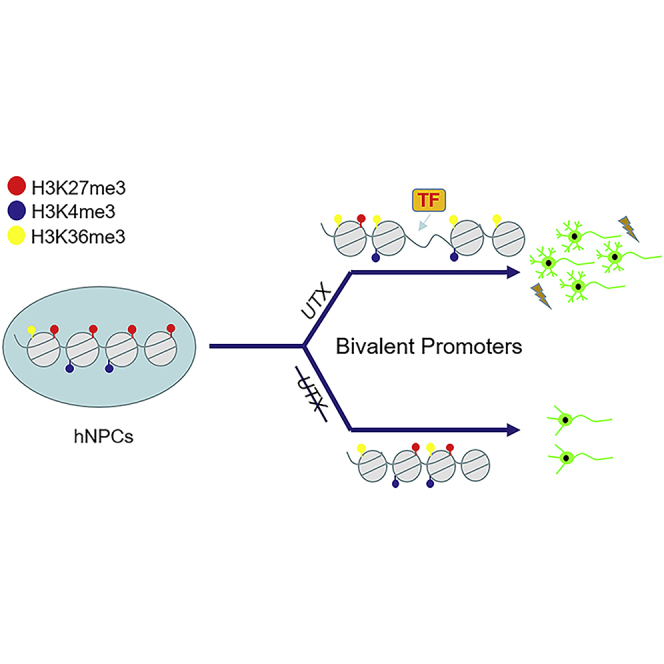
Summary
UTX, a H3K27me3 demethylase, plays an important role in mouse brain development. However, so little is known about the function of UTX in human neural differentiation and dendritic morphology. In this study, we generated UTX-null human embryonic stem cells using CRISPR/Cas9, and differentiated them into neural progenitor cells and neurons to investigate the effects of UTX loss of function on human neural development. The results showed that the number of differentiated neurons significantly reduced after loss of UTX, and that the dendritic morphology of UTX KO neurons tended to be simplified. The electrophysiological recordings showed that most of the UTX KO neurons were immature. Finally, RNA sequencing identified dozens of differentially expressed genes involved in neural differentiation and synaptic function in UTX KO neurons and our results demonstrated that UTX regulated these critical genes by resolving bivalent promoters. In summary, we establish a reference for the important role of UTX in human neural differentiation and dendritic morphology.
Key words: UTX, bivalent promoters, dendritic morphology, neural differentiation
Graphical Abstract
Highlights
-
•
Loss of UTX in hESCs reduces their neural differentiation potential
-
•
The dendritic morphology of UTX KO neurons tends to be simplified
-
•
UTX regulates human neural development depending on its demethylation
-
•
UTX regulates the expression of genes by resolving bivalent promoters
In this article, Liu and colleagues show that the deletion of UTX in hESCs leads to the decline of their ability to differentiate into neurons, and the decrease of neurite complexity of neurons. UTX regulates dozens of genes involved in neural differentiation and synaptic function by resolving bivalent promoters depending on its demethylation.
Introduction
Epigenetic regulations, which include DNA methylation, genomic imprinting, RNA editing, and histone modification, have been reported playing critical roles in almost every biological process (Bird, 2002; Jenuwein and Allis, 2001; Tsankova et al., 2007). Trimethylation of histone H3 at lysine 27 (H3K27me3) is believed to be a repressive epigenetic mark for maintaining transcriptional gene silencing (Stewart-Morgan et al., 2020). In mammals, the dynamic steady-state levels of di-methylation and tri-methylation of histone H3 lysine 27 (H3K27me2/3) are mainly regulated by the balance between the methyltransferase polycomb repressor complex 2 (PRC2) and the histone demethylases UTX (also known as KDM6A) and JMJD3 (also known as KDM6B) (Agger et al., 2007; Lan et al., 2007; Margueron and Reinberg, 2011; Swigut and Wysocka, 2007). Trimethylated histone H3 lysine 27 (H3K27me3) is critical for wound repair, skeleton growth, and cell fate determination (Ezhkova et al., 2011; Wei et al., 2009; Zhang et al., 2015). Meanwhile, the JMJC domain protein UTX acts as an H3K27-specific demethylases that remove this H3K37me mark, enabling the activation of genes involved in cancer, germ cell epigenetic reprogramming, and inflammation (Grasso et al., 2012; Kruidenier et al., 2012; Mansour et al., 2012).
H3K27me3 is a mark of gene repression, while H3K4me3 is a mark of gene activation. These two marks denoting opposite gene expression states co-localize at many differentiation-specific gene promoters in stem cells to form bivalent promoters in which they are located at different H3 tails in nucleosomes (Blanco et al., 2020; Minoux et al., 2017). The H3K4 methyltransferase mixed-lineage leukemia 2 (MLL2; also known as KMT2B) is important for the formation of H3K4me3 in bivalent domains, and the H3K4 methyltransferase MLL1 plays a redundant role in depositing H3K4me3 to generate bivalent domains (Denissov et al., 2014; Hu et al., 2013). Besides, the H3K27 methyltransferase complex PRC2 and H3K4 methyltransferases SET1A and SET1B are associated with the generation of bivalency (Voigt et al., 2013). However, we still know very little about which histone methylation modifier is responsible for the resolution of bivalent domains into active monovalent states during human neural differentiation.
Previous studies have demonstrated that the regulation of H3K27me3 is absolutely indispensable for the development and function of the mammalian nervous system (Aldiri et al., 2017; Ayata et al., 2018; Henriquez et al., 2013; Liu et al., 2017). For example, Ezh2, a histone methyltransferase of PRC2, is essential for controlling the rate at which cortical progenitor cells proliferate and differentiate into lineage cells, and loss of Ezh2 results in a removal of the repressive mark of H3K27me3 in cortical progenitor cells and a decline in terminally differentiated neurons (Pereira et al., 2010). During neocortical development, the polycomb group complex restricts neurogenic competence of neural progenitor cells (NPCs) and promotes the fate transition of NPCs from neurogenic to astrogenic (Hirabayashi et al., 2009). UTX is broadly expressed in various regions of the mouse brain (Xu et al., 2008), and its mutations are associated with Kabuki syndrome whose patients have developmental delay and intellectual disability (Miyake et al., 2013). In addition, UTX has also been demonstrated to be the promotional transcription factor of Pten by demethylating H3K27me3 at the Pten promoter to control neurogenesis in mice (Lei and Jiao, 2018). However, some other findings suggest that the H3K27me demethylase activity of UTX is dispensable for the development of the nervous system. For instance, Kabuki causative point mutations upstream of the JmjC domain do not destroy UTX demethylation (Shpargel et al., 2017). In addition, deletion of UTX does not affect global H3K27me levels in neural crest cells (Shpargel et al., 2017). Therefore, whether the regulation mechanism of UTX in neurodevelopment depends on its demethylase function remains to be investigated.
In our previous study, we found that specific deletion of UTX in the mouse forebrain results in aberrant dendrite complexity and abnormal synaptic plasticity, and H3K27me3 level increased in the hippocampus of UTX knockout (KO) mice (Tang et al., 2017). However, the role and mechanism of UTX in human neural morphogenesis and development have not yet been elucidated. In this study, we investigated the role of UTX in neural progenitors and neurons differentiated from human embryonic stem cells (hESCs). We found that UTX was upregulated upon neural differentiation of hESCs, and loss of UTX in hESCs led to the decline in their differentiation potential into neurons, the decrease of neurite complexity, and the defect of electrophysiological function. Finally, we provided evidence showing that UTX regulated human neural differentiation and dendritic morphology of neurons by resolving bivalent promoter dependent on its H3K27 demethylase activity.
Results
UTX Is Enriched in Human NPCs during Differentiation of hESCs into Neurons
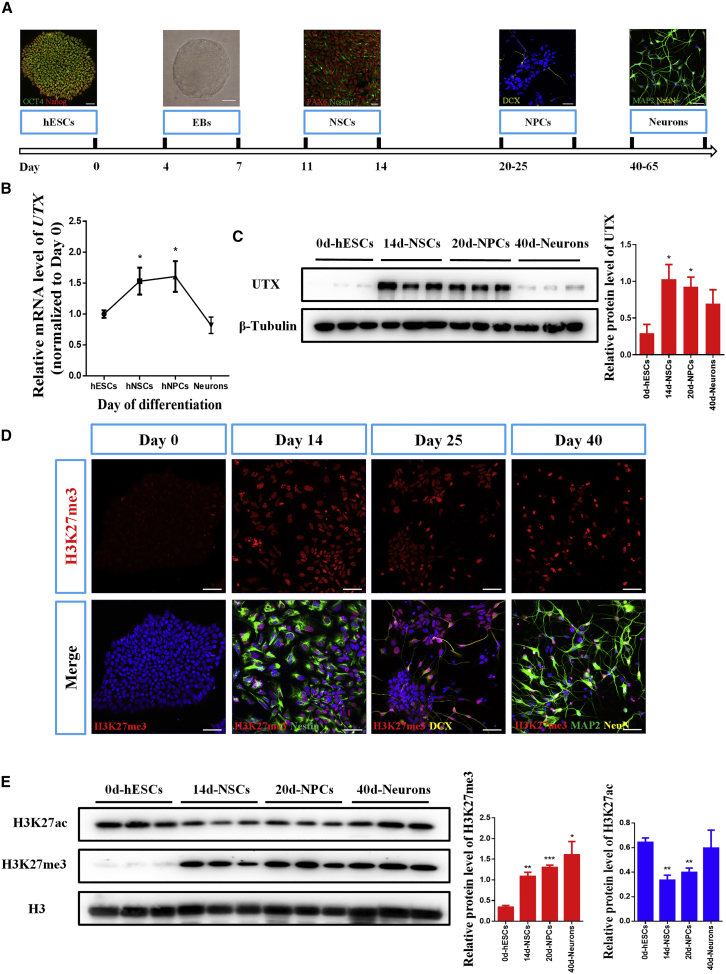
To investigate the expression of UTX and H3K27me3 during human neural differentiation, we utilized an in vitro culture system to differentiate hESCs into human neural stem cells (hNSCs), human neural progenitor cells (hNPCs), and neurons (Figure 1A). Real-time RT-PCR analysis showed that the mRNA expression of UTX was not abundant in hESCs, but was increased in hNSCs and hNPCs (Figure 1B). Consistent with this, higher protein levels of UTX in hNSCs and hNPCs were also detected by western blotting (Figure 1C). Although UTX is known to have histone demethylase activity, both immunofluorescence and western blotting assays demonstrated that the expression levels of both H3K27me3 and UTX were also significantly increased during the neural differentiation of hESCs (Figures 1D and 1E). Meanwhile, the expression of H3K27ac was declined by almost 30% in hNSCs and hNPCs (Figure 1E). These results suggested that UTX might play an important role in neural differentiation of hESCs.
Figure 1.
UTX Is Upregulated in Earlier Stages of Neural Differentiation of hESCs
(A) Schematic illustration of neural differentiation of hESCs. hESCs, human embryonic stem cells; EB, embryoid bodies; hNSCs, human neural stem cells; hNPCs, human neural progenitor cells. Scale bar, 50 μm.
(B) Real-time PCR analysis showing higher mRNA levels of UTX in earlier stages of neural differentiation of hESCs. GAPDH is used as an internal control for real-time PCR.
(C) Western blotting indicating upregulated protein levels of UTX at earlier stages during neural differentiation of hESCs. β-Tubulin is used as a loading control.
(D) Representative images of H3K27me3 immunostaining at different stages of neural differentiation of hESCs. DAPI, a nuclei marker; Nestin, an NSC marker; DCX, an NPCs marker; MAP2, a mature neuron marker. Scale bar, 50 μm.
(E) Western blotting showing protein levels of H3K27me3 and H3K27ac upon neural differentiation of hESCs. Histone 3 was used as a loading control.
Results are presented as mean ± SEM; n ≥ 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
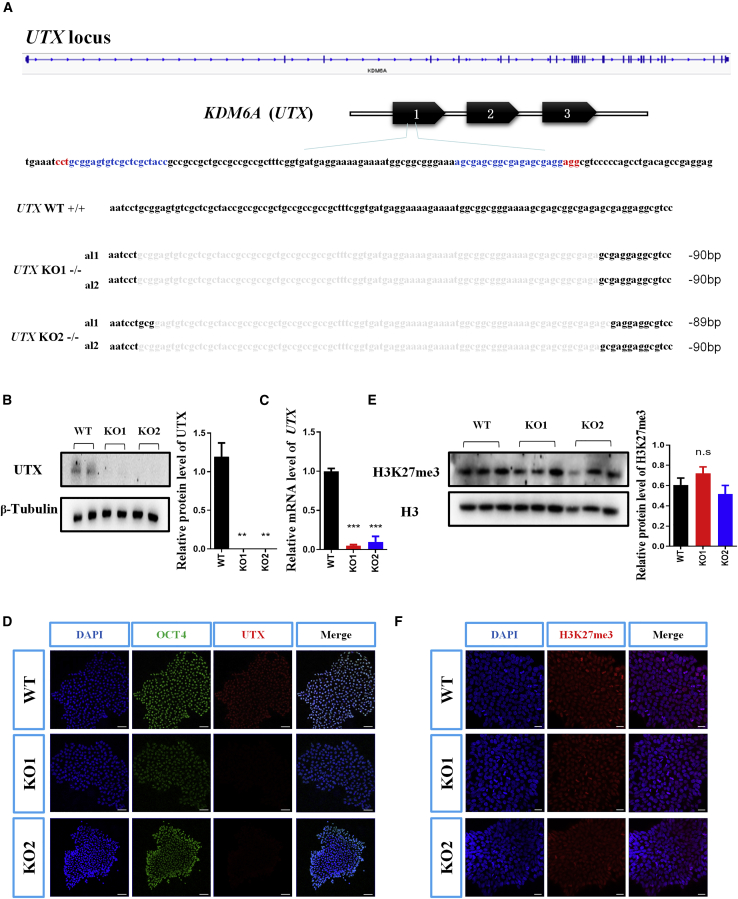
The Expression of H3K27me3 Is Not Altered in UTX KO hESCs
To explore the function of UTX in neural differentiation of hESCs, we conducted CRISPR/Cas9-mediated gene editing to knock out UTX in hESCs. We successfully established two UTX-null hESC clones (KO1, KO2) (Figure 2A). Two specific single-guide RNA (sgRNAs) targeting exon 1 of UTX were designed (as shown in Figure 2A and at the Optimized CRISPR Design website http://crispr.mit.edu) (Cong et al., 2013). Off-target effect was not detected in six potential genes that were predicted by the Optimized CRISPR Design tool, indicating the specificity of the selected sgRNAs (Figure S1A).
Figure 2.
Generation and Characterization of UTX Knockout hESCs
(A) CRISPR sgRNA sequences and mutations in two UTX knockout clones (KO1, KO2). The sgRNA sequences are labeled in blue and PAM recognition sequences are highlighted in red. Light-colored sequences indicating the deletion of allele 1 (al1) and allele 2 (al2). WT, wild type.
(B) Western blotting demonstrating that UTX protein is undetectable in both UTX KO clones.
(C) Real-time PCR analysis confirming the mutation of UTX in both UTX KO clones.
(D) Representative images of UTX immunostaining of UTX KO hESCs. Scale bar, 20 μm.
(E) Western blotting indicating normal protein expression of H3K27me3 in UTX KO clones. Histone 3 was used as a loading control.
(F) Representative images of H3K27me3 immunostaining showing its unaltered expression in UTX KO clones. Results are presented as mean ± SEM; n = 3 independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001.
To confirm the deletion of UTX in hESCs, we performed western blotting, RT-PCR, and immunofluorescent staining analyses of UTX KO clones. Compared with wild-type (WT) cells, we observed a complete loss of UTX protein both in KO1 and KO2 hESCs (Figures 2B and 2D). Consistent with this, the mRNA expression of UTX was almost undetectable in UTX KO hESCs (Figure 2C). Next, we examined the expression of H3K27me3 in UTX KO hESCs, and found that H3K27me3 was unchanged upon the deletion of UTX in hESCs (Figures 2E and 2F). Besides, the expression of JMJD3, another H3K27me3 demethylase, was not changed in UTX KO hESCs (Figure S1B) and two UTX knockout cell lines had a normal karyotype (Figure S1C). In summary, these observations supported the idea that UTX might play roles independent of its histone lysine demethylase activity in hESCs.
Deletion of UTX Has No Effects on Pluripotency and Self-Renewal of hESCs
To confirm the pluripotency status of the established UTX KO hESC clones, we examined the expression of several markers for stemness and cell proliferation. We observed similar expression levels of pluripotency markers in UTX WT and KO cell lines by immunofluorescence staining, qPCR, and western blotting (Figures S2A–S2C). UTX WT and KO cells also displayed similar proliferation potentials by 5-bromo-2′-deoxyuridine (BrdU) incorporation and PH3S10 immunofluorescent staining analyses (Figures S3A and S3B). These results suggested that UTX deletion did not affect pluripotency or self-renewal of hESCs.
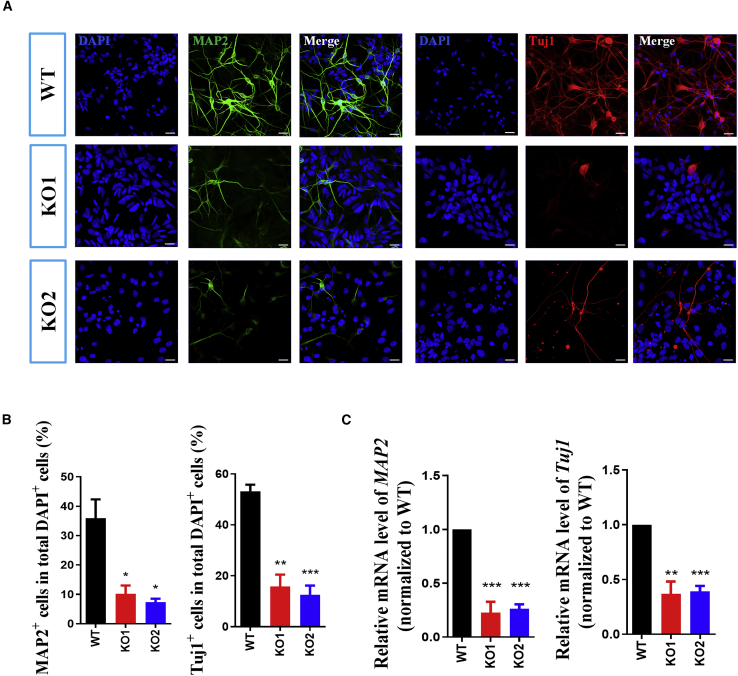
UTX Is Required for Neural Differentiation of hESCs
We next induced hESCs to differentiate toward hNSCs to investigate the function of UTX during early neural differentiation of hESCs. At day 14 after neural differentiation, both UTX WT and KO hNSCs could express the typical neural progenitor-specific markers PAX6 and Nestin, with no discernible difference in the percentage of PAX6+ or the fluorescence of Nestin+ cells (Figures S4A–S4C). RT-PCR results then confirmed that both UTX WT and KO hNSCs had the same mRNA expression levels of PAX6 and Nestin (Figure S4E).
Quantification of BrdU+ cells after pulse labeling indicated that UTX loss of function had no obvious effect on the proportion of BrdU+ cells compared with WT hNSCs (Figures S4A and S4D). However, at day 40 after neural differentiation, we observed decreased numbers of MAP2+ neurons and Tuj1+ neurons in both UTX KO clones (Figures 3A and 3B), indicating that UTX loss of function decreased neural differentiation efficiency of hNSCs. Moreover, our RT-PCR analysis showed a significant decrease in mRNA expression levels of MAP2 and TUJ1 in UTX KO cells compared with WT control at day 40 after neural differentiation of hESCs (Figure 3C), further proving that deletion of UTX inhibited neural differentiation of hESCs.
Figure 3.
UTX Loss-of-function Inhibits Neural Differentiation of hESCs
(A) Immunostaining of neuron markers MAP2 and Tuj1 at day 40 of neural differentiation. Scale bar, 20 μm.
(B) Quantitative analysis showing reduced percentages of MAP2+ and Tuj1+ neurons derived from UTX KO hESCs.
(C) qRT-PCR analysis showing that the expression levels MAP2 and Tuj1 were significantly lower in UTX KO cells at day 40 of neural differentiation. Results are presented as mean ± SEM; n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
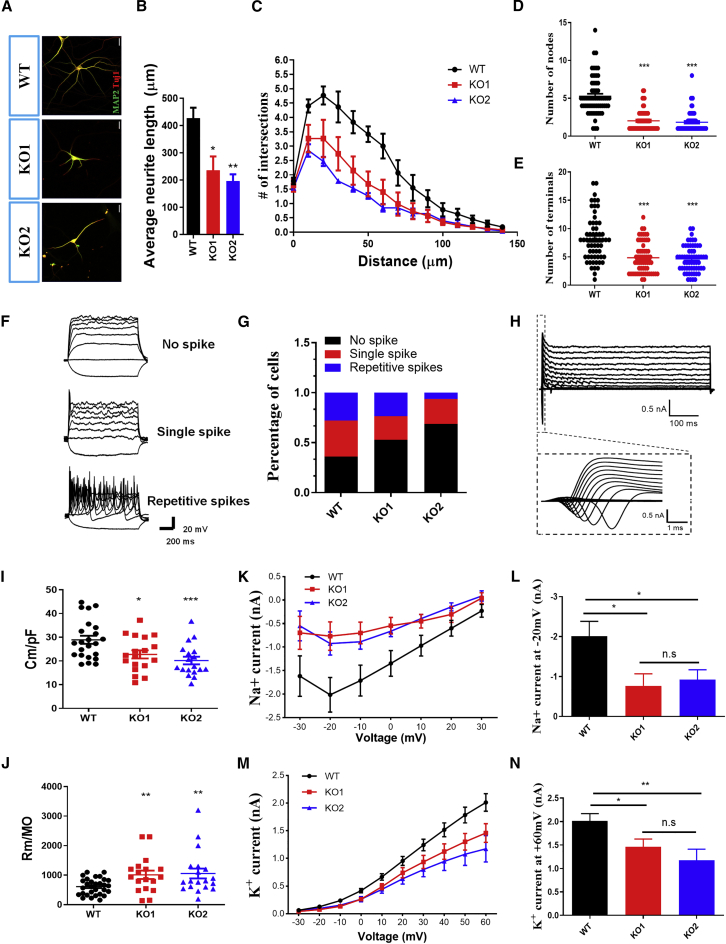
Loss of UTX Results in Decreased Neurite Complexity and Electrophysiological Defects in hESC-Derived Neurons
We next determined whether the loss of UTX affected neurite growth of hESC-derived neurons. Morphology analysis showed that UTX KO hESC-derived neurons exhibited a significant reduction in neurite complexity with decrease in total neurite length, and numbers of nodes and ends, compared with WT neurons at day 40 after neural differentiation (Figures 4A–4E).
Figure 4.
Loss of UTX Results in Decreased Neurite Complexity and Electrophysiological Defects of hESC-Derived Neurons
(A) Representative images of neurons derived from UTX WT and KO hESCs at day 40 of neural differentiation.
(B) Sholl analysis showing that total neurite length reduced in UTX KO hESC-derived neurons (60 neurons in every group in 3 independent experiments).
(C–E) Compared with WT group, UTX KO hESC-derived neurons exhibited decreased dendritic complexity as shown in reduced numbers of intersections (C), nodes (D), and ends (E).
(F) Representative traces of membrane potential responding to step depolarization by current injection steps from −10 pA to +60 pA in 10-pA increments. Membrane potential was current-clamped at around -65 mV. Representative traces were displayed by WT neurons.
(G) Quantification of the neuron maturity by recorded AP firing patterns at day 60 after differentiation (more than 15 neurons in every group have been recorded).
(H) Representative traces of whole-cell currents in voltage-clamp mode. Cells were held at −70 mV. Step depolarization from −80 to +60 mV at 10-mV intervals was delivered. Inset shows Na+ currents.
(I) Quantification of membrane capacitance in neurons at 60 days after differentiation. Error bars indicate ±SEM. ∗p < 0.05; a two-tailed t test was performed.
(J) Quantification of membrane resistance in neurons at 60 days after differentiation. ∗p < 0.05.
(K and L) Averaged current-voltage relationship (I-V curves) for Na+ currents, recorded from hESC-derived neurons. ns, not significant; ∗p < 0.05, ∗∗p < 0.01.
(M and N) Averaged current-voltage relationship (I-V curves) for K+ currents, recorded from hESC-derived neurons. ns, not significant; ∗p < 0.05, ∗∗p < 0.01.
To further examine the effect of UTX loss of function on the maturity of hESC-derived neurons, we performed electrophysiological recordings of hESC-derived neurons in each group using whole-cell patch recordings at day 60 (ASV 0 nM, n = 28; 50 nM, n = 20; 100 nM, n = 26; 1,000 nM) (Figure 4F). We found that, in the WT group, 27.7% of the recorded mature action potential (AP) firing properties with faster and consistent AP velocity, and 36.1% fire single or fewer spikes of APs, 36.1% of the recorded cells fail to show spikes of APs (Figure 4G). As expected, the proportion of immature neurons and no-spike cells increased in both UTX KO1 (52.9%) and KO2 (68.7%) cell lines, again suggesting that loss of UTX affects the maturity of hESC-derived neurons (Figure 4G). Moreover, membrane resistance (Figure 4I), membrane capacitance (Figure 4J), and sodium/potassium current (Figures 4K–4N) exhibited downward trends in UTX KO hESC-derived neurons compared with the control cells, indicating that hESC-derived neurons lacking UTX have impaired electrophysiological features.
Loss of UTX Leads to an Abnormal Transcription Profile in hESC-Derived Neurons
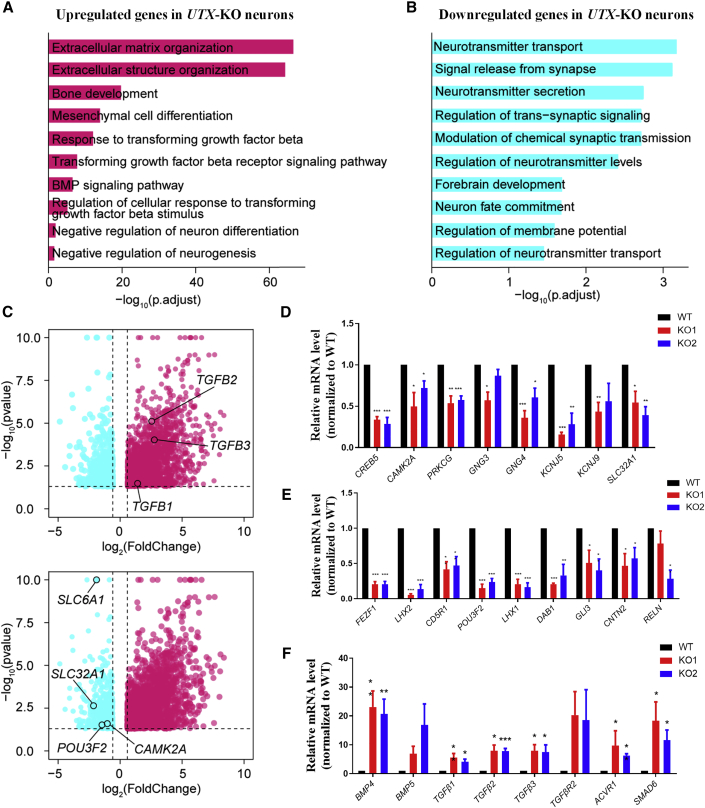
To understand the molecular mechanisms underlying the decreased neurite complexity and electrophysiological defects in UTX KO hESC-derived neurons, we performed RNA sequencing (RNA-seq) of hESC-derived neurons at day 40 after neural differentiation. RNA-seq data analysis showed that the transcription profile of UTX KO neurons was significantly different from that of control cells (Figure S5A). The clustering analysis and principal-component analysis proved clear discrimination between the UTX KO group and the WT group, indicating that neurons displayed distinctly gene expression pattern under the loss of UTX (Figures S5B and S5C). A total of 954 downregulated genes and 2,611 upregulated genes were identified in UTX KO neurons (Figure S5A). Gene ontology analysis showed that upregulated genes in UTX KO neurons were enriched for several cellular biological processes, including extracellular matrix organization, bone development and mesenchymal cell differentiation, and transforming growth factor β (TGF-β) signaling pathway (Figure 5A). Meanwhile, downregulated genes in UTX KO neurons were enriched for neurotransmitter transport, signal release from synapse, and neuron fate commitment (Figure 5B). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis further demonstrated that many downregulated genes in UTX KO neurons contributed to glutamatergic, dopaminergic, GABAergic, or cholinergic synapse pathway enrichment (Figures S5D and S5E). In contrast, KEGG analysis showed that the upregulated genes in UTX KO neurons were enriched in the TGF-β signaling pathway (Figures S5D and S5E), a well-known pathway which regulates a diverse range of biological processes during development and adult tissue homeostasis. Finally, we validated the differential expression of 25 genes which were randomly selected from genes positively participated in synaptic function (Figure 5D), neural development (Figure 5E), or TGF-β signaling pathway (Figure 5F) using qPCR. These results suggest that UTX is a critical regulator for human neural development and synaptic function.
Figure 5.
UTX KO Neurons Have an Abnormal Gene Expression Profile that Is Related to the Development and Synaptic Function
(A) Gene ontology terms enriched in genes upregulated in UTX KO-derived neurons.
(B) Gene ontology terms enriched in genes downregulated in UTX KO-derived neurons.
(C) Volcano plot of downregulated genes (blue) and upregulated genes (red) in UTX KO-derived neurons. Genes associated with TGF-β signaling pathway (TGF-β1, TGF-β2, TGF-β3) and neural function (SLC6A1, SLC32A1, POU3F2, CAMK2A) are indicated.
(D) qPCR analysis verified that genes positively participated in synapse-related pathways were significantly downregulated in UTX KO neurons.
(E) qPCR analysis verified that genes positively participated in neural development were significantly downregulated in UTX KO neurons.
(F) qPCR analysis verified that genes positively participated in TGF-β signaling pathway were significantly upregulated in UTX KO neurons. Results are presented as mean ± SEM; n = at least 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. FC, fold change.
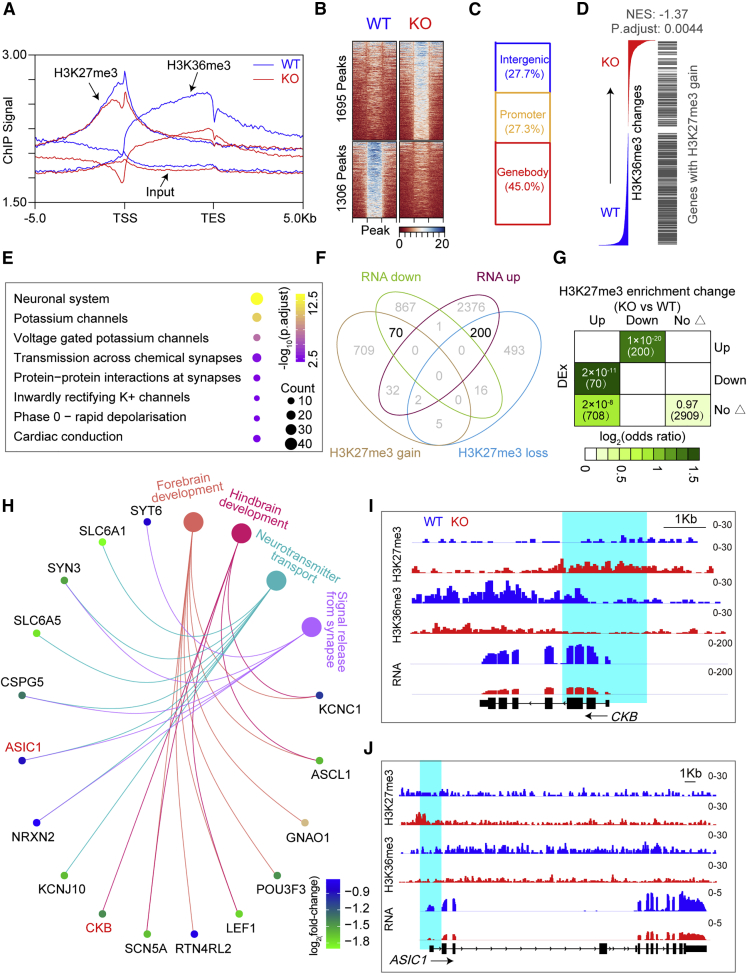
UTX KO Results in H3K27me3 Accumulation and H3K36me3 Reduction in hESC-Derived Neurons
To better understand the regulatory mechanism of UTX in hESC-derived neurons, we performed chromatin immunoprecipitation sequencing (ChIP-seq) to analyze the abundance of histone modifications in gene promoters. Meta-analysis of ChIP-seq data showed that both UTX KO and WT neurons displayed a broad distribution of H3K27me3 and H3K36me3 in promoters (±5 kb from the transcription start site [TSS]), and there was a slight reduction in H3K27me3, but an obvious reduction in H3K36me3 at the TSS regions (Figures 6A and S6A). We then focused on 1,695 increased peaks of H3K27me3 enrichment (Figure 6B), and found these increased peaks most distributed in gene bodies and promoters (Figure 6C). Furthermore, gene set enrichment analysis showed these H3K27me3-gained regions tended to lose H3K36me3 enrichment (Figures 6D and S6B). Reactome pathway enrichment analysis indicated these gained H3K27me3 peaks were mostly associated with neuronal system, voltage-gated potassium channel, and transmission across chemical synapses, and so on (Figure 6E). To further explore the regulatory mechanism of UTX, we performed conjoint data analysis of Chip-seq and RNA-seq, and found that there were 70 downregulated genes from RNA-seq analysis that gained H3K27me3 enrichment after the loss of UTX (Figure 6F). Moreover, odds ratio analysis and BETA plot of combined computational analysis showed that the decreased expression of these genes was obviously correlated with the gain of H3K27me3 (Figures 6G and S6C). Gene ontology enrichment analysis of these 70 genes showed that they are involved in forebrain development, hindbrain development, neurotransmitter transport, and signal release from synapse (Figure 6H). CKB and ASIC1 are important for neural development and synaptic function (Inoue et al., 2004; Wemmie et al., 2003), and a reduced H3K36me3 enrichment and an increased H3K27me3 enrichment at promoters of these two genes were detected in UTX KO neurons compared with that of WT neurons. Therefore, our results demonstrated that UTX regulated transcription of genes associated with neural development and synaptic function depending on its demethylase.
Figure 6.
The Gain of H3K27me3 Associates with the Reduction of Transcription in UTX KO Neurons
(A) Average profiles of histone marks and input within ±5 kb of gene body of all GENCODE annotated genes.
(B) Heatmaps of H3K27me3 peaks divided into increased enrichment (1,695 peaks) and decreased enrichment (1,306 peaks) group before and after UTX knockout separately.
(C) Bar chart showing distribution of gained H3K27me3 peaks caused by UTX knockout at annotated genomic regions.
(D) Gene set enrichment analysis of association between changes of H3K27me3 and H3K36me3 changes.
(E) Reactome pathway enrichment analysis of genes with gained H3K27me3.
(F) Venn diagram of a combined comparison of differentially expressed genes (RNA-up, upregulated genes; RNA-down, downregulated genes) and genes with different H3K27me3 enrichment (H3K27me3 gain, genes with increased H3K27me3 enrichment; H3K27me3 loss, genes with decreased H3K27me3 enrichment).
(G) Odds ratio analysis of overlapping genes displaying differential histone H3K27me3 enrichment versus differential gene expression (DEx), insert numbers indicate respective p values for associations, with the number of genes overlapping in parentheses.
(H) Gene ontology enrichment analysis of these 70 genes with increased H3K27me3 enrichment and downregulated expression. Displayed terms are all with an “p.adjust” < 0.05.
(I and J) Genome-browser view at CKB gene (I) and ASIC1 (J) of different sequencing datasets.
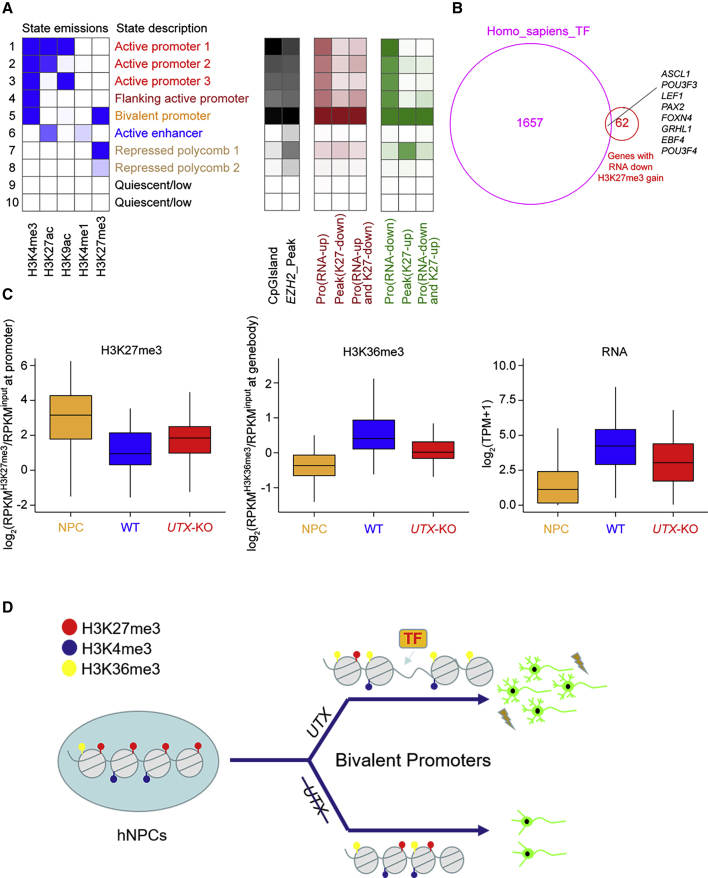
UTX Acts as a Demethylase in Resolution and Activation of Bivalent Promoters during the Neural Differentiation of hESCs
Because H3K27 methyltransferase complex PRC2 was associated with the generation of bivalency (Jadhav et al., 2016), we further assessed the effect of UTX loss on bivalent gene expression. Firstly, we used ChromHMM to model the chromatin states in NPCs, which included a bivalent promoter in the fifth row (Figure 7A, left), and analyzed the distribution of all promoters of differentiated genes after the loss of UTX. Because EZH2 and CpG islands have been proven to be enriched in the bivalent promoters (Beguelin et al., 2016; Mantsoki et al., 2015), we used them as positive controls. Interestingly, we found that different expressed genes between UTX KO and WT neurons, which could be divided into six groups: (1) RNA-up, (2) K27-down, (3) RNA-up and K27-down, (4) RNA-down, (5) K27-up, and (6) RNA-down and K27-up. Promoters of these six groups all belonged to bivalent promoters compared with control groups (Figure 7A, right). As the combination of transcription factors is also a key step in activating bivalent promoters, we then compared the 70 differentiated genes which belonged to RNA-down and K27-up groups with the Homo sapiens TF database, and found that 8 critical TFs were intersected, including ASCL1, POU3F3, and POU3F4, which are important for neural differentiation and morphogenesis (Figure 7B). We then compared the H3K27me3 changes of bivalent promoters of these 70 genes in UTX KO neurons and NPCs. Our results showed that the H3K27me3 enrichment decreased when UTX WT NPCs differentiated to neurons; however, the H3K27me3 enrichment level increased in differentiated UTX KO neurons (Figure 7C). The H3K36me3 enrichments in gene bodies of these 70 genes were also decreased in UTX KO neurons compared with WT neurons. These results indicated that UTX was involved in the resolution and activation of bivalent promoters during the neural differentiation of hESCs.
Figure 7.
UTX Resolves Bivalent Promoters for the Induction of Lineage-Specific Genes during Human Neural Differentiation
(A) Heatmap showing distribution of indicated groups of genomic regions on the chromatin states in NPCs modeled using ChromHMM (pro, promoters).
(B) Venn diagram of a combined comparison of 70 genes with downregulated expression and H3K27me3 gain with TFs.
(C) Left panel, boxplots showing changes of promoter H3K27me3 level of different conditions of 70 genes with downregulated expression and H3K27me3 gain; middle panel, boxplots showing changes of gene body H3K36me3 level of these genes; right panel, boxplots showing gene expression changes of these genes. (ChIP-seq and RNA-seq data of neural progenitor cell originated from H9 were downloaded from ENCODE [https://www.encodeproject.org/, biosample: ENCBS044KWE] and re-analyzed.)
(D) A hypothetical model of UTX as a bivalency-resolving demethylase necessary for neural differentiation of hESCs.
Discussion
Aberrant neural differentiation and dendritic morphogenesis in neurons are pervasive in many developmental brain disorders (Kulkarni and Firestein, 2012; Moretti et al., 2006). Deletion of UTX results in increased anxiety-like behaviors and impaired spatial learning and memory in mice, and UTX deficiency in the hippocampus leads to reduced long-term potentiation and amplitude of miniature excitatory postsynaptic current, aberrant dendrite development, and defective synapse formation (Tang et al., 2017). In this paper, we show that UTX regulates neural differentiation and dendritic morphology during the differentiation of hESCs into neurons by resolving bivalent promoters, partially through association with transcription factors. Deletion of UTX makes some bivalent promoters of critical genes associating with neural development to increase H3K27me3 and to reduce H3K36me3 to inhibit their transcription, which eventually causes defects in human neural differentiation, dendritic morphogenesis, and electrophysiological function (Figure 7D).
In the process of neurogenesis, we find that H3K27me3 levels were unchanged in the hESCs after the loss of UTX, and the differentiation potential of UTX KO hESCs to hNPCs is not affected. This finding supports the idea that maintaining the level of H3K27me3 may be important for the early stage of neurogenesis (Shan et al., 2020).
There is a great debate on how UTX regulates gene transcription. Given the role of UTX in removal of repressive H3K27me3 to establish transcriptionally permissive chromatin, its demethylase activity is required for activating gene expression in T cells (Manna et al., 2015), osteoblasts (Yang et al., 2015), and myoblasts (Faralli et al., 2016). A previously published study reports that human 53BP1 contains a UTX-binding site and the 53BP1-UTX interaction is required to upregulate key neurodevelopmental genes during the differentiation of hESCs into neurons (Yang et al., 2019). In the present study, we provide more evidence to demonstrate that UTX not only regulates the transition of hNPCs to neurons, but also affects neural morphogenesis and synaptic function by resolving bivalent promoters. UTX has been reported necessary for the resolution and activation of numerous retinoic acid (RA)-inducible bivalent genes during the RA-driven differentiation of mouse embryonic stem cells (Dhar et al., 2016). Consistent with this, we also demonstrate that UTX plays an important role in human neural differentiation and neural morphology by regulating bivalent promoters.
Importantly, we find that UTX deficiency in hESC-derived neurons also causes transcriptional activation of dozens of genes, i.e., TGF-β signaling pathway-related genes, including BMP4, TGF-β1, TGF-β2, TGF-β3, and ACVR1, which negatively regulate neuronal morphogenesis (Nakashima et al., 2018), and the inhibition of the TGF-β signaling pathway is important for the differentiation of hESCs to neurons (Morizane et al., 2011). How these genes are activated after the loss of UTX still needs to be studied. Interestingly, EZH2 is downregulated in UTX-null hESC-derived neurons, we speculate that EZH2 reduction may be responsible for the de-repression of upregulated genes during neural differentiation of UTX-null hESCs. For a better understanding of the complex regulatory mechanisms mediated by UTX, future studies are necessary to characterize chromatin structure and epigenetic modification by a higher resolution of both RNA and DNA sequencing.
Experimental Procedures
Maintenance and Differentiation of hESCs
The human H9 (WA09) ESC line was obtained from Dr. Baoyang Hu at the Institute of Zoology, Chinese Academy of Sciences. H9 cells were cultured on Matrigel (BD, New Jersey, USA) using TeSR-E8 (STEMCELL Technologies, Vancouver, BC, Canada) and passaged with EDTA (100 μM). Neural differentiation of the H9 cells was initiated by the formation of embryoid bodies (EBs) as described previously with minor modifications (Jiang et al., 2013). In brief, hESC colonies were dissociated with Accutase (BD) and resuspended as single cells in E8 medium with 10 μM Rock inhibitor (Y27632, Selleck, Shanghai, China). To produce EBs from hESCs, hESC medium was replaced with N2 medium (DMEMF/12, 1× N2, 1× NEAA) at day 3 and the medium was changed every other day. To obtain NPCs derived from hESCs, the EBs were plated at day 7 on Matrigel-coated plates with 1 μg/mL laminin (Life Technologies, Carlsbad, CA, USA) and cultured in N2 medium. NPCs in the form of rosettes were manually picked at day 14 and expanded as neurospheres in N2B27 medium (DMEM/F12, 1× N2, 1× B27, 0.2 μM RA, and 20 ng/mL fibroblast growth factor 2) until day 20. For neuronal differentiation, the dissociated NPCs at day 20 were plated onto poly-L-ornithine/laminin (50 μg/mL)-coated coverslips, and cultured in neuronal differentiation medium (DMEM/F12, 1× N2, 1× B27, 10 ng/mL brain-derived neurotrophic factor, 10 ng/mL GDNF, 1 mM dibutyryl-cyclic AMP, and 200 nM ascorbic acid).
Construction of the RNA-Guided CRISPR/Cas9 Vector
Two specific guide RNA sequences (sgRNAs) targeting exon 1 of UTX were designed at the Optimized CRISPR Design website (http://crispr.mit.edu) (Cong et al., 2013). Then, the two guide RNA sequences were respectively annealed and cloned to the PX330-GFP-U6 plasmid (a gift from Dr. Haoyi Wang at the Institute of Zoology, Chinese Academy of Sciences) to generate all-in-one expression vector containing two sgRNA expression cassettes.
CRISPR/Cas9-Mediated KO of UTX in hESCs
H9 cells were dissociated into single cells using Accutase at 37°C for 5 min. Cells (2 × 106) were electroporated with 10 μg UTX double-guide RNA expression plasmid using Human Stem Cell Nucleofector kit 2 (Lonza) with program CM115 in a Nucleofector II device. Cells were resuspended in hESC medium supplemented with 10 μM Rock inhibitor (Y-27632) and re-plated on Matrigel-coated plates. After 36 h of culture, GFP-positive cells were selected with flow cytometry and reseeded as single cells. Approximately 2 weeks later, colonies were singly picked and expanded for genotype sequencing.
Histone Extraction
Histones were isolated from cells by following a standard acid extraction protocol (Shechter et al., 2007). In brief, the cells were suspended with hypotonic lysis buffer and blown into single cells. Then the cells were shaken at 4°C for 30 min and centrifuged at 10,000 × g for 10 min at 4°C. Supernatant was discarded and the nuclei pellet was resuspended in 0.4 N H2SO4. Then the nucleic solution was centrifuged and supernatant was collected, and 132 μL tricarboxylic acid (TCA) was added drop by drop to histone solution and the tube was inverted several times to mix the solutions (the final concentration of TCA was 33%). The precipitate was centrifuged, collected and washed with acetone twice. Finally, histone was dissolved with 4 M urea.
Proliferation Analysis of Cultured NPCs
To examine the proliferation ability of NPCs, EBs at day 7 of culture were plated on Matrigel-coated coverslips. After 7 days of differentiation, 5 μM BrdU (Sigma-Aldrich, St. Louis, MO, USA) was added to the culture medium for 3.5 h. NPCs were then fixed with 4% paraformaldehyde for 20 min at room temperature, and finally washed with 1× PBS, followed by BrdU immunostaining.
In Vitro Analysis of Dendritic Morphology
For quantitative analysis of dendrites, neurons at day 40 in culture were fixed with 4% paraformaldehyde and then washed with PBS in vitro. Fixed neurons were blocked and permeabilized with 2% normal goat serum/0.1% Triton X-100 for 1 h at room temperature. Neurons were incubated with anti-Map2 antibody (cat. no. 822501, BioLegend, 1:1,000) overnight at 4°C, and then incubated with secondary antibody for 1 h at room temperature. Images of the dendrites were acquired using a Zeiss LSM710 confocal microscope with a 40× lens. Dendritic branches were traced, and their lengths were calculated using the Simple Neurite Tracer plugin of Fiji.
Western Blot Analysis
Cell pellets were lysed in RIPA lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with protease inhibitor (Roche Applied Science), and samples were then quantified using a BCA protein assay kit (Thermo Fisher Scientific). After electrophoresis, proteins were transferred onto nitrocellulose membranes. The following primary antibodies were used: UTX (cat no. GTX121246, GeneTex, 1:2,000), JMJD3 (cat no. GTX124222, GeneTex 1:1,000), Oct-3/4 (cat no. sc-5279 Santa Cruz 1:1,000), and NANOG (cat no. 14295-1 Proteintech 1:1,000).
Electrophysiological Recordings
Whole-cell current and clamp recordings were performed at 22°C in artificial cerebral spinal fluid, bubbled with 95% O2 and 5% CO2. The extracellular fluid consisted of: 124 mM NaCl, 3.3 mM KCl, 2.4 mM MgSO4, 1.2 mM KH2PO4, 26 mM NaHCO3, 2.5 mM CaCl2, and 10 mM glucose (pH 7.4). Borosilicate glass electrodes (resistance 6–10 MΩ) were filled with an intracellular solution containing 135 mM potassium gluconate, 7 mM NaCl, 10 mM HEPES, 2 mM MgATP, 0.3 mM Na2GTP, and 2 mM MgCl2, adjusted to pH 7.4 with KOH. Cell visualization and patch pipette micromanipulation were performed by video-microscopy, using a 40× water-immersion objective mounted on an upright microscope equipped with infrared differential interference contrast (Nikon, Eclipse fn1, Japan). Intracellular membrane electrical potentials were recorded in current-clamp mode, using a Multi-clamp 700B amplifier (Molecular Devices, Palo Alto, CA, USA). For voltage-clamp recordings, cells were held at −70 mV. All compounds were obtained from Sigma (St. Louis, MO). Data were digitized at 10 kHz with a 2-kHz low-pass filter. Data processing and analysis were performed using Clampfit 10.6 (Axon Instruments).
RNA Isolation and qRT-PCR
Total RNA was extracted with TRIzol (Invitrogen, New York, UK) and then was reverse-transcribed into cDNA using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (cat no. 20200725 TRAN). Real-time qPCR was performed using the Super Real PreMix Plus (SYBR Green I) (Takara) in a 20-μL reaction system on the Roche LightCycler@480II. ACTB was used as endogenous control to normalize the RNA content of samples. The primers we used were listed in Table S1. The relative expression level of each mRNA was analyzed by the ΔΔCT method. All experiments were repeated at least in triplicate.
Statistical Analysis
Experiments were conducted in at least three biological replicates for each group. For statistical analyses, unpaired two-tailed Student's t tests (two groups) or ANOVA (three or more groups) were performed using GraphPad Prism 6.0 software. All data were presented as mean ± SEM. Differences of p < 0.05 were considered statistically significant.
Data and Code Availability
The RNA-seq and ChIP-seq data presented in this manuscript are accessible through GEO series accession number GEO: GSE152448.
Author Contributions
C.-M.L., Q.-Y.T., and Z.-Q.T., conceived and designed the study, collected, assembled, analyzed, and interpreted the data, wrote the manuscript, and approved the final manuscript. S.-F.Z., S.-K.D., C.L., Y.-Y.W., and H.-Z.D. collected and assembled the data.
Acknowledgments
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16010300) and grants from the National Key Research and Development Program of China Project (2016YFA0101402 and 2018YFA0108001), and the National Science Foundation of China (91753140and 81771224).
Published: July 16, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.06.015.
Contributor Information
Zhao-Qian Teng, Email: tengzq@ioz.ac.cn.
Chang-Mei Liu, Email: liuchm@ioz.ac.cn.
Supplemental Information
References
- Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Aldiri I., Xu B.S., Wang L., Chen X., Hiler D., Griffiths L., Valentine M., Shirinifard A., Thiagarajan S., Sablauer A. The dynamic epigenetic landscape of the retina during development, reprogramming, and tumorigenesis. Neuron. 2017;94:550. doi: 10.1016/j.neuron.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayata P., Badimon A., Strasburger H.J., Duff M.K., Montgomery S.E., Loh Y.H.E., Ebert A., Pimenova A.A., Ramirez B.R., Chan A.T. Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci. 2018;21:1049. doi: 10.1038/s41593-018-0192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beguelin W., Teater M., Gearhart M.D., Calvo Fernandez M.T., Goldstein R.L., Cardenas M.G., Hatzi K., Rosen M., Shen H., Corcoran C.M. EZH2 and BCL6 cooperate to assemble CBX8-BCOR complex to repress bivalent promoters, mediate germinal center formation and lymphomagenesis. Cancer Cell. 2016;30:197–213. doi: 10.1016/j.ccell.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Blanco E., Gonzalez-Ramirez M., Alcaine-Colet A., Aranda S., Di Croce L. The bivalent genome: characterization, structure, and regulation. Trends Genet. 2020;36:118–131. doi: 10.1016/j.tig.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S., Hofemeister H., Marks H., Kranz A., Ciotta G., Singh S., Anastassiadis K., Stunnenberg H.G., Stewart A.F. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141:526–537. doi: 10.1242/dev.102681. [DOI] [PubMed] [Google Scholar]
- Dhar S.S., Lee S.H., Chen K., Zhu G., Oh W., Allton K., Gafni O., Kim Y.Z., Tomoiga A.S., Barton M.C. An essential role for UTX in resolution and activation of bivalent promoters. Nucleic Acids Res. 2016;44:3659–3674. doi: 10.1093/nar/gkv1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E., Lien W.H., Stokes N., Pasolli H.A., Silva J.M., Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faralli H., Wang C.C., Nakka K., Benyoucef A., Sebastian S., Zhuang L.N., Chu A., Palii C.G., Liu C.Y., Camellato B. UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J. Clin. Invest. 2016;126:1555–1565. doi: 10.1172/JCI83239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso C.S., Wu Y.M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez B., Bustos F.J., Aguilar R., Becerra A., Simon F., Montecino M., van Zundert B. Ezh1 and Ezh2 differentially regulate PSD-95 gene transcription in developing hippocampal neurons. Mol. Cell Neurosci. 2013;57:130–143. doi: 10.1016/j.mcn.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y., Suzki N., Tsuboi M., Endo T.A., Toyoda T., Shinga J., Koseki H., Vidal M., Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hu D., Garruss A.S., Gao X., Morgan M.A., Cook M., Smith E.R., Shilatifard A. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1093–1097. doi: 10.1038/nsmb.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Ueno S., Fukuda A. Interaction of neuron-specific K+-Cl− cotransporter, KCC2, with brain-type creatine kinase. FEBS Lett. 2004;564:131–135. doi: 10.1016/S0014-5793(04)00328-X. [DOI] [PubMed] [Google Scholar]
- Jadhav U., Nalapareddy K., Saxena M., O'Neill N.K., Pinello L., Yuan G.C., Orkin S.H., Shivdasani R.A. Acquired tissue-specific promoter bivalency is a basis for PRC2 necessity in adult cells. Cell. 2016;165:1389–1400. doi: 10.1016/j.cell.2016.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jiang P., Chen C., Wang R., Chechneva O.V., Chung S.H., Rao M.S., Pleasure D.E., Liu Y., Zhang Q., Deng W. hESC-derived Olig2+ progenitors generate a subtype of astroglia with protective effects against ischaemic brain injury. Nat. Commun. 2013;4:2196. doi: 10.1038/ncomms3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruidenier L., Chung C.W., Cheng Z., Liddle J., Che K., Joberty G., Bantscheff M., Bountra C., Bridges A., Diallo H. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni V.A., Firestein B.L. The dendritic tree and brain disorders. Mol. Cell Neurosci. 2012;50:10–20. doi: 10.1016/j.mcn.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Lan F., Bayliss P.E., Rinn J.L., Whetstine J.R., Wang J.K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lei X., Jiao J. UTX affects neural stem cell proliferation and differentiation through PTEN signaling. Stem Cell Reports. 2018;10:1193–1207. doi: 10.1016/j.stemcr.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.C., Wu X.W., Zhang H.Y., Pfeifer G.P., Lu Q. Dynamics of RNA polymerase II pausing and bivalent histone H3 methylation during neuronal differentiation in brain development. Cell Rep. 2017;20:1307–1318. doi: 10.1016/j.celrep.2017.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna S., Kim J.K., Bauge C., Cam M., Zhao Y.M., Shetty J., Vacchio M.S., Castro E., Tran B., Tessarollo L. Histone H3 Lysine 27 demethylases Jmjd3 and Utx are required for T-cell differentiation. Nat. Commun. 2015;6:8152. doi: 10.1038/ncomms9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour A.A., Gafni O., Weinberger L., Zviran A., Ayyash M., Rais Y., Krupalnik V., Zerbib M., Amann-Zalcenstein D., Maza I. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–413. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- Mantsoki A., Devailly G., Joshi A. CpG island erosion, polycomb occupancy and sequence motif enrichment at bivalent promoters in mammalian embryonic stem cells. Sci. Rep. 2015;5:16791. doi: 10.1038/srep16791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M., Holwerda S., Vitobello A., Kitazawa T., Kohler H., Stadler M.B., Rijli F.M. Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science. 2017;355:eaal2913. doi: 10.1126/science.aal2913. [DOI] [PubMed] [Google Scholar]
- Miyake N., Mizuno S., Okamoto N., Ohashi H., Shiina M., Ogata K., Tsurusaki Y., Nakashima M., Saitsu H., Niikawa N. KDM6A point mutations cause Kabuki syndrome. Hum. Mutat. 2013;34:108–110. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- Moretti P., Levenson J.M., Battaglia F., Atkinson R., Teague R., Antalffy B., Armstrong D., Arancio O., Sweatt J.D., Zoghbi H.Y. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J. Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane A., Doi D., Kikuchi T., Nishimura K., Takahashi J. Small-molecule inhibitors of bone morphogenic protein and activin/nodal signals promote highly efficient neural induction from human pluripotent stem cells. J. Neurosci. Res. 2011;89:117–126. doi: 10.1002/jnr.22547. [DOI] [PubMed] [Google Scholar]
- Nakashima H., Tsujimura K., Irie K., Ishizu M., Pan M., Kameda T., Nakashima K. Canonical TGF-beta signaling negatively regulates neuronal morphogenesis through TGIF/Smad complex-mediated CRMP2 suppression. J. Neurosci. 2018;38:4791–4810. doi: 10.1523/JNEUROSCI.2423-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J.D., Sansom S.N., Smith J., Dobenecker M.W., Tarakhovsky A., Livesey F.J. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. U S A. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y., Zhang Y., Zhao Y., Wang T., Zhang J., Yao J., Ma N., Liang Z., Huang W., Huang K. JMJD3 and UTX determine fidelity and lineage specification of human neural progenitor cells. Nat. Commun. 2020;11:382. doi: 10.1038/s41467-019-14028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D., Dormann H.L., Allis C.D., Hake S.B. Extraction, purification and analysis of histones. Nat. Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- Shpargel K.B., Starmer J., Wang C., Ge K., Magnuson T. UTX-guided neural crest function underlies craniofacial features of Kabuki syndrome. Proc. Natl. Acad. Sci. U S A. 2017;114:E9046–E9055. doi: 10.1073/pnas.1705011114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Morgan K.R., Petryk N., Groth A. Chromatin replication and epigenetic cell memory. Nat. Cell Biol. 2020;22:361–371. doi: 10.1038/s41556-020-0487-y. [DOI] [PubMed] [Google Scholar]
- Swigut T., Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- Tang G.B., Zeng Y.Q., Liu P.P., Mi T.W., Zhang S.F., Dai S.K., Tang Q.Y., Yang L., Xu Y.J., Yan H.L. The histone H3K27 demethylase UTX regulates synaptic plasticity and cognitive behaviors in mice. Front Mol. Neurosci. 2017;10:267. doi: 10.3389/fnmol.2017.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N., Renthal W., Kumar A., Nestler E.J. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Voigt P., Tee W.W., Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T.Y., Watford W.T. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie J.A., Askwith C.C., Lamani E., Cassell M.D., Freeman J.H., Welsh M.J. Acid-sensing ion channel 1 is localized in brain regions with high synaptic density and contributes to fear conditioning. J. Neurosci. 2003;23:5496–5502. doi: 10.1523/JNEUROSCI.23-13-05496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Deng X., Watkins R., Disteche C.M. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J. Neurosci. 2008;28:4521–4527. doi: 10.1523/JNEUROSCI.5382-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Okamura H., Teramachi J., Haneji T. Histone demethylase Utx regulates differentiation and mineralization in osteoblasts. J. Cell Biochem. 2015;116:2628–2636. doi: 10.1002/jcb.25210. [DOI] [PubMed] [Google Scholar]
- Yang X., Xu B., Mulvey B., Evans M., Jordan S., Wang Y.D., Pagala V., Peng J., Fan Y., Patel A. Differentiation of human pluripotent stem cells into neurons or cortical organoids requires transcriptional co-regulation by UTX and 53BP1. Nat. Neurosci. 2019;22:362–373. doi: 10.1038/s41593-018-0328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Xu L., Xu L., Xu Q., Li D., Yang Y., Karsenty G., Chen C.D. JMJD3 promotes chondrocyte proliferation and hypertrophy during endochondral bone formation in mice. J. Mol. Cell Biol. 2015;7:23–34. doi: 10.1093/jmcb/mjv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and ChIP-seq data presented in this manuscript are accessible through GEO series accession number GEO: GSE152448.