Summary
Recent studies suggest that metabolic regulation may improve differentiation of cardiomyocytes derived from induced pluripotent stem cells (iPSCs). AMP-activated protein kinase (AMPK) is a master regulator of metabolic activities. We investigated whether AMPK participates in iPSC-derived cardiomyocyte differentiation. We observed that AMPK phosphorylation at Thr172 increased at day 9 but then decreased after day 11 of differentiation to cardiomyocytes. Inhibition of AMPK with compound C significantly reduced mRNA and protein expression of cardiac troponins TNNT2 and TNNI3. Moreover, sustained AMPK activation using AICAR from days 9 to 14 of differentiation increased mRNA and protein expression of both TNNT2 and TNNI3. AICAR decreased acetylation of histone 3 at Lys9 and 56 and histone 4 at Lys16 (known target sites for nuclear-localized sirtuins [SIRT1, SIRT6]), suggesting that AMPK activation enhances sirtuin activity. Sustained AMPK activation during days 9–14 of differentiation induces sirtuin-mediated histone deacetylation and may enhance cardiomyocyte differentiation from iPSCs.
Keywords: iPSC-derived cardiomyocytes, AMPK, sirtuin, histone acetylation
Highlights
-
•
iPSC-derived cardiomyocytes transiently increased AMPK phosphorylation at Thr172
-
•
Chemical inhibition of AMPK with compound C decreased TNNI3 and TNNT2 expression
-
•
Sustained activation of AMPK using AICAR increased expression of TNNT2 and TNNI3
-
•
AICAR decreased acetylation of histones H3 (at Lys9 and Lys56) and H4 (at Lys16).
Lee and colleagues demonstrate that sustained activation of AMPK enhances differentiation of iPSC-derived cardiomyocytes. Sustained AMPK activation decreased histone acetylation at known target sites for nuclear-localized sirtuins, suggesting that AMPK activation enhances sirtuin activity. AMPK-induced sirtuin-mediated deacetylation of histone proteins may regulate chromatin accessibility and enhance cardiomyocyte differentiation.
Introduction
The application of stem cell-derived cardiomyocytes for cardiac disease modeling, drug screening and potential cell therapies is highly dependent on production of differentiated cardiomyocytes. However, cardiomyocytes derived from embryonic or induced pluripotent stem cells (ESCs or iPSCs, respectively) do not fully differentiate with existing protocols (Yang et al., 2014). Despite various strategies, iPSC-derived cardiomyocytes are metabolically, structurally, and functionally immature, with molecular mechanisms of cardiomyocyte differentiation incompletely defined (Scuderi and Butcher, 2017; Yang et al., 2014). Successful translation of cell therapies for cardiovascular disease may require improving methods to differentiate stem cell-derived cardiomyocytes.
The mammalian heart relies on carbohydrate metabolism via glycolysis during early development in utero (Yuan and Braun, 2017). After birth, metabolic changes, including exposure to higher oxygen levels and initiation of enteral nutrition affect the early regenerative capability of cardiomyocytes and differentiation (Yuan and Braun, 2017). The first meal of the newborn is enriched in lipids from maternal milk (colostrum) and accelerates a metabolic switch from carbohydrate to lipid metabolism (Piquereau and Ventura-Clapier, 2018), leading to upregulation of genes involved in fatty acid uptake to provide cells with required energy (Sim et al., 2015). This switch is necessary to establish the highly oxidative metabolism of the postnatal heart and provide increased ATP to meet demand, facilitating cardiomyocyte maturation (Yuan and Braun, 2017). Relative availability of carbohydrate and fatty acid substrates affects the cellular metabolic phenotype (Wanet et al., 2015). The metabolic shift is accompanied by increased mitochondrial number and activity to help differentiation and maturation during heart development, with mitochondria occupying 20%–40% of the adult myocyte volume (Yang et al., 2014). Thus, evidence supports the role of metabolism in cardiac growth and maturation.
Regulation of AMP-activated protein kinase (AMPK) during heart failure is well studied (Arad et al., 2007); however, the role of AMPK in cardiac development is not well understood. AMPK is a heterotrimeric enzyme that regulates metabolism by enhancing fatty acid uptake, glycolysis, glucose uptake, and autophagy (Arad et al., 2007). AMPK is activated when the AMP/ATP ratio increases, triggering AMPK to help the cell to produce energy (Zaha and Young, 2012). Each AMPK molecule is comprised of a catalytic α and regulatory β and γ subunit. The α1β1γ1 complex is ubiquitous, whereas α2β2γ2 is found primarily in the heart in humans (Arad et al., 2007). Mice with deletion of AMPKα1 or AMPKα2 are viable, but AMPKα1/α2 double deletion causes embryonic lethality at ~10.5 days (Viollet et al., 2009). Prolonged AMPK activation increases expression of fatty acid transporters in cardiomyocytes (Chabowski et al., 2006). Moreover, AMPK activation enhances NAD+ abundance and the NAD/NADH ratio which enhances NAD+-dependent type III deacetylase SIRT1 (silent information regulator of transcription 1) activity (Canto et al., 2009). Phosphorylation of AMPK occurs via one of two known AMPK kinases (AMPKKs) in the heart: the tumor suppressor kinase, LKB1, and a calmodulin-dependent protein kinase, CaMKK (Arad et al., 2007). LKB1 is deactivated by deacetylation of LKB1 at lysine 48 by SIRT1 (Lan et al., 2008); thus the sirtuin family of deacetylases may both be activated by AMPK and also provide negative feedback to regulate AMPK.
The sirtuin family of proteins includes a group of class III lysine deacetylases that regulate various intracellular processes, including metabolism (Alcendor et al., 2004; Bao and Sack, 2010), oxidative stress, apoptosis (Alcendor et al., 2004; Motta et al., 2004), chromatin condensation (Jing and Lin, 2015), and the cell cycle (Sasaki et al., 2006). There are seven known sirtuins that act in cellular regulation in humans (Li and Kazgan, 2011). Sirtuins are localized in different compartments, with SIRT1, 6, and 7 located mainly in the nucleus, SIRT2 located mainly in the cytosol and also shuttled in the nucleus, and SIRT3, 4, and 5 located in the mitochondria (Herskovits and Guarente, 2013). Activation of SIRT is dependent on NAD+ (Imai et al., 2000). Among the seven mammalian sirtuins, SIRT1, 2, 6, and 7 are demonstrated to have important epigenetic roles (Jing and Lin, 2015). SIRT1 regulates chromatin structure by deacetylating histone lysines (H4K16, H3K9, H3K14, H4K8, and H4K12) (Imai et al., 2000; Jing and Lin, 2015; Vaquero et al., 2004) or indirectly regulating other epigenetic enzymes, including histone acetyltransferase (CBP/P300) and methyltransferases (Benetti et al., 2007; Yamagoe et al., 2003). SIRT2 deletion increases H4K16 acetylation (Serrano et al., 2013), and SIRT2 inhibition leads to hyperacetylation at H3K18 and H3K56 (Das et al., 2009; Eskandarian et al., 2013). SIRT6 has also been shown to be a weak deacetylase but also can play a role in genome stability (Liao and Kennedy, 2016).
In this study, we tested the hypothesis that AMPK regulates differentiation of cardiomyocytes from iPSCs. In addition, we evaluated whether changes in AMPK activation alters NAD+ levels and subsequent SIRT1 activation to affect chromatin accessibility during differentiation of iPSC-derived cardiomyocytes.
Results
AMPK Undergoes Transient Phosphorylation during Early iPSC-Derived Cardiomyocyte Differentiation
Recent studies revealed the importance of metabolic pathways in cardiomyocyte maturation (Horikoshi et al., 2019; Hu et al., 2018; Marchiano et al., 2019; Yang et al., 2019). AMPK is a master regulator of metabolism, yet its role during differentiation of iPSC-derived cardiomyocytes is not defined. To perform this study, we used a well-established iPSC-derived cardiomyocyte differentiation protocol in three human iPSC lines (BJRiPS-A, Gibco episomal iPSC line, UCSD142i-86-1) (Lian et al., 2013) and harvested cells at different differentiation time points (days 0, 2, 4, 7, 9, 11, and 14) (Figure 1A, non-cropped images shown in Figures S3–S5 for all westerns). We observed that AMPK was hyperphosphorylated at days 9–11 of differentiation with a decrease in AMPK phosphorylation at later time points in each of the three iPSC lines tested (Figure 1B); this was repeated in multiple batches in each cell line and a similar transient activation of AMPK was seen consistently (Figure 1H shows a different batch in the Gibco cell line). Also, AMPK remains hypophosphorylated at later time points to at least day 21 of differentiation (Figure S1A). The three iPSC lines tested showed similar time courses for mRNA (Figure 1C) and protein (Figure 1B) expression of selected cardiomyocyte markers (TNNT2, NKX2.5, and GATA4) (Figure 1B). PRKAA1 (AMPK-α1) had decreased mRNA expression at days 9–11 versus day 7 but PRKAA2 (AMPK-α2) mRNA expression was significantly increased on days 9–11 and increased sharply at day 14 versus day 7 (Figures 1D and 1E, respectively).
Figure 1.
Transient Activation of AMPK during Early iPSC-Derived Cardiomyocyte Differentiation
(A) Cardiomyocyte differentiation protocol.
(B) Western analysis of phospho-AMPK and AMPK at baseline on days 0, 2, 4, 7, 9, 11, and 14 of differentiation in Gibco, BJRiPS-A, and UCSD cell lines.
(C) Time course qPCR of sarcomere genes (TNNT2, NKX2-5, and GATA4) in untreated iPSC-derived cardiomyocytes at baseline on days 7, 9, 11, 14, and 16, n = 3 independent experiments (I.E.).
(D and E) Time course qPCR for (D) PRKAA1 and (E) PRKAA2, n = 3 I.E.
(F) Western analysis of phospho-LKB1 at days 7, 9, 11, and 14.
(G) Time course qPCR for LKB1, n = 3 I.E.
(H) Endogenous LKB1 immunoprecipitated from days 4, 7, 9, 11, and 14 and probed for acetyl lysine. Data shown from untreated Gibco iPSC-derived cardiomyocytes unless noted otherwise.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 one-way ANOVA.
AMPK can be phosphorylated at Thr172 by either LKB1 or calmodulin-dependent protein kinase kinase (CaMKK) in the heart (Arad et al., 2007). LKB1 appears to be the primary upstream kinase of AMPK in the heart (Sakamoto et al., 2006). We observed transient LKB1 phosphorylation between days 9 and 11 of differentiation, similar to the time course of AMPK phosphorylation, suggesting that AMPK phosphorylation occurs via LKB1 (Figure 1F, mRNA expression of LKB1 shown in Figure 1B). SIRT1-mediated deacetylation of LKB1 enhances its activity (Lan et al., 2008). Western analysis demonstrated that SIRT1 protein levels decreased with time (Figure 1H), coinciding with increased acetylation of LKB1 (Figure 1H). These data demonstrate transient AMPK activation during cardiomyocyte differentiation from iPSCs, with LKB1 activation likely mediating hyperphosphorylation of AMPK.
Antagonism of AMPK Inhibits iPSC-Derived Cardiomyocyte Differentiation
To determine if transient activation of AMPK regulates iPSC-derived cardiomyocyte differentiation, we inhibited AMPK using the selective small-molecule inhibitor compound C (10 μM) (Liu et al., 2014) versus vehicle (DMSO) for 5 days from ~1 day after onset of beating (Figure 2A). Compound C decreased expression of phosphorylated AMPK as well as expression of TNNT2 and TNNI3 by western analysis (Figures 2B–2D). We observed a similar significant decrease in the mean fluorescence intensity (MFI) of TNNT2 by flow cytometry (Figure 2E) and mRNA expression of TNNT2 and TNNI3 (Figure 2F). Compound C also decreased mRNA expression of MYH7 and CD36, with no change in cardiac transcription factors (TFs), GATA4, NKX2.5, or MEF2 (Figure S1B). Compound C administration unexpectedly increased mRNA expression of the ion channel, KCNJ2, and glucose transporter, GLUT1, with no change in expression GLUT4 (Figure S1B).
Figure 2.
Antagonism of AMPK Inhibits iPSC-Derived Cardiomyocyte Differentiation
(A) Schematic depicting timing of compound C treatment.
(B) Western analysis of phospho-AMPK, TNNT2, and TNNI3, GAPDH as loading control, n = 5 I.E.
(C) Densitometry of TNNI3 bands by western analysis, n = 3; ∗∗∗p < 0.001 by t test.
(D) Densitometry of TNNT2 band by western analysis, n = 3 independent experiments (I.E.); ∗∗p < 0.01 by t test.
(E) Mean fluorescence intensity (MFI) of TNNT2-Alexa Fluor 647 of TNNT2+ cells. n = 3 I.E.; ∗p < 0.05 by unpaired t test.
(F) qPCR of TNNT2 and TNNI3, n = 3 I.E.; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001 one-way ANOVA.
(G) Oxygen consumption rate (OCR) normalized to cell number versus time, Seahorse Mito Stress Test, n = 20 wells/group.
(H and I) (H) Maximum OCR and (I) respiratory reserve capacity normalized to baseline of control, ∗p < 0.05 t test.
(J) Extracellular acidification rate (ECAR) normalized to cell number versus time, Seahorse Mito Stress Test, n = 20 wells/group.
All data are from Gibco-CMs treated with DMSO (control) or compound C for 5 days; sample numbers indicate biological replicates per group.
Cardiomyocytes have high mitochondrial density which facilitates generation of ATP (Piquereau et al., 2013), and AMPK promotes mitochondrial respiration (Wang et al., 2019). Using the Seahorse Mito Stress Test to evaluate mitochondrial function (oxygen consumption rate [OCR] profile shown in Figure 2G), inhibition of AMPK with compound C decreased normalized maximal OCR (Figure 2H) and respiratory reverse capacity (Figure 2I) and decreased extracellular acidification rate (ECAR) (Figure 2J). These results suggest that the transient activation of AMPK is necessary for cardiomyocyte differentiation from iPSCs.
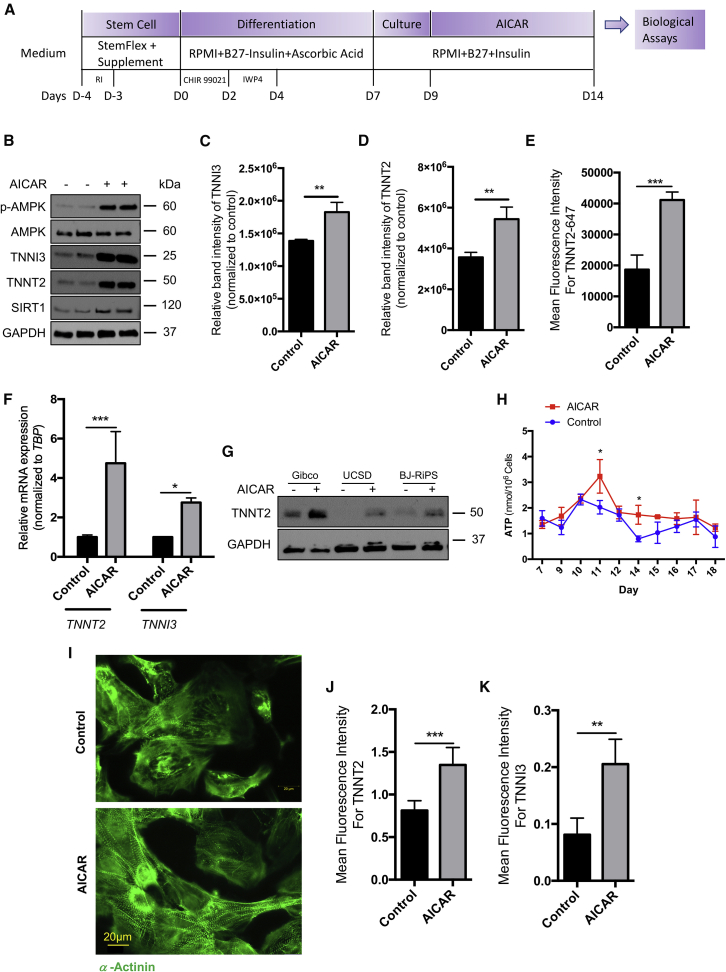
Sustained AMPK Activation Improves iPSC-Derived Cardiomyocyte Differentiation
Because inhibition of AMPK impaired iPSC-derived cardiomyocyte differentiation, we hypothesized that sustaining AMPK phosphorylation beyond day 11 of differentiation might enhance cardiomyocyte differentiation. To test this hypothesis, we treated cardiomyocytes ~1 day after onset of beating for 5 days with AICAR (AMPK activator) (Kim et al., 2016) (Figure 3A). AICAR-treated cells significantly increased mRNA (Figure 3F) and protein expression of TNNT2 and TNNI3 (Figures 3B–3D, western analysis; Figure 3E, MFI of TNNT2 by flow cytometry; Figures 3I–3K, MFI of TNNT2 and TNNI3 by image-based analysis). We observed increased TNNT2 protein expression in three different iPSC lines treated with AICAR (Figure 3G). Moreover, AICAR treatment significantly increased mRNA and protein expression of the cardiac TFs GATA4 and NKX2.5 (Figures S1C [western] and 1D [qPCR]) and mRNA expression of MYH7 and MYH6 (Figure S1C). ATP levels were also higher at multiple time points after initiation of AICAR (Figure 3H), suggesting that sustained activation of AMPK improved the metabolic phenotype of cardiomyocytes. Arguably, mature cardiomyocytes would be expected to have eventual downregulation of GATA4, NKX2-5, and MYH6 so AMPK activation may facilitate mid-late differentiation but not later maturation. AMPK activation also led to a significant decrease in KCNJ2 (Kir 2.1) expression, consistent with the results we observed with compound C administration (Figure S1C). While we did not measure the Ik1 current in this study, these results would suggest that AMPK activation may have a detrimental effect on electrical maturation, contrary to the effects on sarcomere proteins and metabolic phenotype. We also note that the change in medium composition starting on day 7 of differentiation (with the removal of ascorbic acid and addition of insulin) could drive the changes seen on AMPK phosphorylation at days 9–11. However, even when insulin was not added to the medium on day 7 (and ascorbic acid was continued), we still observed a significant increase in TNNT2 expression (Figure S1E, with no change in %TNNT2+ cells with AICAR in Figure S1F).
Figure 3.
Persistent AMPK Activation Improves iPSC-Derived Cardiomyocyte Differentiation
(A) Protocol depicting timing of AICAR treatment.
(B) Western analysis of phospho-AMPK, TNNI3, TNNT2, and SIRT1.
(C and D) (C) TNNI3 and (D) TNNT2 western densitometry analyses, n = 3 independent experiments (I.E.); ∗∗p < 0.001 t test.
(E) MFI of TNNT2-Alexa Fluor 647 of TNNT2+ cells, n = 3 I.E.; ∗∗∗p < 0.001 t test.
(F) qPCR TNNI3 and TNNT2, n = 3 I.E.; ∗∗∗p < 0.001 one-way ANOVA.
(G) Western analysis of TNNT2 in Gibco, UCSD, and BJRiPS.
(H) ATP concentration nmol/106 cells per time point, n = 3 per time point I.E.; ∗p < 0.05 two-way ANOVA.
(I) Confocal imaging of cells stained with α-actinin (green).
(J and K) (J) MFI of TNNT2 of TNNT2+ cells or (K) MFI of TNNI3 of TNNI3+ cells, n = 5 wells/group; ∗∗∗p < 0.001 by unpaired t test.
All data are from Gibco-CMs treated with DMSO (control) or compound C for 5 days unless noted otherwise; sample numbers indicate biological replicates per group.
Because AMPK can affect cell proliferation, we performed cell-cycle analysis by flow cytometry in beating cardiomyocytes at day 16 of differentiation in cells treated with 1 or 10 mM of AICAR for 5 days (Figure S1G). We observed an increase in the %TNNT2+ cardiomyocytes in G1 and a significant decrease in the %TNNT2+ cardiomyocytes in G2 (Figure S1G). We also found a significant decrease in the number of EdU+ cells with AMPK activation (Figures S1H and S1I). These data from TNNT2+ cardiomyocytes suggest that AICAR activity directly regulates iPSC-derived cardiomyocyte differentiation, but we cannot exclude a possible role of non-cardiomyocytes (TNNT2− cells account for ~20% of total cells, Figure S1F) in differentiation and maturation. AMPK can also increase apoptosis (Kuznetsov et al., 2011), and we found that 10 mM but not 1 mM AICAR significantly increased the percentage of cells in sub-G1 (Figure S1G) (representing cells with DNA fragmentation seen during apoptosis [Huang et al., 2005]). We also found that AICAR significantly increased mRNA expression of BCL2-like 1 (BCL2L1) (Figure S2A), which can have either pro- and anti-apoptotic effects due to alternative splicing (Boise et al., 1993).
Removal of AICAR from the medium after 5 days led to dephosphorylation of AMPK by 48 h (Figure S2B), while sustained treatment with AICAR beyond 5 days led to sustained phosphorylation of AMPK (Figure S2C). Paradoxically, prolonged activation of AMPK beyond day 14 of differentiation led to a decrease in TNNT2 expression at later time points (Figure S2C), suggesting that additional mechanisms may be required to maintain the effects of AMPK at later time points. Therefore, we chose to focus on molecular changes that occur during the narrow window of cardiomyocyte differentiation between days 7 and 16 for the remainder of this study.
Activation of AMPK May Enhance iPSC-Derived Cardiomyocyte Metabolic Maturation
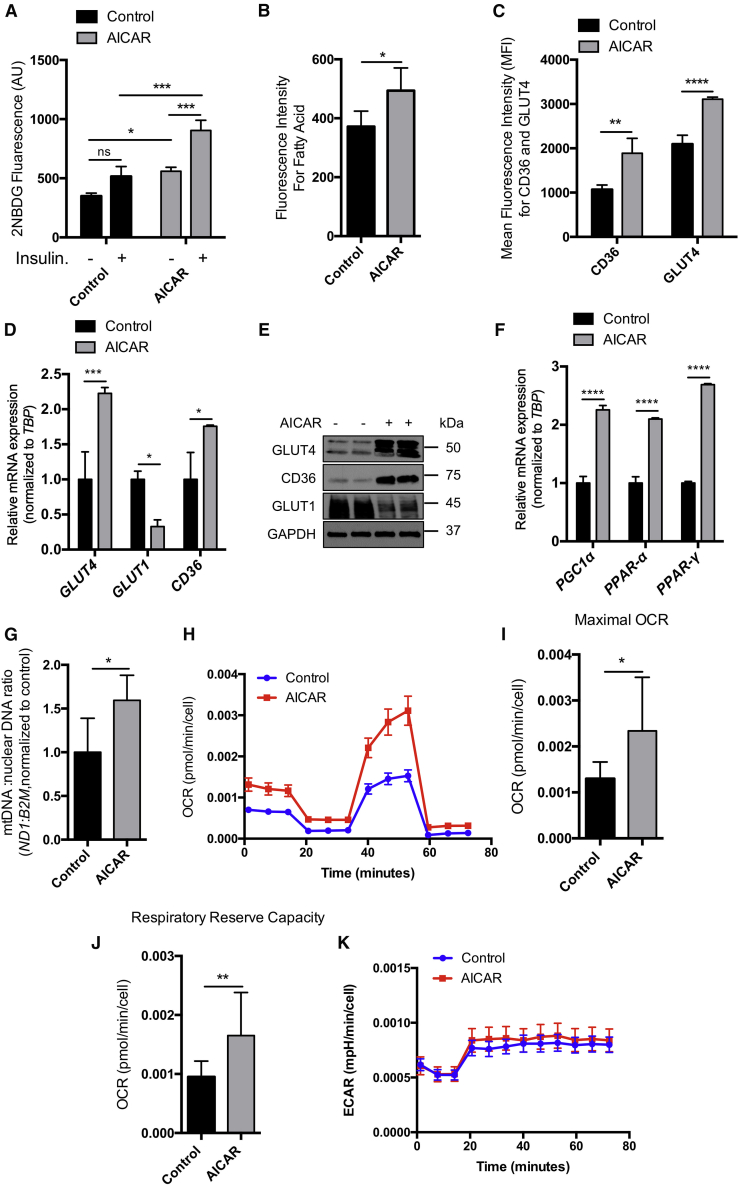
Considering the importance of AMPK in the regulation of glucose metabolism and fatty acid oxidation (Ofir et al., 2008), the relative abundance of ATP could play an important role in cardiomyocyte differentiation. It has recently been shown that fatty acids enhance iPSC-derived cardiomyocyte maturation by increasing mitochondrial activity (Yang et al., 2019). In addition, iPSC-derived cardiomyocytes express high levels of GLUT1 and low levels of GLUT4 compared with primary cardiomyocytes (Bowman et al., 2019). GLUT4 and CD36 are the major glucose and fatty acid transporters in the heart, respectively (Holloway et al., 2009; Shao and Tian, 2015). However, GLUT4 is predominantly upregulated in the postnatal and adult heart (Liu et al., 1992) and is the major mediator of insulin-stimulated glucose uptake in the heart (Laybutt et al., 1997). AMPK also increases gene expression of GLUT4 to facilitate glucose uptake (Zheng et al., 2001), and activation of AMPK in HepG2 cells leads to increased expression of CD36 (Choi et al., 2017). Regional variation in GLUT4 expression has been described in the heart (Maria et al., 2015), thus further investigation should be performed to determine whether AMPK has similar effects on atrial, nodal, and ventricular cardiomyocytes. Interestingly, we observed a significant decrease in mRNA expression of the atrial TF, COUP-TFII, with AICAR treatment, while we saw no difference in HCN4 (pacemaker ion channel) or IRX4 (ventricular marker) mRNA levels (Figure S2D) or MYL2 protein expression (Figure S2E).
To evaluate glucose uptake with AMPK activation, we performed 2NBDG (fluorescent glucose analog) uptake in the presence and absence of insulin. Our data showed that AICAR significantly increased 2NBDG uptake under basal conditions as well as in insulin-treated cells (Figure 4A). AICAR also significantly increased fatty acid uptake (Figure 4B). AICAR-treated cells significantly increased membrane recruitment of GLUT4 and CD36 (Figure 4C). AICAR increased mRNA (Figure 4D) and protein (Figure 4E) expression of SLC2A4 (GLUT4) and CD36 while decreasing SLC2A1 (GLUT1) expression (Figures 4D and 4E). AICAR also increased mRNA expression of PPARGC1a (PGC1α), which regulates mitochondrial biogenesis (Irrcher et al., 2008) (Figure 4F). We also observed significantly increased PPARA (PPARα) and PPARG (PPARγ) mRNA levels in AICAR-treated cells versus control, suggesting that AICAR treatment increased fatty acid β-oxidation (Figure 4F). AICAR treatment also significantly increased the mitochondrial DNA to nuclear DNA ratio (Figure 4G). Cardiomyocytes treated with AICAR had a significantly increased OCR (Figure 4I) and respiratory reserve capacity (Figure 4J), suggesting that these cells are metabolically more active (Figure 4H). Interestingly, AICAR did not change ECAR (Figure 4K), suggesting that glucose utilization is unchanged despite an increase in the OCR. Thus, while AMPK activation may increase oxidative phosphorylation, it may not inhibit glycolysis, consistent with our finding that GLUT4 expression increased. Overall, these data suggest that sustaining activation of AMPK may enhance iPSC-derived cardiomyocyte metabolic maturation through upregulation of fatty acid oxidation pathways, although it may also increase glycolysis.
Figure 4.
Activation of AMPK Enhances iPSC-Derived Cardiomyocyte Metabolic Maturation
(A) 2-NBDG uptake measured in control and AICAR-treated Gibco-CMs in the presence and absence of insulin, n = 6 independent experiments (I.E.); ∗p < 0.05 two-way ANOVA.
(B) Fluorescence fatty acid uptake, n = 6 I.E.; ∗p < 0.05 t test.
(C) MFI of CD36-Alexa Fluor 594 and GLUT4-Alexa Fluor 488, n = 3 I.E.; ∗∗p < 0.001, ∗∗∗∗p < 0.0001 two-way ANOVA.
(D) mRNA levels of CD36, GLUT4, and GLUT1 by qPCR, n = 3 I.E.; ∗p < 0.05 one-way ANOVA.
(E) Western analysis of GLUT4, CD36, and GLUT1.
(F) qPCR for PPARGC1A, PPARA, and PPARG. n = 3 I.E.; ∗∗∗∗p < 0.0001 one-way ANOVA.
(G) Mitochondrial (ND1) to nuclear (B2M) DNA ratio, n = 3 I.E.; ∗p < 0.05 unpaired t test.
(H) OCR normalized to cell number versus time, Seahorse Mito Stress Test, n = 20 wells/group.
(I and J) (I) Maximum OCR and (J) respiratory reserve capacity normalized to baseline of control, ∗p < 0.05 unpaired t test. (K) ECAR normalized to cell number versus time, Seahorse Mito Stress Test, n = 20 wells/group.
All data shown from Gibco-CMs treated with DMSO (control) or AICAR for 5 days unless noted otherwise; sample numbers indicate biological replicates.
AMPK Activation Increased NAD/NADH and Reduced Lysine Acetylation through Sirtuins
To understand how sustained activation of AMPK can affect NAD+ levels and consequently activate sirtuins, we evaluated NAD+ levels in control and AICAR-treated cells and found significant increases in NAD+ levels from days 9 to 16 of differentiation with AMPK activation (Figure 5A). To determine whether AMPK-mediated iPSC-derived cardiomyocyte differentiation acts via sirtuins, cells were treated with nicotinamide (NAM) (Bonkowski and Sinclair, 2016) (known sirtuin class 3 histone deacetylases inhibitor) and trichostatin A (TSA) (inhibitor of class I and class II histone deacetylases) (Vanhaecke et al., 2004). We observed a significant decrease in MFI of TNNT2 in NAM-treated cells compared with control by flow cytometry, while no change was observed in TSA-treated cells (Figure 5B). Similar findings were observed by western analysis with NAM-treated cells with decreased expression of TNNT2 and TNNI3 (Figure 5C), while no change was observed in TSA-treated cells. Pan-hyperacetylation was also observed in NAM-treated but not TSA-treated cells (Figures 5C and 5D, respectively).
Figure 5.
AMPK Activation Increases NAD/NADH and Reduces Lysine Acetylation through Sirtuins
(A) NAD to NADH ratio with DMSO or AICAR, n = 3/time point independent experiments (I.E.); ∗∗∗p < 0.001 two-way ANOVA.
(B) MFI of TNNT2-Alexa Fluor 647 of TNNT2+ cells for DMSO, NAM and TSA, n = 3/group I.E.; ∗∗p < 0.001, ∗∗∗∗p < 0.0001 one-way ANOVA.
(C) Western analysis for TNNI3, TNNT2, and total acetylation in control and NAM-treated cells.
(D) Immunoblot images of TNNI3, TNNT2, and total acetylation with control and TSA.
(E) Western analysis of control, NAM, AICAR, and AICAR + NAM-treated cells for phospho-AMPK, TNNI3, and TNNT2.
(F) mRNA levels of TNNT2 and TNNI3 in DMSO, AICAR, NAM, and NAM + AICAR, n = 3 I.E.; ∗p < 0.05 one-way ANOVA.
(G) Western analysis for sirtuin target histone deacetylation lysine in control and AICAR-treated cells.
(H) Representative immunoblot analysis for sirtuin histone target deacetylation lysine in control and compound C-treated cells.
All data shown from Gibco-CMs after indicated treatment; sample numbers indicate biological replicates.
To further evaluate whether AMPK-mediated sirtuin activation improves differentiation of cardiomyocytes, we treated cells with AICAR, NAM, and NAM + AICAR. We observed that the AICAR-mediated increase in TNNI3 and TNNT2 was reduced in AICAR + NAM cells by western analysis (Figure 5E). In addition, we observed a trend toward decreased acetylation in cells treated with AICAR and AICAR + NAM compared with control or those treated with NAM alone (Figure 5E). Similarly, mRNA expression of TNNT2 was significantly decreased in NAM + AICAR-treated cells compared with AICAR (Figure 5F). SIRT1 acetylation sites, H3K9 and H4K16, and the SIRT6 acetylation site, H3K56, had decreased acetylation in AICAR-treated cells (Figure 5G), while compound C increased acetylation of these sites (Figure 5H). To examine expression of SIRT1, SIRT2, and SIRT6 we performed qPCR and found that SIRT1 expression was significantly increased at day 11 while SIRT2 mRNA levels after day 4 of differentiation significantly increased, and SIRT6 mRNA levels on days 7, 9, and 11 were significantly increased and followed by a decrease (Figure S2F).
Sustained AMPK Activation in iPSC-Derived Cardiomyocytes Changes Gene Expression through Chromatin Accessibility
We hypothesized that the transcriptome change associated with NAM and AICAR treatment was mediated through changes in chromatin accessibility. To investigate the global epigenomic landscape of iPSC-derived cardiomyocyte samples, an assay for transposase-accessible chromatin using sequencing (ATAC-seq) to detect chromatin accessibility (Buenrostro et al., 2013) was performed. We calculated motif accessibility scores (Schep et al., 2017) and found that ATAC-seq data from NAM and AICAR conditions showed high correlation (R = 0.97 and 1.00) between biological replicates, indicating that ATAC-seq could reliably and reproducibly measure chromatin accessibility in these samples. We focused on cardiac- or metabolic-related TFs and calculated their motif scores (Schep et al., 2017). We observed global depletion of chromatin accessibility at MEF2A, HGMA1, and MYOD1 TF motif binding sites and enrichment at GATA4, NKX2-5, HNF4, and SP3 TF motif binding sites upon AICAR treatment (Figures 6A and 6B). GATA4 and NKX2-5 TFs are known to control cardiac biogenesis and maintenance pathways (Kohli et al., 2011). We found significantly increased chromatin accessibility around TNNT2 and MYH6/7 loci with AICAR alone compared with NAM alone, consistent with changes in gene expression (Figure 6C) (Koshiba-Takeuchi et al., 2016). Histone acetylation is associated with increased transcriptional activity (euchromatin) and deacetylation of histones leads to gene silencing (heterochromatin) (Verdone et al., 2005). High DAPI intensity is a marker of heterochromatin, while euchromatin displays weaker DAPI fluorescence (Gorisch et al., 2005). We observed significantly increased DAPI intensity by flow cytometry and imaging in AICAR-treated samples versus control (Figures 6D and 6E), suggesting that AICAR-mediated histone deacetylation may increase heterochromatin and gene expression profiling. Taken together, these data show that AMPK activation affects chromatin accessibility, which may enhance cardiomyocyte differentiation through transcription.
Figure 6.
Persistent AMPK Activation in iPSC-Derived Cardiomyocytes Changes Gene Expression through Chromatin Accessibility
(A) Heatmap of motif scores on cardiac- or metabolic-related transcription factors.
(B) Volcano plots showing differential motif scores of cardiac and metabolic transcription factors.
(C) Comparison of chromatin accessibility between NAM only (top two replicates) and AICAR only (bottom two replicates) to control at TNNT2, TNNI1, TNNI3, MYH6, and MYH7 loci. The peak heights are normalized to the ACTB locus.
(D) MFI of DAPI in control and AICAR-treated Gibco-CMs, n = 5 wells with 10 fields/well; ∗∗∗∗p < 0.0001 unpaired t test.
(E) MFI by flow cytometry for DAPI in control and AICAR, n = 3 I.E.; ∗∗p < 0.001 unpaired t test.
(F) Unsupervised hierarchical clustering of cardiac gene expression from NanoString analysis, n = 3 independent biological replicates.
(G) Volcano plots showing differential gene expression analysis of cardiac genes.
To evaluate whether transcriptional changes were consistent with the ATAC-seq data observed, we utilized the NanoString Metabolic Pathways Panel with an additional 30 cardiac custom genes selected to provide a multiplex analysis of genes associated with metabolism and cardiomyocyte phenotype in cells treated with AICAR or NAM. With hierarchical clustering and principal-component analysis, we observed clustering by AICAR- and NAM-treated samples at the level of all gene pathways in the panel, including in selected cardiac genes (Figure 6F), with significantly increased mRNA levels of TNNT2, TNNI3, NPPA, ACTN2, and ATP2A2 in AICAR-treated cells compared with NAM-treated cells (Figures 6G, S2G, and S2H).
A schematic summarizing the major findings in this study is presented in Figure 7. We propose that AMPK activation via AICAR increased recruitment of GLUT4 and CD36 to the membrane and increased mitochondrial activity. In addition, hyperactivation of AMPK led to increased NAD/NADH which may mediate sirtuin activation and histone modification.
Figure 7.
Schematic of AMPK Activation Inducing Increased Expression of GLUT4 and CD36
Glucose and fatty acid uptake also increased NAD+ levels which activates sirtuins, leading to chromatin remodeling.
Discussion
Current protocols to differentiate cardiomyocytes from pluripotent stem cells generate immature cardiomyocytes with fetal-like phenotype (Yang et al., 2014). Fetal cardiomyocytes rely predominantly on aerobic glycolysis until undergoing a postnatal shift to β-oxidation of fatty acids to fulfill a high demand for ATP production (Hu et al., 2018). In this study, we tested the hypothesis that sustained AMPK activation facilitates cardiomyocyte differentiation via sirtuin activation, leading to altered histone acetylation, chromatin accessibility, and gene expression. AMPK activation transiently increased in early differentiation. AMPK activation was accompanied with hyperactivation of AMPK kinase LKB1. Inhibition of AMPK reduced expression of sarcomere components (TNNI3 and TNNT2) and OCR, suggesting that AMPK activation is required for normal iPSC-derived cardiomyocyte differentiation. We also observed that sustained AMPK activation with AICAR treatment significantly increased TNNI3 and TNNT2 protein and gene expression, and that these changes were accompanied with increased ATP levels. These effects appear to be confined to a narrow time window, as 10 days (instead of 5 days) of AICAR treatment appeared to have a detrimental effect on TNNT2 expression, thus we focused our study on evaluation of metabolic and molecular effects during the 7- to 16-day window of differentiation. AMPK activation leads to increased fatty acid uptake and inhibition of fatty acid synthesis by facilitating more fatty acid transporters to the membrane and increasing fatty acid β-oxidation (Arad et al., 2007). Sustained AMPK activation led to increased glucose and fatty acid uptake as well as increased mRNA and protein expression of GLUT4 and CD36 with increased OCR. In previous literature, fatty acid treatment of human pluripotent stem cell-derived cardiomyocytes led to hyperphosphorylation of AMPK, which ultimately increased cardiomyocyte maturation (Horikoshi et al., 2019; Yang et al., 2019). Moreover, mammalian target of rapamycin (mTOR) inhibition promotes iPSC-derived cardiomyocyte maturation through enhancing a quiescent state (Garbern et al., 2020). mTOR inhibition is also known to be regulated by AMPK activation (Agarwal et al., 2015). Here, activation of AMPK increased NAD/NADH levels and led to hyperactivation of sirtuins.
These data demonstrate that AICAR treatment reduced acetylation of sirtuin target sites (H3K9, H3K56, and H4K16) while AMPK inhibition led to hyperacetylation of the same target sites. Histone acetylation increases chromatin accessibility, thus these data suggest that NAD-mediated sirtuin activation may participate in iPSC-derived cardiomyocyte differentiation (Jing and Lin, 2015). ATAC-seq data showed that chromatin accessibility of cardiac transcription factors are affected by AICAR and NAM treatment, which supports the hypothesis that AMPK modifies the epigenetic state through sirtuin activation. These results were also supported by gene expression data which confirmed that AMPK activation not only increased expression of metabolic genes but also increased expression of cardiac transcription factors, such as GATA4 and NKX2-5, and sarcomere components, including TNNT2 and TNNI3. Several epigenetic mechanisms, including DNA methylation, histone methylation, and histone acetylation, are essential to control expression during cardiac development (Chang and Bruneau, 2012; Wu and Sun, 2006). DNA methylation plays an important role in mouse heart development and maturation, which is mediated by methyltransferase activity during the first 2 weeks (Sim et al., 2015). SIRT1 alters methylation of histones via recruitment and activation of histone methyltransferase (Jing and Lin, 2015). Sirtuins also directly deacetylate histones, transcription factors, and coregulators thereby altering gene expression through changes in chromatin accessibility (Bosch-Presegue and Vaquero, 2015; Gorisch et al., 2005; Jing and Lin, 2015; Sarikhani et al., 2018a, 2018b). These results demonstrate that sustained activation of AMPK in iPSC-derived cardiomyocytes during early differentiation led to epigenetic changes mediated by sirtuins. These changes may help prepare cardiomyocytes for the high demand for ATP post-differentiation.
Experimental Procedures
Additional details on experimental procedures can be found in Supplemental Experimental Procedures.
Cell Lines
Human iPSC lines used in this study include the Gibco episomal-derived iPSC line, BJ RiPS-A cell line from Harvard Stem Cell Institute, and UCSD142i-86-1 iPSC line. Cells were maintained in StemFlex (Thermo Fisher Scientific).
Differentiation of iPSC-Derived Cardiomyocytes
Differentiation was performed in a 12-well plate, with cells at 80% confluency used for differentiation (day 0). Cells were treated with CHIR99021 (6 μM) in RPMI + B27 minus insulin for 48 h (days 0–2), then with IWP4 (5 μM) for 48 h (days 2–4). On day 7, the medium was changed to RPMI + B27 with insulin (Lian et al., 2013). Batches of cells that started beating before day 9 with 70% TNNT2+ cells were used in this study. Beating cardiomyocytes were treated 1 day after onset of beating with 1 mM AICAR, 10 μM compound C, 10 mM nicotinamide (NAM), 10 nM trichostatin A (TSA), or vehicle (DMSO) for 5 days and used for the assay.
Quantitative Reverse Transcriptase PCR
Quantitative PCR (qPCR) was carried out using the iTaq Universal SYBR Green Supermix (Bio-Rad) with the primers listed in Table S1.
Western Analysis, Immunoprecipitation, Flow Cytometry, and Immunocytochemistry
Primary and secondary antibodies used in this study are shown in Table S2. Western blot band intensity was quantified using ImageJ software (NIH). Flow cytometry data were acquired via a BD LSRII instrument and analyzed using FlowJo software. Immunostained cells were visualized using a confocal microscope (Celldiscoverer7, Zeiss).
Metabolic Assays
The Glucose Uptake Cell-Based Assay Kit (Cayman), NAD/NADH Assay Kit (Abcam), ATP Assay Kit (Abcam), and the Free Fatty Acid Uptake Kit (Abcam) were used after cardiomyocyte treatment with DMSO or AICAR (1 mM) according to the manufacturer's instructions. For the glucose uptake and free fatty acid uptake assays, pre-treated cardiomyocytes were dissociated then plated at a density of 50,000 cells per well in a 96-well plate 1 day before assay. For the NAD/NADH and ATP assays, cells were collected at each time point then snap frozen in liquid nitrogen until use.
Seahorse Mito Stress Test
Cardiomyocytes were differentiated as described above then treated with DMSO, AICAR (1 mM), or compound C (10 nM) and seeded (20,000–30,000 cells) on Geltrex-coated Seahorse XFe96 cell culture microplates. The XF Cell Mito Stress Test Kit was used according to the manufacturer's instructions and OCR and ECAR were measured in the Seahorse XFe96 Analyzer. Cell density was determined using a CyQUANT assay (Thermo) for normalization.
ATAC-Seq Library Preparation and Data Analysis
ATAC-seq experiments were performed using a modified protocol (Xu et al., 2019). In brief, 50,000 cells were used for library preparation. The libraries were amplified for 9 cycles, purified with Zymo DNA Clean and Concentrator, and sequenced on a NextSeq 500 using a 75-cycle high output V2 Kit (read 1, 38 cycles; index 1, 8 cycles; read 2, 38 cycles). Raw reads were trimmed and aligned to the human reference genome (hg19) with Bowtie2 aligner (Langmead and Salzberg, 2012) using the option -X2000. The MACS2 peak caller (Zhang et al., 2008) (version 2.1.1) was used to call accessible regions of open chromatin regions (ATAC-seq peaks). TF motif scores were calculated using chromVAR (Schep et al., 2017).
Accession of ATAC-Seq Data
The ATAC-seq data generated from this study have been deposited in NCBI and are accessible through series accession number GEO: GSE152265.
NanoString Analysis
RNA was prepared for analysis using the NanoString nCounter Metabolic Pathways Panel with an additional 30 cardiac-related genes included as a custom spike-in (NanoString Technologies) according to the manufacturer's instructions.
Quantification and Statistical Analysis
Data analysis was performed by t test, one-way analysis of variance (ANOVA), and two-way ANOVA (GraphPad Prism). NanoString data were evaluated using nSolver 4.0 software with the Advanced Analysis Module 2.0. Open source R, Zen, Columbus, and ImageJ software were used to analyze confocal images and ImageJ was used for densitometric analysis.
Author Contributions
M.S. designed and performed the majority of the experiments, performed data analysis, prepared figures, and wrote the initial manuscript draft. J.C.G. helped design and perform the experiments and contributed to manuscript writing. R.S. performed the experiments and prepared figures. S.M. and J.D.B. performed and analyzed the ATAC-seq data. J.C., G.K., G.O.E., and A.A. assisted in performing the experiments. R.T.L. designed the experiments and contributed to manuscript writing.
Conflicts of Interest
R.T.L. is a co-founder, scientific advisory board member, and private equity holder of Elevian, Inc. Elevian also provides sponsored research support to the Lee Laboratory.
Acknowledgments
The authors thank the Harvard Center for Biological Imaging for use of their microscopy center and technical support and the Flow Cytometry Core in the Department of Stem Cell and Regenerative Biology. This study was supported by a grant from the National Heart, Lung, and Blood Institute (HL151684) and the Harvard Stem Cell Institute. J.C.G. was supported by an NIH T32 Fellowship (HL007572) and the American Academy of Pediatrics Children’s Heart Foundation Research Fellowship Award. J.D.B. and S.M. acknowledge support by the Allen Distinguished Investigator Program through the Paul G. Allen Frontiers Group and from the NIH New Innovator Award (DP2).
Published: July 9, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.06.012.
Supplemental Information
References
- Agarwal S., Bell C.M., Rothbart S.B., Moran R.G. AMP-activated protein kinase (AMPK) control of mTORC1 is p53- and TSC2-independent in pemetrexed-treated carcinoma cells. J. Biol. Chem. 2015;290:27473–27486. doi: 10.1074/jbc.M115.665133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcendor R.R., Kirshenbaum L.A., Imai S., Vatner S.F., Sadoshima J. Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ. Res. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- Arad M., Seidman C.E., Seidman J.G. AMP-activated protein kinase in the heart: role during health and disease. Circ. Res. 2007;100:474–488. doi: 10.1161/01.RES.0000258446.23525.37. [DOI] [PubMed] [Google Scholar]
- Bao J., Sack M.N. Protein deacetylation by sirtuins: delineating a post-translational regulatory program responsive to nutrient and redox stressors. Cell Mol. Life Sci. 2010;67:3073–3087. doi: 10.1007/s00018-010-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R., Gonzalo S., Jaco I., Schotta G., Klatt P., Jenuwein T., Blasco M.A. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 2007;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boise L.H., Gonzalez-Garcia M., Postema C.E., Ding L., Lindsten T., Turka L.A., Mao X., Nunez G., Thompson C.B. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- Bonkowski M.S., Sinclair D.A. Slowing ageing by design: the rise of NAD(+) and sirtuin-activating compounds. Nat. Rev. Mol. Cell. Biol. 2016;17:679–690. doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Presegue L., Vaquero A. Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. FEBS J. 2015;282:1745–1767. doi: 10.1111/febs.13053. [DOI] [PubMed] [Google Scholar]
- Bowman P.R.T., Smith G.L., Gould G.W. GLUT4 expression and glucose transport in human induced pluripotent stem cell-derived cardiomyocytes. PLoS One. 2019;14:e0217885. doi: 10.1371/journal.pone.0217885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C., Elliott P.J., Puigserver P., Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabowski A., Momken I., Coort S.L., Calles-Escandon J., Tandon N.N., Glatz J.F., Luiken J.J., Bonen A. Prolonged AMPK activation increases the expression of fatty acid transporters in cardiac myocytes and perfused hearts. Mol. Cell Biochem. 2006;288:201–212. doi: 10.1007/s11010-006-9140-8. [DOI] [PubMed] [Google Scholar]
- Chang C.P., Bruneau B.G. Epigenetics and cardiovascular development. Annu. Rev. Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- Choi Y.J., Lee K.Y., Jung S.H., Kim H.S., Shim G., Kim M.G., Oh Y.K., Oh S.H., Jun D.W., Lee B.H. Activation of AMPK by berberine induces hepatic lipid accumulation by upregulation of fatty acid translocase CD36 in mice. Toxicol. Appl. Pharmacol. 2017;316:74–82. doi: 10.1016/j.taap.2016.12.019. [DOI] [PubMed] [Google Scholar]
- Das C., Lucia M.S., Hansen K.C., Tyler J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskandarian H.A., Impens F., Nahori M.A., Soubigou G., Coppee J.Y., Cossart P., Hamon M.A. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science. 2013;341:1238858. doi: 10.1126/science.1238858. [DOI] [PubMed] [Google Scholar]
- Garbern J.C., Helman A., Sereda R., Sarikhani M., Ahmed A., Escalante G.O., Ogurlu R., Kim S.L., Zimmerman J.F., Cho A. Inhibition of mTOR signaling enhances maturation of cardiomyocytes derived from human-induced pluripotent stem cells via p53-induced quiescence. Circulation. 2020;141:285–300. doi: 10.1161/CIRCULATIONAHA.119.044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorisch S.M., Wachsmuth M., Toth K.F., Lichter P., Rippe K. Histone acetylation increases chromatin accessibility. J. Cell Sci. 2005;118:5825–5834. doi: 10.1242/jcs.02689. [DOI] [PubMed] [Google Scholar]
- Herskovits A.Z., Guarente L. Sirtuin deacetylases in neurodegenerative diseases of aging. Cell Res. 2013;23:746–758. doi: 10.1038/cr.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway G.P., Jain S.S., Bezaire V., Han X.X., Glatz J.F., Luiken J.J., Harper M.E., Bonen A. FAT/CD36-null mice reveal that mitochondrial FAT/CD36 is required to upregulate mitochondrial fatty acid oxidation in contracting muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009;297:R960–R967. doi: 10.1152/ajpregu.91021.2008. [DOI] [PubMed] [Google Scholar]
- Horikoshi Y., Yan Y., Terashvili M., Wells C., Horikoshi H., Fujita S., Bosnjak Z.J., Bai X. Fatty acid-treated induced pluripotent stem cell-derived human cardiomyocytes exhibit adult cardiomyocyte-like energy metabolism phenotypes. Cells. 2019;8:1095. doi: 10.3390/cells8091095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Linders A., Yamak A., Correia C., Kijlstra J.D., Garakani A., Xiao L., Milan D.J., van der Meer P., Serra M. Metabolic maturation of human pluripotent stem cell-derived cardiomyocytes by inhibition of HIF1alpha and LDHA. Circ. Res. 2018;123:1066–1079. doi: 10.1161/CIRCRESAHA.118.313249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Halicka H.D., Traganos F., Tanaka T., Kurose A., Darzynkiewicz Z. Cytometric assessment of DNA damage in relation to cell cycle phase and apoptosis. Cell Prolif. 2005;38:223–243. doi: 10.1111/j.1365-2184.2005.00344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong C.M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Irrcher I., Ljubicic V., Kirwan A.F., Hood D.A. AMP-activated protein kinase-regulated activation of the PGC-1alpha promoter in skeletal muscle cells. PLoS One. 2008;3:e3614. doi: 10.1371/journal.pone.0003614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H., Lin H. Sirtuins in epigenetic regulation. Chem. Rev. 2015;115:2350–2375. doi: 10.1021/cr500457h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yang G., Kim Y., Kim J., Ha J. AMPK activators: mechanisms of action and physiological activities. Exp. Mol. Med. 2016;48:e224. doi: 10.1038/emm.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli S., Ahuja S., Rani V. Transcription factors in heart: promising therapeutic targets in cardiac hypertrophy. Curr. Cardiol. Rev. 2011;7:262–271. doi: 10.2174/157340311799960618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba-Takeuchi K., Morita Y., Nakamura R., Takeuchi J.K. Combinatorial functions of transcription factors and epigenetic factors in heart development and disease. In: Nakanishi T., Markwald R.R., Baldwin H.S., Keller B.B., Srivastava D., Yamagishi H., editors. Etiology and Morphogenesis of Congenital Heart Disease: From Gene Function and Cellular Interaction to Morphology. Springer; 2016. pp. 295–303. [Google Scholar]
- Kuznetsov J.N., Leclerc G.J., Leclerc G.M., Barredo J.C. AMPK and Akt determine apoptotic cell death following perturbations of one-carbon metabolism by regulating ER stress in acute lymphoblastic leukemia. Mol. Cancer Ther. 2011;10:437–447. doi: 10.1158/1535-7163.MCT-10-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Cacicedo J.M., Ruderman N., Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J. Biol. Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybutt D.R., Thompson A.L., Cooney G.J., Kraegen E.W. Selective chronic regulation of GLUT1 and GLUT4 content by insulin, glucose, and lipid in rat cardiac muscle in vivo. Am. J. Physiol. 1997;273:H1309–H1316. doi: 10.1152/ajpheart.1997.273.3.H1309. [DOI] [PubMed] [Google Scholar]
- Li X., Kazgan N. Mammalian sirtuins and energy metabolism. Int. J. Biol. Sci. 2011;7:575–587. doi: 10.7150/ijbs.7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X., Hsiao C., Kamp T.J., Palecek S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat. Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C.Y., Kennedy B.K. SIRT6, oxidative stress, and aging. Cell Res. 2016;26:143–144. doi: 10.1038/cr.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.L., Olson A.L., Moye-Rowley W.S., Buse J.B., Bell G.I., Pessin J.E. Expression and regulation of the human GLUT4/muscle-fat facilitative glucose transporter gene in transgenic mice. J. Biol. Chem. 1992;267:11673–11676. [PubMed] [Google Scholar]
- Liu X., Chhipa R.R., Nakano I., Dasgupta B. The AMPK inhibitor compound C is a potent AMPK-independent antiglioma agent. Mol. Cancer Ther. 2014;13:596–605. doi: 10.1158/1535-7163.MCT-13-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchiano S., Bertero A., Murry C.E. Learn from your elders: developmental biology lessons to guide maturation of stem cell-derived cardiomyocytes. Pediatr. Cardiol. 2019;40:1367–1387. doi: 10.1007/s00246-019-02165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maria Z., Campolo A.R., Lacombe V.A. Diabetes alters the expression and translocation of the insulin-sensitive glucose transporters 4 and 8 in the atria. PLoS One. 2015;10:e0146033. doi: 10.1371/journal.pone.0146033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta M.C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Ofir M., Arad M., Porat E., Freimark D., Chepurko Y., Vidne B.A., Seidman C.E., Seidman J.G., Kemp B.E., Hochhauser E. Increased glycogen stores due to gamma-AMPK overexpression protects against ischemia and reperfusion damage. Biochem. Pharmacol. 2008;75:1482–1491. doi: 10.1016/j.bcp.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Piquereau J., Caffin F., Novotova M., Lemaire C., Veksler V., Garnier A., Ventura-Clapier R., Joubert F. Mitochondrial dynamics in the adult cardiomyocytes: which roles for a highly specialized cell? Front Physiol. 2013;4:102. doi: 10.3389/fphys.2013.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquereau J., Ventura-Clapier R. Maturation of cardiac energy metabolism during perinatal development. Front Physiol. 2018;9:959. doi: 10.3389/fphys.2018.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Zarrinpashneh E., Budas G.R., Pouleur A.C., Dutta A., Prescott A.R., Vanoverschelde J.L., Ashworth A., Jovanovic A., Alessi D.R. Deficiency of LKB1 in heart prevents ischemia-mediated activation of AMPKalpha2 but not AMPKalpha1. Am. J. Physiol. Endocrinol. Metab. 2006;290:E780–E788. doi: 10.1152/ajpendo.00443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikhani M., Maity S., Mishra S., Jain A., Tamta A.K., Ravi V., Kondapalli M.S., Desingu P.A., Khan D., Kumar S. SIRT2 deacetylase represses NFAT transcription factor to maintain cardiac homeostasis. J. Biol. Chem. 2018;293:5281–5294. doi: 10.1074/jbc.RA117.000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikhani M., Mishra S., Desingu P.A., Kotyada C., Wolfgeher D., Gupta M.P., Singh M., Sundaresan N.R. SIRT2 regulates oxidative stress-induced cell death through deacetylation of c-Jun NH2-terminal kinase. Cell Death Differ. 2018;25:1638–1656. doi: 10.1038/s41418-018-0069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Maier B., Bartke A., Scrable H. Progressive loss of SIRT1 with cell cycle withdrawal. Aging Cell. 2006;5:413–422. doi: 10.1111/j.1474-9726.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- Schep A.N., Wu B., Buenrostro J.D., Greenleaf W.J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods. 2017;14:975–978. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuderi G.J., Butcher J. Naturally engineered maturation of cardiomyocytes. Front. Cell Dev. Biol. 2017;5:50. doi: 10.3389/fcell.2017.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L., Martinez-Redondo P., Marazuela-Duque A., Vazquez B.N., Dooley S.J., Voigt P., Beck D.B., Kane-Goldsmith N., Tong Q., Rabanal R.M. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013;27:639–653. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao D., Tian R. Glucose transporters in cardiac metabolism and hypertrophy. Compr. Physiol. 2015;6:331–351. doi: 10.1002/cphy.c150016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim C.B., Ziemann M., Kaspi A., Harikrishnan K.N., Ooi J., Khurana I., Chang L., Hudson J.E., El-Osta A., Porrello E.R. Dynamic changes in the cardiac methylome during postnatal development. FASEB J. 2015;29:1329–1343. doi: 10.1096/fj.14-264093. [DOI] [PubMed] [Google Scholar]
- Vanhaecke T., Papeleu P., Elaut G., Rogiers V. Trichostatin A-like hydroxamate histone deacetylase inhibitors as therapeutic agents: toxicological point of view. Curr. Med. Chem. 2004;11:1629–1643. doi: 10.2174/0929867043365099. [DOI] [PubMed] [Google Scholar]
- Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Verdone L., Caserta M., Di Mauro E. Role of histone acetylation in the control of gene expression. Biochem. Cell Biol. 2005;83:344–353. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- Viollet B., Athea Y., Mounier R., Guigas B., Zarrinpashneh E., Horman S., Lantier L., Hebrard S., Devin-Leclerc J., Beauloye C. AMPK: lessons from transgenic and knockout animals. Front. Biosci. (Landmark Ed.) 2009;14:19–44. doi: 10.2741/3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanet A., Arnould T., Najimi M., Renard P. Connecting mitochondria, metabolism, and stem cell fate. Stem Cells Dev. 2015;24:1957–1971. doi: 10.1089/scd.2015.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., An H., Liu T., Qin C., Sesaki H., Guo S., Radovick S., Hussain M., Maheshwari A., Wondisford F.E. Metformin improves mitochondrial respiratory activity through activation of AMPK. Cell Rep. 2019;29:1511–1523.e5. doi: 10.1016/j.celrep.2019.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Sun Y.E. Epigenetic regulation of stem cell differentiation. Pediatr. Res. 2006;59:21R–25R. doi: 10.1203/01.pdr.0000203565.76028.2a. [DOI] [PubMed] [Google Scholar]
- Xu H., Ding J., Porter C.B.M., Wallrapp A., Tabaka M., Ma S., Fu S., Guo X., Riesenfeld S.J., Su C. Transcriptional atlas of intestinal immune cells reveals that neuropeptide alpha-CGRP modulates group 2 innate lymphoid cell responses. Immunity. 2019;51:696–708.e9. doi: 10.1016/j.immuni.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagoe S., Kanno T., Kanno Y., Sasaki S., Siegel R.M., Lenardo M.J., Humphrey G., Wang Y., Nakatani Y., Howard B.H. Interaction of histone acetylases and deacetylases in vivo. Mol. Cell. Biol. 2003;23:1025–1033. doi: 10.1128/MCB.23.3.1025-1033.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Pabon L., Murry C.E. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2014;114:511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Rodriguez M.L., Leonard A., Sun L., Fischer K.A., Wang Y., Ritterhoff J., Zhao L., Kolwicz S.C., Jr., Pabon L. Fatty acids enhance the maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Reports. 2019;13:657–668. doi: 10.1016/j.stemcr.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Braun T. Multimodal regulation of cardiac myocyte proliferation. Circ. Res. 2017;121:293–309. doi: 10.1161/CIRCRESAHA.117.308428. [DOI] [PubMed] [Google Scholar]
- Zaha V.G., Young L.H. AMP-activated protein kinase regulation and biological actions in the heart. Circ. Res. 2012;111:800–814. doi: 10.1161/CIRCRESAHA.111.255505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., MacLean P.S., Pohnert S.C., Knight J.B., Olson A.L., Winder W.W., Dohm G.L. Regulation of muscle GLUT-4 transcription by AMP-activated protein kinase. J. Appl. Physiol. (1985) 2001;91:1073–1083. doi: 10.1152/jappl.2001.91.3.1073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.