Abstract
Background
Signal-regulatory-protein beta 1 (SIRPB1) is a signal-regulatory-protein member of the immunoglobulin superfamily and is capable of modulating receptor tyrosine kinase-coupled signaling. Copy-number variations (CNVs) at the SIRPB1 locus were previously reported to associate with prostate cancer aggressiveness in patients, however the role of SIRPB1 in prostate carcinogenesis is unknown.
Methods
FISH and laser capture microdissection coupled with qPCR was utilized to determine SIRPB1 gene amplification and mRNA expression in prostate cancer specimens. The effect of knockdown of SIRPB1 by RNAi in PC3 prostate cancer cells on cell growth in colony formation assays and cell mobility in wound-healing, transwell assays and cell cycle analysis was determined. Overexpression of SIPRB1 in C4–2 prostate cancer cells on cell migration, invasion, colony formation and cell cycle progression and tumor take rate in xenografts was also determined. Western blot assay of potential downstream SIRPB1 pathways was also performed.
Results
SIRPB1 gene amplification was detected in up to 37.5% of prostate cancer specimens based on in silico analysis of several publicly available datasets. SIRPB1 gene amplification and overexpression was detected in prostate cancer specimens. Knockdown of SIRPB1 significantly suppressed cell growth in colony formation assays and cell mobility. SIRPB1 knockdown also induced cell cycle arrest during the G0/G1 phase and an enhancement of apoptosis. Conversely, overexpression of SIPRB1 in C4–2 prostate cancer cells significantly enhanced cell migration, invasion, colony formation and cell cycle progression and increased C4–2 xenograft tumor take rate in nude mice. Finally, this study presented evidence for SIRPB1 regulation of Akt phosphorylation and showed that Akt inhibition could abolish SIRPB1 stimulation of prostate cancer cell proliferation.
Conclusions
These results suggest that SIRPB1 is a potential oncogene capable of activating Akt signaling to stimulate prostate cancer proliferation and could be a biomarker for patients at risk of developing aggressive prostate cancer.
Keywords: SIRPB1, prostate cancer, Akt signaling, FISH, qPCR, C4–2, PC3
Introduction
Signal regulatory proteins (SIRPs) are cell surface receptors, each containing three extracellular Ig-like domains, and are classified in human as SIRPA, SIRPB, and SIRPG. SIRPA, SIRPB and SIRPG have high sequence similarity and similar extracellular regions, but different cytoplasmic regions (1–6). SIRPA appears to act as a potential tumor suppressor (7–10). The growth suppression of SIRPA appears to require its long cytoplasmic region (110–113 amino acid) containing tyrosine residues that resemble inhibitory immunoreceptor tyrosine-based inhibition motifs (ITIMs), which recruit the phosphatase SH2-domain-containing protein SHP-1 and SHP-2 in vivo (11). A recent study found that SHP-2 suppression resulted in blockade of SIRPA-mediated inhibition of anchorage-independent growth in rat fibroblasts (12). SIRPA inhibited anchorage-independent growth of v-Src-transformed cells by eliciting anoikis, and SHP-2 was required for this effect (7). The same group also found that SIRPA/SHP-2 signaling induced anoikis in human breast carcinoma cells with activated c-Src (7). These studies suggest a critical role of the long cytoplasmic tail in SIRPA suppression of cell growth. In contrast to SIRPA, SIRPB has a short 6 amino acid cytoplasmic domain that lacks signaling motifs. However, SIRPB has a charged amino acid residue in the transmembrane domain which could interact with immunoreceptor tyrosine-based activation motif (ITAM)-bearing molecules such as Dap12 (DNAX-activating protein of molecular mass 12 kilodaltons (kDa) (13). SIRPB1 is a disulphide-linked dimer, while SIRPA and SIRPG are monomeric proteins, suggesting that these proteins have different ligand-binding topology (14). In addition to SIRPB1, two additional isoforms, SIRPB2 and SIRPB3 can be transcribed from the same gene (15,16). The role of SIRPB1 in carcinogenesis and cell growth has not been previously reported.
Prostate cancer is the most common cancer and the second leading cause of cancer death in men in the United States, with predicted 174,650 new cases and 31,620 deaths in 2019 (17). Identification of the genes and molecular mechanisms involved in prostate cancer development and progression remain incompletely understood. One frequently utilized approach to identify genes playing important roles in prostate cancer is genome-wide copy-number variation (CNV) analysis. This approach was used previously to identify several CNVs in genes that were significantly associated with aggressive prostate tumors (18). Jin and colleagues observed that one CNV (CNP2454, a 32.3 kb deletion polymorphism at 20p13), affecting all three isoforms of SIRPB1, was significantly associated with prostate cancer aggressiveness in 448 aggressive and 500 non-aggressive patients recruited from Johns Hopkins Hospital (JHH1) [odds ratio (OR) = 1.30, 95% confidence interval (CI): 1.01–1.68; P = 0.045]. Using the best tagging SNP for CNP2454, rs2209313, this association was confirmed in both JHH1 and additional 2895 aggressive and 3094 nonaggressive cases (18). CNP2454 (rs2209313) was significantly associated with an increased risk of aggressive PC (OR = 1.17, 95% CI: 1.07–1.27; P = 2.75 × 10−4). However, CNV at SIRPB1 locus in prostate cancer patients suggests, but does not prove, a functional role of SIRPB1 in prostate cancer.
In the present study, we analyzed CNV and mRNA expression of SIRPB1 in publicly available datasets through the cBioPortal (19,20) and in human prostate cancer specimens obtained through the University of Pittsburgh Biospecimen Core. We investigated the function of SIRPB1 in prostate cancer cell lines using siRNA knockdown and overexpression approaches coupled with clonogenic assay, wound healing assay, transwell migration assay, fluorescence activated cell sorting (FACS) analysis of cell cycle and apoptosis, and xenograft tumor model. Furthermore, we explored potential signaling pathways regulated by SIRPB1 in prostate cancer cells.
Material and Methods
Data Analysis Using cBioPortal
The cBioPortal for Cancer Genomics site (http://cbioportal.org) (19,20) was used to determine the alteration frequency and disease progression-free survival for SIRPB1 in several publicly available prostate cancer data sets.
Fluorescence In situ Hybridization (FISH)
Treatment naïve human prostate cancer tissue specimens were obtained from the UPMC Hillman Cancer Center’s Tissue and Research Pathology Services/Pitt Biospecimen Core under approval by the University of Pittsburgh Institutional Review Board. Specimens included prostate tumor and normal adjacent tissue sections from 20 prostate cancer patients (11 patients with Gleason 4+3 tumors and 9 patients with Gleason 4+5 tumors). FISH analysis was performed by the UPMC Hillman Cancer Center’s Tissue and Research Pathology Services/Pitt Biospecimen Core following a standardized protocol for clinical specimens by a cytogenetic histotechnologist (M.M.) in the In Situ Laboratory. Formalin-fixed, paraffin-embedded tissues were serially sectioned at 4 μm intervals. An H&E stained section from each specimen was prepared and reviewed by a board-certified genitourinary pathologist (R. Dhir) to identify and mark one ‘normal’ area and one ‘tumor’ area for further analysis. Slides were baked for one hour to overnight at 56°C (Oven Range: 55–65°C). Sections were deparaffinized in xylene twice for 10 minutes each, immersed twice in 100% ethanol, and then pretreated (1N sodium thiocyanate). Slides were digested in pepsin solution (0.5 mg/mL in 0.9% sodium chloride) followed by drying. Dual-color FISH was then performed using SIRBP1 20q13 Spectrum Orange FISH probe (Empire Genomics, Buffalo, New York, USA) and Vysis CEP20 Spectrum Green FISH Probe (Abbott Molecular Inc., Des Plaines, IL, USA). FISH analysis was performed as described (21). The slides and probe were denatured in 70% formamide at 75°C for 5 minutes and through 70%, 85%, and 100% alcohols for 2 minutes each before hybridization. A 10 μl volume of probe mixture was applied to the target area, then a 22 mm square coverslip was placed over the probe in order to allow the probe to spread evenly under the coverslip. Hybridization was performed using a Dako Hybridizer (Dako, Santa Clara, CA). After denaturing, the coverslip was sealed with rubber cement. Slides were incubated overnight at 37°C in a humidified chamber. Post-hybridization washes were performed in 2x SSC/0.3% IGEPAL for 2 minutes at 72°C. Rubber cement was gently removed by pulling on the sealant with forceps, and slide was immersed in post hybridization buffer at room temperature to float off the coverslip. Slides were air-dried in the dark and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Analysis was performed using a Leica Biosystems (CytoVision FISH Capture and Analysis Workstation, Buffalo Grove, IL, USA). Only individual and well-delineated cells were counted; overlapping cells were excluded from the analysis.
A total of 60 cells were scored for each case, at least 30 non-overlapping cells from the marked normal area and 30 cells from the tumor area. Nuclei with weak intensity, non-specificity, noisy background or insufficient counterstain to determine the nuclear border were not enumerated. Signals were rejected if signals were non-uniform (>25%), auto-fluorescence was high or nuclear resolution was poor. The number of gene signals (orange) and centromere signals (green) were counted in each cell on a standard worksheet. Two images per probe are maintained for each case. A ratio of gene signals to centromere signals was calculated for each of the marked areas, as well as signal to nucleus ratio (SNR) and centromere to nucleus ratio (CNR). Hyperploidy cells include nuclei with greater than two green signals per cell. Monosomy cells include nuclei with one orange signal and one green signal. A board-certified genitourinary pathologist (R.D.) confirmed results through review of counts and image analysis and confirmed tumor was counted.
Laser-Capture Microdissection (LCM) and Quantitative Polymerase Chain Reaction (qPCR)
A second set of 20 human prostate cancer tissue specimens was obtained from the University of Pittsburgh Biospecimen Core for LCM coupled with qPCR analysis. Specimens included prostate tumor and normal adjacent tissue sections from 1 prostate cancer patient with Gleason 3+3, 7 patients with Gleason 3+4, 2patients with Gleason 4+3, 1 patient with Gleason 3+5, 6 patients with Gleason 4+5, and 3 patients with Gleason 5+4 tumors. Prostate cancer cells and adjacent normal glandular cells were isolated by LCM using a Leica LMD6000 Microsystems microscope (Wetzlar, Germany) equipped with an HV-D20P Hitachi (Tokyo, Japan) color camera and Leica Laser Microdissection V 6.3 imaging software (Wetzlar, Germany). LCM and qPCR were performed as described previously (22). Captured individual tissue specimens were lysed, and RNA isolation, reverse transcription, and qPCR were performed using CellsDirect One-Step qRT-PCR Kit (Invitrogen, Carlsbad, CA, USA). Gene-specific primers and Taqman probes cross exon/exon junctions and were designed (Table S1). Probes contained FAM fluorophore and TAMRA quencher. qPCR was performed on ABI Step-One Plus (Applied Biosystems, Foster City, CA, USA) and data were analyzed by ΔCp (crossing point) method as R=2[Cp sample − Cp control] to generate the relative expression ratio (R) of each target gene relative to ACTB (22).
Cell Culture and Plasmids
PC3, LNCaP and 22Rv1cells (23–25) were purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA). The C4–2 cell line (26) was a gift from Leland W.K. Chung (Cedars-Sinai, Los Angeles, CA, USA). C4–2, PC3, LNCaP, and 22Rv1 cells were cultured in RPMI-1640 (#10–040-CV, Corning, Corning, NY, USA) media supplemented with 10% FBS (#10099–141, Thermo Fisher Scientific, Waltham, MA, USA). All cell lines were maintained at low passage (<6 months) after thawing from master vials (ATCC) subjected to short tandem repeat (STR) profiling of polymorphic loci with a >80% match criteria for cell line authentication. pCMV6-AC-GFP-SIRPB1 (#RG211765) and pCMV6-AC-GFP-vector (#PS100010) were purchased from OriGene Technologies (Rockville, MD, USA). C4–2 cells were transduced with pCMV6-AC-GFP-SIRPB1 or pCMV6-AC-GFP-vector using PolyJet In Vitro DNA Transfection Reagent (#SL100688, SignaGen Laboratories, Rockville, MD, USA), according to the manufacturer’s instructions. At 72 hours post-transfection, the cells were selected with 1 mg/mL of neomycin (#10131035, Thermo Fisher Scientific, Waltham, MA, USA) for at least 1 week, and sorted by flow cytometry to isolate the stably transfected GFP or GFP-SIRPB1 C4–2 cells. C4–2 cells were stimulated with increasing doses (0.1 – 100 nM) of dihydrotestosterone (DHT) for 48 hours before western blot analysis to determine whether SIRPB1 expression was sensitive to androgens.
RNAi Knockdown
For transient knockdown, PC3 cells were transfected with control siRNA (#D-001810–10-50, GE Healthcare Dharmacon, Inc., Lafayette, CO, USA) or two different siRNAs targeting SIRPB1 using Dharma FECT siRNA transfection reagent (T-2002–03, GE Healthcare Dharmacon, Inc.) in six-well plates. The final concentration of siRNA SIRPB1 (siSIRPB1–1 or siSIRPB1–2) or siRNA Control (si-Control) was 50 nM. For the mock control (Mock), cells were treated with the transfection reagent only. At 72 hours after transfection or treatment, the above Mock, si-Control, siSIRPB1–1, and siSIRPB1–2 PC3 cells were used for further analysis. The sequences of siRNAs specific for SIRPB1 are listed in Supplemental Table 2 (Integrated DNA Technologies, Coralville, IA, USA).
Extraction of total RNA and qPCR analysis in cell line studies
Total RNAs were isolated from cultured cells using Trizol reagent (#15596026, Thermo Fisher Scientific) and then reverse transcribed to generate cDNA using GoScript reverse transcription system (#A5000, Promega, Madison, WI, USA). The cDNA samples were analyzed by the TB GreenTM AdvantageR qPCR premix (#639676, Takara, Kusatsu, Shiga Prefecture, Japan). Gene-specific primers crossing exon/exon junctions were designed (Supplemental Table S3) (Integrated DNA Technologies) and qPCR was performed on ABI Step-One Plus (Applied Biosystems, Foster City, CA) and data were analyzed the same as described above.
Western Immunoblotting
Harvested cells were lysed in RIPA buffer (#0278, Sigma-Aldrich Corp., St. Louis, MO, USA) supplemented with proteinase inhibitors and Phosphatase Inhibitor Cocktail 2 (#P5726, Sigma-Aldrich Corp) and Phosphatase Inhibitor Cocktail 3 (#P0044, Sigma-Aldrich Corp.). The lysates were resolved by SDS-PAGE and transferred to Whatman Western polyvinylidene difluoride (PVDF) membrane (#1620177, Millipore Sigma, Burlington, MA, USA), followed by incubation with antibodies listed in Supplemental Table 4. Immunoblot band intensities were measured by Image Lab Software 6.0.1 (Bio-Rad, Hercules, CA, USA).
Clonogenic assay
The Mock, si-Control, si-SIRPB1–1 and si-SIRPB1–2 treated PC3 cells were counted and reseeded at 1×104 cells per 100 mm dishes in complete RPMI 1640 media. The stable GFP C4–2 or GFP-SIRPB1 C4–2 cells were cultured with 1×103 cells in 100 mm dishes in complete RPMI 1640 media with 1,000 μg/ml G418 (#10131035,Thermo Fisher,Waltham,MA,USA) GFP C4–2 or GFP-SIRPB1 C4–2 cells were cultured by plating 370 cells per 60 mm dish and then treated with indicated doses of AKT inhibitor MK 2206 dihydrocholoride (#A3010, APExBIO, Houston, TX, USA). Media were replenished every 3–4 days in clonogenic assays. After 10 or 14 days, colonies were photographed and washed with phosphate buffered saline (PBS), fixed in methanol, stained with crystal violet, and the numbers or size of colonies were counted and measured using the Image-Pro Software (Media Cybernetics, Inc., Rockville, MD, USA).
Scratch motility (wound-healing) assay
PC3 cells were cultured at a concentration of 5×105 cells per well in six-well plates and transfected with Mock, si-Control, siSIRPB1–1 and siSIRPB1–2 when the cells reached 80% confluency. The PC3 cells formed a confluent monolayer at 72 hours post-transfection. The monolayer was scratched with a sterile pipette tip thenwashed with PBS to remove any floating cells. Cells were cultured with serum-free medium and imaged at 0, 15 and 24 hours after wounding. Images were acquired with a Nikon microscope and processed using Image-Pro. The distance traveled by the cells was determined by measuring the wound area at different time points and then subtracting it from the wound width at time 0. The values obtained were expressed as a migration percentage, setting the gap width at 0 hours as 0%.
Transwell migration assay
The Mock, si-Control, siSIRPB1–1 and siSIRPB1–2 PC3 cells were serum starved in serum free media for 15 hours before migration assay. Cell migration was assayed using chambers with noncoated 8 μM pore size transwell membranes (#3422, Corning) in 24-well plates as previously (27). Cells (5×104) were suspended in 400 μl of serum-free medium and loaded into the upper chamber. The lower chamber was filled with 750 μl of medium supplemented with 20% fetal bovine serum (FBS) that was used as a chemoattractant. GFP or GFP-SIRPB1 C4–2 cells were serum starved in 1% FBS media for 5 hours. Cells (1×105) were suspended in 400 μl of 1% FBS media and loaded into the upper chamber. The lower chamber was filled with 750 μl of medium supplemented with 20% FBS that was used as a chemoattractant. Following 22 hours of incubation, the transwell inserts were removed and the nonmigrating cells on the upper surface were removed with a cotton swab. The cells on the lower surface of the membrane were fixed in cold 100% methanol for 5 minutes, air-dried, and stained with DAPI and photographed. The cells were counted using Image-Pro Software (Media Cybernetics, Inc.)
Cell-cycle analysis by FACS
The Mock, si-Control, siSIRPB1–1 and siSIRPB1–2 PC3 cells were trypsinized, washed in PBS and processed using a BD Cycletest Plus DNA Kit (#340242, BD Biosciences, San Jose, CA, USA) at 72 hours after transfection, according to manufacturer’s instruction. C4–2 stable GFP or GFP-SIRPB1 cells were processed similarly. A total of 10,000 events were collected for each sample using a BD Accuri Flow Cytometer (BD Biosciences), and data were analyzed using the ModFit LT 5.0 Software (Verity Software House, Topsham, ME, USA).
Apoptosis analysis by FACS
The Mock, si-Control, siSIRPB1–1 and siSIRPB1–2 PC3 cells were processed using an Annexin V-PE apoptosis detection kit (#559763, BD Biosciences) 72-hour post-transfection, according to manufacturer’s instruction. GFP or GFP-SIRPB1 C4–2 cells were processed using an Annexin V-APC apoptosis detection kit (#550475, BD Biosciences). The above cells were analyzed total ten thousand events by BD Accuri flow cytometer (BD Biosciences), and data were analyzed using the FlowJo 7.6.1 program software (FlowJo, LLC., Ashland, OR, USA).
Tumor xenograft study
Immunodeficient nude mice were maintained in accordance with the Institutional Animal Care and Use Committee of Guangxi Medical University. A total of 2×106 GFP or GFP-SIRPB1 stably-transfected C4–2 cells resuspended in 100 μl 25% Matrigel were injected subcutaneously into the flanks of 6-week-old nude male mice (n=24 per group). Mice were followed for two weeks to determine whether or not palpable xenograft tumors were established.
Statistical analysis
Statistical analysis was performed using Prism 6.0 software (Graphpad Software, Inc., San Diego, CA, USA). Data were shown as mean ± SD and analyzed using student t-test or one-way ANOVA or Fisher and a statistically significant difference was considered at P<0.05.
Results
Status of SIRPB1 gene in prostate cancer specimens and cell lines
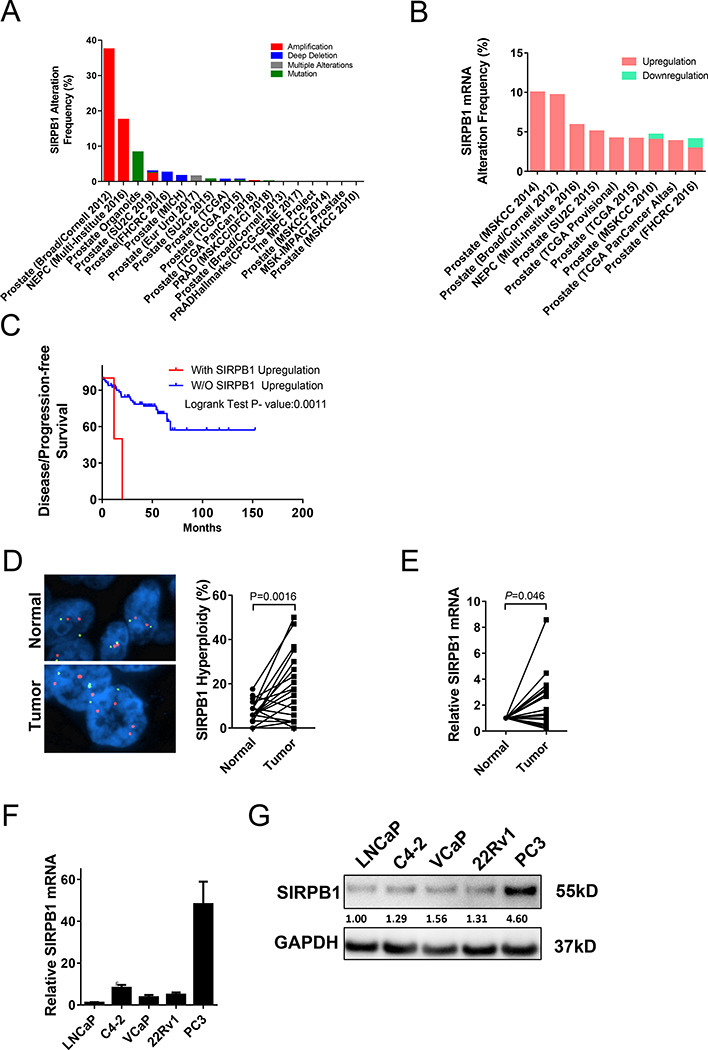
To evaluate the potential alterations of the SIRPB1 gene in prostate cancer, we performed a limited in silico analysis of several prostate cancer datasets available through the cBioportal database (28–34). At the time of analysis, there were 18 studies with a total of 5,170 samples available for query. SIRPB1 amplification, deep deletion, and mutation were detected in several prostate cancer datasets. SIRPB1 gene amplification was identified in four of the studies and ranged from 0.2 to 37.5% in prostate cancer specimens, with the highest incidence in the Broad/Cornell study from 2012 (Fig. 1A) (28). In other studies, alterations ranged from (0–17.54%) (Fig. 1A). mRNA expression data was available in 9 out of the 20 available datasets. SIRPB1 gene mRNA upregulation was increased up to 10% (1/10) (Fig. 1B). Down-regulation of SIRPB1 mRNA was identified in two of the studies (~1%). In the MSKCC 2010 dataset (34), SIRPB1 gene mRNA upregulation was significantly associated with poor disease or progression-free survival (P=0.0011) (Fig. 1C). Although this was not a comprehensive analysis of all publicly available datasets, alteration of SIRPB1 was identified in several of the cBioPortal for Cancer Genomics datasets in addition to the finding of an association between CNV at the SIRPB1 locus with prostate cancer aggressiveness in the JHH1 study (18).
Fig. 1.
SIRPB1 gene alteration (A) mRNA upregulation (B) and disease-free survival (C) in prostate cancer specimens from publicly available genomic datasets from the cBioportal for Prostate Cancer Genomics. Data expressed as a percentage of total specimens. D. Representative FISH images of prostate cancer tissue sections show more frequent amplification in tumor cells (lower panel) than adjacent normal cells (upper panel). SIRPB1 gene was amplified in tumor cells significantly more frequently compared with adjacent normal cells (P=0.0016). E. Comparative quantification analysis of SIRPB1 mRNA expression levels in tumor cells and paired adjacent normal cells. Values are shown as ddCt fold induction P=0.0305. The right panel shows incidence rate of SIRPB1 upregulation between tumor and normal cells. F. qPCR analysis of SIRPB1 expression in prostate cancer cell lines. Data are presented as mean ± SD. G. Western blot analysis of SIRPB1 expression in prostate cancer cell lines.
To further assess the frequency of genomic alterations of SIRPB1, fluorescence in situ hybridization (FISH) analysis was performed on radical prostatectomy tissue sections from 20 individual patients obtained through the University of Pittsburgh Pitt Biospecimen Core. All analyzed tissue sections had prostate cancer cells and adjacent normal glandular cells. FISH signals were imaged and quantified (Fig. 1D), showing that the SIRPB1 gene hyperploidy in prostate cancer cells was more frequent than in adjacent normal glandular cells (P = 0.0016). SIRPB1 gene amplification was detected in 70% (14/20) of the prostate cancer specimens compared to normal adjacent tissues. The average hyperploidy (%) in normal adjacent tissues was 7.317% compared to an average of 20.36% in tumor tissue. Of the 20 patients we examined, 4 had hyperploidy in normal adjacent areas > 10%. Two of these patients had Gleason scores of 4+3, and two had Gleason scores of 4+5. Previously reported FISH analyses identified a low incidence (0 to 20%) of chromosomal abnormalities in BPH tissues (35,36). We were unable to identify any previous studies showing whether chromosomal abnormalities were more or less frequent in normal tissues adjacent to prostate cancer compared to BPH tissues.
We next performed LCM coupled with qPCR to determine SIRPB1 mRNA expression in prostate tumor cells and adjacent normal epithelial cells in a separate cohort of 20 prostate cancer patients. SIRPB1 mRNA expression was 1.95-fold higher in prostate cancer cells compared to paired adjacent normal cells (P=0.046) and SIRPB1 mRNA upregulation was increased in 65%(13/20) of prostate cancer specimens (Fig. 1E). We also examined the expression of SIRPB1 in prostate cancer cell lines using both qPCR and western blot analyses. Both SIRPB1 mRNA and protein were expressed in all assayed prostate cancer cell lines, with the highest expression levels detected in PC3 cells (Fig. 1F, G). Stimulation of C4–2 cells with DHT increased endogenous SIRPB1 expression (Supplemental Fig. S1), suggesting that the elevation of SIRPB1 in AR-negative cells was unlikely due to a lack of androgen signaling.
Knockdown of SIRPB1 inhibited PC3 prostate cancer cell proliferation and migration in vitro
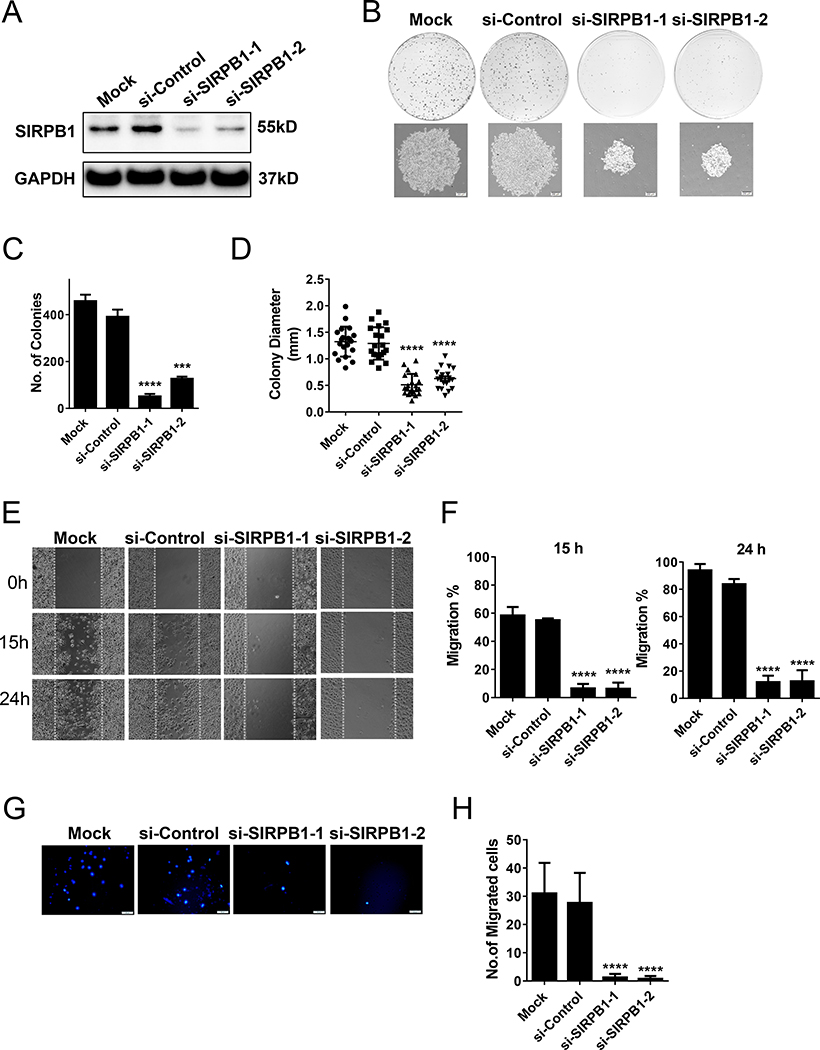
To characterize the function of SIRPB1 in prostate cancer cells, we performed knockdown experiments using two siRNAs targeting different sequences of SIRPB1 mRNA.PC3 cells were used as a model since SIRPB1 expression was the highest in PC3 as compared to LNCaP, C4–2, VCaP and 22Rv1 cell lines (see Fig. 1G). Western blot confirmed reduction of SIRPB1 protein levels by both siRNAs as compared to mock control or control siRNA samples (Fig. 2A). In the PC3 colony formation assay, both SIRPB1 RNAi caused significant reduction in the colony number and size (Fig. 2B,C,D). The wound-healing assay (Fig. 2E) showed that compared to si-Control or Mock, siSIRPB1–1 or siSIRPB1–2 treated cells exhibited considerably slower migration at 15 and 24 hours. Quantification of wound closure showed that siSIRPB1–1 and siSIRPB1–2 cells closed about 6.25% and 6.57% of the original wound after 15 hours, respectively. Conversely, si-Control or Mock cells closed about 55.04% and 58.38% of the wound after 15 and 24 hours, respectively (Fig. 2F). After 24 hours, compared with siSIRPB1–1 and siSIRPB1–2 cells, the gaps in si-Control and Mock cells were almost closed (83.76–93.94% vs. 11.93–12.51%). Furthermore, transwell migration assays revealed that SIRPB1 knockdown significantly blocked prostate cancer cells migration (Fig. 2G). The mean migrated number of siSIRPB1–1 or siSIRPB1–2 cells (1.25 and 0.75, respectively) was approximately one-thirtieth of si-Control (27.6) or Mock cells (31) (Fig. 2H).
Fig. 2.
Effect of SIRPB1 knockdown on prostate cancer cell proliferation and migration in vitro. A. Immunoblot of SIRPB1 protein expression in PC3 cells transfected with control (si-Control) and two different SIRPB1 siRNAs (siSIRPB1–1, siSIRPB1–2) and mock control. B. PC3 cells transfected with control (si-Control), SIRPB1 siRNA (siSIRPB1–1, siSIRPB1–2) or mock control and reseeded in 100 mm dishes in complete media. Media were replenished every 3 days and the cells were stained with crystal violet after 10 or 14 days. The size of cells (upper) and the total number of cells (lower) were quantitated. C. Quantification of colony number. D. Quantification of colony diameter. E. Scratch assay was performed at 72 h post-transfection and images were captured at the time points indicated. F. Quantification of gap closure at 15 h and 24 h. G. Boyden chamber transwell migration assay of PC3 cells following siRNA knockdown and after 22 h with 20% FBS as chemoattractant. H. Quantification of number of migrated cells. Images are representative three independent experiments. Data are presented as mean ± SD. ***P<0.001, ****P<0.0001 compared with si-Control groups.
Knockdown of SIRPB1 induced PC3 cell cycle arrest in G1 phase and enhanced apoptosis
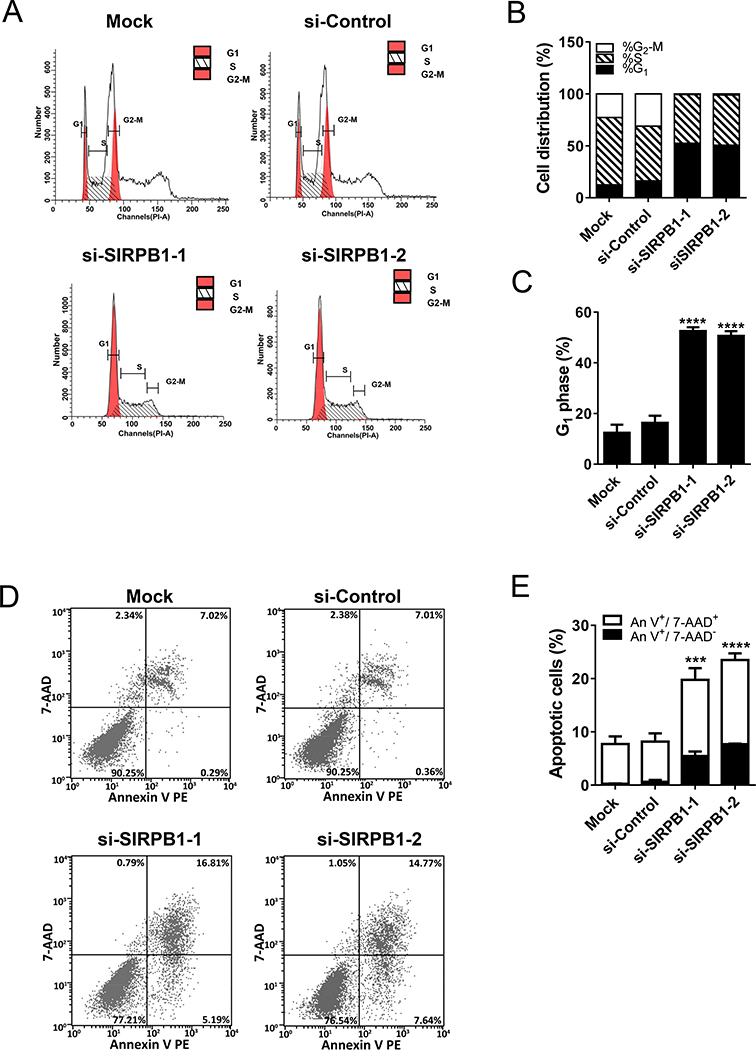
To investigate whether SIRPB1 inhibited prostate cancer cell growth through modulating cell cycle progression, we used siRNA knockdown coupled with flow cytometry in the PC3 model. In Fig. 3A,B, & C, knockdown of SIRPB1 increased the percentage of cells in G1-phase compared with si-Control (50.65–52.51% vs. 12.40–16.32%). To more accurately assess the consequences of SIRPB1 knockdown on cell apoptosis, the early (Annexin V+7AAD-) and late (Annexin V+7AAD+) apoptotic cells were measured. Compared with si-Control or Mock, SIRPB1 knockdown cells exhibited a dramatic increase in both early (5.19–7.64% vs. 0.29–0.36%) and late apopotic cells (14.77–16.81% vs. 7.01–7.02%) (Fig. 3D,E).
Fig. 3.
PC3 cells transfected with control or SIRPB1 siRNA or mock control as indicated were analyzed 72 h post-transfection by flow cytometry. A. Cell number versus propidium iodide fluorescence intensity. B. Percentage of cells in each cell-cycle phase estimated from flow cytometry analysis. C. Percentage of cells in G1 phase. D. Apoptotic cells of PC3 cells detected by flow cytometry. E. Percentage of early (Annexin V+/7AAD-) and late apoptotic PC3 cells (Annexin V+/7AAD-) based on flow cytometry analysis. Images are representative three independent studies. Data are presented as mean ± SD. ***P<0.001, and ****P<0.0001 when the data were compared to the control groups.
SIRPB1 overexpression in C4–2 prostate cancer cells promoted cell proliferation and migration in vitro and tumor formation in vivo
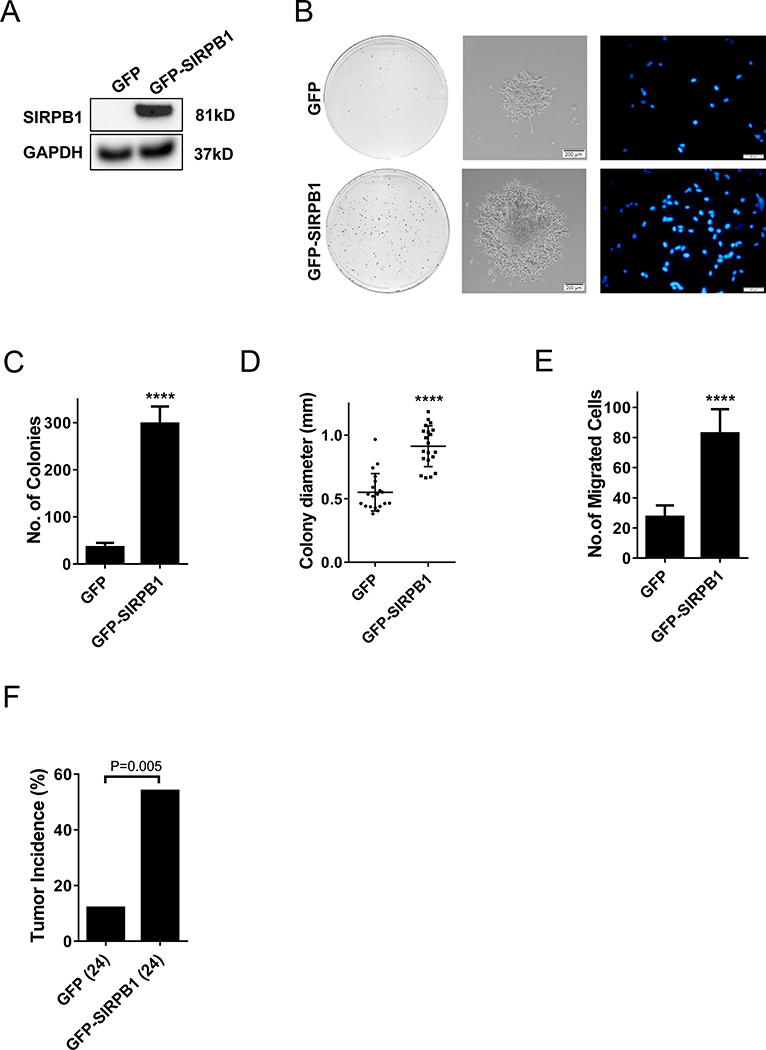
To further evaluate the role of SIRPB1 in prostate cancer, we investigated the effect of SIRPB1 overexpression in prostate cancer cells. The C4–2 cell line was used as a model to study the effect of SIRPB1 overexpression because C4–2 cells express SIRPB1 at a level similar to the other AR-positive prostate cancer cell lines and much lower than in PC3 cells. pCMV6-AC-GFP-SIRPB1 and the empty vector were transfected into C4–2 cells. Stably transfected GFP-SIRPB1 C4–2 cells were selected using FACS sorting. GFP C4–2 stably transfected cells were prepared in parallel as the control. The expression of GFP-SIRPB1 in the stably transfected cells was confirmed by immunoblotting (Fig. 4A). To observe the effect of SIRPB1 overexpression on C4–2 cells, GFP-SIRPB1 C4–2 cells was evaluated by colony formation assay, along with the GFP C4–2 cells as the control. As shown in Fig 4B (left panels) and 4C,GFP-SIRPB1 C4–2 cells formed much more and larger colonies as compared to the GFP C4–2 cells. To determine if SIRPB1 overexpression enhanced cell migration, we employed the transwell chamber-based assay to assess the migration of GFP-SIRPB1 C4–2 and GFP C4–2 cells. As shown in Fig. 4B (right panels), the number of the GFP-SIRPB1 C4–2 cells migrated through the diaphragm membrane was higher than that of the GFP C4–2 control cells. Quantitative analysis showed a 3-fold increase in the migrated GFP-SIRPB1 C4–2 cells as compared to the migrated GFP C4–2 control cells (82 vs. 27) (Fig. 4E). Together, these results indicated that SIRPB1 overexpression could stimulate prostate cancer cell proliferation and migration in vitro.
Fig. 4.
A. Immunoblot of SIRPB1 protein expression in C4–2 cells transfected with GFP-SIRPB1 (GFP-SIRPB1) or GFP vector (GFP). B. The above C4–2 cells were cultured in 100 mm dishes in complete media. Media were replenished every 3 days and the cells were stained with crystal violet after 10 or 14 days in colony formation assay (left panels, center panel) and Boyden chamber transwell migration assay (right panels). Migrated cells were photographed after 22 h culture with 20% FBS as chemoattractant. C. Quantification of colony number. D. Quantification of colony diameter. E. Quantification of migrated cells in transwell assay. F. Tumor incidence in mice 15 days after inoculation of C4–2 cells stably expressing GFP-SIRPB1 or GFP. Data presented are mean ± SD. ****P<0.0001.
We next tested if SIRPB1 overexpression could affect prostate cancer cells in vivo in nude mice. GFP-SIRPB1 C4–2 or GFP C4–2 cells were injected subcutaneously into the right flanks of nude mice (n=24 per group). Two weeks after the injection, tumor take rate of xenografts from GFP-SIRPB1 C4–2 cells (13/24) was significantly higher than the GFP C4–2 control group (3/24) (P = 0.005) (Fig. 4F). This result suggested that overexpression of SIRPB1 in human prostate cancer cells could promote tumor growth in vivo.
SIRPB1 overexpression induces cell proliferation via Akt activation
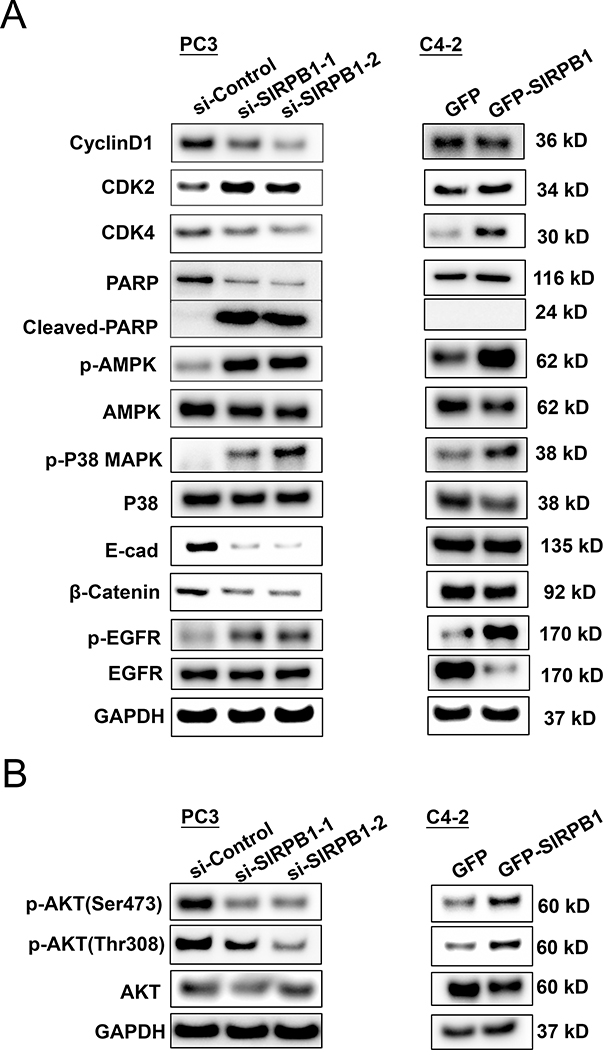
To explore the mechanism of SIRPB1 action in prostate cancer cells, we tested whether knockdown or overexpression of SIRPB1 could modulate proteins involved in cell growth and motility including Cyclin D1, CDK2, CDK4, PARP, AMPK, P38, E-cadherin, β-catenin, and EGFR (Fig. 5A). The differential expression of genes involved in cell cycle, cell division, cell development and cell junction has been reported in prostate cancer (37). Cyclin D1, E-cadherin and β-catenin were downregulated by SIRPB1 knockdown in PC3 cells and slightly downregulated by GFP-SIRPB1 overexpression in C4–2 cells. Thus, the expression levels of these molecules did not correlate with the expression levels of SIRPB1 in PC3 and C4–2 cell lines, suggesting that the effect of SIRBP1 on these molecules could be either inhibitory or stimulatory depending upon the cellular context. Since E-cadherin was down-regulated, we also checked whether N-cadherin expression was altered in response to SIRPB1 knockdown in PC3 cells. Interestingly, N-cadherin expression was slightly up-regulated in the PC3 cell line (Supplemental Fig. S2), further suggesting that SIRPB1 could play a role in several different pathways. The effect of SIRPB1 knockdown or overexpression on CDK2 expression was weak, suggesting CDK2 may not be a major target of SIRPB1. Knockdown of SIRPB1 inhibited CDK4 expression while overexpression of SIRPB1 induced CDK4 expression, suggesting that SIRPB1 may modulate key cell-cycle regulatory proteins such as CDK4 to induce cell cycle progression. Western blot analysis also showed that knockdown of SIRPB1 induced PARP cleavage, which is a marker for apoptosis. This result is consistent with apoptosis inducing potential of SIRPB1in prostate cancer cells (see Fig. 3). Interestingly, phosphorylation of AMPK, P38, and EGFR was significantly increased by both SIRPB1 knockdown in PC3 and GFP-SIRBP1 overexpression in C4–2 cells. Thus, the activation of these signaling molecules can happen when cell growth is either inhibited by SIRPB1 knockdown or stimulated by SIRPB1 overexpression, suggesting that activation of these signaling molecules may not be associated with SIRPB1 regulation of cell growth.
Fig. 5.
Effect of SIRPB1 knockdown or overexpression on expression and/or phosphorylation of some signaling molecules. A. Knockdown of SIRPB1 PC3 cells or overexpression SIRPB1 C42 cells were harvested and lysed for the detection of the indicated protein expression by Western blot analysis. GAPDH was run for each individual experiment, representative results from at least two experimental replicates are shown. B. Knockdown SIRPB1 downregulated Akt phosphorylation, while overexpression of SIRPB1 upregulated Akt phosphorylation. Experiments were performed at least twice and results shown are representative.
Akt is known to play an important role in regulating prostate cancer cell proliferation (38,39), and p-Akt (ser473) has been correlated with an increase in prostate cancer recurrence (40,41). Thus, we also tested whether knockdown or overexpression of SIRPB1 could modulate Akt and Akt phosphorylation. SIRPB1 knockdown in PC3 cells resulted in a reduction of both p-Akt (Ser473) and p-Akt (Thr308) (Fig. 5B), while SIRPB1 overexpression in C4–2 cells enhanced p-Akt levels (Fig. 5B). Thus, we hypothesized that SIRPB1 could regulate cell proliferation via Akt activation and could promote malignant transformation and prostate cancer progression.
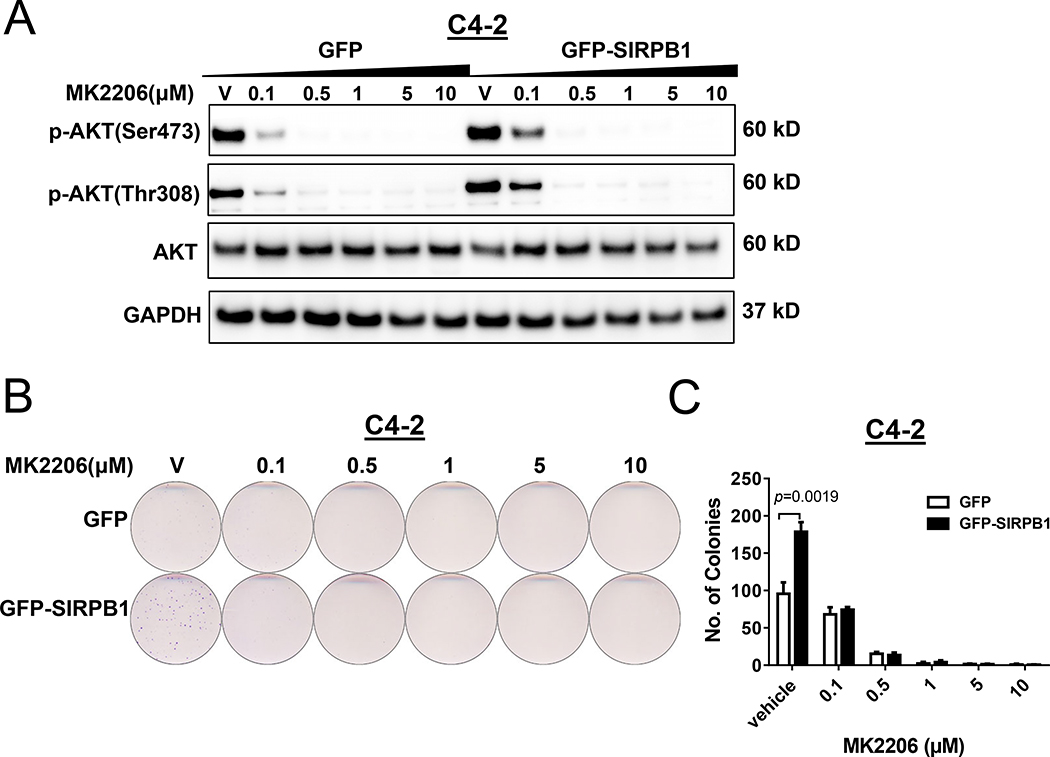
To test this hypothesis that SIRPB1 induced proliferation via activation of the Akt pathway, we first examined whether Akt inhibitor MK2206 could suppress cell proliferation in GFP-SIRPB1 C4–2 cells. As expected, treatment with MK2206 for 72 hours caused a dose-dependent suppression of Akt phosphorylated at Ser473 and/or Thr308 in both GFP- and GFP-SIRPB1 C4–2 cells (Fig. 6A). Importantly, GFP-SIRPB1 stimulated proliferation of C4–2 cells was nullified when the cells were treated with Akt inhibitor MK2206 (Fig. 6B & C). These results suggest that prostate cancers with SIRPB1 up-regulation may be susceptible to Akt inhibition.
Fig. 6.
SIRPB1 affected cell proliferation by Akt activation. A. Effects of the Akt inhibitor MK2206 on Akt phosphorylation (p-Akt) in GFP-SIRPB1 C4–2 cells. GFP or GFP-SIRPB1 C4–2 cells were treated with MK2206 at the indicated concentrations for 72 h, and cell lysates were subjected to Western blotting. B. Clonogenic assay evaluated the effect of long-term exposure to MK2206 in C4–2 cells. Stable GFP or GFP-SIRPB1 C4–2 cells were cultured in 60 mm dishes in complete media with indicated doses of Akt inhibitor MK 2206 dihydrocholoride. Media were replenished every 3–4 days and the cells were stained with crystal violet after 10 to 20 days. C. Quantification of colony number. Data presented are mean ± SD. Western blot experiments were performed at least twice and the clonogenic assay was performed a minimum of three times. Results shown are representative.
Discussion
SIRPB1 was identified in a genome-wide copy number variation analysis of prostate cancer (18), suggesting the potential involvement of SIRPB1 in prostate cancer development and progression. Here, we present evidence for SIRPB1 as a potential oncogene in prostate cancer. In silico analysis of several public databases available through the cBioPortal showed SIRPB1 gene amplification in a subset of prostate cancer specimens as well as mutation, deep deletion and dysregulation of mRNA expression in several specimens. FISH and LCM-coupled with qPCR provided further evidence for SIRPB1 gene amplification and overexpression in prostate cancer specimens compared to normal adjacent prostate. SIRPB1 knockdown in PC3 prostate cancer cells profoundly inhibited cell growth and motility. In contrast, SIRBP1 overexpression in C4–2 cells enhanced cell growth and motility. Cell cycle analysis showed SIRPB1 knockdown induced cell cycle arrest and SIRPB1 overexpression stimulated cell cycle progression. Furthermore, overexpression of SIRPB1 enhanced tumor take rate in the C4–2 xenograft tumor model. These findings suggest that SIRPB1 plays an important growth-stimulatory role in prostate cancer progression.
Based on the literature and this study, SIRPB1 and SIRPA1 function very differently, with SIRPA1 inhibiting growth and SIRPB1 promoting growth. SIRPA1 was reported to act as a tumor suppressor in rat embryonic fibroblast cells (12). Decreased expression of SIRPA1 has been reported in breast cancer (42), and transfection of SIRPA1 into the glioblastoma cell line U87MG negatively impacted several cancer-associated characteristics (8). In contrast, our studies suggest that SIRPB1 acts as an oncogene in prostate cancer cells. The major structural difference between SIRPB1 and SIRPA is that SIRPA has a cytoplasmic domain whereas SIRPB1 does not (Reviewed in (11)). Transfection of an SIRPA mutant with most of the cytoplasmic region deleted remarkably induced cell growth in NIH3T3 cells when compared to the control cells transfected with full-length SIRPA or the empty vector. This suggests that the cytoplasmic domain of SIRPA is essential for tumor suppression and the SIRPA mutant lacking the cytoplasmic domain is oncogenic. The cytoplasmic portion of SIRPB1 consists of only 6 amino acids, which is structurally similar to the SIRPA mutant lacking the cytoplasmic domain (Reviewed in (11)). Thus, the presence of the cytoplasmic domain in SIRPA, but not in SIRPB1, is likely responsible for the major functional difference between SIRPA and SIRPB1.
SIRPB1 knockdown down-regulated CDK4, while SIRPB1 overexpression increased CDK4. CDK4/6 is thought to play a significant role in prostate cancer cell growth and proliferation prompting the development of CDK inhibitors as potential therapies. The pan-CDK inhibitor flavopiridol was associated with significant toxicity and adverse effects in prostate cancer patients (43–45). Selective inhibition of CDK4/6 has been shown to decrease prostate cancer cell proliferation by promoting G1 cell cycle arrest and delayed growth in prostate cancer cell xenografts (46) suggesting that CDK4/6 activity is important in prostate cancer cell proliferation.
Our studies also showed that SIRPB1 could activate the Akt signaling pathway by enhancing Akt phosphorylation. Inhibition of SIRPB1-stimulated cell growth by Akt inhibitor MK2206 suggests that Akt signaling is a major pathway mediating SIRPB1 stimulation of prostate cancer cell growth. This finding has potential clinical relevance. Patients with SIRPB1 overexpression in prostate cancer may be more sensitive to therapeutic agents targeting Akt. We recognize that SIRPB1 could induce other signaling pathways such as CDK4 to stimulate cell proliferation. Therapeutic agents targeting CDK4 signaling pathways may be also suitable for patients with SIRPB1 overexpression.
Our studies suggested a potential oncogenic function for SIRPB1 in prostate cancer. FISH analysis of clinical prostate cancer specimens showed hyperploidy of SIRPB1 locus in prostate cancer cells as compared to the non-malignant prostatic cells. Also, LCM-coupled qPCR analysis showed up-regulation of SIRPB1 mRNA in prostate cancer specimens. However, CNV analysis identified CNP2454 as a 32.3 kb deletion polymorphism of SIRPB1 at 20p13 that significantly associated with aggressiveness of prostate cancer (18). This deletion polymorphism in the SIRPB1 gene may impact three different isoforms, SIRPB1, SIRPB2, and SIRPB3, differently. Our studies did not address functional roles of SIRPB2 and SIRPB3, because we were not able to detect SIRPB2 and SIRPB3 expression in prostate cancer cells using the existing antibodies (Data not shown). The impact of the deletion polymorphism may be different from siRNA knockdown of SIRPB1 in prostate cancer cells. Also, the deletion polymorphism of SIRPB1 gene may affect prostate cancer cells indirectly. For example, SIRPB1 appears to play an important role in immune cells (11) and its deletion in immune cells may lead to changes in microenvironment and subsequently impact on prostate carcinogenesis. How deletion polymorphism of SIRPB1 gene is associated with aggressive prostate cancer remains unclear.
In summary, our studies presented evidence for SIRPB1 as an important factor promoting prostate cancer cell growth and motility, functioning as a potential oncogene in the prostate. Future studies will be needed to determine whether SIRPB1 overexpression can serve as a biomarker to identify aggressive prostate cancers and to elucidate the mechanism by which SIRPB1 activates the Akt signaling pathway and other signaling pathways, such as CDK4. Defining SIRPB1 signaling pathways may lead to novel approaches to target SIRPB1 in prostate cancer.
Supplementary Material
Supplemental Fig. S1. Androgen regulation of endogenous SIRPB1 in GFP C4-2 cells. GFP C4-2 cells were stimulated with DHT (0.1 – 100 nM) for 48 h, and cell lysates were subjected to Western blotting.
Supplemental Fig. S2. Effect of SIRPB1 knockdown on N-cadherin. Knockdown of SIRPB1 PC3 cells were harvested and lysed for the detection of the N-cadherin protein expression by Western blot analysis. GAPDH served as a loading control. Experiment was performed at least twice and results shown are representative.
Acknowledgements
This work was supported in part by NIH P50 CA180995 (WJC), R01 CA186780 (ZW), R50 CA211242 (LEP), GXNSF 2018GXNSFBA050019 (QS), NSFC 81773146 (YL), NSFC 81130046 (JZ), and the Enhancement Foundation for Young Teachers in Guangxi Universities 2019KY0132 (QS). This project used the Hillman Cytometry Facility and the Tissue and Research Pathology Services Pitt Biospecimen Core that are supported in part by award P30CA047904.
Footnotes
Competing Interests
The authors indicate no potential conflicts of interest
Additional disclosure: The first author, Dr. Song, who performed the majority of this study in Pittsburgh, is a Visiting Scholar to our institution from Guangxi Medical University in China, where Prof. Yi Lu (lyu3@sustech.edu.cn) is her supervisor. Therefore, Dr. Lu was included as a co-author in this article; however, Dr. Lu has no conflict of interest for this study, and was involved only in the final approval process of the manuscript.
References
- 1.Fujioka Y, Matozaki T, Noguchi T, Iwamatsu A, Yamao T, Takahashi N, Tsuda M, Takada T, Kasuga M. A novel membrane glycoprotein, SHPS-1, that binds the SH2-domain-containing protein tyrosine phosphatase SHP-2 in response to mitogens and cell adhesion. Molecular and cellular biology 1996;16(12):6887–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohnishi H, Kubota M, Ohtake A, Sato K, Sano S. Activation of protein-tyrosine phosphatase SH-PTP2 by a tyrosine-based activation motif of a novel brain molecule. The Journal of biological chemistry 1996;271(41):25569–25574. [DOI] [PubMed] [Google Scholar]
- 3.Kharitonenkov A, Chen Z, Sures I, Wang H, Schilling J, Ullrich A. A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature 1997;386(6621):181–186. [DOI] [PubMed] [Google Scholar]
- 4.Veillette A, Thibaudeau E, Latour S. High expression of inhibitory receptor SHPS-1 and its association with protein-tyrosine phosphatase SHP-1 in macrophages. The Journal of biological chemistry 1998;273(35):22719–22728. [DOI] [PubMed] [Google Scholar]
- 5.Ichigotani Y, Matsuda S, Machida K, Oshima K, Iwamoto T, Yamaki K, Hayakawa T, Hamaguchi M. Molecular cloning of a novel human gene (SIRP-B2) which encodes a new member of the SIRP/SHPS-1 protein family. Journal of human genetics 2000;45(6):378–382. [DOI] [PubMed] [Google Scholar]
- 6.Seiffert ER, Simons EL. Astragalar morphology of late Eocene anthropoids from the Fayum Depression (Egypt) and the origin of catarrhine primates. Journal of human evolution 2001;41(6):577–606. [DOI] [PubMed] [Google Scholar]
- 7.Hara K, Senga T, Biswas MH, Hasegawa H, Ito S, Hyodo T, Hirooka Y, Niwa Y, Goto H, Hamaguchi M. Recovery of anoikis in Src-transformed cells and human breast carcinoma cells by restoration of the SIRP alpha1/SHP-2 signaling system. Cancer research 2011;71(4):1229–1234. [DOI] [PubMed] [Google Scholar]
- 8.Wu CJ, Chen Z, Ullrich A, Greene MI, O’Rourke DM. Inhibition of EGFR-mediated phosphoinositide-3-OH kinase (PI3-K) signaling and glioblastoma phenotype by signal-regulatory proteins (SIRPs). Oncogene 2000;19(35):3999–4010. [DOI] [PubMed] [Google Scholar]
- 9.Yan HX, Wang HY, Zhang R, Chen L, Li BA, Liu SQ, Cao HF, Qiu XH, Shan YF, Yan ZH, Wu HP, Tan YX, Wu MC. Negative regulation of hepatocellular carcinoma cell growth by signal regulatory protein alpha1. Hepatology 2004;40(3):618–628. [DOI] [PubMed] [Google Scholar]
- 10.Qin JM, Yan HX, Liu SQ, Wan XW, Zeng JZ, Cao HF, Qiu XH, Wu MC, Wang HY. Negatively regulating mechanism of Sirpalpha1 in hepatocellular carcinoma: an experimental study. Hepatobiliary & pancreatic diseases international : HBPD INT 2006;5(2):246–251. [PubMed] [Google Scholar]
- 11.Barclay AN, Brown MH. The SIRP family of receptors and immune regulation. Nature reviews Immunology 2006;6(6):457–464. [DOI] [PubMed] [Google Scholar]
- 12.Machida K, Matsuda S, Yamaki K, Senga T, Thant AA, Kurata H, Miyazaki K, Hayashi K, Okuda T, Kitamura T, Hayakawa T, Hamaguchi M. v-Src suppresses SHPS-1 expression via the Ras-MAP kinase pathway to promote the oncogenic growth of cells. Oncogene 2000;19(13):1710–1718. [DOI] [PubMed] [Google Scholar]
- 13.Lahoud MH, Proietto AI, Gartlan KH, Kitsoulis S, Curtis J, Wettenhall J, Sofi M, Daunt C, O’Keeffe M, Caminschi I, Satterley K, Rizzitelli A, Schnorrer P, Hinohara A, Yamaguchi Y, Wu L, Smyth G, Handman E, Shortman K, Wright MD. Signal regulatory protein molecules are differentially expressed by CD8- dendritic cells. Journal of immunology 2006;177(1):372–382. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Soto I, Tong Q, Chin A, Buhring HJ, Wu T, Zen K, Parkos CA. SIRPbeta1 is expressed as a disulfide-linked homodimer in leukocytes and positively regulates neutrophil transepithelial migration. J Biol Chem 2005;280(43):36132–36140. [DOI] [PubMed] [Google Scholar]
- 15.van Beek EM, Cochrane F, Barclay AN, van den Berg TK. Signal regulatory proteins in the immune system. Journal of immunology 2005;175(12):7781–7787. [DOI] [PubMed] [Google Scholar]
- 16.van den Berg TK, van Beek EM, Buhring HJ, Colonna M, Hamaguchi M, Howard CJ, Kasuga M, Liu Y, Matozaki T, Neel BG, Parkos CA, Sano S, Vignery A, Vivier E, Wright M, Zawatzky R, Barclay AN. A nomenclature for signal regulatory protein family members. Journal of immunology 2005;175(12):7788–7789. [DOI] [PubMed] [Google Scholar]
- 17.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 18.Jin G, Sun J, Liu W, Zhang Z, Chu LW, Kim ST, Sun J, Feng J, Duggan D, Carpten JD, Wiklund F, Gronberg H, Isaacs WB, Zheng SL, Xu J. Genome-wide copy-number variation analysis identifies common genetic variants at 20p13 associated with aggressiveness of prostate cancer. Carcinogenesis 2011;32(7):1057–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pradhan D, Roy S, Quiroga-Garza G, Cieply K, Mahaffey AL, Bastacky S, Dhir R, Parwani AV. Validation and utilization of a TFE3 break-apart FISH assay for Xp11.2 translocation renal cell carcinoma and alveolar soft part sarcoma. Diagn Pathol 2015;10:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Malley KJ, Dhir R, Nelson JB, Bost J, Lin Y, Wang Z. The expression of androgen-responsive genes is up-regulated in the epithelia of benign prostatic hyperplasia. Prostate 2009;69(16):1716–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Investigative urology 1979;17(1):16–23. [PubMed] [Google Scholar]
- 24.Horoszewicz JS, Leong SS, Chu TM, Wajsman ZL, Friedman M, Papsidero L, Kim U, Chai LS, Kakati S, Arya SK, Sandberg AA. The LNCaP cell line--a new model for studies on human prostatic carcinoma. Progress in clinical and biological research 1980;37:115–132. [PubMed] [Google Scholar]
- 25.Sramkoski RM, Pretlow TG 2nd, Giaconia JM, Pretlow TP, Schwartz S, Sy MS, Marengo SR, Rhim JS, Zhang D, Jacobberger JW. A new human prostate carcinoma cell line, 22Rv1. In vitro cellular & developmental biology Animal 1999;35(7):403–409. [DOI] [PubMed] [Google Scholar]
- 26.Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int J Cancer 1994;57(3):406–412. [DOI] [PubMed] [Google Scholar]
- 27.Qiu X, Pascal LE, Song Q, Zang Y, Ai J, O’Malley KJ, Nelson JB, Wang Z. Physical and Functional Interactions between ELL2 and RB in the Suppression of Prostate Cancer Cell Proliferation, Migration, and Invasion. Neoplasia 2017;19(3):207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, MacDonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA, Garraway LA. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012;44(6):685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, Tomlins SA, Nanus DM, Tagawa ST, Van Allen EM, Elemento O, Sboner A, Garraway LA, Rubin MA, Demichelis F. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nature medicine 2016;22(3):298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, Beltran H, Abida W, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel EA, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis F, Robinson BD, Schiffman M, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161(5):1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, Wongvipat J, Kossai M, Ramazanoglu S, Barboza LP, Di W, Cao Z, Zhang QF, Sirota I, Ran L, MacDonald TY, Beltran H, Mosquera JM, Touijer KA, Scardino PT, Laudone VP, Curtis KR, Rathkopf DE, Morris MJ, Danila DC, Slovin SF, Solomon SB, Eastham JA, Chi P, Carver B, Rubin MA, Scher HI, Clevers H, Sawyers CL, Chen Y. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014;159(1):176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, Etzioni R, Bolouri H, Montgomery B, White T, Lucas JM, Brown LG, Dumpit RF, DeSarkar N, Higano C, Yu EY, Coleman R, Schultz N, Fang M, Lange PH, Shendure J, Vessella RL, Nelson PS. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nature medicine 2016;22(4):369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ, Chinnaiyan AM, Tomlins SA. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487(7406):239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyauchi T, Nagayama T, Maruyama K. [Chromosomal abnormalities in carcinoma and hyperplasia of the prostate]. Nihon Hinyokika Gakkai Zasshi 1992;83(1):66–74. [DOI] [PubMed] [Google Scholar]
- 36.Lieber MM. DNA ploidy in prostate cancer: potential measurement as a surrogate endpoint biomarker. J Cell Biochem Suppl 1994;19:246–248. [PubMed] [Google Scholar]
- 37.Fan S, Liang Z, Gao Z, Pan Z, Han S, Liu X, Zhao C, Yang W, Feng W. Identification of the key genes and pathways in prostate cancer. Oncol Lett 2018;16(5):6663–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao H, Dupont J, Yakar S, Karas M, LeRoith D. PTEN inhibits cell proliferation and induces apoptosis by downregulating cell surface IGF-IR expression in prostate cancer cells. Oncogene 2004;23(3):786–794. [DOI] [PubMed] [Google Scholar]
- 39.Murillo H, Huang H, Schmidt LJ, Smith DI, Tindall DJ. Role of PI3K signaling in survival and progression of LNCaP prostate cancer cells to the androgen refractory state. Endocrinology 2001;142(11):4795–4805. [DOI] [PubMed] [Google Scholar]
- 40.Ayala G, Thompson T, Yang G, Frolov A, Li R, Scardino P, Ohori M, Wheeler T, Harper W. High levels of phosphorylated form of Akt-1 in prostate cancer and non-neoplastic prostate tissues are strong predictors of biochemical recurrence. Clin Cancer Res 2004;10(19):6572–6578. [DOI] [PubMed] [Google Scholar]
- 41.Kreisberg JI, Malik SN, Prihoda TJ, Bedolla RG, Troyer DA, Kreisberg S, Ghosh PM. Phosphorylation of Akt (Ser473) is an excellent predictor of poor clinical outcome in prostate cancer. Cancer Res 2004;64(15):5232–5236. [DOI] [PubMed] [Google Scholar]
- 42.Oshima K, Machida K, Ichigotani Y, Nimura Y, Shirafuji N, Hamaguchi M, Matsuda S. SHPS-1: a budding molecule against cancer dissemination. Cancer research 2002;62(14):3929–3933. [PubMed] [Google Scholar]
- 43.Senderowicz AM, Headlee D, Stinson SF, Lush RM, Kalil N, Villalba L, Hill K, Steinberg SM, Figg WD, Tompkins A, Arbuck SG, Sausville EA. Phase I trial of continuous infusion flavopiridol, a novel cyclin-dependent kinase inhibitor, in patients with refractory neoplasms. J Clin Oncol 1998;16(9):2986–2999. [DOI] [PubMed] [Google Scholar]
- 44.Liu G, Gandara DR, Lara PN Jr., Raghavan D, Doroshow JH, Twardowski P, Kantoff P, Oh W, Kim K, Wilding G. A Phase II trial of flavopiridol (NSC #649890) in patients with previously untreated metastatic androgen-independent prostate cancer. Clin Cancer Res 2004;10(3):924–928. [DOI] [PubMed] [Google Scholar]
- 45.Thomas JP, Tutsch KD, Cleary JF, Bailey HH, Arzoomanian R, Alberti D, Simon K, Feierabend C, Binger K, Marnocha R, Dresen A, Wilding G. Phase I clinical and pharmacokinetic trial of the cyclin-dependent kinase inhibitor flavopiridol. Cancer Chemother Pharmacol 2002;50(6):465–472. [DOI] [PubMed] [Google Scholar]
- 46.Comstock CE, Augello MA, Goodwin JF, de Leeuw R, Schiewer MJ, Ostrander WF Jr., Burkhart RA, McClendon AK, McCue PA, Trabulsi EJ, Lallas CD, Gomella LG, Centenera MM, Brody JR, Butler LM, Tilley WD, Knudsen KE. Targeting cell cycle and hormone receptor pathways in cancer. Oncogene 2013;32(48):5481–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. S1. Androgen regulation of endogenous SIRPB1 in GFP C4-2 cells. GFP C4-2 cells were stimulated with DHT (0.1 – 100 nM) for 48 h, and cell lysates were subjected to Western blotting.
Supplemental Fig. S2. Effect of SIRPB1 knockdown on N-cadherin. Knockdown of SIRPB1 PC3 cells were harvested and lysed for the detection of the N-cadherin protein expression by Western blot analysis. GAPDH served as a loading control. Experiment was performed at least twice and results shown are representative.