Abstract
Accumulating evidence indicates that aberrant regulation of metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1), a long noncoding RNA (lncRNA), plays a vital role in tumorigenesis. However, its association with breast cancer has not been systematically evaluated. In the current study, a meta-analysis was conducted to clarify the association between MALAT-1 and the prognosis and clinicopathological features of breast cancer. Relevant literature published in several databases was searched. Hazard ratio (HR) and odds ratio (OR) with 95% confidence interval (CI) were calculated to evaluate the effect of MALAT-1 expression on the survival outcomes and clinicopathological features of breast cancer. A total of 12 studies involving 4106 patients were identified. Pooled HR demonstrated that elevated MALAT-1 expression significantly predicted unfavorable overall survival (HR = 2.06, 95% CI: 1.66–2.56, P<0.0001) in patients with breast cancer. Subgroup analysis stratified by cancer type, sample size, and method of variance analysis also showed statistically significant associations. Additionally, the HR of patients with up-regulated MALAT-1 expression concerning disease-free survival (DFS), recurrence-free survival (RFS), and disease-specific survival (DSS) was 1.91 (95% CI: 1.53–2.39, P<0.0001). Further, elevated MALAT-1 expression was positively correlated with the progesterone receptor (PR) status (OR = 1.47, 95% CI: 1.18–1.82). Thus, MALAT-1 is a promising biomarker for predicting survival outcomes in patients with breast cancer.
Keywords: breast cancers, Long noncoding RNA, MALAT-1, prognosis
Introduction
Breast cancer is the most common malignancy affecting females, and although substantial progress in terms of its management has been made, it still remains the major cause of cancer-related deaths among females worldwide due to recurrence and metastasis [1,2]. Breast cancer is a heterogeneous disease that exhibits considerable variability in its prognostic patterns and treatment response [3,4]. Molecular biomarkers have been identified as promising candidates and may not only predict the biological behavior and clinical outcome but also contribute to the improvement of treatment protocols [5–7].
Long noncoding RNAs (lncRNAs), with a length of more than 200 nucleotides, do not encode proteins and were previously regarded as ‘transcriptional noise’ or ‘junk RNA’; however, a growing number of studies have indicated that lncRNAs are key regulators that influence various biological processes [8,9]. LncRNAs have been found to be consistently dysregulated and closely related to carcinogenesis and tumor progression in many cancers including liver, pancreas, colon, lung, prostate, and breast cancers [10]. In particular, several lncRNAs are considered to be promising prognostic markers in cancer [11,12].
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1), approximately 8000 nucleotides in length, is one of the most conserved lncRNAs and is extremely abundant and ubiquitously expressed in different tissues [13]. MALAT-1 was initially recognized as a metastasis-associated gene that might predict the potential metastasis of non-small-cell lung cancer [14]. There is accumulating evidence that dysregulated MALAT-1 is closely associated with the development of various tumors. For example, MALAT-1 can promote cell invasion and migration by serving as a competitive endogenous RNA (ceRNA) to attenuate the function of miR-200c-3p/ZEB1 in pancreatic ductal adenocarcinoma [15]. MALAT-1 has also been shown to induce epithelial–mesenchymal transition through the miR-204/ZEB2 axis in breast cancer [16]. A previous meta-analysis has shown that the up-regulation of MALAT-1 is significantly correlated with the prognosis and metastasis of cancers [17]. Nevertheless, the prognostic role of MALAT-1 in breast cancer is poorly characterized, with no systematic evaluation conducted to date [18,19]. Therefore, we conducted a systematic review to investigate the associations of MALAT-1 with clinicopathological features and survival of patients with breast cancer.
Materials and methods
Identification
We carefully searched international databases (Web of Science, PubMed, and Embase) to identify studies referring to MALAT-1 in breast cancer. We used the following search terms or key words ‘MALAT-1 OR MALAT1 OR metastasis-associated lung adenocarcinoma transcript 1’ and ‘breast or mammary’ and ‘tumor OR cancer OR carcinoma OR neoplasm.’ The last update of our research was conducted on 22 January 2020.
Inclusion and exclusion criteria
Studies fulfilling the following criteria were regarded as eligible studies: (1) the expression of MALAT-1 was measured in either tumor tissue or serum; (2) patients in the study were diagnosed with breast cancer; (3) the relationship between MALAT-1 expression and clinicopathological features or prognosis in breast cancer patients was investigated; and (4) odds ratios (ORs) or hazard ratios (HRs) and 95% confidence interval (CI) were recorded or sufficient published information was provided to calculate them. Studies meeting any of the following criteria were excluded: (1) important information including clinicopathological parameters and survival outcomes was not available; (2) studies were in the form of reviews, letters, comments, or conference abstracts; (3) studies were published in a language other than English; and (4) studies that reprocessed data from public databases.
Quality assessment and data extraction
Two investigators (Y.y.W. and Y.j.Z.) independently assessed all potentially eligible studies based on the Newcastle–Ottawa Scale (NOS) [20]. Articles with NOS ≥ 6 were regarded as high-quality studies. Any discrepancy was resolved by discussion. We extracted the following required information: (1) first author and publication year; (2) patient characteristics (country, ethnicity, sample size, type of specimen, histological grade, stage of disease, hormone receptor including progesterone receptor (PR) and estrogen receptor (ER) status, human epidermal growth factor receptor 2 (HER2) status, lymphoid node status, and follow-up); (3) detection methods of MALAT-1 and cut-off value; and (4) HRs and the corresponding 95% CI of up-regulated MALAT-1 for overall survival (OS), metastasis-free survival (MFS), disease-specific survival (DSS), disease-free survival (DFS), or recurrence-free survival (RFS). We also utilized Engauge Digitizer (version 4.1) to estimate the HRs based on the graphical survival plots when HRs were not directly available in the study [21]. We also corresponded with relevant original authors for additional data required for meta-analysis.
Breast cancer patient cohort analysis based on public datasets
The gene expression profile of GSE19615 containing chemotherapy-treated breast cancer samples was obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The genome expression data of breast cancer samples from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) were also downloaded. MALAT-1 expression levels were compared between the two groups according to whether the included patients received chemotherapy. Kaplan–Meier survival analysis was then employed to compare the survival outcomes between groups. Given the vital roles of immune infiltration in the tumorigenesis and metastasis, we applied the CIBERSORT algorithm to determine the association between MALAT-1 and infiltrating immune cells from gene expression profiles. The correlation was evaluated using the Pearson’s correlation coefficient; an estimated P-value of <0.05 was considered statistically significant.
Statistical analysis
HRs and their associated 95% CIs for OS, MFS, DFS, DSS, and RFS were used to evaluate the prognostic effect of MALAT-1 in breast cancer. An HR > 1 and 95% CI not including 1 suggested a significant association between poor prognosis and elevated MALAT-1 expression. Cochran’s Q test and P-values were applied to find the heterogeneity among studies. If the calculation suggested significant heterogeneity (I2 ≥ 50% and/or P≤0.05), the random-effects model was utilized. Otherwise, the fixed-effects model was more appropriate. ORs and their corresponding 95% CIs were calculated to determine the association of MALAT-1 with the clinicopathological variables of breast cancer. We used a sensitivity analysis to evaluate the stability of the results. The possibility of publication bias was also tested through Egger’s test and Begg’s funnel plot. All statistical analyses were performed using the Stata 15.0 software.
Results
Summary of the included studies
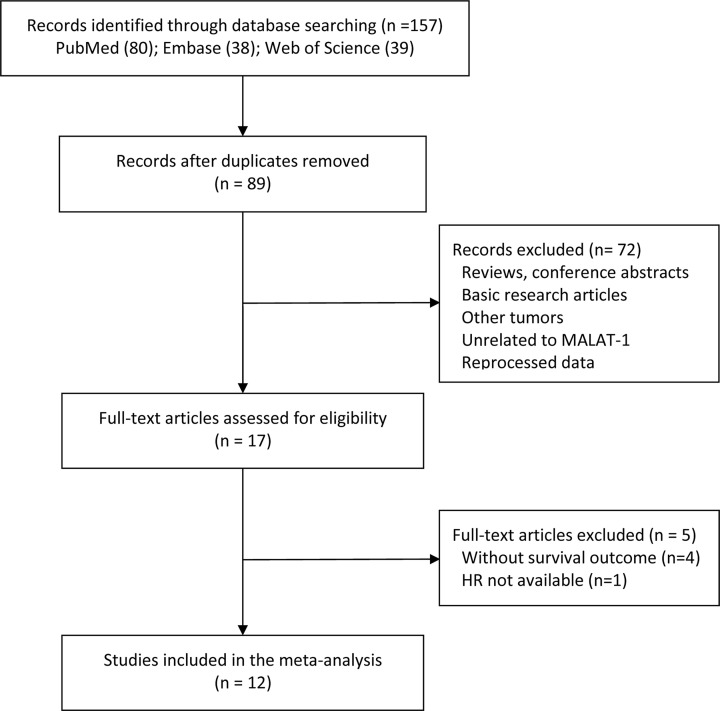
Based on the Web of Science, PubMed, and Embase databases, we initially retrieved 157 papers, 68 of which were excluded due to duplication. After careful screening of the titles and abstracts, 17 potentially eligible articles were reviewed in detail. Due to the lack of key information regarding survival outcomes, 5 of the 17 articles were subsequently excluded; hence, we finally included a total of 12 articles in the meta-analysis [16,18,19,22–30]. Figure 1 summarizes the details of the literature selection procedure.
Figure 1. Flow chart depicting the literature selection procedure in this meta-analysis.
A total of 4186 patients from China, France, and the United States were investigated, with a minimum and maximum sample size of 43 and 1992 patients, respectively. MALAT-1 expression levels were assessed using reverse transcriptase polymerase chain reaction (RT-PCR) in all studies, with 11 studies employing cancerous tissue and 1 using serum specimens [26]. The cut-off estimates for MALAT-1 expression, including the median, fold change, and expression level, were inconsistent among these studies. Elevated MALAT-1 expression levels in breast cancer were reported in most studies, except in one study that reported the down-regulation of MALAT-1 [18]. Among the different cancer subtypes, Jadaliha et al. found that MALAT-1 expression was consistently higher in ER-positive patients than in those with the triple-negative breast cancer (TNBC) subtype [24]. They also suggested that TNBC subtype cells express relatively lower levels of MALAT-1 than other subtypes of breast cancer cells. There were two studies which focused only on TNBC [22,28]. Of these, Huang et al. divided all patients into groups based on receptor status, with ER-positive or -negative groups [23], and Jadaliha et al. did not report a negative result in all patients but mentioned the survival outcomes in the lymph node-negative TNBC and HER2-positive breast cancer subsets [24]. Eight of all included studies described the clinicopathological features of breast cancer [16,18,19,22,23,25,26,30]. Of the 12 articles, 2 studies only described the survival outcomes of breast cancer, without providing necessary data to obtain HR [18,25]. HR was reported directly in four studies [19,23,24,27] and estimated indirectly for the remaining six studies [16,22,26,28–30]. Table 1 shows the detailed information of the included studies.
Table 1. Main characteristics of the studies included in the present meta-analysis.
| Author’s name | Year | Country | Number | Type | Cut-off | Follow-up (months) | Sample | Detection method | Expression level | HR | Survival outcomes | Multivariate analysis | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jin [22] | 2015 | China | 139 | TNBC | median | 55 | tissue | RT-PCR | ↑ | SC | OS | No | 7 |
| Xu [18] | 2015 | China | 135 | Mixed | NR | NR | tissue | RT-PCR | ↓ | NR | NR | No | 6 |
| Huang [23] | 2016 | China | 204 | ER+ and ER− | 75% expression | 65 | tissue | RT-PCR | ↑ | Reported | RFS | No | 8 |
| Jadaliha [24] | 2016 | U.S.A. | 1992 | TNBC and Her2+ | NR | NR | tissue | RT-PCR | ↑ | Reported | DSS | No | 6 |
| Meseure [25] | 2016 | France | 446 | Mixed | 3.02-fold | NR | tissue | RT-PCR | ↑ | NR | MFS | No | 7 |
| Miao [26] | 2016 | China | 78 | Mixed | median | 60 | tissue | RT-PCR | ↑ | SC | DFS | No | 8 |
| Wang [16] | 2017 | China | 118 | Mixed | NR | 50 | tissue | RT-PCR | ↑ | SC | OS | No | 7 |
| Zhang [27] | 2017 | China | 178 | Mixed | 3.48 | NR | serum | RT-PCR | ↑ | Reported | OS | Yes | 7 |
| Zuo [28] | 2017 | China | 43 | TNBC | median | 60 | tissue | RT-PCR | ↑ | SC | OS | No | 7 |
| Li [29] | 2018 | China | 258 | Mixed | NR | 23 | tissue | RT-PCR | ↑ | SC | RFS | No | 7 |
| Wang [19] | 2018 | China | 509 | Mixed | median | 82.8 | tissue | RT-PCR | ↑ | Reported | OS/RFS | Yes | 7 |
| Zhang [30] | 2018 | China | 86 | Mixed | median | NR | tissue | RT-PCR | ↑ | SC | OS | No | 6 |
Abbreviations: ER+, ER positive; ER−, ER negative; Her2+, Her2 positive; NM, not mentioned; NR, not reported; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; SC, survival curve.
Association between MALAT-1 expression and OS
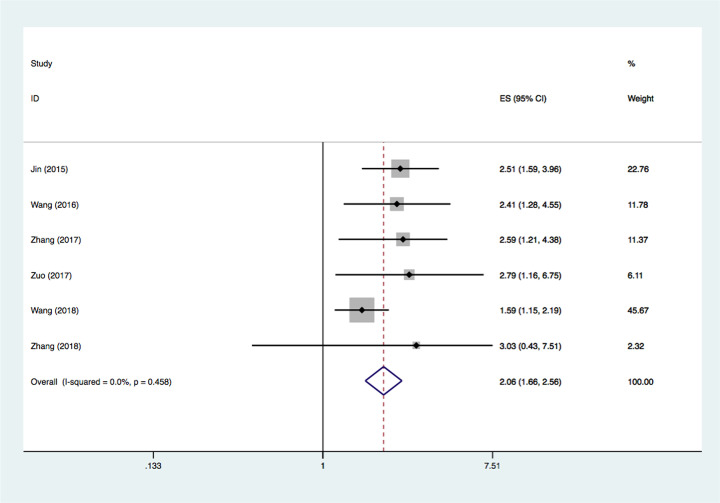
Six studies evaluated the connection between MALAT-1 expression levels and OS [16,19,22,27,28,30]; no statistically significant heterogeneity was observed among the six studies (I2 = 0.0%, P=0.458). Thus, the fixed-effects model was applied. Collectively, the results indicated that increased MALAT-1 expression in breast cancer was related to a worse OS (HR = 2.06, 95% CI: 1.66–2.56, P<0.0001) (Figure 2). Next, we performed subgroup analysis according to cancer type, sample size, and the method of variance analysis. As shown in Table 2, the subgroup analysis for cancer type suggested that MALAT-1 up-regulation was a strong prognostic biomarker in both patients with TNBC (HR = 1.88, 95% CI = 1.45‒2.44, P<0.0001) and in those with mixed type breast cancer (HR = 2.57, 95% CI = 1.71‒3.85, P<0.0001). Next, we performed subgroup analysis according to the sample size, with a threshold for 100 patients. The results showed that the association of MALAT-1 dysregulation and OS was not significantly influenced by sample size (n≥100: HR = 2.00, 95% CI = 1.59‒2.51, P<0.0001; n<100: HR = 2.86, 95% CI = 1.35‒6.04, P=0.006). Similarly, pooled results of subgroup analysis according to the analysis method revealed that elevated MALAT-1 expression was significantly associated with worse OS in both the multivariate (HR = 1.75, 95% CI = 1.31‒2.34, P<0.0001) and univariate groups (HR = 2.55, 95% CI = 1.83‒3.55, P<0.0001).
Figure 2. Forest plots of studies evaluating the association between MALAT-1 expression and OS in patients with breast cancer.
Table 2. Results of subgroup analyses of pooled HRs for OS with elevated MALAT-1 expression.
| Categories | Number of studies | Number of patients | Pooled HR (95% CI) | Heterogeneity | |
|---|---|---|---|---|---|
| I2 (%) | P-value | ||||
| Cancer type | |||||
| Mixed | 4 | 891 | 2.57 (1.71, 3.85) | 0.4 | 0.834 |
| TNBC | 2 | 182 | 1.88 (1.45, 2.44) | 0 | 0.39 |
| Sample size | |||||
| ≥100 | 4 | 944 | 2.00 (1.59, 2.51) | 22.2 | 0.277 |
| <100 | 2 | 128 | 2.85 (1.35, 6.04) | 0 | 0.923 |
| Analysis method | |||||
| Univariate | 4 | 386 | 2.55 (1.83, 3.55) | 0 | 0.988 |
| Multivariate | 2 | 687 | 1.75 (1.31, 2.34) | 43.4 | 0.184 |
Association between MALAT-1 expression and DFS/RFS/DSS
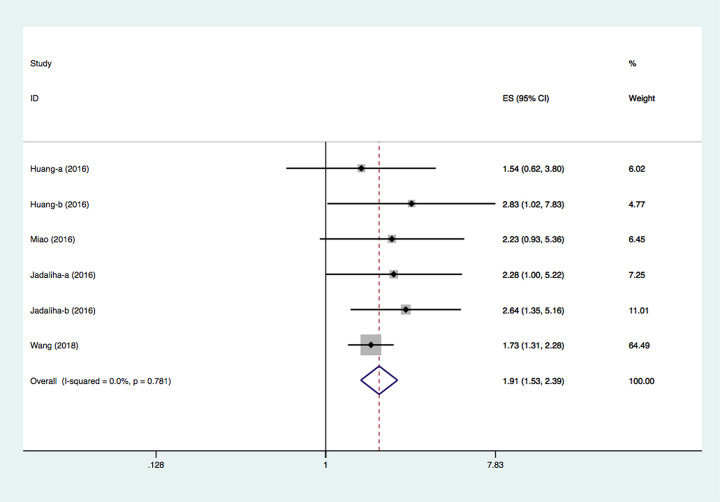
Six studies investigated the connection between MALAT-1 expression levels and DFS/MFS/DSS/RFS [19,23–26,29]. Li et al.’s study suggested that low MALAT-1 expression predicted favorable RFS in their cohort (HR = 0.0093, 95% CI: 0.0013‒0.0637, P<0.001) [29]. Their data were insufficient for quantitative analysis but were still included in the qualitative analysis. Meseure et al.’s study suggested that the impact of MALAT-1 expression level on MFS was not significant. Unfortunately, this study did not provide useful data for HR calculation [25]. Eventually, a total of four studies compromising six groups of data reported the prognostic significance of MALAT-1 for DFS, RFS, and DSS. The pooled HR was 1.91 (95% CI: 1.53–2.39, P<0.0001, using the fixed-effects model) (Figure 3). On account of the limited number of studies, a subgroup analysis for DFS/RFS/DSS was not performed.
Figure 3. Forest plots of studies evaluating the association between MALAT-1 expression and DFS/RFS/DSS in patients with breast cancer.
Association between MALAT-1 expression and clinicopathological parameters
We also evaluated the relationship between the expression level of MALAT-1 and the clinicopathological features by pooling the clinicopathological data. As shown in Table 3, we found that high expression of MALAT-1 was significantly associated with PR status (OR = 1.47, 95% CI = 1.18–1.82, P=0.0006, fixed-effects model). Unlike the PR status, the ER status did not display a significant correlation with MALAT-1 expression, as there was great heterogeneity (OR = 1.37, 95% CI = 0.85‒2.22, P=0.19, random-effects model). In addition, a significant correlation was not found at TNM stage (OR = 1.27, 95% CI = 0.56‒2.87, P=0.57, random-effects model), tumor size (OR = 0.95, 95% CI = 0.69–1.31, P=0.75, fixed-effects model), distant metastasis (OR = 1.17, 95% CI = 0.27– 5.15, P=0.83, random-effects model), lymph node metastasis (OR = 0.86, 95% CI = 0.41– 1.82, P=0.7, random-effects model), histological grade (OR = 0.90, 95% CI = 0.47–1.71, P=0.74, random-effects model), and HER2 status (OR = 1.10, 95% CI = 0.81–1.48, P=0.56, fixed-effects model).
Table 3. Meta-analyses of MALAT-1 expression with respect to clinicopathologic parameters.
| Variables | Number of studies | Number of patients | Model | OR (95% CI) | P-value | Heterogeneity (I2, P-value) |
|---|---|---|---|---|---|---|
| TNM stage (III/IV vs. I/II) | 5 | 902 | Random | 1.27 (0.56, 2.87) | 0.57 | 82%, 0.0002 |
| Tumor size (>2 vs <2 cm) | 6 | 639 | Fixed | 0.95 (0.69, 1.31) | 0.75 | 0.0%, 0.58 |
| Distant metastasis (positive vs. negative) | 4 | 386 | Random | 1.17 (0.27, 5.15) | 0.83 | 89%, <0.00001 |
| Lymph node metastasis (positive vs. negative) | 4 | 862 | Random | 0.86 (0.41, 1.82) | 0.7 | 77%, 0.004 |
| Histological grade (III vs. I/II) | 5 | 937 | Random | 0.90 (0.47, 1.71) | 0.74 | 74%, 0.004 |
| ER status (positive vs. negative) | 8 | 1380 | Random | 1.38 (0.85, 2.22) | 0.19 | 72.0%, 0.0008 |
| PR status (positive vs. negative) | 8 | 1400 | Fixed | 1.47 (1.18, 1.82) | 0.0006 | 0.0%, 0.44 |
| HER2 status (positive vs. negative) | 6 | 1007 | Fixed | 1.10 (0.81, 1.48) | 0.56 | 0.0%, 0.84 |
Sensitivity analysis and publication bias
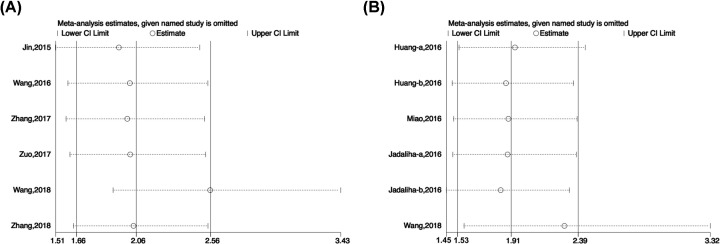
We performed sensitivity analysis to examine the stability of the meta-analysis. When any one study was sequentially omitted, no individual study dominantly affected the overall HR for OS (Figure 4A) or DFS, RFS, or DSS (Figure 4B).
Figure 4. Sensitivity analysis of the pooled HRs of MALAT-1 expression.
(A) OS; (B) DFS/RFS/DSS.
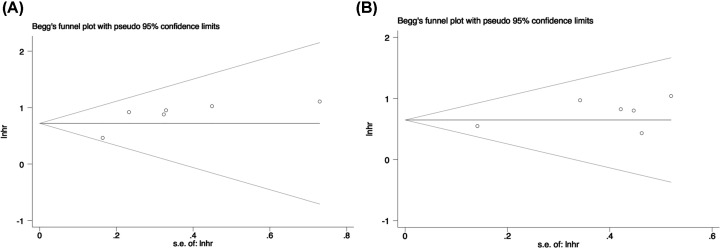
Egger’s test and Begg’s funnel plot were used to test publication bias. As shown in Figure 5A,B, the funnel plot demonstrated basic symmetrical shape in OS studies (Egger’s test P=0.071) and in DFS/RFS/DSS studies (Egger’s test P=0.142). Hence, there was no noticeable publication bias in the current meta-analysis.
Figure 5. Funnel plot of publication bias analysis.
(A) OS; (B) DFS/RFS/DSS.
Breast cancer patient cohort analysis based on public datasets
We also checked MALAT-1 expression levels in chemotherapy-treated patients based on publicly available microarray datasets. Our analysis showed that the expression level of MALAT-1 was markedly lower in the chemotherapy-treated groups than that in the non-chemotherapy group (Supplementary Figure S1). Indeed, MALAT-1 could not predict prognosis in these groups (data not shown). These findings suggested that the prognostic value of MALAT-1 for patients receiving chemotherapy requires further investigation.
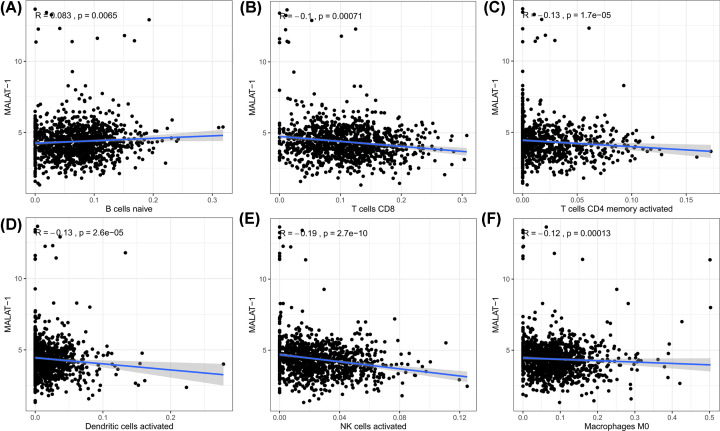
To further explore the possible reason why MALAT-1 is responsible for poor survival in patients with breast cancer, we analyzed the association between MALAT-1 and tumor infiltrating immune cells based on TCGA dataset. As shown in Figure 6, MALAT-1 expression levels showed a significant positive correlation with immune infiltration by B cells (R = 0.083, P=0.0065) and a negative association with CD8+ T cells (R = −0.1, P=0.00071), CD4+ T cells (R = −0.13, P=1.7e−05), dendritic cells (R = −0.13, P<2.6e−05), NK cells (R = −0.19, P=2.7e−10), and macrophages M0 (R = −0.12, P=0.00013). These findings strongly implied that MALAT-1 may be implicated in immune infiltration in patients with breast cancer.
Figure 6. Correlation between MALAT-1 and infiltrating immune cells.
(A) association between MALAT-1 and B cells; (B) association between MALAT-1 and CD8+ T cells; (C) association between MALAT-1 and CD4+ T cells; (D) association between MALAT-1 and dendritic cells; (E) association between MALAT-1 and NK cells; (F) association between MALAT-1 and macrophages M0.
Discussion
MALAT-1, also referred to as noncoding nuclear-enriched abundant transcript 2, was initially identified to promote metastasis in non-small-cell lung cancer [13,14]. To date, aberrant expression of MALAT-1 has been found in multiple cancers, such as ovarian, breast, colorectal, and bladder cancers [19,31–33]. Some studies have shown that elevated MALAT-1 expression levels contribute to breast cancer carcinogenesis, whereas a few studies have demonstrated that MALAT-1 may serve as a tumor-suppressing gene [34,35]. Hence, the role of MALAT-1 and its influence on survival outcomes in patients with breast cancer remain controversial. In addition, although previously published reviews and meta-analyses have reported that MALAT-1 could function as a potential prognostic biomarker in cancers, no studies have focused on its prognostic significance in breast cancer. Here, we pooled published data to highlight the prognostic and clinical value of MALAT-1 in breast cancer. It is well-known that breast cancer is a highly heterogeneous disease. Based on the heterogeneity of ER and HER2 expression, breast cancer can be classified into three main subtypes: luminal (luminal A and luminal B), HER2-positive, and TNBC. These subtypes display distinct histological features, molecular etiologies, and clinical behaviors [5,6]. Our comprehensive meta-analysis of 4186 cases from 12 cohorts showed that high MALAT-1 expression levels in patients with breast cancer were observed in most studies, with no obvious subtype or cell specificity. Patients with breast cancer having elevated MALAT-1 expression levels displayed worse survival outcomes (both OS and DFS/RFS/DSS). The association between MALAT-1 and poor prognosis in patients with breast cancer was consistent in most of the original studies as well as in the reprocessed data obtained from TCGA database [36]. The biological behavior of MALAT-1 in cancer may explain this correlation. First, MALAT-1 is involved in pre-mRNA alternative splicing through interactions with serine- and arginine-rich proteins [37,38]. Second, MALAT-1 participates in transcriptional regulation. MALAT-1 actively interacts with the 3′ end of the gene body and overlaps with H3K36me2 peaks, a marker of active transcriptional elongation, indicating its role in gene expression [13]. Third, MALAT-1 can function as a post-transcriptional regulator of gene expression through a ‘ceRNA’ mechanism. MALAT-1 utilizes miRNA-responsive elements in miRNA sequences as a ‘language’ to communicate with mRNAs and pseudogenes, thereby resulting in phenotypic alterations such as cell invasion and metastasis [16,39]. Multiple studies have suggested that MALAT-1 plays oncogenic roles in breast cancer. For instance, MALAT-1 modulates cdc42 expression by sponging miR-1 in cell lines, thereby triggering migration and invasion [40]. MALAT-1 can promote angiogenesis by interacting with miR-145 [41]. MALAT-1 also contributes to the maintenance of stem cell-like phenotypes in breast cancer cells by regulating self-renewal-associated factors [42]. Moreover, MALAT-1 exerts a crucial role in tumor progression and metastasis in both TNBC and luminal cells [23,28]. Immune cell infiltration and the tumor microenvironment have been verified to play essential roles in the initiation and progression of cancers [43,44]. Low levels of immune-infiltrating cells in the tumor microenvironment may confer worse prognosis in breast cancer [45,46]. In our study, high MALAT-1 expression levels were associated with low immune cell infiltration (e.g., CD4+ and CD8+ T cells), which may explain the correlation between MALAT-1 and poor prognosis in patients with breast cancer. Taken together, the extensive range of functions of MALAT-1 enables it to predict survival outcomes in patients with breast cancer.
Notably, our results suggested that high MALAT-1 expression levels were significantly associated with PR status (OR = 1.47, 95% CI = 1.18–1.82). ER status also had a tendency for correlation with MALAT-1 expression, albeit not significant (P>0.05) due to the evident heterogeneity. Previous studies have shown that MALAT-1 regulates the effect of 17β-estradiol treatment on breast cell lines [47] and is associated with ER and its target genes. Moreover, MALAT-1 might confer tamoxifen resistance by affecting the ER signaling pathway [23,48]. These studies demonstrate that MALAT-1 is closely related to hormone receptor status in breast cancer, which is consistent with our results. No significant association was observed between MALAT-1 expression and advanced clinical parameters, such as distant metastasis, lymph node metastasis, and TNM stage. This negative result seems reasonable, given the limited number of enrolled patients as well as the diverse enrollment criteria among studies. In addition, Kim et al. [34] and Kwok et al. [35] recently demonstrated a metastasis-suppressing rather than a metastasis-promoting role of MALAT-1 in breast cancer. Moreover, multiple molecular networks, and not MALAT-1 alone, contribute to tumor development. Therefore, further studies should be conducted to identify the function of MALAT-1 in breast cancer and verify the association between clinical parameters and MALAT-1 expression.
Although the prognostic value of MALAT-1 was identified in this meta-analysis, there are a few limitations of the study. First, marked heterogeneity existed owing to the difference in methodology, such as in the determination of cut-off values, sample selection, RT-PCR primer sets, and statistical analyses. We applied the random-effects model to minimize the influence of these differences when heterogeneity was large. Second, HR may have been overestimated because only two studies provided multivariate HR estimate for survival outcomes. In addition, the HR value extracted from the survival curve might cause slight error for prognosis. Third, we should carefully consider the preference for publications with significant results, as studies showing significant results are easier to publish than negative ones. Notably, Messeur et al. [25] did not report the negative result regarding MALAT-1 and breast cancer prognosis, whereas Jadaliha et al. [24] and Zuo et al. [28] only reported significant results for specific groups. Fourth, the sample size of some included studies was small (<100), and smaller studies could obtain higher effect size as well as heterogeneity compared with larger studies. Finally, language bias could also have been implicated as the selected studies were only published in English.
In conclusion, our study indicated that elevated MALAT-1 expression levels are predictive of unfavorable prognosis, not only for OS but also DFS, RFS, and DSS in breast cancer. Therefore, it could potentially function as a prognostic biomarker for patients with breast cancer. Furthermore, our results demonstrated that the level of MALAT-1 tended to correlate with hormone receptor status. Nevertheless, due to the limitations mentioned above, additional comprehensive, well-designed multicenter studies should be performed to confirm and strengthen our findings regarding the prognostic role of MALAT-1 in breast cancer.
Supplementary Material
Abbreviations
- ceRNA
competitive endogenous RNA
- CI
confidence interval
- DFS
disease-free survival
- DSS
disease-specific survival
- ER
estrogen receptor
- HER2
human epidermal growth factor receptor 2
- HR
hazard ratio
- H3K36me2
Histone H3 dimethylation at lysine 36
- lncRNA
long noncoding RNA
- MALAT-1
metastasis-associated lung adenocarcinoma transcript 1
- MFS
metastasis-free survival
- NK
natural killer
- NOS
Newcastle–Ottawa Scale
- OR
odds ratio
- OS
overall survival
- PR
progesterone receptor
- RFS
recurrence-free survival
- RT-PCR
reverse transcriptase polymerase chain reaction
- TCGA
The Cancer Genome Atlas
- TNBC
triple-negative breast cancer
- TNM
Tumor Node Metastasis
- ZEB
zinc-finger E-box-binding homeobox
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Nature Science Foundation of Zhejiang Province [grant numbers LQ19H060002, LQ19H160041]; and the Medical and Health Science and Technology Project of Zhejiang Province [grant numbers 2018KY089, 2020KY143].
Author Contribution
Y.y.W., Y.j.Z., and S.z.Z. conceived and designed the research. Y.y.W., Y.j.Z., K.m.H., and J.l.Q. searched the literature and analyzed the data. Y.y.W. and Y.j.Z. wrote the manuscript. Y.H. and M.q.Z. proofread the manuscript. S.z.Z. finally revised the manuscript. Y.y.W. and Y.j.Z. contributed equally to this work as first authors.
References
- 1.Allemani C., Matsuda T., Di Carlo V. et al. (2018) Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 391, 1023–1075 10.1016/S0140-6736(17)33326-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D. and Jemal A. (2019) Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 3.Slamon D.J., Clark G.M., Wong S.G. et al. (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182 10.1126/science.3798106 [DOI] [PubMed] [Google Scholar]
- 4.Prat A., Parker J.S., Fan C. and Perou C.M. (2012) PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res. Treat. 135, 301–306 10.1007/s10549-012-2143-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slamon D.J., Leyland-Jones B., Shak S. et al. (2001) Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 10.1056/NEJM200103153441101 [DOI] [PubMed] [Google Scholar]
- 6.Perou C.M., Sorlie T., Eisen M.B. et al. (2000) Molecular portraits of human breast tumours. Nature 406, 747–752 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 7.Prat A., Lluch A., Albanell J. et al. (2014) Predicting response and survival in chemotherapy-treated triple-negative breast cancer. Br. J. Cancer 111, 1532–1541 10.1038/bjc.2014.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinn J.J. and Chang H.Y. (2016) Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- 9.Jandura A. and Krause H.M. (2017) The New RNA World: growing evidence for long noncoding RNA functionality. Trends Genet. 33, 665–676 10.1016/j.tig.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 10.Huarte M. (2015) The emerging role of lncRNAs in cancer. Nat. Med. 21, 1253–1261 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 11.Chen X., Yang Y., Cao Y. et al. (2018) lncRNA PVT1 identified as an independent biomarker for prognosis surveillance of solid tumors based on transcriptome data and meta-analysis. Cancer Manag. Res. 10, 2711–2727 10.2147/CMAR.S166260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Wen W., Zhao S. et al. (2015) Prognostic role of HOTAIR in four estrogen-dependent malignant tumors: a meta-analysis. Onco Targets Ther. 8, 1471–1482 10.2147/OTT.S84687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X., Hamblin M.H. and Yin K.J. (2017) The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 14, 1705–1714 10.1080/15476286.2017.1358347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji P., Diederichs S., Wang W. et al. (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 10.1038/sj.onc.1206928 [DOI] [PubMed] [Google Scholar]
- 15.Zhuo M., Yuan C., Han T. et al. (2018) A novel feedback loop between high MALAT-1 and low miR-200c-3p promotes cell migration and invasion in pancreatic ductal adenocarcinoma and is predictive of poor prognosis. BMC Cancer 18, 1032 10.1186/s12885-018-4954-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Zhou Y., Yang Z. et al. (2017) MiR-204/ZEB2 axis functions as key mediator for MALAT1-induced epithelial-mesenchymal transition in breast cancer. Tumour Biol. 39, 1010428317690998, 10.1177/1010428317690998 [DOI] [PubMed] [Google Scholar]
- 17.Li J., Cui Z., Li H. et al. (2018) Clinicopathological and prognostic significance of long noncoding RNA MALAT1 in human cancers: a review and meta-analysis. Cancer Cell Int. 18, 109 10.1186/s12935-018-0606-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu S., Sui S., Zhang J. et al. (2015) Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int. J. Clin. Exp. Pathol. 8, 4881–4891 [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z., Katsaros D., Biglia N. et al. (2018) High expression of long non-coding RNA MALAT1 in breast cancer is associated with poor relapse-free survival. Breast Cancer Res. Treat. 171, 261–271 10.1007/s10549-018-4839-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang S. and Kleijnen J. (2010) Quality assessment tools for observational studies: lack of consensus. Int. J. Evid. Based Healthc. 8, 247 10.1111/j.1744-1609.2010.00195.x [DOI] [PubMed] [Google Scholar]
- 21.Tierney J.F., Stewart L.A., Ghersi D., Burdett S. and Sydes M.R. (2007) Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8, 16 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin C., Yan B., Lu Q., Lin Y. and Ma L. (2016) Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumour Biol. 37, 7383–7394 10.1007/s13277-015-4605-6 [DOI] [PubMed] [Google Scholar]
- 23.Huang N.S., Chi Y.Y., Xue J.Y. et al. (2016) Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget 7, 37957–37965 10.18632/oncotarget.9364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jadaliha M., Zong X., Malakar P. et al. (2016) Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 7, 40418–40436 10.18632/oncotarget.9622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meseure D., Vacher S., Lallemand F. et al. (2016) Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br. J. Cancer 114, 1395–1404 10.1038/bjc.2016.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao Y., Fan R., Chen L. and Qian H. (2016) Clinical significance of long non-coding RNA MALAT1 expression in tissue and serum of breast cancer. Ann. Clin. Lab. Sci. 46, 418–424 [PubMed] [Google Scholar]
- 27.Zhang P., Luo Q., Hu T. and Yan D. (2017) Serum MALAT1 expression predicts clinical outcome in breast cancer patients receiving cyclophosphamide based treatment. Int. J. Clin. Exp. Med. 10, 7624–7635 [Google Scholar]
- 28.Zuo Y., Li Y., Zhou Z., Ma M. and Fu K. (2017) Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed. Pharmacother. 95, 922–928 10.1016/j.biopha.2017.09.005 [DOI] [PubMed] [Google Scholar]
- 29.Li Z., Xu L., Liu Y. et al. (2018) LncRNA MALAT1 promotes relapse of breast cancer patients with postoperative fever. Am. J. Transl. Res. 10, 3186–3197 [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P., Zhou H., Lu K. et al. (2018) Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. Onco Targets Ther. 11, 291–299 10.2147/OTT.S155134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y., Xu X., Lv H. et al. (2016) The long noncoding RNA MALAT-1 is highly expressed in ovarian cancer and induces cell growth and migration. PLoS ONE 11, e0155250 10.1371/journal.pone.0155250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P.L., Zhang X., Wang H.Y. et al. (2017) MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol. Cancer Ther. 16, 739–751 10.1158/1535-7163.MCT-16-0591 [DOI] [PubMed] [Google Scholar]
- 33.Li C., Cui Y., Liu L.F. et al. (2017) High expression of long noncoding RNA MALAT1 indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Clin. Genitourin. Cancer 15, 570–576 10.1016/j.clgc.2017.05.001 [DOI] [PubMed] [Google Scholar]
- 34.Kim J., Piao H.L., Kim B.J. et al. (2018) Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50, 1705–1715 10.1038/s41588-018-0252-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwok Z., Roche V., Chew X., Fadieieva A. and Tay Y. (2018) A non-canonical tumor suppressive role for the long non-coding RNA MALAT1 in colon and breast cancers. Int. J. Cancer 143, 668–678 10.1002/ijc.31386 [DOI] [PubMed] [Google Scholar]
- 36.Zheng L., Zhang Y., Fu Y. et al. (2019) Long non-coding RNA MALAT1 regulates BLCAP mRNA expression through binding to miR-339-5p and promotes poor prognosis in breast cancer. Biosci. Rep. 39, 1–12 10.1042/BSR20181284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tripathi V., Ellis J.D., Shen Z. et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tripathi V., Shen Z., Chakraborty A. et al. (2013) Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 9, e1003368 10.1371/journal.pgen.1003368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomson D.W. and Dinger M.E. (2016) Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 17, 272–283 10.1038/nrg.2016.20 [DOI] [PubMed] [Google Scholar]
- 40.Chou J., Wang B., Zheng T. et al. (2016) MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem. Biophys. Res. Commun. 472, 262–269 10.1016/j.bbrc.2016.02.102 [DOI] [PubMed] [Google Scholar]
- 41.Huang X., Xia Y., He G., Zheng L., Cai Y., Yin Y. et al. (2018) MALAT1 promotes angiogenesis of breast cancer. Oncol. Rep. 40, 2683–2689 [DOI] [PubMed] [Google Scholar]
- 42.Zeng L., Cen Y. and Chen J. (2018) Long non-coding RNA MALAT-1 contributes to maintenance of stem cell-like phenotypes in breast cancer cells. Oncol. Lett. 15, 2117–2122 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Sharma P., Hu-Lieskovan S., Wargo J. and Ribas A. (2017) Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato T., Noma K., Ohara T., Kashima H., Katsura Y., Sato H. et al. (2018) Cancer-associated fibroblasts affect intratumoral CD8(+) and FoxP3(+) T cells via IL6 in the tumor microenvironment. Clin. Cancer Res. 24, 4820–4833 10.1158/1078-0432.CCR-18-0205 [DOI] [PubMed] [Google Scholar]
- 45.Bense R.D., Sotiriou C., Piccart-Gebhart M.J., Haanen J., van Vugt M., de Vries E.G.E. et al. (2017) Relevance of tumor-infiltrating immune cell composition and functionality for disease outcome in breast cancer. J. Natl. Cancer Inst. 109, 10.1093/jnci/djw192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tawfik O., Kimler B.F., Karnik T. and Shehata P. (2018) Clinicopathological correlation of PD-L1 expression in primary and metastatic breast cancer and infiltrating immune cells. Hum. Pathol. 80, 170–178 10.1016/j.humpath.2018.06.008 [DOI] [PubMed] [Google Scholar]
- 47.Zhao Z., Chen C., Liu Y. and Wu C. (2014) 17β-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem. Biophys. Res. Commun. 445, 388–393 10.1016/j.bbrc.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 48.Sherene L., Benjamin H.-K., Christine D. et al. (2007) Definition of clinically distinct molecular subtypes in estrogen receptor–positive breast carcinomas through genomic grade. J. Clin. Oncol. 25, 1239–1246 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.