Abstract
Background
Castration-resistant prostate cancer can develop resistance to enzalutamide because of androgen receptor (AR) point mutations, AR overexpression, constitutively active AR splice variants, and/or elevated intratumoral androgen synthesis. The point mutation ARF876L was reported to be stimulated, instead of inhibited, by enzalutamide, thus contributing to enzalutamide resistance. We have recently developed JJ-450 as a novel AR antagonist with potential to treat enzalutamide-resistant CRPC.
Methods
We employed several assays to determine the impact of JJ-450 and enzalutamide on prostate cancer cell lines expressing GFP-ARF876L. These assays include a prostate-specific antigen (PSA) enhancer/promoter-based luciferase assay to determine AR transcriptional activity, a quantitative real-time polymerase chain reaction (qPCR) assay, and western blot to detect expression of AR target genes at the mRNA and protein level, fluorescence microscopy to show AR subcellular localization, and a 5-bromo-2'-deoxyuridine (BrdU) assay to measure prostate cancer cell proliferation.
Results
As expected, enzalutamide inhibited wild-type (wt) AR but not ARF876L transcriptional activity in the luciferase assay. In contrast, JJ-450 inhibited both wt-AR and ARF876L transcriptional activity to a similar extent. Also, enzalutamide retarded androgen-induced nuclear import of GFP-AR, but not GFP- ARF876L, whereas JJ-450 retarded nuclear import of both GFP-AR and GFP- ARF876L. To further evaluate JJ-450 inhibition of ARF876L, we stably transfected C4–2 cells separately with GFP-AR or GFP- ARF876L. Enzalutamide inhibited endogenous AR-target gene expression in C4–2-GFP-ARWT, but not in the C4–2-GFP- ARF876L subline, whereas JJ-450 inhibited AR-target gene expression in both C4–2 sublines. More importantly, enzalutamide inhibited proliferation of C4–2-GFP-ARWT, but not of the C4–2-GFP- ARF876L subline, whereas JJ-450 inhibited proliferation of both C4–2 sublines.
Conclusion
JJ-450 inhibits enzalutamide-resistant ARF876L mutant nuclear translocation and function. Our findings suggest that JJ-450 and its analogs should be further developed to provide a potential new approach for the treatment of enzalutamide-resistant CRPC.
Keywords: CRPC, JJ-450, prostate cancer, LNCaP, C4–2, PC3
Introduction
Prostate cancer is the most commonly diagnosed non-cutaneous malignancy and second leading cause of cancer-related death among men in the United States (1). Androgen deprivation therapy (ADT) remains the standard treatment for patients with metastatic prostate cancer. Despite the initial favorable response to ADT, almost all patients will inevitably progress to lethal castration-resistant prostate cancer (CRPC) within 1–2 years after the initiation of ADT (2,3). Many studies have contributed to the uncovering of potential mechanisms leading to CRPC, including androgen receptor (AR) overexpression, AR point mutations, constitutively active AR splice variants, and increased intratumoral androgen synthesis (4,5).
Enzalutamide (also called MDV3100), a second generation AR antagonist, has been shown to prolong the survival of CRPC patients for several months via multiple mechanisms, including blocking androgen binding to AR, attenuating activated AR nuclear translocation and reducing binding of AR to DNA (6–8). Unfortunately, the treatment is not curative because of acquired resistance after the administration of this FDA-approved drug. There is an urgent need to develop new AR antagonists for patients with enzalutamide resistance.
Point mutations in receptors represent a key mechanism leading to compromised receptor-targeted therapy (9). For instance, the non-sense mutation T790M in the epidermal growth factor receptor (EGFR) is considered to confer resistance to tyrosine kinase inhibitors like gefitinib in non-small-cell lung cancer (10), and D538G substitution in estrogen receptor can greatly decrease the sensitivity to tamoxifen in breast cancer (11). In prostate cancer, AR point mutations, especially in the ligand-binding domain (LBD), are a major cause for the resistance to first-generation AR antagonist such as hydroxyflutamide and bicalutamide. AR mutants such as T877A can utilize hydroxyflutamide as an agonist (12). Similarly, the AR mutant W741C was reported to be resistant to bicalutamide. These changes in the LBD result in a broadened ligand specificity for AR transcriptional activation such that antagonists could act as agonists. One of the benefits of enzalutamide over the first-generation AR antagonists is that it can exhibit full antagonist activity to these AR mutants.
To explore the potential mechanisms leading to the acquired resistance of prostate tumors to enzalutamide, Korpal and colleagues have identified a missense F876L mutation in the AR of the LNCaP cell line subjected to prolonged exposure to enzalutamide in vitro (13). Almost at the same time, by employing a similar method, Joseph et al. also found the same mutation capable of conferring resistance to both enzalutamide and ARN-509 (13,14). More importantly, the ARF876L mutation was detectable in circulating, cell-free DNA from CRPC patients resistant to enzalutamide (15). Further work demonstrated that enzalutamide was an agonist for the ARF876L mutant. The switch of enzalutamide from antagonist to agonist is likely due to a repositioning of helix H12 of the mutant AR which results in a more agonist-like conformation compatible with co-activator recruitment (16).
Recently, our lab developed a novel AR antagonist JJ-450 with the potential to inhibit AR variants which lack LBD (17), suggesting that it may have different target site(s) from the current AR antagonists, such as bicalutamide and enzalutamide which target LBD. Thus, we hypothesize that JJ-450 can overcome enzalutamide resistance caused by the ARF876L mutant. To test this hypothesis, we investigated the effect of JJ-450 on transiently and stably transfected GFP-ARF876L in prostate cancer cells, along with controls.
Materials and Methods
Cell culture
LNCaP, PC3, HEK293 prostate cancer cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). C4–2 was kindly provided by Dr. Leland WK Chung (Cedars-Sinai Medical Center, Los Angeles, CA). If not specifically stated, all cells were maintained at 37 °C in a 5% CO2 incubator, grown in medium supplemented with 10% FBS (Atlanta Biologicals, Flowery Branch, GA), L-glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 mg/ml). LNCaP, PC3 and C4–2 were grown in RPMI 1640 (Corning, Corning, NY); HEK293 were grown in Dulbecco’s modified Eagle’s medium (Lonza, Basel, Switzerland). Cells in experiments using the synthetic androgen methyltrienolone (R1881) were cultured in corresponding phenol-red free medium supplemented with 5% charcoal-stripped serum (CSS) 24 hours prior to transfection. The genetic identity of C4–2 cell lines was authenticated in 2016 using DNA fingerprinting by examining microsatellite loci in a multiplex PCR (AmpFlSTR Identifiler PCR Amplification Kit, Applied Biosystems, Foster City, CA) by the University of Pittsburgh Cell Culture and Cytogenetics Facility. Cell lines were confirmed as mycoplasma free by PCR testing.
Plasmids
The expression vector pEGFP-C1 (Clontech, Mountain View, CA) was used to generate GFP-ARWT as described previously (18). Renilla luciferase reporter (pRL-TK) was purchased from Promega (Madison, WI). PSA luciferase reporter vector (pPSA6.1-Luc) was a kind gift provided by Dr. Marianne Sadar (19). The plasmids were double CsCl gradient purified for transfection using PolyJet DNA In Vitro Transfection Reagent (SignaGen Laboratories, Rockville, MD) according to manufacturer’s protocols.
Site-directed mutagenesis and Sanger Sequencing
The single-amino acid substitution Phe 876 to Leu was introduced into the GFP-ARWT expression vector to generate a GFP-ARF876L vector using the Q5 site-directed mutagenesis kit (NEB, Ipswich, MA). Two overlapping primers for generating the point mutation in AR were designed using the NEB online design software, NEBaseChanger™, with the forward primer consisting of 5’-GCTGCATCAGCTCATTTGAC-3’ and the reverse primer consisting of 5’-TCTCTCGCAATAGGTGC-3’. We performed the mutagenesis PCR following the manufacturer’s instruction, with the following steps: 1) initial denaturation at 98 °C for 30 seconds, 2) 25 cycles of 98 °C for 10 seconds, 64 °C for 20 seconds and 72 °C for 3.5 minutes, and 3) final extension at 72 °C for 2 minutes. The GFP-ARF876L expression vector was sequence verified by Genewiz (South Plainfield, NJ).
Stably transfected LNCaP and C4–2 sublines
LNCaP and C4–2 cells were stably transfected as described previously (20). Briefly, C4–2 or LNCaP cells at 70–80% confluency were transfected with 1 μg GFP-ARWT or GFP-ARF876L in 6-well plates using PolyJet™ In Vitro DNA Transfection Reagent (SignaGen) according to the manufacturer’s protocol. Twenty-four hours after transfection, half of the cells from each well were reseeded in 10 cm dishes containing 10 ml regular RPMI 1640 medium. After another 24 hours, medium was replaced with RPMI1640 containing 1000 μg/ml G418 (for C4–2) or 500 μg/ml (for LNCaP) for selection of stably transfected cells. The selection medium was changed every week until the colonies formed were about 1 mm in diameter. Single colonies were transferred individually to separate wells in a 96-well plate. As the colonies were expanded, each was reseeded into a 48-well, followed by a 12-well, then a 10 cm dish. The C4–2 GFP-ARWT, C4–2 GFP-ARF876L and LNCaP GFP-ARF876L stable sublines were monitored using fluorescence microscopy (Nikon Eclipse TS100, Prior Scientific Lumen 200 Fluorescence Illumination System, Tokyo, Japan) and confirmed by western blotting. Unfortunately, we were not able to generate LNCaP-GFP-ARWT, because LNCaP-GFP-ARWT cells did not propagate.
Luciferase assay
PC3 or HEK293 cells were seeded into 24-well plates and transfected with GFP-ARWT or GFP-ARF876L together with pPSA6.1-Luc and pRL-TK with the ratio 10:10:1, in 5% charcoal-stripped FBS (cFBS), phenol-red free medium 16 hours before treatment with the indicated compounds. Twenty-four hours after the treatment, a luciferase assay was performed as described previously (21). Briefly, cells were lysed and luciferase activity was assayed using a Dual-Luciferase Reporter Assay kit (Promega) and measured using LMax II Microplate Reader (Molecular Devices, San Jose, CA) following the manufacturer’s recommendations. The firefly luciferase activity was normalized to Renilla luciferase activity.
Western blot analysis
Cell lysates were prepared in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, 1% NP-40, 0.1% SDS, 0.5% Sodium Deoxycholate, pH 8.0) containing protease inhibitor cocktails (P8340, Sigma-Aldrich, St. Louis, MO). Cell lysates were boiled in SDS loading buffer, size fractionated by SDS-PAGE, and transferred onto a PVDF membrane (BioRad). The membranes were blocked for 1 hour at room temperature in TBST buffer containing 5% non-fat milk, then incubated overnight with the following primary antibodies: anti-AR mouse monoclonal (1:1000, sc-7305, Santa Cruz Biotechnology, Dallas, TX ), anti-PSA goat polyclonal (1:500, sc-7638, Santa Cruz Biotechnology), anti-PARP rabbit monoclonal (1:1000, # 9532S, Cell Signaling Technology, Danvers, MA), anti-cleaved caspase 3 rabbit monoclonal (1:1000, #9664S, Cell Signaling Technology) and anti-GAPDH mouse monoclonal (1:10000, sc-47724 HRP, Santa Cruz Biotechnology) antibodies. After three washes in TBST, membranes were incubated with secondary antibody (sc-2004, sc-2005, or sc-2354) diluted in blocking buffer for 1 hour at room temperature. Following three washes in TBST, the secondary antibody was detected by Enhanced Chemiluminescent (ECL) using an ECL kit (BioRad Laboratories, Hercules, CA) and signals on the membranes were visualized using a ChemiDoc Imaging System (Bio-Rad).
Quantitative real-time PCR assays
Total RNA was isolated using the RNeasy Mini kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol, then 500 ng RNA was reverse transcribed using the PrimeScript RT Reagent Kit (TaKaRa, Mountain View, CA). All reactions were performed in triplicate on an ABI StepOnePlus™ system (Applied Biosystems, Foster City, CA) using SYBR Advantage qPCR Premix (TaKaRa). The mRNA levels of genes of interest were normalized to GAPDH level and then compared to the control group individually. The following primers were used for amplification: PSA‐F (forward): 5’-AGGCCTTCCCTGTACACCAA-3’, PSA-R (reverse): 5’-GTCTTGGCCTGGTCATTTCC-3’, TMPRSS2-F: 5’-CTGCCAAGGTGCTTCTCATT-3’, TMPRSS2-R: 5’-CTGTCACCCTGGCAAGAATC-3’, GAPDH-F: 5’-CTCCTCACAGTTGCCATGTA-3’, and GAPDH-R: 5’-GTTGAGCACAGGGTACTTTATTG-3’.
Analysis of GFP-ARWT and GFP-ARF876L subcellular localization
GFP-ARWT or GFP-ARF876L was transiently transfected into PC3 cells at >60% confluence in 5% cFBS, phenol-red free medium for 16 hours, followed by indicated treatment for 24 hours. The subcellular localization of GFP fusion proteins was imaged using fluorescence microscopy. Nuclei were stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (Sigma-Aldrich). Cytoplasmic localization of GFP-ARWT or GFP-ARF876L in transfected cells was defined as GFP fluorescence in the cytoplasm being more intense relative to the nuclei. Nuclear localization of GFP-ARWT or GFP-ARF876L was defined as GFP fluorescence in the nuclei being more intense than in the cytoplasm. Even distribution was defined as GFP fluorescence evenly distributed between the nucleus and cytoplasm in transfected cells. Quantification of subcellular localization of GFP-tagged fusion proteins was determined by counting at least 200 transfected cells per well in each experiment. All experiments were performed in triplicate and repeated at least twice.
BrdU Assay
Cells seeded in 24-well plates were treated as indicated for 48 hours. The cells were subsequently cultured in the presence of 10 μM 5-bromo-2′-deoxyuridine (BrdU) for 2 hours and then fixed by Carnoy’s fixative (3:1 methanol: glacial acetic acid) for 20 minutes at −20 °C. After treatment with 2 M HCl at 37 °C for 1 hour, cells were washed with 0.1 M boric acid, then incubated with 3% hydrogen peroxide for 20 minutes at 37 °C, followed by blocking with 3% BSA for 1 hour at 37 °C. Cells were then incubated with anti-BrdU antibody (sc-51514, Santa Cruz Biotechnology) overnight at 4 °C followed by CY3-labeled goat anti-mouse secondary antibody (A10521, Life Technologies, Carlsbad, CA) for 1 hour at 37 °C. Nuclei were stained with 1 μM SYTOX Green (S7020, Life technologies) for 15 minutes at room temperature.
Colony formation assay
Stably transfected cells were plated at a density of about 500 cells per 60 mm dish. Cells were treated with DMSO, 10 μM enzalutamide (Enz), or 10 μM JJ-450. The media containing indicated concentration of compounds was replenished every 5 days for 10 days. The colonies were then rinsed with PBS and fixed with 4% paraformaldehyde (PFA) for 20 minutes. After another two washes with PBS to remove remaining PFA, fixed cells were stained with 0.5% crystal violet solution for 20 minutes. ImageJ was used to count colony area following a standard protocol (22).
MTS assay
LNCaP GFP-ARF876L subline cells were plated into 96-well plate at a density of 3000 cells/well and treated with indicated compounds in complete medium for 72 hours prior to the MTS assay. The experiment was carried out using CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) (G3580, Promega) according to the manufacturer’s protocol.
Statistical analysis
All the data were analyzed statistically using Student’s t-test (two groups) or one-way ANOVA followed by the LSD post hoc test (more than two groups). *P<0.05, **P<0.01, ***P<0.001 are determined as significance. Value was presented as the mean ± standard deviation (SD) by GraphPad Prism software 7.0 (GraphPad Software, CA, USA).
Results
Compound JJ-450 inhibits ARF876L transcriptional activity
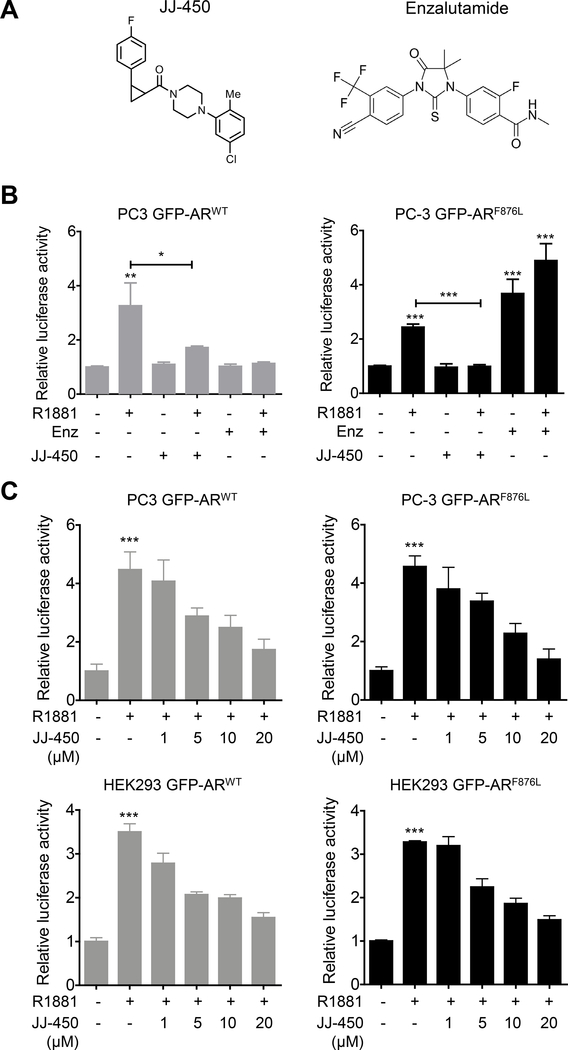
Compound JJ-450 was recently identified as a novel AR antagonist capable of inhibiting both full-length AR and AR variants function (17). This suggested the possibility that JJ-450 may target AR in domain(s) other than LBD, unlike the current clinical AR antagonists that target LBD. JJ-450 is a mixture of 2 stereoisomers, (–)-JJ-450 and (+)-JJ-450 (chemical structure is shown in Figure 1A). One mechanism leading to failure of enzalutamide administration is a single amino-acid substitution Phe 876 to Leu in LBD of AR (13,14). Enzalutamide acts as an agonist, instead of an antagonist, for mutant ARF876L (13,14). However, whether JJ-450 will experience the same switch for ARF876L is not clear. To address this question, we first examined whether JJ-450 could stimulate ARF876L transcriptional activity. As expected, 10 μM enzalutamide stimulated ARF876L transcriptional activity to a comparable level induced by 0.1nM R1881, while it effectively inhibited androgen-stimulated ARWT transcriptional activity (Figure 1B). However, we did not observe a similar antagonist to agonist switch in JJ-450 when AR was mutated to be ARF876L (Figure 1B). To further determine the efficacy of JJ-450 on ARF876L, we performed a luciferase assay using two AR-negative cell lines, PC3 and HEK293. The results showed that there was no obvious difference between ARWT and ARF876L in their response to androgens, since 1 nM R1881 could induce similar levels of activation, about 4.5-fold in PC3 cells and 3.5-fold in HEK293 cells. In the luciferase assays, JJ-450 suppressed both ARWT and ARF876L transcriptional activity effectively in a dose-dependent fashion (Figure 1C).
Figure 1.
Compound JJ-450 inhibits ARF876L mediated transcriptional activity. (A) Structures of JJ-450 and enzalutamide, respectively. (B) JJ-450 does not experience an antagonist to agonist switch in ARF876L. PC3 Cells were transiently transfected with GFP-ARWT or GFP-ARF876L, pPSA6.1-Luc, and pRL-TK as described in materials and methods. Transfected cells were subsequently treated with the following: 1) DMSO vehicle, 2) 0.1 nM R1881, 3) 10 μM Enz, 4) 0.1 nM R1881+10 μM Enz, 5) 10 μM JJ-450, and 6) 0.1 nM R1881+10 μM JJ-450 for an additional 24 h prior to luciferase assays. (C) Compound JJ-450 inhibits R1881-induced ARF876L mediated transcriptional activity in a dose dependent manner. PC3 Cells or HEK293 were transfected as described in materials and methods, then treated with 1 nM R1881 combined with the indicated dose of JJ-450 for 24 h before luciferase assay. All data were normalized to Renilla luciferase expression, then compared to the control group and expressed as mean ± SD. *P<0.05; **P<0.01; ***P < 0.001. All experiments were performed in triplicate and repeated. Enz, enzalutamide.
Compound JJ-450 partially retards GFP-ARF876L nuclear translocation upon androgen stimulation
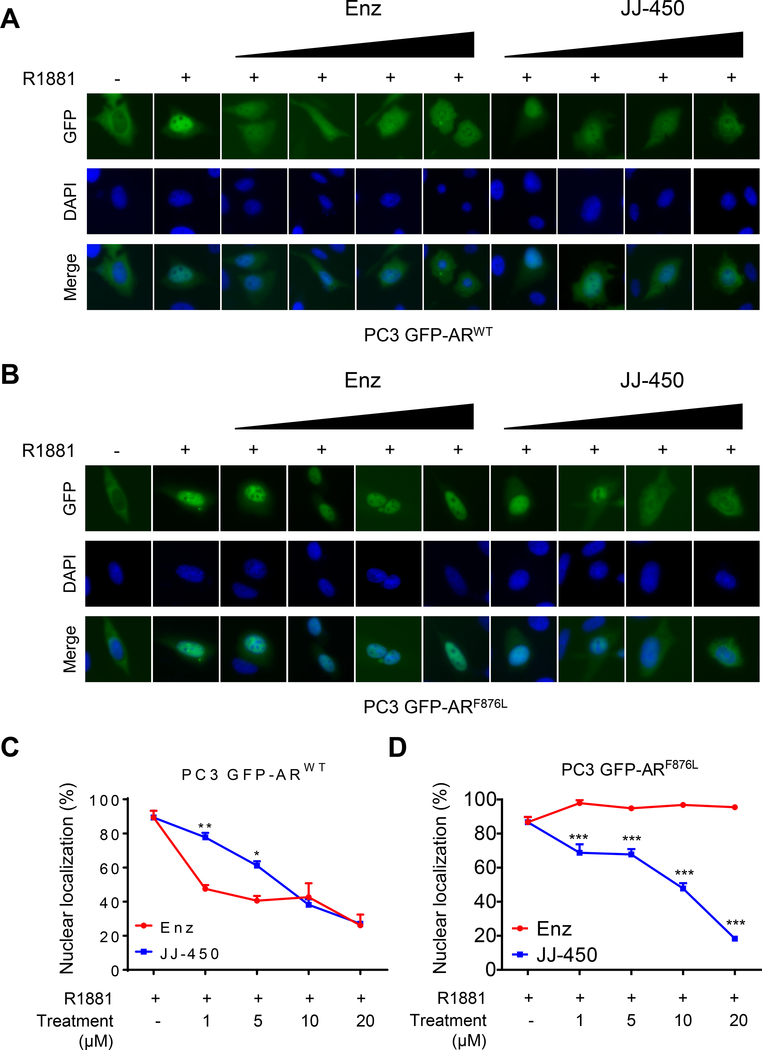
Nuclear localization is a prerequisite for AR transcriptional activity. We thus tested whether JJ-450 can affect GFP-ARF876L nuclear translocation. Consistent with a previous finding (23), GFP-ARWT in PC3 cells showed predominant cytoplasmic localization under androgen-free condition (Figure 2A). GFP-ARF876L in transfected PC3 cells also displayed similar cytoplasmic localization in the absence of androgen (Figure 2B). GFP-ARWT and GFP-ARF876L were imported into nuclei in the presence of 0.1 nM R1881, with about 90% transfected cells exhibiting predominant nuclear localization of GFP-ARWT or GFP-ARF876L 24 hours after R1881 treatment (Figure 2). When transfected cells were treated with 0.1 nM R1881 combined with different concentrations of enzalutamide (1–20 μM) or JJ-450 (1–20 μM), GFP-ARWT translocation into nuclei was inhibited by enzalutamide or JJ-450. At low dosages (1 μM, 5 μM), enzalutamide appeared to be more potent than JJ-450 in the inhibition of R1881-induced GFP-ARWT nuclear import. Consistent with a previous report (13), enzalutamide did not inhibit R1881-induced GFP-ARF876L nuclear import. Instead, enzalutamide slightly enhanced R1881-induced GFP-ARF876L nuclear import in transfected cells. In contrast, JJ-450 inhibited R1881-induced GFP-ARF876L nuclear import in a dose-dependent fashion, which was similar to its inhibition of GFP-ARWT nuclear import.
Figure 2.
Compound JJ-450 partially retarded ARF876L nuclear translocation upon R1881 stimulation. PC3 cells were transiently transfected with GFP-ARWT (A) or GFP-ARF876L (B) in androgen-free conditions for 16 h, then treated with 0.1 nM R1881 or 0.1 nM R1881 combined with an increasing dose (1–20 μM) of Enz or JJ-450 for an additional 24 h. The subcellular localization of GFP-ARWT was visualized using fluorescent microscopy. The nuclei were stained with DAPI (blue). (C) Quantification of cells exhibiting nuclear localization in PC3 transfected with GFP-ARWT(A) or GFP-ARF876L (B) was carried out as describe in material and methods. Data are expressed as mean ± SD. *P<0.05; **P<0.01; ***P < 0.001. Enz, enzalutamide.
Compound JJ-450 downregulates endogenous AR target gene expression
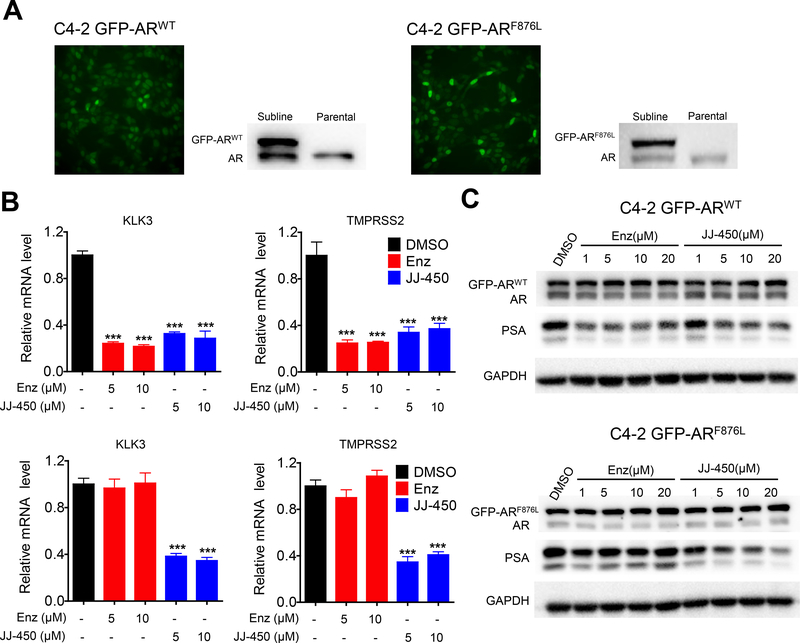
To further characterize the effect of JJ-450 of inhibiting prostate cancer cells bearing mutant ARF876L, we established three sublines stably expressing GFP-ARF876L or GFP-ARWT, namely, C4–2 GFP-ARF876L, C4–2 GFP-ARWT, and LNCaP GFP-ARF876L. We attempted, but were unable, to establish another stable cell line LNCaP GFP-ARWT as a control cell line. For unknown reasons, the GFP-ARWT-transfected LNCaP cells did not grow when we tried to establish sublines. The stable sublines were verified by fluorescence microscopy and western blot analysis (Figure 3A). Considering that enzalutamide was reported to overcome castration resistance caused by AR overexpression (24), exogenous AR overexpression in these sublines should not lead to resistance to enzalutamide. qPCR analysis showed the expression of two canonical AR target genes, KLK3 (25) and TMPRSS2 (26), was inhibited by enzalutamide or JJ-450 in the C4–2 GFP-ARWT subline (Figure 3B). However, the expression of these two target genes was inhibited by JJ-450, but not enzalutamide, in C4–2 GFP-ARF876L subline (Figure 3B). Western blot analysis of PSA protein showed similar results that JJ-450 inhibited PSA expression in both C4–2 GFP-ARWT and C4–2 GFP-ARF876L sublines whereas enzalutamide inhibited PSA expression in the C4–2 GFP-ARWT, but not the C4–2 GFP-ARF876L subline (Figure 3C). In the LNCaP GFP-ARF876L subline, JJ-450, but not enzalutamide, inhibited the expression of PSA (Figure S1A).
Figure 3.
Compound JJ-450 downregulated endogenous AR target gene expression. (A) Generation of stable GFP-ARWT and GFP-ARF876L sublines. Fluorescence microscopy and western blot analysis confirmed GFP-ARWT and GFP-ARF876L expression in the sublines. AR antibody detected both endogenous AR (lower band) and transfected GFP-tagged AR (upper band) in western blotting analyses. (B) qPCR analysis of KLK3 and TMPRSS2 mRNA expression in C4–2 GFP-ARWT subline (upper panels) and C4–2 GFP-ARF876L subline (lower panels) following treatment with DMSO, 5 μM Enz, 10 μM Enz, 5 μM JJ-450 or 10 μM JJ-450 in complete medium for 24 h. GAPDH was used to normalize expression. Data represent mean ± SD; ***P < 0.001. (C) Western blot analysis of PSA protein level in two sublines treated with an increasing dose of Enz or JJ-450 for 24 h in complete medium. Enz, enzalutamide.
Compound JJ-450 inhibits ARF876L-mediated proliferation but does not induce apoptosis
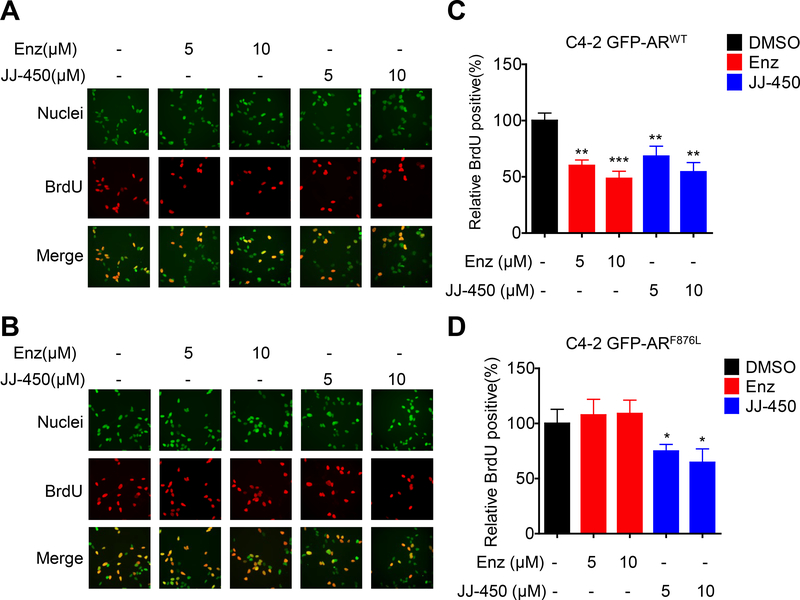
Since JJ-450 inhibited GFP-ARF876L activity, we next tested its influence on proliferation of cells bearing GFP-ARF876L. In BrdU incorporation assays, JJ-450 inhibited proliferation of both C4–2 GFP-ARWT and C4–2 GFP-ARF876L sublines (Figure 4). In contrast, enzalutamide suppressed proliferation of the C4–2 GFP-ARWT, but not the C4–2 GFP-ARF876L subline (Figure 4A&B). In the LNCaP GFP-ARF876L subline, JJ-450, but not enzalutamide, inhibited cellular growth in the MTS assay (Figure S1B).
Figure 4.
Compound JJ-450 inhibits ARF876L-mediated cell proliferation. Representative images show BrdU staining in C4–2 GFP-ARWT (A) or C4–2 GFP-ARF876L (B) sublines. C4–2 GFP-ARWT or C4–2 GFP-ARF876L stable sublines were treated with DMSO, 5 μM Enz, 10 μM Enz, 5 μM JJ-450 or 10 μM JJ-450 in complete medium for 48 h prior to BrdU assay as described in materials and methods. Upper panel shows nuclear staining with SYTOX Green (green), middle panel shows BrdU-positive nuclei (red), lower panel shows merged images of SYTOX and BrdU staining. (C) and (D) show quantification of BrdU-positive cells percentages of C4–2 GFP-ARWT and C4–2 GFP-ARF876L subline, respectively. Data represent mean ± SD, relative to the control. *P<0.05; **P<0.01; ***P < 0.001. Enz, enzalutamide.
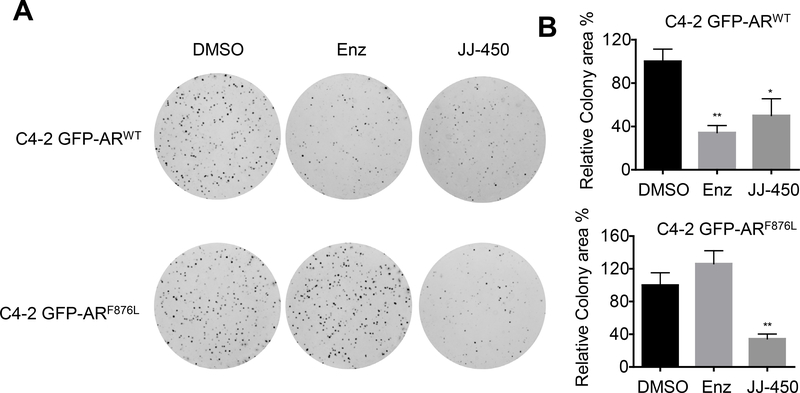
To further test the effects of JJ-450 on cells bearing GFP-ARF876L, we performed colony formation assays. Consistent with the BrdU results, JJ-450 inhibited colony formation of both C4–2 GFP-ARWT and C4–2 GFP-ARF876L sublines. In contrast, enzalutamide inhibited colony formation of the C4–2 GFP-ARWT, but not the C4–2 GFP-ARF876L subline (Figure 5A&B).
Figure 5.
Compound JJ-450 suppresses ARF876L-mediated clonogenic ability. (A) Representative images of C4–2 GFP-ARWT (upper panel) and C4–2 GFP-ARF876L (lower panel) subline colonies. Cells were treated with DMSO, 10 μM Enz, 10 μM JJ-450 in complete RPMI 1640 and plated for colony formation assays. The media were changed every 5 d for 10 d. (B) Quantitative analysis of colony formation in (A). Colony formation assay was carried out in triplicate and repeated. Data represent mean ± SD, relative to the control. *P<0.05; **P<0.01. Enz, enzalutamide.
We also evaluated if JJ-450 and/or enzalutamide could affect cell death in C4–2 GFP-ARWT and C4–2 GFP-ARF876L sublines. The level of PARP and caspase-3 cleavage are known markers of apoptosis (27,28). The C4–2 GFP-ARWT and C4–2 GFP-ARF876L sublines treated with 5 nM docetaxel, which induces apoptosis in prostate cancer cells (29), were included as positive controls. Figure 6 showed that neither JJ-450 nor enzalutamide induced apoptosis in both C4–2 GFP-ARWT and C4–2 GFP-ARF876L sublines.
Figure 6.
Compound JJ-450 does not induce apoptosis in C4–2 GFP-ARWT and C4–2 GFP-ARF876L sublines. (A) Western blot analysis of cleaved-PARP and cleaved-caspase3 in C4–2 GFP-ARWT. Cells were treated with DMSO, 10 μM Enz, 10 μM JJ-450 or 5 nM docetaxel for 48 h prior to western blot analysis. (B) Western blot analysis of cleaved-PARP and cleaved-caspase 3 in C4–2 GFP-ARF876L. Cells were treated the same as in (A). Results are representative of three separate experiments. Enz, enzalutamide.
Discussion
AR mutation is an important mechanism leading to enzalutamide resistance in CRPC. ARF876L is a well-characterized enzalutamide-resistant mutant AR (13,14). Small molecules capable of inhibiting enzalutamide-resistant AR mutants would have potential as new therapeutics for the treatment of enzalutamide-resistant CRPC. The results in the present paper provide evidence that JJ-450, a novel AR inhibitor, can effectively inhibit androgen-induced nuclear import and activation of ARF876L.
JJ-450 inhibition of enzalutamide-resistant mutant ARF876L suggests that the mechanism of JJ-450 action is different from enzalutamide. Enzalutamide competitively inhibits AR via blocking androgen binding to the LBD. Enzalutamide binding to AR inhibits androgen-induced AR nuclear localization and AR recruitment to DNA. Our previous studies showed that the parental compound of JJ-450, IMTPPE, could inhibit NAR, an AR deletion construct lacking LBD, suggesting that JJ-450 and related compounds can inhibit AR independent of LBD (21). Thus, it is likely that mutations in the LBD, such as F876L, should not affect the JJ-450 inhibition of AR. This expectation is supported by inhibition of ARF876L by JJ-450 in the current study. However, the mechanism for inhibition of AR by JJ-450 is not yet clear and the binding site(s) of JJ-450 on AR remains to be determined. Defining the mechanism of JJ-450 inhibition of AR will facilitate our understanding of how JJ-450 can effectively inhibit ARF876L.
JJ-450 is a small molecule capable of inhibiting enzalutamide-resistant AR mutant ARF876L. ASC-J9, an AR degradation enhancer, was reported to inhibit ARF876L via induction of the mutant AR degradation (30). Compound 30, which was developed by optimizing AR ligand-binding efficiency of aryloxy tetramethylcyclobutyl lead compounds, inhibited enzalutamide-resistant resistant MR49F prostate cancer cells (31,32). Darolutamide (ODM-201), a potent AR antagonist targeting LBD, can also inhibit ARF876L (33). The mechanisms of AR inhibition by the above small molecules appear to be very different: with JJ-450 being capable of inhibiting AR independent of LBD, ASC-J9 being an effective AR degrader, and compound 30 and darolutamide targeting the LBD. Additionally, small molecules such as EPI-001 should inhibit ARF876L by targeting the N-terminal domain (NTD) of AR (34). Enzalutamide resistance could be overcome by alternate small molecules which function by different mechanisms of action. The availability of these small molecules offers the potential for new therapeutic approaches for CRPC expressing enzalutamide-resistant AR.
We have tried to grow LNCaP GFP-ARF876L xenograft tumors in immunodeficient mice. Although LNCaP GFP-ARF876L tumors were established in castrated mice, most of them stopped growing after reaching ~0.1 ml in size. This prevented us from testing the efficacy of JJ-450 in these tumors. The inability of establishing LNCaP GFP-ARF876L tumors was also observed by Korpal and colleagues (13). One possible reason for the inability of LNCaP GFP-ARF876L tumors to grow may be due to the excessive expression of GFP-ARF876L. Another possibility is that LNCaP GFP-ARF876L cells may undergo phenotype drifting during the stable selection of the subline.
In summary, our studies demonstrate that JJ-450 can inhibit enzalutamide-resistant ARF876L mutant nuclear translocation and function. These findings suggest that JJ-450 and its analogs should be further developed to provide a potential new approach for the treatment of enzalutamide-resistant CRPC.
Supplementary Material
Supplemental Figure S1. The effect of compound JJ-450 on LNCaP GFP-ARF876L sublines. (A) Western blot analysis of PSA level. LNCaP GFP-ARF876L subline cells were treated at indicated concentrations of enzalutamide (Enz) or JJ-450 in complete medium for 24 h before harvesting for western analysis. (B) MTS assay of LNCaP GFP-ARF876L subline cells. The cells were treated with indicated doses of Enz or JJ-450 for 72 h prior to the MTS assay. Data represent mean ± SD relative to the control group. The experiment was repeated twice. ***P < 0.001.
Acknowledgements
We are grateful to Wei Chen, Taber Maskrey, and Mingming Zhong for technical support. This work was funded in part by DOD Award W81XWH-16–1-0659 and R50 CA211242 (L.E.P.) as well as by UPMC Enterprises. This project used the UPMC Hillman Cancer Center Animal Facility which was supported in part by NCI award P30 CA047904. Special thanks to the China Scholarship Council and the Third Xiangya Hospital of Central South University for supporting Zeyu Wu’s living expense and training fees in University of Pittsburgh.
Footnotes
Disclosure: Z.W., J.B.N., and P.W. are co-inventors named in US Patent #9,708,276 on JJ-450, held by the University of Pittsburgh. The remaining authors have no competing interests. The first author, Zeyu Wu, performed the experiments in Pittsburgh and is a Visiting Scholars to University of Pittsburgh from Central South University Xiangya School of Medicine, China.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Zegarra-Moro OL, Schmidt LJ, Huang H, Tindall DJ. Disruption of androgen receptor function inhibits proliferation of androgen-refractory prostate cancer cells. Cancer Res 2002;62(4):1008–1013. [PubMed] [Google Scholar]
- 3.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat Med 2004;10(1):33–39. [DOI] [PubMed] [Google Scholar]
- 4.Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi OP. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet 1995;9(4):401–406. [DOI] [PubMed] [Google Scholar]
- 5.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 2015;15(12):701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, Wasielewska T, Welsbie D, Chen CD, Higano CS, Beer TM, Hung DT, Scher HI, Jung ME, Sawyers CL. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009;324(5928):787–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, Armstrong AJ, Flaig TW, Flechon A, Mainwaring P, Fleming M, Hainsworth JD, Hirmand M, Selby B, Seely L, de Bono JS, Investigators A. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367(13):1187–1197. [DOI] [PubMed] [Google Scholar]
- 8.Beer TM, Tombal B. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 2014;371(18):1755–1756. [DOI] [PubMed] [Google Scholar]
- 9.Jin J, Wu X, Yin J, Li M, Shen J, Li J, Zhao Y, Zhao Q, Wu J, Wen Q, Cho CH, Yi T, Xiao Z, Qu L. Identification of Genetic Mutations in Cancer: Challenge and Opportunity in the New Era of Targeted Therapy . Front Oncol 2019;9:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352(8):786–792. [DOI] [PubMed] [Google Scholar]
- 11.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, Fizazi K, Mainwaring P, Piulats JM, Ng S, Carles J, Mulders PF, Basch E, Small EJ, Saad F, Schrijvers D, Van Poppel H, Mukherjee SD, Suttmann H, Gerritsen WR, Flaig TW, George DJ, Yu EY, Efstathiou E, Pantuck A, Winquist E, Higano CS, Taplin ME, Park Y, Kheoh T, Griffin T, Scher HI, Rathkopf DE, Investigators C-A-. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 2013;368(2):138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bohl CE, Miller DD, Chen J, Bell CE, Dalton JT. Structural basis for accommodation of nonsteroidal ligands in the androgen receptor. J Biol Chem 2005;280(45):37747–37754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korpal M, Korn JM, Gao X, Rakiec DP, Ruddy DA, Doshi S, Yuan J, Kovats SG, Kim S, Cooke VG, Monahan JE, Stegmeier F, Roberts TM, Sellers WR, Zhou W, Zhu P. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov 2013;3(9):1030–1043. [DOI] [PubMed] [Google Scholar]
- 14.Joseph JD, Lu N, Qian J, Sensintaffar J, Shao G, Brigham D, Moon M, Maneval EC, Chen I, Darimont B, Hager JH. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov 2013;3(9):1020–1029. [DOI] [PubMed] [Google Scholar]
- 15.Azad AA, Volik SV, Wyatt AW, Haegert A, Le Bihan S, Bell RH, Anderson SA, McConeghy B, Shukin R, Bazov J, Youngren J, Paris P, Thomas G, Small EJ, Wang Y, Gleave ME, Collins CC, Chi KN. Androgen Receptor Gene Aberrations in Circulating Cell-Free DNA: Biomarkers of Therapeutic Resistance in Castration-Resistant Prostate Cancer. Clin Cancer Res 2015;21(10):2315–2324. [DOI] [PubMed] [Google Scholar]
- 16.Balbas MD, Evans MJ, Hosfield DJ, Wongvipat J, Arora VK, Watson PA, Chen Y, Greene GL, Shen Y, Sawyers CL. Overcoming mutation-based resistance to antiandrogens with rational drug design. Elife 2013;2:e00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, Wang D, Johnson JK, Pascal LE, Takubo K, Avula R, Chakka AB, Zhou J, Chen W, Zhong M, Song Q, Ding H, Wu Z, Chandran UR, Maskrey TS, Nelson JB, Wipf P, Wang Z. A novel small molecule targets androgen receptor and its splice variants in castration-resistant prostate cancer. Mol Cancer Ther 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saporita AJ, Zhang Q, Navai N, Dincer Z, Hahn J, Cai X, Wang Z. Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem 2003;278(43):41998–42005. [DOI] [PubMed] [Google Scholar]
- 19.Sadar MD. Androgen-independent induction of prostate-specific antigen gene expression via cross-talk between the androgen receptor and protein kinase A signal transduction pathways. J Biol Chem 1999;274(12):7777–7783. [DOI] [PubMed] [Google Scholar]
- 20.Dong J, Wu Z, Wang D, Pascal LE, Nelson JB, Wipf P, Wang Z. Hsp70 Binds to the Androgen Receptor N-terminal Domain and Modulates the Receptor Function in Prostate Cancer Cells. Mol Cancer Ther 2019;18(1):39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masoodi KZ, Eisermann K, Yang Z, Dar JA, Pascal LE, Nguyen M, O’Malley K, Parrinello E, Feturi FG, Kenefake AN, Nelson JB, Johnston PA, Wipf P, Wang Z. Inhibition of Androgen Receptor Function and Level in Castration-Resistant Prostate Cancer Cells by 2-[(isoxazol-4-ylmethyl)thio]-1-(4-phenylpiperazin-1-yl)ethanone. Endocrinology 2017;158(10):3152–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzman C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One 2014;9(3):e92444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saporita AJ, Ai J, Wang Z. The Hsp90 inhibitor, 17-AAG, prevents the ligand-independent nuclear localization of androgen receptor in refractory prostate cancer cells. Prostate 2007;67(5):509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung ME, Ouk S, Yoo D, Sawyers CL, Chen C, Tran C, Wongvipat J. Structure-activity relationship for thiohydantoin androgen receptor antagonists for castration-resistant prostate cancer (CRPC). J Med Chem 2010;53(7):2779–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, Hood L, Lin B. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A 2002;99(18):11890–11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, Hood L, Nelson PS. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res 1999;59(17):4180–4184. [PubMed] [Google Scholar]
- 27.Boulares AH, Yakovlev AG, Ivanova V, Stoica BA, Wang G, Iyer S, Smulson M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem 1999;274(33):22932–22940. [DOI] [PubMed] [Google Scholar]
- 28.Porter AG, Janicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ 1999;6(2):99–104. [DOI] [PubMed] [Google Scholar]
- 29.Mediavilla-Varela M, Pacheco FJ, Almaguel F, Perez J, Sahakian E, Daniels TR, Leoh LS, Padilla A, Wall NR, Lilly MB, De Leon M, Casiano CA. Docetaxel-induced prostate cancer cell death involves concomitant activation of caspase and lysosomal pathways and is attenuated by LEDGF/p75. Mol Cancer 2009;8:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R, Lin W, Lin C, Li L, Sun Y, Chang C. ASC-J9((R)) suppresses castration resistant prostate cancer progression via degrading the enzalutamide-induced androgen receptor mutant AR-F876L. Cancer Lett 2016;379(1):154–160. [DOI] [PubMed] [Google Scholar]
- 31.Toren P, Kim S, Cordonnier T, Crafter C, Davies BR, Fazli L, Gleave ME, Zoubeidi A. Combination AZD5363 with Enzalutamide Significantly Delays Enzalutamide-resistant Prostate Cancer in Preclinical Models. Eur Urol 2015;67(6):986–990. [DOI] [PubMed] [Google Scholar]
- 32.Kuruma H, Matsumoto H, Shiota M, Bishop J, Lamoureux F, Thomas C, Briere D, Los G, Gleave M, Fanjul A, Zoubeidi A. A novel antiandrogen, Compound 30, suppresses castration-resistant and MDV3100-resistant prostate cancer growth in vitro and in vivo. Mol Cancer Ther 2013;12(5):567–576. [DOI] [PubMed] [Google Scholar]
- 33.Fizazi K, Albiges L, Loriot Y, Massard C. ODM-201: a new-generation androgen receptor inhibitor in castration-resistant prostate cancer. Expert Rev Anticancer Ther 2015;15(9):1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Mol E, Fenwick RB, Phang CT, Buzon V, Szulc E, de la Fuente A, Escobedo A, Garcia J, Bertoncini CW, Estebanez-Perpina E, McEwan IJ, Riera A, Salvatella X. EPI-001, A Compound Active against Castration-Resistant Prostate Cancer, Targets Transactivation Unit 5 of the Androgen Receptor. ACS Chem Biol 2016;11(9):2499–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. The effect of compound JJ-450 on LNCaP GFP-ARF876L sublines. (A) Western blot analysis of PSA level. LNCaP GFP-ARF876L subline cells were treated at indicated concentrations of enzalutamide (Enz) or JJ-450 in complete medium for 24 h before harvesting for western analysis. (B) MTS assay of LNCaP GFP-ARF876L subline cells. The cells were treated with indicated doses of Enz or JJ-450 for 72 h prior to the MTS assay. Data represent mean ± SD relative to the control group. The experiment was repeated twice. ***P < 0.001.