No two individuals are alike as we contain different genetic codes that vary from person-to-person and make us unique. However, increasing evidence shows that within the same individual the genome also differs from cell-to-cell. This phenomenon is known as somatic mosaicism, and it results from the accumulation of somatic mutations that inevitably occur during embryonic development and over the course of an individual’s lifetime. Indeed, recent advances in DNA sequencing methodology reveal that somatic mutations are much more prevalent in normal cells than we initially thought. In most cases, these mutations are phenotypically neutral and there are no consequences on the cell or organism. If the mutation confers a deleterious phenotype, that cell may be lost from the organism over time as it competes with normal cells. In contrast, a ‘driver’ gene mutation can confer a fitness advantage that leads to the clonal expansion of the cell that harbours the mutation. While it is well-established that the accumulation of fitness-conferring mutations can contribute to the development of cancer, recent evidence suggests that they are remarkably prevalent in the normal tissues of non-symptomatic individuals. Furthermore, it is becoming increasingly apparent that these aberrant clonal expansions can be harbingers of non-cancerous disease development in elderly individuals. Here, we summarize recent findings that document the extents of somatic mosaicism in different tissues and discuss the provocative idea that these somatic mutations can impact various age-associated diseases including cardiovascular disease.
Mosaicism throughout the body
Advances in technology associated with DNA sequencing and the preparation of single-cell libraries have allowed Lodato et al.1,2 to assess the degree of genomic heterogeneity among post-mitotic neurons of the brain. Using computational algorithms to minimize false-positive results, it was found that the number of somatic single nucleotide variants (sSNV) in neurons increases according to the subject’s age with an estimated mutation accumulation rate of 20 sSNV per year per single neuron. The analysis of DNA base-pair substitutions revealed enrichment in the C > T ‘clock-like’ signature that is associated with the aging process, supporting the concept that neurons accumulate somatic mutations in an age-dependent manner. Neurons from patients with the progeroid neurodegenerative disorders xeroderma pigmentosum and Cockayne syndrome, that are caused by defects in DNA damage repair, showed greater accumulation of sSNV. Interestingly, somatic mutations were enriched in the coding regions of genes, indicating that they arise from errors during the transcriptional processes. In particular, mutations in aged neurons were found to be enriched in genes involved in neurological diseases, such as SCN1A (seizure disorder) and SLC12A2 (schizophrenia), suggesting that the accumulation of somatic mutations could affect neuron function and contribute to age-related cognitive decline.
In tissues with proliferative cells, such as skin and oesophagus, genomic mosaicism can give rise to multiple competing clonal events that have been described as a ‘patchwork of thousands of evolving clones’.3 Martincorena et al.3 performed targeted-exome sequencing of 74 cancer genes in multiple biopsies from sun-exposed eyelid epidermises that were removed during plastic surgery. While the tissues were physiologically and histologically normal, it was found that somatic mutations in cancer driver genes were frequent, averaging 140 presumptive driver gene mutations per square centimetre of skin. These mutations occurred in NOTCH1, NOTCH2, NOTCH3, TP53, FAT1, and RBM10 genes that are associated with squamous cell carcinomas. Sunlight-induced damage is probably a large contributor to this high burden of somatic mutations in aged skin, and this was reflected by the signature of nucleotide substitutions in the DNA. These clonal mutations can propagate as cells divide, generating numerous unique patches of epidermal cells that harbour these mutations. This age-dependent increase in the pool of mutant cells can increase the probability of additional mutations and lead to an increased chance of uncontrolled cell proliferation (i.e. cancer development). However, it was found that the largest epidermal patches had mutations in FGFR3, a gene that is associated with benign seborrhoeic keratosis rather than malignancy. In a subsequent study of normal oesophagus, Martincorena et al.4 showed that there was a similar age-dependent accumulation of somatic mutations to what was found in sun-exposed skin. Unexpectedly, the analysis of mutant clones revealed a higher density of cancer driver mutations in the oesophagus than in the skin, although the number of mutations per cell in normal oesophagus is <10% of sun-exposed skin. In a related study, Yokoyama et al.5 showed that lifestyle risk factors for oesophageal carcinoma (excessive alcohol consumption and smoking) were associated with increased clonal events within the normal epithelium, suggesting the existence of tissue-specific selective pressure on the development of these clones. Overall, it is paradoxical that cells of the skin and oesophagus that harbour these oncogenic mutations at high levels maintain their normal physiological function and do not necessarily give rise to cancer.
The clonal expansion of mutant cells has also been documented in non-cancerous liver. Zhu et al.6 performed whole-exome analyses of cancer genes in the cirrhotic liver from 82 patients and found that the number and volume of clones carrying mutations in PKD1, PPARGC1B, KMT2D, and ARID1A genes increased with liver damage. Subsequent functional analyses in mice indicated that these mutations facilitated liver regeneration in models of injury, suggesting that these clonal mutations can potentially be adaptive rather than maladaptive.
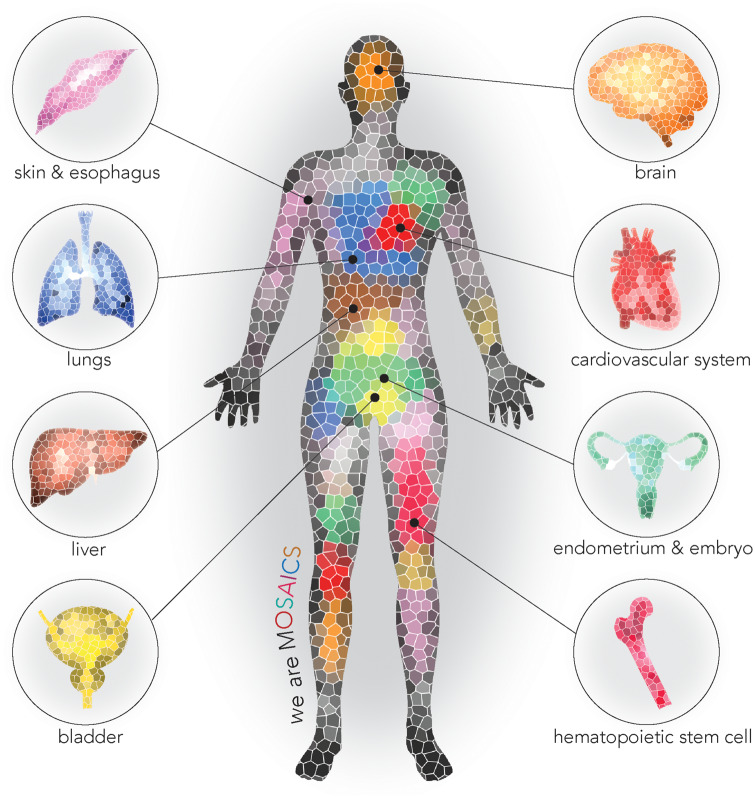
More recently, a study by Yizhak et al.7 utilized RNA sequence data from the Genotype-Tissue Expression Project to assess the occurrence of mutational clones in 29 healthy tissues, including heart and blood vessels, from approximately 500 individuals. Mutational clones were found in nearly every individual. The mutational burden increased with age and the proliferative capacity of the tissue, suggesting that mutations accumulate with time and the number of cell divisions. This study further illustrates the widespread nature of somatic mosaicism and its age-dependence (Figure 1).
Figure 1.
We are mosaics. Recent studies have documented somatic mosaicism and aberrant clonal expansions in multiple cell types and tissues.
Mosaicism in blood: Clonal haematopoiesis and cardiovascular disease
Mosaicism in leucocytes has been well studied due to the accessibility of this material. Clonal haematopoiesis describes the expansion of blood cells derived from dominant haematopoietic stem cell clones. This condition is prevalent in elderly individuals with a normal haematologic phenotype. While the mutational landscape of clonal haematopoiesis has only partially been deciphered, some of these clonal expansions can be attributed to somatic mutations in driver genes that are recurrently mutated in blood malignancies. These driver genes include epigenetic regulators (TET2, DNMT3A, and ASXL1), spliceosome components (SF3B1 and SRSF2), signalling proteins (JAK2), and DNA damage response molecules (TP53 and PPM1D).8–10 The reported prevalence of clonal haematopoiesis in the population is variable due to differences in detection methods and the lack of a consensus definition for this condition. For example, the designation of clonal haematopoiesis with indeterminate potential, or CHIP, refers to individuals who harbour a haematologic malignancy-associated somatic mutation, present at an allele frequency ≥2% in white blood cells, yet they do not meet the criteria for a haematologic malignancy. Focusing exclusively on CHIP genes, it was estimated that 10% of persons over the age of 70 years exhibited these clonal expansions.8 However, non-biased analyses of clonal events deduced from deep whole exome or whole genome sequencing indicate that individuals classified by the confines of the CHIP designation represent a minority of those who exhibit clonal events in their haematopoietic system. For example, a whole genome sequence analysis of >11 000 Icelanders revealed that >50% of individuals over the age of 85 years showed evidence of clonal haematopoiesis, and only a small fraction of these clonal events could be attributed to mutations in known driver genes.10
Studies show that clonal haematopoiesis is associated with an increased risk of all-cause mortality regardless of whether it is associated with a presumptive driver gene or the mechanism is unknown.8–10 While there is a marked increase in the frequency of haematological cancer in individuals with clonal haematopoiesis, which is to be expected, the major cause of the increased mortality in these populations appears to be the increase in the risk of cardiovascular diseases including coronary heart disease, stroke, and early-onset myocardial infarction.8,11 More recent studies have associated clonal haematopoiesis with poor prognosis in patients diagnosed with chronic heart failure and in patients who are treated for aortic stenosis.12,13 Experimental studies with Tet2 and Dnmt3a candidate driver genes in mice support the concept that clonal haematopoiesis can causally contribute to atherogenesis and heart failure progression, illustrating a new mechanism of cardiovascular disease that involves the dysregulation of cytokine expression by the mutant clones.14–16
The case for mosaicism in cardiovascular tissues
The concept that somatic mutations in the cardiovascular system are associated with its pathology is not new. In 1973, Benditt and Benditt17 examined human atherosclerotic plaques for X-chromosome inactivation patterns, the same type of analysis that led to the discovery of clonal haematopoiesis. The Benditt and Benditt study provided evidence that plaques were monoclonal in origin, and they hypothesize that the mechanism was mutational events in smooth muscle cells (SMC). These findings may have a bearing on the interpretation of recent studies that have used lineage-restricted confetti mice to demonstrate that specific cell clones expand in vascular lesions.18–20 It is possible that atherosclerosis and other vascular lesions share features with cancer; i.e. that the accumulation of somatic mutations over time contributes to disease progression. Similar to what has been described for skin, mutant SMC with different phenotypic properties may expand forming a patchwork of clones within the blood vessel wall. Consistent with this notion, the work of Yizhak et al.7 has documented the expansion of mutant clones in cardiovascular tissues.7 Specifically, they detected 518 mutations in blood vessels from 287 individuals (and 256 mutations in the hearts of 138 individuals) with allele frequencies ≥2%. Thus, following exposure to injurious stimuli, these mutant cells may preferentially expand within the lesion, and atherosclerotic plaque progression and stability could be influenced by the nature of the clonal mutations that they harbour.
Concluding remarks
No cell can avoid the accumulation of somatic mutations over time. Advances in DNA sequencing methodology have shown that somatic mutations are remarkably prevalent and ubiquitous. In proliferative tissues, it is common to find mutations in particular driver genes that give rise to clonal events that accumulate with age, and this process is under the control of distinct mutational landscapes in different cell types. In these situations, the clonal expansion amplifies the genomes that harbour these mutations, yet this condition has no impact on overall cell number within the tissue and there is no evidence of malignant transformation. Accumulating evidence suggests that clonal haematopoiesis is a newly recognized cardiovascular disease risk factor that is prevalent in the elderly. Since aging is the single greatest risk factor for cardiovascular disease, greater efforts should be made to understand the epidemiological and mechanistic connections among aging, somatic mutations, and cardiovascular disease development. In this regard, it would be of interest to delineate the somatic mutation landscape of other cell types in the human cardiovascular system, such as cardiac fibroblasts, tissue-resident tissue immune cells, and vascular endothelial cells. The expansion and functional consequences of these mutant clones can be addressed by the development of murine genetic systems where the gene of interest is conditionally manipulated in a portion of cells.
Funding
This work was funded by National Institutes of Health grants HL138014, HL139819, HL141256, and HL142650 to K. Walsh and by American Heart Association Postdoctoral Fellowship 17POST33670076 and the Kanae Foundation for the Promotion of Medical Science to S. Sano.
Conflict of interest: none declared.
References
- 1. Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, Lee S, Chittenden TW, D’Gama AM, Cai X, Luquette LJ, Lee E, Park PJ, Walsh CA.. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 2015;350:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lodato MA, Rodin RE, Bohrson CL, Coulter ME, Barton AR, Kwon M, Sherman MA, Vitzthum CM, Luquette LJ, Yandava CN, Yang P, Chittenden TW, Hatem NE, Ryu SC, Woodworth MB, Park PJ, Walsh CA.. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 2018;359:555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martincorena I, Roshan A, Gerstung M, Ellis P, Loo PV, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, Stebbings L, Menzies A, Widaa S, Stratton MR, Jones PH, Campbell PJ.. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348:880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall M, Cagan A, Murai K, Mahbubani K, Stratton MR, Fitzgerald RC, Handford PA, Campbell PJ, Saeb-Parsy K, Jones PH.. Somatic mutant clones colonize the human esophagus with age. Science 2018;362:911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yokoyama A, Kakiuchi N, Yoshizato T, Nannya Y, Suzuki H, Takeuchi Y, Shiozawa Y, Sato Y, Aoki K, Kim SK, Fujii Y, Yoshida K, Kataoka K, Nakagawa MM, Inoue Y, Hirano T, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Nishikawa Y, Amanuma Y, Ohashi S, Aoyama I, Horimatsu T, Miyamoto S, Tsunoda S, Sakai Y, Narahara M, Brown JB, Sato Y, Sawada G, Mimori K, Minamiguchi S, Haga H, Seno H, Miyano S, Makishima H, Muto M, Ogawa S.. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 2019;565:312–317. [DOI] [PubMed] [Google Scholar]
- 6. Zhu M, Lu T, Jia Y, Luo X, Gopal P, Li L, Odewole M, Renteria V, Singal AG, Jang Y, Ge K, Wang SC, Sorouri M, Parekh JR, MacConmara MP, Yopp AC, Wang T, Zhu H.. Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell 2019;177:608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yizhak K, Aguet F, Kim J, Hess JM, Kubler K, Grimsby J, Frazer R, Zhang H, Haradhvala NJ, Rosebrock D, Livitz D, Li X, Arich-Landkof E, Shoresh N, Stewart C, Segre AV, Branton PA, Polak P, Ardlie KG, Getz G.. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 2019;364:eaaw0726.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, Higgins JM, Moltchanov V, Kuo FC, Kluk MJ, Henderson B, Kinnunen L, Koistinen HA, Ladenvall C, Getz G, Correa A, Banahan BF, Gabriel S, Kathiresan S, Stringham HM, McCarthy MI, Boehnke M, Tuomilehto J, Haiman C, Groop L, Atzmon G, Wilson JG, Neuberg D, Altshuler D, Ebert BL.. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, Purcell SM, Svantesson O, Landén M, Höglund M, Lehmann S, Gabriel SB, Moran JL, Lander ES, Sullivan PF, Sklar P, Grönberg H, Hultman CM, McCarroll SA.. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, Thorgeirsson TE, Sigurdsson A, Gudjonsson SA, Gudmundsson J, Jonasson JG, Tryggvadottir L, Jonsson T, Helgason A, Gylfason A, Sulem P, Rafnar T, Thorsteinsdottir U, Gudbjartsson DF, Masson G, Kong A, Stefansson K.. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 2017;130:742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, Baber U, Mehran R, Fuster V, Danesh J, Frossard P, Saleheen D, Melander O, Sukhova GK, Neuberg D, Libby P, Kathiresan S, Ebert BL.. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017;377:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dorsheimer L, Assmus B, Rasper T, Ortmann CA, Ecke A, Abou-El-Ardat K, Schmid T, Brune B, Wagner S, Serve H, Hoffmann J, Seeger F, Dimmeler S, Zeiher AM, Rieger MA.. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol 2019;4:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mas-Peiro S, Hoffmann J, Fichtlscherer S, Dorsheimer L, Rieger MA, Dimmeler S, Vasa-Nicotera M, Zeiher AM.. Clonal haematopoiesis in patients with degenerative aortic valve stenosis undergoing transcatheter aortic valve implantation. Eur Heart J 2020;41:933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, Wu C-L, Sano S, Muralidharan S, Rius C, Vuong J, Jacob S, Muralidhar V, Robertson AAB, Cooper MA, Andres V, Hirschi KK, Martin KA, Walsh K.. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 2017;355:842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sano S, Oshima K, Wang Y, MacLauchlan S, Katanasaka Y, Sano M, Zuriaga MA, Yoshiyama M, Goukassian D, Cooper MA, Fuster JJ, Walsh K.. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1β/NLRP3 inflammasome. J Am Coll Cardiol 2018;71:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sano S, Oshima K, Wang Y, Katanasaka Y, Sano M, Walsh K.. CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. Circ Res 2018;123:335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benditt EP, Benditt JM.. Evidence for a monoclonal origin of human atherosclerotic plaques. Proc Natl Acad Sci U S A 1973;70:1753–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chappell J, Harman JL, Narasimhan VM, Yu H, Foote K, Simons BD, Bennett MR, Jorgensen HF.. Extensive proliferation of a subset of differentiated, yet plastic, medial vascular smooth muscle cells contributes to neointimal formation in mouse injury and atherosclerosis models. Circ Res 2016;119:1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jacobsen K, Lund MB, Shim J, Gunnersen S, Fuchtbauer E-M, Kjolby M, Carramolino L, Bentzon JF.. Diverse cellular architecture of atherosclerotic plaque derives from clonal expansion of a few medial SMCs. JCI Insight 2017;2:e95890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Manavski Y, Lucas T, Glaser SF, Dorsheimer L, Gunther S, Braun T, Rieger MA, Zeiher AM, Boon RA, Dimmeler S.. Clonal expansion of endothelial cells contributes to ischemia-induced neovascularization. Circ Res 2018;122:670–677. [DOI] [PubMed] [Google Scholar]