatogonia, JMJD1A and JMJD1B are highly expressed and the H3K9me2 mark is removed to extremely low levels in a JMJD1-dependent manner. This H3K9 hypomethylation status is critical for upregulation of SSC genes and the subsequent transition of prospermatogonia to spermatogonia after birth. In JMJD1A/JMJD1B-depleted spermatogonia, H3K9me2 is hypermethylated, resulting in low SSC gene expression and perturbed differentiation of prospermatogonia.
We previously demonstrated that the GLP/G9a complex is responsible for global H3K9me2 in embryonic stem cells (Tachibana et al., 2002, 2005). Recently, we reported that JMJD1A/JMJD1B contributed to transcriptional activation by demethylating H3K9me2 marks deposited by GLP/G9a (Kuroki et al., 2018). We speculate that JMJD1A/JMJD1B-mediated H3K9 demethylation may also counteract GLP/G9a complex-mediated H3K9 methylation in spermatogonia development. In support of this, the GLP/G9a complex is expressed in postnatal spermatogonia (Tachibana et al., 2007). Alternatively, Suv39h1 and Eset H3K9 methyltransferases are candidate enzymes that might counteract JMJD1A/JMJD1B-mediated H3K9 demethylation in prospermatogonia because Suv39h1 (Peters et al., 2001) and Eset (Liu et al., 2014) are expressed in pre-meiotic spermatogonia. To comprehensively examine the role of H3K9 methylation dynamics in spermatogonia development, it is important to identify which H3K9 methyltransferase counteracts JMJD1-mediated H3K9 demethylation.
In this study, we demonstrated partial non-redundancy of JMJD1 isozymes in spermatogenesis. As shown in Figure 3H, 1aΔ/+; 1bΔ/Δ germ cells could differentiate into mature sperm and the corresponding male mice were fertile. In contrast, 1aΔ/Δ; 1bΔ/+ germ cells could not complete meiosis and the corresponding male mice were infertile. These data indicate a predominant function of JMJD1A in postnatal spermatogenesis. Loss of all four Jmjd1a and Jmjd1b alleles resulted in the most severe postnatal male germ cell developmental phenotype, i.e., exhaustion of germ cells before puberty, indicating the redundancy of JMJD1A and JMJD1B in the maintenance of male germ cells. This phenotype was associated with a marked inhibition of first-round spermatogenesis (Figure 4D) and a lack of functional spermatogonia (Figure 4L). Many studies have demonstrated that spermatogonia express specific marker genes related to transcription factors and cell surface receptors. Of these, several transcription factors, including Oct4 (Dann et al., 2008), Id4 (Oatley et al., 2011), Ret (Jain et al., 2004), Gfra1 (Naughton et al., 2006), Plzf (Buaas et al., 2004; Costoya et al., 2004), Etv5 (Morrow et al., 2007), Lhx1 (Oatley et al., 2007), and Bcl6b (Oatley et al., 2006), have important roles in maintaining the male germline. Interestingly, expression levels of these genes were significantly reduced in 1aΔ/Δ; 1bΔ/Δ germ cells (Figure 5F). The decreased expression of multiple spermatogonia-specific transcription factors might synergistically affect the cellular integrity of the male germline in JMJD1A/JMJD1B-depleted germ cells.
DNA methylation levels change dynamically throughout the reproductive cycle (Seisenberger et al., 2013). Several studies, including ours, also reveal that H3K9me2 levels are dynamically regulated during the reproductive cycle. First, we demonstrated that JMJD1A and JMJD1B redundantly control embryogenesis around the peri-implantation period, where JMJD1 isozymes have crucial roles during epiblast development (Kuroki et al., 2018). Second, JMJD1A contributes to sex determination by activating Sry in gonadal somatic cells at E11.5. JMJD1A deficiency resulted in the failure of H3K9 demethylation at the Sry locus and the failure of Sry activation, thereby inducing male-to-female sex reversal (Kuroki et al., 2013). Third, JMJD1A contributed to the elongation step of spermatogenesis by activating genes required for the histone to protamine transition (Liu et al., 2010; Okada et al., 2007). Finally, we show that JMJD1 isozymes substantially contribute to the prospermatogonia to spermatogonia transition. In summary, the dynamic regulation of H3K9me2 plays a critical role in various steps of the reproductive cycle. In addition to H3K9 methylation enzymes, there are many other types of histone K methylating and counteracting demethylating enzymes in mammals (Dillon et al., 2005; Kooistra and Helin, 2012). Therefore, it is likely that histone K methylation marks other than H3K9 methylation are also dynamically regulated during development and cell differentiation. The next important step is, therefore, to identify these methylating/demethylating enzymes and their target loci.
Experimental Procedures
Antibodies
The following antibodies were used were used for immunoblotting. Anti-SOX9 (Merck Millipore, AB5535), anti-HSD3b (TransGenic, KO607), anti-TRA98 (Abcam, ab82527), anti-FLAG (clone M2, Sigma, A8592), anti-VASA (Abcam, ab13840), anti-EPCAM (clone G8.8, eBioscience, no. 14-5791-85), anti-FOXO1 (CST, no. 2880), anti-PLZF (Santa Cruz, H300), anti-c-KIT (BioLegend, no. 105803), anti-JMJD1B (Abvnova, PAB15833), anti-H3K9me1 (Abcam, ab9045), anti-H3K9me2 (clone 6D11) (Kimura et al., 2008), anti-H3K9me3 (clone 2F3) (Kimura et al., 2008), anti-JMJD1A (clone F0618) (Abe et al., 2015), and anti-JMJD1B (Abvnova, PAB15833). Anti-H3K9me2 (Abcam, Ab1220) was used for ChIP analysis.
Mice
For the generation of Jmjd1a2lox and Jmdj1b2lox mice, Jmjd1a and Jmjd1b knockin targeting vectors were constructed using bacterial artificial chromosome recombineering (Copeland et al., 2001). Vectors were introduced into TT2 embryonic stem cells. Chimeric males derived from homologously recombined embryonic stem cell clones were obtained to generate F1 offspring bearing the mutant alleles, which were further backcrossed to C57BL/6 for more than five generations. Jmjd1bFLAG−KI (Kuroki et al., 2018), Vasa-Cre Tg (Gallardo et al., 2007), and Oct4-EGFP Tg mice (Yoshimizu et al., 1999) were described previously. All animal experiments were performed under the animal ethical guidelines of Osaka University, Tokushima University and Kyoto University.
Data and Code Availability
The RNA-seq and ChIP-seq data are deposited in the Gene Expression Omnibus database under accession number GSE148055.
Author Contributions
S. Kuroki and M.T. designed the experiments. S. Kuroki, Y.M., S. Kitano, H.M., and M.T. performed the experiments. S. Kuroki and R.M. analyzed the data. M.F. and Y.S. provided resources. S. Kuroki and M.T. wrote the manuscript. M.T. supervised the project and approved the manuscript.
Acknowledgments
We thank all members of the Tachibana laboratory. This work was supported by JSPS KAKENHI grant numbers JP26250037 (M.T.), JP16H01218 (M.T.), JP16H01409 (M.T.), JP17H06424 (M.T.), JP16K21196 (S. Kuroki), JP20H05364 (S. Kuroki) and JP20H03262 (S. Kuroki); the Funding Program for Next Generation World-Leading Researchers (M.T.); the Takeda Science Foundation (M.T. and S. Kuroki); the Suntory Foundation for life sciences (S. Kuroki); and a Promotion of Science Cooperative Research Grant of the Institute for Enzyme Research, Joint Usage/Research Center, Tokushima University (H.M.). We thank Edanz Group (https://en-author-services.edanzgroup.com/) for editing a draft of this manuscript.
Published: July 16, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.06.013.
Supplemental Information
References
- Abe Y., Rozqie R., Matsumura Y., Kawamura T., Nakaki R., Tsurutani Y., Tanimura-Inagaki K., Shiono A., Magoori K., Nakamura K. JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat. Commun. 2015;6:7052. doi: 10.1038/ncomms8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas F.W., Kirsh A.L., Sharma M., McLean D.J., Morris J.L., Griswold M.D., de Rooij D.G., Braun R.E. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Copeland N.G., Jenkins N.A., Court D.L. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- Costoya J.A., Hobbs R.M., Barna M., Cattoretti G., Manova K., Sukhwani M., Orwig K.E., Wolgemuth D.J., Pandolfi P.P. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- Dann C.T., Alvarado A.L., Molyneux L.A., Denard B.S., Garbers D.L., Porteus M.H. Spermatogonial stem cell self-renewal requires OCT4, a factor downregulated during retinoic acid-induced differentiation. Stem Cells. 2008;26:2928–2937. doi: 10.1634/stemcells.2008-0134. [DOI] [PubMed] [Google Scholar]
- de Rooij D.G. Stem cells in the testis. Int. J. Exp. Pathol. 1998;79:67–80. doi: 10.1046/j.1365-2613.1998.t01-1-00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi K., Nagamatsu G., Miyachi H., Kato Y., Morita S., Kimura H., Kitano S., Hatada I., Saga Y., Tachibana M. Posttranscriptional regulation of histone lysine methyltransferase GLP in embryonic male mouse germ cells. Biol. Reprod. 2013;88:36. doi: 10.1095/biolreprod.112.103572. [DOI] [PubMed] [Google Scholar]
- Dillon S.C., Zhang X., Trievel R.C., Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T., Shirley L., John G.B., Castrillon D.H. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45:413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goertz M.J., Wu Z., Gallardo T.D., Hamra F.K., Castrillon D.H. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J. Clin. Invest. 2011;121:3456–3466. doi: 10.1172/JCI57984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Naughton C.K., Yang M., Strickland A., Vij K., Encinas M., Golden J., Gupta A., Heuckeroth R., Johnson E.M., Jr. Mice expressing a dominant-negative Ret mutation phenocopy human Hirschsprung disease and delineate a direct role of Ret in spermatogenesis. Development. 2004;131:5503–5513. doi: 10.1242/dev.01421. [DOI] [PubMed] [Google Scholar]
- Kimura H., Hayashi-Takanaka Y., Goto Y., Takizawa N., Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- Kooistra S.M., Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Kubo N., Toh H., Shirane K., Shirakawa T., Kobayashi H., Sato T., Sone H., Sato Y., Tomizawa S., Tsurusaki Y. DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. BMC Genomics. 2015;16:624. doi: 10.1186/s12864-015-1833-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki S., Matoba S., Akiyoshi M., Matsumura Y., Miyachi H., Mise N., Abe K., Ogura A., Wilhelm D., Koopman P. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science. 2013;341:1106–1109. doi: 10.1126/science.1239864. [DOI] [PubMed] [Google Scholar]
- Kuroki S., Nakai Y., Maeda R., Okashita N., Akiyoshi M., Yamaguchi Y., Kitano S., Miyachi H., Nakato R., Ichiyanagi K. Combined loss of JMJD1A and JMJD1B reveals critical roles for H3K9 demethylation in the maintenance of embryonic stem cells and early embryogenesis. Stem Cell Reports. 2018;10:1340–1354. doi: 10.1016/j.stemcr.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhou S., Liao L., Chen X., Meistrich M., Xu J. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J. Biol. Chem. 2010;285:2758–2770. doi: 10.1074/jbc.M109.066845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Brind'Amour J., Karimi M.M., Shirane K., Bogutz A., Lefebvre L., Sasaki H., Shinkai Y., Lorincz M.C. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2014;28:2041–2055. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow C.M., Hostetler C.E., Griswold M.D., Hofmann M.C., Murphy K.M., Cooke P.S., Hess R.A. ETV5 is required for continuous spermatogenesis in adult mice and may mediate blood testes barrier function and testicular immune privilege. Ann. N. Y. Acad. Sci. 2007;1120:144–151. doi: 10.1196/annals.1411.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C.K., Jain S., Strickland A.M., Gupta A., Milbrandt J. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol. Reprod. 2006;74:314–321. doi: 10.1095/biolreprod.105.047365. [DOI] [PubMed] [Google Scholar]
- Oatley J.M., Avarbock M.R., Telaranta A.I., Fearon D.T., Brinster R.L. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc. Natl. Acad. Sci. U S A. 2006;103:9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J.M., Avarbock M.R., Brinster R.L. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J. Biol. Chem. 2007;282:25842–25851. doi: 10.1074/jbc.M703474200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley M.J., Kaucher A.V., Racicot K.E., Oatley J.M. Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol. Reprod. 2011;85:347–356. doi: 10.1095/biolreprod.111.091330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Scott G., Ray M.K., Mishina Y., Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- Peters A.H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schofer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Schrans-Stassen B.H., van de Kant H.J., de Rooij D.G., van Pelt A.M. Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology. 1999;140:5894–5900. doi: 10.1210/endo.140.12.7172. [DOI] [PubMed] [Google Scholar]
- Seisenberger S., Peat J.R., Hore T.A., Santos F., Dean W., Reik W. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20110330. doi: 10.1098/rstb.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Nozaki M., Takeda N., Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 2007;26:3346–3359. doi: 10.1038/sj.emboj.7601767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Pereira L.A., Nozaki M., Tsuchida J., Sawada K., Mori H., Nishimune Y. A germ cell-specific nuclear antigen recognized by a monoclonal antibody raised against mouse testicular germ cells. Int. J. Androl. 1997;20:361–366. doi: 10.1046/j.1365-2605.1998.00080.x. [DOI] [PubMed] [Google Scholar]
- Ueda J., Tachibana M., Ikura T., Shinkai Y. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J. Biol. Chem. 2006;281:20120–20128. doi: 10.1074/jbc.M603087200. [DOI] [PubMed] [Google Scholar]
- Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Sukeno M., Nakagawa T., Ohbo K., Nagamatsu G., Suda T., Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development. 2006;133:1495–1505. doi: 10.1242/dev.02316. [DOI] [PubMed] [Google Scholar]
- Yoshimizu T., Sugiyama N., De Felice M., Yeom Y.I., Ohbo K., Masuko K., Obinata M., Abe K., Scholer H.R., Matsui Y. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 1999;41:675–684. doi: 10.1046/j.1440-169x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and ChIP-seq data are deposited in the Gene Expression Omnibus database under accession number GSE148055.