Key Points
HLA disparity quantified by ME is associated with clinical outcomes of haploidentical HSCT.
HLA-DPB1 nonpermissive mismatch in GVH direction is associated with significant relapse protection without increasing the risk of acute GVHD.
Abstract
Haploidentical donors are increasingly used for patients requiring hematopoietic stem cell transplantation (HSCT). Although several factors have been associated with transplant outcomes, the impact of HLA disparity in haploidentical HSCT (haplo-HSCT) remains unclear. We investigated the impact of HLA disparity quantified by mismatched eplets (ME) load of each HLA locus on the clinical outcome of 278 consecutive haploidentical transplants. Here, we demonstrated that the degree of HLA molecular mismatches, at individual HLA loci, may be relevant to clinical outcome in the haplo-HSCT. A significantly better overall survival was associated with higher ME load from HLA-A (hazard ratio [HR], 0.97; 95% confidence interval [CI], 0.95-0.99; P = .003) and class I loci (HR, 0.99; 95% CI, 0.97-0.99; P = .045) in the host-versus-graft direction. The apparent survival advantage of HLA-A ME was primarily attributed to reduced risk in relapse associated with an increase in HLA-A ME load (subdistribution HR, 0.95; 95% CI, 0.92-0.98; P = .004). Furthermore, we have identified an association between the risk of grade 3-4 acute graft-versus-host disease (GVHD) and a higher ME load at HLA-B and class I loci in graft-versus-host (GVH) direction. Additionally, GVH nonpermissive HLA-DPB1 mismatch defined by T-cell epitope grouping was significantly associated with relapse protection (subdistribution HR, 0.19; 95% CI, 0.06-0.59; P = .004) without a concurrent increase in GVHD. These findings indicate that alloreactivity generated by HLA disparity at certain HLA loci is associated with transplant outcomes, and ME analysis of individual HLA loci might assist donor selection and risk stratification in haplo-HSCT.
Visual Abstract
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) is the only curative therapy for many advanced hematologic malignancies and nonmalignant hematologic disorders. With the success in the prophylaxis of graft-versus-host disease (GVHD) and graft rejection,1,2 haploidentical related donors, who share 1 haplotype with recipients, have become widely accepted stem cell sources in clinical practice with comparable clinical outcomes with HLA-matched donor transplants.3-5 The use of haploidentical donors significantly expanded the likelihood of finding a source of hematopoietic stem cells, especially in certain ethnic minority groups.6-8
It has been shown that HLA disparity has a negative effect on transplant outcomes of patients with HLA-mismatched unrelated donors. Compared with 8/8 matched unrelated donor transplant (MUD), a single mismatch at HLA-A, HLA-B, HLA-C, or HLA-DRB1 was associated with ∼10% decrease in 1-year survival and higher incidence of GVHD.9 In the setting of haploidentical HSCT (haplo-HSCT) performed with conventional GVHD prophylaxis, hyperacute reactions in graft-versus-host (GVH) and host-versus-graft (HVG) reactions occurred as a result of strong bidirectional alloreactivity.10,11 However, with the use of posttransplant cyclophosphamide (PTCy) as GVHD prophylaxis, the influence of HLA mismatch in haplo-HSCT appears to be less significant and distinct from the impact seen in the transplantation with unrelated donors. Raiola et al recently studied a relatively large haplo-HSCT cohort and concluded that there is no correlation between the number of mismatched HLA antigens and clinical outcomes.12 However, although the immediate hyperacute reactions may be attenuated, higher disparities at particular HLA loci could perpetuate different alloreactive immune responses. The European Society for Blood and Marrow Transplantation reported recently that there is no influence of a cumulative number of mismatched HLA antigens on clinical outcomes in their haplo-HSCT cohort, yet an association between mismatched HLA-DRB1 and a higher risk of grade 2-4 GVHD was observed.13 Additionally, the molecular mechanisms behind T-cell alloreactivity are complex and presumably determined by permissibility and structural homology of HLA/peptide complexes. A recent study categorized the HLA mismatches into several of supertype groups, which were defined by anchor specificity of the presented peptide on HLA molecules. Compared with the supertype matched group, HLA-B supertype mismatch was associated with an increased risk of grade 2-4 acute GVHD (aGVHD) in mismatched unrelated donor transplants.14 Therefore, a comprehensive study of the structural and functional disparities at individual HLA loci, in both GVH and HVG directions, might shed light on minimizing risks and maximizing the benefit of alloreactive reactions in HLA-haploidentical transplantation.
HLAMatchmaker is a molecular matching algorithm that considers the functional components of epitopes exposed on HLA molecules. As the key determinants, eplets represent distinct configurations of amino acid polymorphisms that could elicit the immune response (Figure 1). HLAMatchmaker program is used to quantitatively determine the degree of mismatch by comparing the eplet repertories between donor and recipient.15,16 It has been demonstrated that less ME load is associated with longer graft survival17 and reduced humoral sensitization in solid organ transplantation.18 Additionally, alloreactive T-cell clones that are specific to certain eplets were identified,19-21 suggesting that HLAMatchmaker could likewise reflect compatibility at many polymorphic residues involved in T-cell receptor interaction.
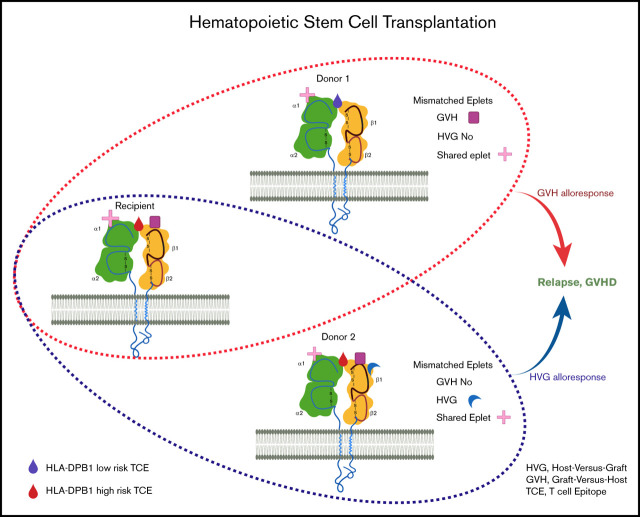
Figure 1.
Simplified illustration of the mismatched eplets (ME) between recipient and different donors. HLA class I molecules from recipient and 2 donors are shown with 1 membrane-spanning α (α) chain and 1 light chain, beta2-microglobulin (β2m). Each specific HLA molecule consists of a set of eplets on the surface, represented by different shapes. Compared with donor 1, there are more MEs that are only present in the recipients, resulting in 2 MEs in the GVH direction and 0 mismatch in HVG direction. Similarly, donor 2 has extra eplets compared with recipient eplet repertories and the net result will be 3 MEs in the HVG direction and 0 mismatch in GVH direction. MEs could, therefore, be completely different between different haploidentical donors. Additionally, with 1 donor, MEs recognized from the donor perspective (GVH) could be significantly different from that seen from the recipient’s perspective (HVG). The provoked immune response might be consequently differed, depending on the number of ME, the direction of ME, or even the specific types of ME.
Solomon et al recently reported a positive survival advantage associated with HLA-DPB1 nonpermissive mismatch defined by T-cell epitope (TCE) grouping in PTCy-based haplo-HSCT, which was attributed to a significant reduction in relapse rate without a parallel increase in the development of GVHD.22 A recent study in unrelated donor transplantation demonstrated that HLA-DPB1 nonpermissive mismatches in GVH direction were associated with an increased risk of aGVHD.23 Concurrently, a reduced risk for disease progression was observed with HLA-DPB1 mismatches.24,25 Consequently, whether the directionality of nonpermissive mismatches affects the risk of relapse or GVHD in haplo-HSCT needs to be clarified.
In this study, we aimed to comprehensively assess the ability of HLAMatchmaker algorithm in predicting the risks or benefits associated with HLA molecular disparity in haplo-HSCT. In addition, we investigated the effect of HLA-PDB1 nonpermissive mismatches, either in the HVG or GVH direction, on clinical outcomes in a relatively large cohort of patients receiving haplo-HSCT at our institution.
Methods
Patients and transplant characteristics
Our cohort included 278 consecutively treated patients, 18 years of age or older, with hematologic malignancies who underwent first or second haplo-HSCT at The University of Texas MD Anderson Cancer Center between July 2009 and January 2015. The majority of patients received T cell–replete bone marrow graft (n = 247, 90%) and all received PTCy-based GVHD prophylaxis with tacrolimus and mycophenolate, as previously described by our group.26 Donors were first-degree relatives who share 1 haplotype match with the recipients. All patients with a significant level (mean fluorescence intensity >2000) of donor-specific anti-HLA antibodies (DSA) received desensitization treatment before transplant per institutional protocol.27 Comorbidities before transplant were evaluated using the Hematopoietic Cell Transplant-Comorbidity Index (HCT-CI).28 Hematological malignancies were risk-stratified using the refined Disease Risk Index (DRI).29 Clinical and laboratory data were collected via electronic medical records. All patients provided written informed consent for transplant following the Declaration of Helsinki. The study protocol was approved by the institutional review board of The University of Texas MD Anderson Cancer Center.
HLA typing and ME analysis
Patients eligible for this study had donor and recipient HLA typing performed at the HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DRB3/4/5, HLA-DQA, HLA-DQB1, and HLA-DPB1 loci using sequenced-based typing methods at high resolution.23 For patients or donors who had no DQA typing in our record (∼15% of the cohort), inferred typing was assigned based on HLA-DR-DQ association at http://www.allelefrequencies.net/. Alleles reported with National Marrow Donor Program codes were deduced to the most common allele in the coded string. ME load at each HLA locus and total ME of class I or class II loci were quantified by using HLAMatchmaker module incorporated in HLA Fusion software v4.4. The analysis was performed in both GVH and HVG directions. The software identified antibody-verified eplets (ME-Ab) with informative HLA antibodies and theoretically predicted eplets (ME-ALL) based on the crystalized HLA molecule models.16 Eplet repertoires are listed in HLA Epitope Registry (http://www.epitopes.net/downloads.html). Inter-locus eplets were assigned as a given eplet identified in 2 or more loci within class I or class II molecules. While comparing eplets between donor and recipient to identify the mismatched eplets, the inter-locus eplets mismatched on 1 locus but matched on another locus were removed to truly reflect the molecular disparity.
HLA-DPB1 permissiveness and KIR benefit
HLA-DPB1 mismatches were classified into permissive and nonpermissive mismatches based on TCE algorithms (version 2.0) at the IPD-IMGT/HLA Web site (https://www.ebi.ac.uk/ipd/imgt/hla/dpb.html).30 As previously described,23 the vectors of HLA-DPB1 mismatches, either in the GVH or HVG direction, were assigned. Transplantations were therefore classified into 4 groups: (1) HLA-DPB1 matched; (2) HLA-DPB1 permissive mismatched; (3) nonpermissive mismatches in HVG direction; and (4) nonpermissive mismatches in GVH direction. Ligand-ligand model31 was used to predict natural killer (NK) cell alloreactivity based on the high-resolution HLA typing of the donor and recipient. Briefly, killer immunoglobulin receptor (KIR) ligand HLA-C and HLA-B molecules were grouped into 3 major categories (C1, C2, Bw4) based on the specific amino acid sequence that defines specific KIR ligand binding (https://www.ebi.ac.uk/ipd/kir/ligand.html). The NK cell alloreactivity in the GVH direction was assigned when the recipient lacked at least 1 of the HLA ligands that were present in the donor. This theoretical prediction of KIR benefit has been commonly used in haplo-HSCT studies.32,33
Clinical endpoints and statistical methods
Clinical endpoints included overall survival (OS), progression-free survival (PFS), relapse, nonrelapse mortality (NRM), acute and chronic GVHD, neutrophil engraftment, and platelet engraftment. All outcomes were measured from the time of stem cell infusion. OS was based on death from any cause. PFS events included death or relapse. NRM was defined as death without a previous relapse. Patients without an event were censored at the time of last contact. Time to neutrophil engraftment was defined as the first of 3 consecutive days with an absolute neutrophil count exceeding 0.5 × 103/µL. Time to platelet engraftment was defined as the first day of a platelet count >20 × 103/µL without transfusion support for 7 consecutive days. Cumulative incidence with competing risks method was used to calculate aGVHD, chronic GVHD, NRM, relapse, and engraftment. For NRM, relapse was the competing risk, and for relapse, the competing risk was NRM. For acute and chronic GVHD, death without the event and relapse were the competing risks, whereas death without engraftment was considered a competing risk for engraftment. Associations between survival outcomes (OS and PFS) and each HLA locus ME number were determined using univariable and multivariable Cox proportional hazards regression models, whereas the impact of the number of HLA locus ME on relapse, NRM, and acute and chronic GVHD was determined using univariable and multivariable proportional subdistribution hazards regression model. HLA locus ME associated with P < .10 by the univariable analysis were included in the multivariable analysis along with other potential prognostic factors including sex, age, HCT-CI, DRI, transplant protocol, conditioning regimen intensity, donor relation, stem cell source, and ABO match. Given that ME-Ab and ME-ALL at the same locus are highly associated and overlapped, only 1 factor with the lowest P value was chosen and included in multivariable analysis. All variables of interest were tested for the proportional hazard assumption and interaction terms. All tests were 2-sided. The type 1 error rate was fixed at 0.05. No adjustments for multiple testing were made. Stata statistical software (SE 13, Stata Corp LP, College Station, TX) was used for statistical analyses.
Results
Patients characteristics and HLA disparity quantified by ME
The median age of this cohort was 48 years (range, 18-72 years). The clinical characteristics are summarized in Table 1. Acute myeloid leukemia/myelodysplastic syndrome was the main indication for haplo-HSCT (163 patients, 58.6%). One hundred and thirty-four patients had high- (40.7%) or very high-risk diseases (7.6%) based on the DRI. Myeloablative conditioning was used in approximately one-half of the patients. Fifty-one percent of the patients had HCT-CI score ≥3. CMV serostatus was nonreactive in 13% of the recipients. NK cell alloreactivity was present in 28% of transplants. HLA-DPB1 nonpermissive mismatches, either in GVH or HVG directions, were noted in one-quarter of the transplantations. In our cohort, the NRM at 1 and 3 years was 25.9% and 29.7% and relapse rate at 1 and 3 years was 24.1% and 27.4%, respectively. PFS and OS at 3 years were 42.9% and 47.4%.
Table 1.
Patient demographics and transplant characteristics
| No. of patients | % | |
|---|---|---|
| Age at transplant, y | ||
| Median (range) | 48 (18-72) | |
| 18-60 | 211 | 75.9 |
| >60 | 67 | 24.1 |
| Female patients | 122 | 43.9 |
| Disease type | ||
| AML/MDS | 163 | 58.6 |
| Others | 115 | 41.4 |
| ALL | 43 | 15.5 |
| CML/MPNs | 32 | 11.5 |
| Lymphoma | 19 | 6.8 |
| Hodgkin | 11 | 4.0 |
| CLL | 9 | 3.2 |
| Myeloma | 1 | 0.4 |
| HCT-CI score | ||
| Median (range) | 3 (0-9) | |
| ≥3 | 142 | 51.1 |
| DRI | ||
| Low | 27 | 9.7 |
| Intermediate | 117 | 42.1 |
| High | 113 | 40.7 |
| Very high | 21 | 7.6 |
| Donor-recipient sex | ||
| Female donor to male recipient | 54 | 19.4 |
| Other recipient-donor sex combination | 225 | 80.6 |
| Conditioning regimen intensity | ||
| MAC | 144 | 51.8 |
| NMA/RIC | 134 | 48.2 |
| Recipient CMV serostatus | ||
| Reactive | 241 | 86.7 |
| Nonreactive | 36 | 12.9 |
| Nondetermined | 1 | 0.4 |
| Donor-recipient ABO match | ||
| Match | 193 | 69.4 |
| Minor mismatch | 45 | 16.2 |
| Major mismatch | 37 | 13.3 |
| Bidirectional mismatch | 3 | 1.1 |
| Hematopoietic cell source | ||
| PB | 31 | 11.2 |
| BM | 247 | 88.9 |
| NK cell alloreactivity | ||
| No | 201 | 72.3 |
| Yes | 77 | 27.7 |
| HLA-DPB1 match/mismatch | ||
| Match | 55 | 19.8 |
| Permissive mismatch | 155 | 55.8 |
| Nonpermissive mismatch in GVH direction | 39 | 14.0 |
| Nonpermissive mismatch in HVG direction | 29 | 10.4 |
| Presence of DSAs | ||
| Yes | 27 | 9.6 |
| No | 252 | 90.4 |
| Median follow-up (range), mo | 16.9 (0.2-101.9) | |
| Median follow-up of 131 survivors (range), mo | 41.3 (4.7-101.9) |
AML, acute myeloid leukemia; BM, bone marrow; CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; MAC, myeloablative conditioning; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm; NMA, nonmyeloablative; PB, peripheral blood; RIC, reduced-intensity conditioning.
Both ME-Ab and ME-ALL load were quantified on each locus in the HVG or GVH direction (supplemental Table 1). The median of the total number of class I ME and class II ME in HVG direction was 24 (range, 16-32) and 21 (range, 11-31), respectively. The median of the total number of class I ME and class II ME in the GVH direction was 26 (range, 17-35) and 23 (range, 14-32), respectively. No correlation was identified between the number of ME in HVG direction vs that in GVH direction (supplemental Figure 1A,C), suggesting that the ME load recognized from the donor perspective is different from that recognized from the recipient perspective. In contrast, the number of theoretical ME-ALL and ME-Ab at 1 specific locus was highly and positively correlated. For instance, the linear regression between ME-ALL and ME-Ab at HLA-A locus and class I were analyzed, the r2 value was 0.78 and 0.80, respectively (supplemental Figure 1B,D). Similar to a recent study in solid organ transplantation,34 no significant differences in clinical impact was recognized between antibody-verified ME and theoretical ME in the present study.
HLA-A ME in HVG direction is associated with an improved OS, PFS, and reduced risk of relapse
We observed a significant association between higher ME at HLA-DRB3/4/5 in HVG direction and an inferior OS on the univariable analysis (hazard ratio [HR], 1.03; 95% confidence interval [CI], 1.00-1.05; P = .025) (Table 2). After adjusting for potential confounders, the association remained significantly in multivariable analysis (HR, 1.03; 95% CI, 1.00-1.05; P = .038) (Table 2). ME load at HLA-A locus and class I in the HVG direction was associated with a better survival on the univariable analysis (Table 2). After adjusting for other patients, disease, and transplant characteristics, an increased HLA-A ME load in the HVG direction remained significantly associated with better OS with a 3% mortality reduction for each ME (HR, 0.97; 95% CI, 0.95-0.99; P = .003). For transplants with lower ME load at HLA-A locus, a 3-year OS rate was only 39%, significantly lower than 54.3% seen in transplants with a higher number of ME at HLA-A locus (Figure 2A). Consistently, the risk of mortality decreased significantly when the total number of ME from class I increased on the multivariable analysis (HR, 0.99; 95% CI, 0.97-0.99; P = .047) (Table 2). Because no association was found between OS and ME from HLA-B or HLA-C, the survival advantage from class I ME is likely attributed to ME from HLA-A locus. Other variables associated with worse OS were poorer HCT-Cl score, high- and very-high DRI, and older age at transplantation (Table 2). No significant influence on OS from ME at other individual locus and total class II loci was detected, indicating the alloimmunity elicited by the mismatches from each HLA locus could be different.
Table 2.
Selected univariable and multivariable analysis for OS
| Factors | HR | 95% CI | P |
|---|---|---|---|
| Univariable analysis for OS | |||
| A-ME Ab HVG | 0.95 | 0.90-1.00 | .053 |
| A-ALL ME HVG | 0.98 | 0.96-0.99 | .024 |
| Class I-Ab ME HVG | 0.98 | 0.95-1.01 | .188 |
| Class I-ALL ME HVG | 0.99 | 0.97-0.99 | .045 |
| DRB345-Ab ME HVG | 1.11 | 1.00-1.23 | .042 |
| DRB345-ALL ME HVG | 1.03 | 1.00-1.05 | .025 |
| DP match/mismatch | |||
| Match | Ref | ||
| Permissive mismatch | 0.71 | 0.47-1.06 | .102 |
| Nonpermissive mismatch GVH | 0.77 | 0.45-1.34 | .368 |
| Nonpermissive mismatch HVG | 0.085 | 0.29-1.08 | .085 |
| Multivariable analysis for OS | |||
| Impact of A-ME HVG | |||
| A-ALL ME HVG (continuous) | 0.97 | 0.95-0.99 | .003 |
| Sex | 1.03 | 0.73-1.45 | .879 |
| Age (continuous) | 1.03 | 1.02-1.05 | <.001 |
| HCT-CI (continuous) | 1.08 | 0.99-1.17 | .064 |
| DRI | |||
| Low | Ref | ||
| Intermediate | 2.08 | 0.92-4.67 | .078 |
| High | 4.04 | 1.81-0.03 | .001 |
| Very high | 6.18 | 2.44-15.61 | <.001 |
| Conditioning regimen intensity | |||
| MAC | Ref | ||
| NMA/RIC | 1.01 | 0.70-1.45 | .950 |
| Hematopoietic cell source | |||
| PB | Ref | ||
| BM | 1.17 | 0.66-2.04 | .598 |
| ABO mismatch | |||
| Match | Ref | ||
| Minor | 0.99 | 0.62-1.57 | .962 |
| Major | 1.74 | 1.09-2.77 | .020 |
| Bidirectional | 1.34 | 0.18-10.06 | .77 |
| Impact of Class I ME HVG | |||
| Class I-All ME HVG (continuous) | 0.99 | 0.97-0.99 | .047 |
| Sex | 1.08 | 0.76-1.51 | .681 |
| Age (continuous) | 1.31 | 1.01-1.05 | <.001 |
| HCT-CI (continuous) | 1.08 | 0.99-1.17 | .054 |
| DRI | |||
| Low | Ref | ||
| Intermediate | 2.24 | 0.99-5.01 | .05 |
| High | 4.10 | 1.83-9.15 | .001 |
| Very high | 6.08 | 2.41-15.32 | <.001 |
| Conditioning regimen intensity | |||
| MAC | Ref | ||
| NMA/RIC | 1.02 | 0.71-1.45 | .927 |
| Hematopoietic cell source | |||
| PB | Ref | ||
| BM | 1.24 | 0.71-2.19 | .445 |
| ABO mismatch | |||
| Match | Ref | ||
| Minor | 1.01 | 0.63-1.60 | .975 |
| Major | 1.71 | 1.07-2.72 | .024 |
| Bidirectional | 1.26 | 0.17-9.43 | .822 |
| Impact of HLA DRB345-ALL ME HVG | |||
| DRB345-All HVG (continuous) | 1.03 | 1.00-1.05 | .038 |
Figure 2.
Influence of ME of HLA-A locus in HVG direction on OS, PFS, and cumulative incidence of relapse. HLA-A ME in HVG direction is associated with superior OS (A) and PFS (B), and reduced risk of relapse (C). HR and SHR represent the impact of HLA-A ME in the HVG direction on outcomes as a continuous variable, adjusted for sex, age, HCT-CI, DRI, transplant protocol, conditioning regimen intensity, donor relation, stem cell source, and ABO match.
Further studies identified the association between better PFS and a higher load of ME at HLA-A and class I in the HVG direction as presented in Table 3 and Figure 2B. As a continuous variable, each class I ME was significantly associated with better PFS with 2% mortality or relapse reduction (HR, 0.98; 95% CI, 0.97-0.99; P = .039), and each ME in HLA-A was significantly associated better PFS with 4% mortality or relapse reduction on the multivariable analysis (HR, 0.96; 95% CI, 0.05-0.98; P < .001). The survival advantage of HLA-A ME load was primarily attributed to a reduced risk in relapse, which progressively decreased with an increase in the number of A-ME (subdistribution hazard ratio [SHR], 0.95; 95% CI, 0.92-0.98; P = .004) (Figure 2C; Table 4). The 3-year cumulative incidence of relapse was 21.6% in the higher A-ME group compared with 34.3% seen in the lower A-ME group (Figure 2C). As described previously, the antibody-verified ME was included in the theoretical version of ME and they were highly correlated. Both ME-Ab and ME-ALL at the HLA-A locus were significantly correlated with reduced risk of relapse in our analysis (Table 4). Neither HLA-A ME nor class I ME was significantly associated with NRM (Table 5), whereas an inferior NRM was significantly associated with increased ME at HLA-DRB1 (SHR, 1.04; 95% CI, 1.00-1.09; P = .048) but not with increased ME from class II (SHR, 1.01; 95% CI, 0.99-1.04; P = .053).
Table 3.
Selected univariable and multivariable analysis for PFS
| Factors | HR | 95% CI | P |
|---|---|---|---|
| Univariable analysis for PFS | |||
| A-ME Ab HVG | 0.93 | 0.89-0.98 | .007 |
| A-ALL ME HVG | 0.97 | 0.95-0.99 | .003 |
| Class I-Ab ME HVG | 0.97 | 0.94-1.00 | .091 |
| Class I-ALL ME HVG | 0.09 | 0.97-0.99 | .032 |
| Class II-Ab ME HVG | 1.03 | 0.99-1.07 | .155 |
| Class II-ALL ME HVG | 1.01 | 0.99-1.03 | .050 |
| DP match/mismatch | |||
| Match | Ref | ||
| Permissive mismatch | 0.66 | 0.45-0.96 | .034 |
| Nonpermissive mismatch GVH | 0.66 | 0.39-1.11 | .120 |
| Nonpermissive mismatch HVG | 0.59 | 0.32-1.07 | .084 |
| Multivariable analysis for PFS* | |||
| A-ALL ME HVG (continuous) | 0.96 | 0.05-0.98 | <.001 |
| Class I-ALL ME HVG (continuous) | 0.98 | 0.97-0.99 | .039 |
| Impact of DP match/mismatch | |||
| Match | Ref | ||
| Permissive mismatch | 0.66 | 0.45-0.98 | .040 |
| Nonpermissive mismatch GVH | 0.64 | 0.37-1.10 | .110 |
| Nonpermissive mismatch HVG | 0.66 | 0.36-1.24 | .202 |
| HLA Class II-ALL ME HVG (continuous) | 1.01 | 0.99-1.02 | .107 |
*Impact of HLA eplet mismatch on each locus on PFS was analyzed in separate models, adjusted for sex, age, HCT-CI, DRI, transplant protocol, conditioning regimen intensity, donor relation, stem cell source, and ABO match. Impacts of these covariates are not shown.
Table 4.
Selected univariable and multivariable analysis for relapse
| Factors | SHR | 95% CI | P |
|---|---|---|---|
| Univariable analysis for relapse | |||
| A-ME Ab HVG | 0.92 | 0.85-0.99 | .026 |
| A-ALL ME HVG | 0.96 | 0.93-0.99 | .006 |
| Class I-Ab HVG | 0.99 | 0.94-1.04 | .770 |
| Class I-ALL ME HVG | 0.99 | 0.97-1.02 | .668 |
| Class II-Ab ME HVG | 1.01 | 0.95-1.07 | .723 |
| Class II-ALL ME HVG | 1.00 | 0.98-1.02 | .903 |
| DP match/mismatch | |||
| Match | Ref | ||
| Permissive mismatch | 0.86 | 0.51-1.48 | .601 |
| Nonpermissive mismatch GVH | 0.25 | 0.09-0.76 | .014 |
| Nonpermissive mismatch HVG | 0.54 | 0.22-1.33 | .181 |
| Multivariable analysis for relapse* | |||
| A-ALL ME HVG (continuous) | 0.95 | 0.92-0.98 | .004 |
| Impact of DP match/mismatch | |||
| Match | Ref | ||
| Permissive mismatch | 0.74 | 0.42-1.30 | .302 |
| Nonpermissive mismatch GVH | 0.19 | 0.06-0.59 | .004 |
| Nonpermissive mismatch HVG | 0.54 | 0.19-1.52 | .247 |
*Impact of HLA eplet mismatch on each locus on relapse was analyzed in separate models, adjusted for sex, age, HCT-CI, DRI, transplant protocol, conditioning regimen intensity, donor relation, stem cell source, and ABO match. Impacts of these covariates are not shown.
Table 5.
Selected univariable and multivariable analysis for NRM
| Factors | SHR | 95% CI | P |
|---|---|---|---|
| Univariable analysis for NRM | |||
| B-ME Ab HVG | 0.94 | 0.88-1.01 | .110 |
| B-ALL ME HVG | 0.96 | 0.94-0.99 | .040 |
| DRB1-Ab ME HVG | 1.13 | 1.01-1.26 | .033 |
| DRB1-ALL ME HVG | 1.05 | 1.01-1.09 | .026 |
| Class II-Ab ME HVG | 1.04 | 0.99-1.09 | .157 |
| Class II-ALL ME HVG | 1.02 | 1.00-1.04 | .042 |
| Multivariable analysis for NRM* | |||
| B-ALL ME HVG (continuous) | 0.97 | 0.94-1.01 | .162 |
| DRB1-ALL ME HVG (continuous) | 1.04 | 1.00-1.09 | .048 |
| Class II-ALL ME HVG (continuous) | 1.01 | 0.99-1.04 | .053 |
*Impact of HLA eplet mismatch on each locus on NRM was analyzed in separate models, adjusted for sex, age, HCT-CI, DRI, transplant protocol, conditioning regimen intensity, donor relation, stem cell source, and ABO match. Impacts of these covariates are not shown.
Nonpermissive HLA-DPB1 mismatch in GVH direction is associated with decreased risk of relapse
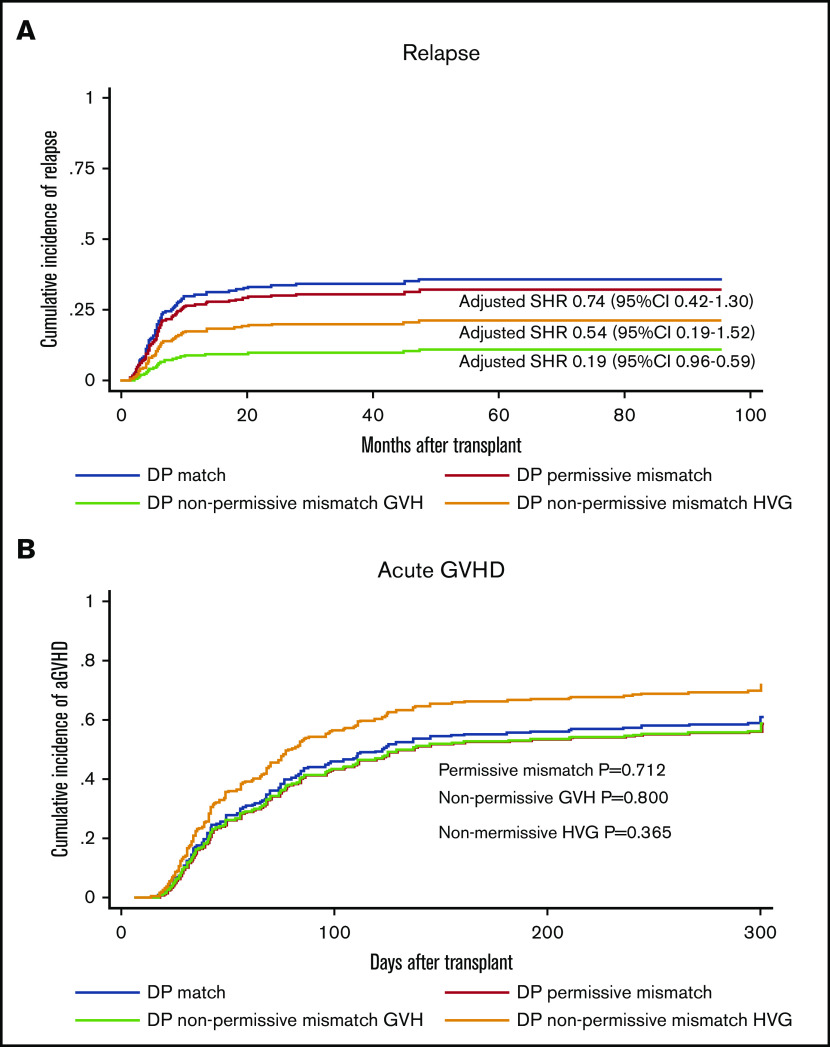
A previous study suggested that HLA-DPB1 disparity might be associated with relapse protection in haplo-HSCT with PTCy.22 We further characterized the relapse protection by separately assessing the nonpermissive mismatches in HVG or GVH directions predicted by TCE grouping. A nonsignificant trend of relapse protection was found with HLA-DP nonpermissive in HVG direction (SHR, 0.54; 95% CI, 0.22-1.33; P = .181) on univariable analysis. In contrast, we observed a significant reduction in the incidence of relapse associated with HLA-DPB1 nonpermissive mismatch in the GVH direction (SHR, 0.25; 95% CI, 0.09-0.76; P = .014). The 3-year cumulative incidence of relapse was 15.3% in the nonpermissive GVH group, compared with 31.3% in HLA-DPB1 matched group (Figure 3A). We further investigated the role of nonpermissive mismatches in either HVG or GVH vectors on multivariable analysis and confirmed that only GVH nonpermissive mismatch was significantly associated with relapse protection (SHR, 0.19; 95% CI, 0.06-0.59; P = .004) (Table 4) without impact on OS (Table 2) or risk of GVHD (Figure 3B). Unlike observed in transplant with MUD donor,23 no significantly higher risk of grade 3-4 aGVHD was observed in nonpermissive GVH group (SHR, 1.00; 95% CI, 0.27-3.74; P = .995) (Table 6) with univariable analysis. On multivariable analysis, the HLA-DPB1 ME in GVH direction was not significantly correlated with a higher risk of aGVHD or grade 3-4 aGVHD (SHR, 0.78; 95% CI, 0.65-1.02; P = .112).
Figure 3.
Influence of HLA-DPB1 mismatch permissive status on relapse and aGVHD. HLA-DPB1 nonpermissive mismatch in GVH direction is associated with relapse protection (A) without increasing the risk of aGVHD (B). SHRs represent the effect of each group on outcomes in comparison with HLA-DPB1 matched group (reference), adjusted for sex, age, HCT-CI, DRI, transplant protocol, conditioning regimen intensity, donor relation, stem cell source, and ABO match.
Table 6.
Selected univariable and multivariable analysis for aGVHD grade 3-4
| Factors | SHR | 95% CI | P |
|---|---|---|---|
| Univariable analysis for aGVHD grade 3-4 | |||
| B-ME Ab GVH | 1.09 | 1.01-1.19 | .034 |
| B-ALL ME GVH | 1.04 | 0.99-1.09 | .052 |
| Class I-Ab ME GVH | 1.06 | 0.99-1.14 | .063 |
| Class I-ALL ME GVH | 1.02 | 0.99-1.06 | .119 |
| DP match/mismatch | |||
| Match | Ref | ||
| Permissive mismatch | 1.12 | 0.45-3.28 | .708 |
| Nonpermissive mismatch GVH | 1.00 | 0.27-3.74 | .995 |
| Nonpermissive mismatch HVG | 1.24 | 0.28-5.41 | .768 |
| Multivariable analysis for aGVHD grade 3-4* | |||
| B-ALL ME GVH (continuous) | 1.05 | 1.01-1.12 | .036 |
| Class I-Ab ME GVH (continuous) | 1.09 | 1.00-1.18 | .041 |
| DPB1-Ab ME GVH (continuous) | 0.78 | 0.65-1.02 | .112 |
*Impact of HLA eplet mismatch on each locus on aGVHD grade 3-4 was analyzed in separate models, adjusted for sex, age, HCT-CI, DRI, transplant protocol, conditioning regimen intensity, donor relation, stem cell source, and ABO match. Impacts of these covariates are not shown.
HLA-B and class I ME in GVH direction is associated with a higher incidence of grade 3-4 aGVHD
On univariable analysis, no significant association was found between a higher load of ME from an individual locus and risk of aGVHD (Table 7). The impact on grade 3-4 aGVHD was further investigated and the significance was noted on multivariable analysis with a significantly higher incidence of grade 3-4 aGVHD in the group with higher HLA-B ME (SHR, 1.05; 95% CI, 1.01-1.12; P = .036) and class I ME in GVH direction (SHR, 1.09; 95% CI, 1.00-1.18; P = .041) (Table 6). As shown in Figure 4A, the 100-day cumulative incidence of grade 3-4 aGVHD in a higher B-ME group was 10% compared with 7% in the group with lower B-ME.
Table 7.
Selected univariable analysis for aGVHD
| SHR | 95% CI | P | |
|---|---|---|---|
| DP match | |||
| Match | Ref | ||
| Permissive mismatch | 0.92 | 0.60-1.41 | .712 |
| Nonpermissive mismatch GVH | 0.93 | 0.55-1.58 | .800 |
| Nonpermissive mismatch HVG | 1.35 | 0.71-2.56 | .365 |
| NK benefit | 0.95 | 0.68-1.34 | .780 |
| A-Ab ME HVG | 1.01 | 0.97-1.06 | .607 |
| A-All ME HVG | 0.99 | 0.98-1.02 | .912 |
| B-Ab ME HVG | 1.02 | 0.97-1.06 | .453 |
| B-ALL ME HVG | 1.01 | 0.99-1.03 | .336 |
| C-Ab ME HVG | 0.95 | 0.86-1.06 | .429 |
| C-ALL ME HVG | 0.99 | 0.97-1.03 | .879 |
| Class I-Ab ME HVG | 1.00 | 0.98-1.04 | .572 |
| Class I-ALL ME HVG | 1.00 | 0.98-1.02 | .720 |
| DRB1-Ab ME HVG | 0.97 | 0.88-1.06 | .491 |
| DRB1-ALL ME HVG | 0.99 | 0.97-1.03 | .844 |
| DRB345-Ab ME HVG | 0.97 | 0.87-1.08 | .591 |
| DRB345-ALL ME HVG | 0.99 | 0.97-1.02 | .625 |
| DPB1-Ab ME HVG | 1.00 | 0.92-1.10 | .950 |
| DPB1-ALL ME HVG | 1.01 | 0.97-1.06 | .538 |
| DQ-Ab ME HVG | 0.99 | 0.92-1.08 | .973 |
| DQ-ALL ME HVG | 1.00 | 0.98-1.03 | .749 |
| Class II-Ab ME HVG | 0.99 | 0.96-1.03 | .797 |
| Class II-ALL ME HVG | 1.00 | 0.99-1.01 | .693 |
| A-Ab ME GVH | 1.00 | 0.96-1.04 | .964 |
| A-ALL ME GVH | 1.00 | 0.98-1.02 | .911 |
| B-Ab ME GVH | 1.01 | 0.96-1.06 | .627 |
| B-ALL ME GVH | 0.99 | 0.97-1.02 | .775 |
| C-Ab ME GVH | 0.99 | 0.90-1.10 | .925 |
| C-ALL ME GVH | 0.98 | 0.94-1.01 | .210 |
| Class I-Ab ME GVH | 1.00 | 0.97-1.03 | .759 |
| Class I-ALL ME GVH | 0.99 | 0.98-1.01 | .571 |
| DRB1-Ab ME GVH | 0.92 | 0.84-1.02 | .112 |
| DRB1-ALL ME GVG | 0.97 | 0.94-1.01 | .144 |
| DRB345-Ab ME GVH | 0.92 | 0.83-1.02 | .118 |
| DRB345-ALL ME GVH | 0.97 | 0.95-1.00 | .079 |
| DPB1-Ab ME GVH | 0.96 | 0.89-1.03 | .233 |
| DPB1-ALL ME GVH | 0.98 | 0.95-1.02 | .306 |
| DQ-Ab ME GVH | 0.99 | 0.92-1.08 | .937 |
| DQ-ALL ME GVH | 1.00 | 0.98-1.03 | .765 |
| Class II-Ab ME GVH | 0.97 | 0.93-1.00 | .086 |
| Class II-ALL ME GVH | 0.99 | 0.98-1.00 | .257 |
NK cell, natural killer cell.
Figure 4.
Influence of HLA-B ME and class I ME in the GVH direction on grade 3-4 aGVHD. HLA-B ME (A) and class I ME (B) in the GVH direction is associated with higher incidence of grade 3-4 aGVHD.SHRs represent the effect of HLA-B ME and class I ME in GVH direction on grade 3-4 aGVHD as a continuous variable, adjusted for sex, age, HCT-CI, DRI, transplant protocol, conditioning regimen intensity, donor relation, stem cell source, and ABO match.
ME load was not significantly associated with engraftment and chronic GVHD
The effect of HLA disparity on engraftment and chronic GVHD was also evaluated. Different from a recent study on HLA mismatch in haplo-HSCT,35 we did not find any association between ME load and neutrophil and platelet engraftment. The association between ME load and chronic GVHD was not identified. No significant impact from NK alloreactivity was noted on OS (HR, 1.13; 95% CI, 0.79-1.61; P = .499), PFS (HR, 1.11; 95% CI, 0.79-1.57; P = .520), or relapse (SHR, 1.20; 95% CI, 0.74-1.94; P = .441).
Discussion
It has been well accepted that a greater HLA disparity is associated with worse clinical outcomes and a higher risk of GVHD in HSCT with mismatched donors receiving conventional GVHD prophylaxis.36,37 However, the immune system seems to respond differently in the setting of haplo-HSCT with GVHD prophylaxis using PTCy regimen. As haplo-HSCT is increasingly used for patients requiring transplantation, it is essential to understand the immunogenicity to alloantigen on the mismatched graft from haploidentical donors. In the present study, with patient cohort receiving PTCy-based GVHD prevention and bone marrow graft, we demonstrated that HLA disparity quantified at molecular mismatch level, and not by the cumulative number of mismatched antigens, could be relevant to the clinical outcome of patients receiving haploidentical hematopoietic cell transplantation.
With the rapid progress in molecular typing and protein modeling, the degree of HLA disparity has recently been assessed at the molecular level in solid organ transplantation to precisely determine HLA alloimmune risk.38,39 Different algorithms of molecular mismatching in predicting immunogenicity have been developed with different focuses, such as the number of mismatched amino acids or physiochemical properties of the amino acid substitution.40 Initially, HLAMatchmaker quantified mismatched epitopes and predicted humoral response along with antibody-mediated rejection. The revised HLAMatchmaker algorithm considers the structural proximity that is accessible by antibodies and includes all 3-dimensional patch of linear or discontinuous polymorphic amino acids within 3 Å radium. It defines both antibody-verified eplets and theoretical eplets based on stereochemical modeling of HLA molecule and antigen-antibody complex. In the present study, we demonstrated that the revised HLAMatchmaker could be used for the assessment of histocompatibility in haplo-HSCT at the molecular level.
Here, we have found that ME load at different loci has different effects on the clinical outcomes of haplo-HSCT. Patients with higher ME load at class I loci and HLA-A locus in the HVG direction was found to be associated with improved OS. Our study also suggested that this survival benefit of A-ME may primarily result from a decreased risk of relapse. Kasamon et al studied a cohort of haplo-HSCT patients receiving bone marrow graft and high-dose PTCy and demonstrated that presence of higher allelic level mismatches (3/4 vs 1/2) was associated with a nonsignificant trend to less relapse, and the higher number of class I mismatches (2/3 vs 0/1) was significantly associated with lower relapse rate.41 Interestingly, further analysis indicated that the significant protective effect was mainly associated with mismatches in the HVG direction. The authors were unable to disentangle the effect from HVG versus that from GVH direction because their mismatches at the antigen level were tightly linked from both vectors. In our study, using ME instead of mismatched antigens/alleles, we showed that the ME at the same locus but from different directions are not correlated, which allowed us to separately assess their effect (supplemental Figure 1). Surprisingly, we found that a lower risk of relapse was significantly associated with HLA disparity in the HVG but not in the GVH direction, indicating that alloreactive eplets present in the HVG vector could be involved in a subsequent immune response. Several clinical studies demonstrated the association between the rejection of donor bone marrow and the regression of hematological tumor without GVHD.42-44 In addition, several studies in animal models (reviewed by Li and Sykes45) suggested that HVG alloresponse possibly induces an antitumor response by enhancing the pro-inflammatory environment and leads to subsequent activation of particular cell subsets.46,47 Fleischhauer et al studied the effect of HLA-DPB1 nonpermissive mismatches on clinical outcomes in bidirectional vs unidirectional groups.48 The proliferative response in mixed lymphocyte reaction assay was significantly higher in the presence of nonpermissive mismatches. Interestingly, there was no significant difference in the levels of response invoked by alloreactivity in the HVG and GVH direction, whereas a significantly higher response was observed in the bidirectional group. They postulated that in response to allogeneic HLA on the responder cells (graft), the stimulator cells (host) release cytokines that would enhance the proliferation of responder cells.48 In this study, the exact mechanism underlying relapse protection by the HVG alloreactivity is unclear and remains to be explored in future studies. Different from what we observed here, a recent study using peripheral blood graft reported that class II ME in the GVH direction was associated with decreased relapse rate in haplo-HSCT.35 This inconsistency may be attributed to different stem cell sources, transplant protocol, disease status, DSA, and different ethnic cohorts. Recent studies in haplo-HSCT with PTCy-based prophylaxis suggested that HLA disparity is associated with reduced relapse without concurrent increased risk of GVHD, likely because of the distinctive ability of PTCy to manipulate allocative T cells and regulatory T cells.49 In our study, the administration of PTCy appears not to completely mitigate the GVHD in response to allogeneic HLA disparity, as the ME load at HLA-B locus and class I in the GVH direction were correlated with a higher risk of developing grade 3-4 aGVHD. The distinctive allorecognition responses to HLA disparity from different loci, may be dictated by the different expression levels of HLA molecules, or specific reactivity of potential effector cells.41,50 The modulation of alloreactive T cells by PTCy adds an extra layer of complexity. Our current observational study is unable to clarify this complex association and selecting the optimal donor could be difficult when both favorable and undesirable factors exist concomitantly. However, our results suggest that donor selection could be optimized based on the specific impact from ME at 1 HLA locus, in particular, the donor with a higher A-ME load might be preferred over other donors for the patients with a higher risk of relapse.
The role of HLA-DPB1 disparity in HSCT with MUD has been reported by several studies.23,24,51,52 HLA-DPB1 mismatches were classified into nonpermissive and permissive mismatches, the latter was defined as the net result of elicited alloreactivity is toward to graft versus leukemia (GVL) with clinically tolerable GVHD effect.53,54 One model predicted the permissiveness using the TCE grouping55 and another model was based on specific allele expression levels of the HLA-DPB1 allele.56 In HSCT with unrelated donors, nonpermissive mismatches are generally associated with increased risk of GVHD that overweighs the GVL benefit and results in an inferior survival.24,57 The study on HSCT with MUD advocated that HLA-DPB1 disparity in HVG vector, in addition to the disparity in GVH direction, might trigger donor effector cells to exert GVHD and GVL effect.53 In our haplo-HSCT cohort, only GVL but not GVHD effect was observed with GVH nonpermissive mismatches of HLA-DPB1, perhaps because of the use of PTCy. Neither HLA-DPB1 nonpermissive mismatch in HVG direction nor ME in HLA-DPB1 was associated with decreased risk of relapse. The HLA-DPB1 disparity predicted by TCE model seems to be more clinically relevant compared with ME load, possibly because TCE classification considers permissiveness based on the functional “thymic selection” with proven T-cell alloreactivity patterns.53
Our study has several limitations including its retrospective nature and a large number of analyses on a relatively small number of patients, which may result in the false-positive potential resulting from lower statistical power. Although it is well accepted that each eplet does not contribute to immunogenicity equally, we were able to study the disparity only by ME load but not by individual mismatched eplets. Studies with a larger cohort of patients are warranted to fully characterize the eplets with a specific impact in haplo-HSCT. Additionally, inferred DQA was used in about 15% of our cohort, which may not be completely accurate. Although the prevalence of DSA in our cohort was relatively low (9.6%), we were unable to exclude the influence of DSAs on transplant outcomes because the majority of patients with DSAs were desensitized before the transplant. In line with a haplo-HSCT study on patients with acute leukemia,58 we found no association between clinical outcomes and KIR mismatches assessed by the ligand–ligand mismatched model. Compared with the gene–gene model that requires KIR gene typing on both recipient and donor, it has been shown that the model we used predicts a considerably lower number of KIR alloreactivity,31 which might underpower the KIR benefit given the low number of occurrences. Despite these limitations, to the best of our knowledge, this study is the largest study of the clinical impact of HLA disparity measured at the molecular level in haplo-HSCT.
In conclusion, molecular HLA disparity at different loci appears to have a different effect on clinical outcomes in haplo-HSCT. Higher ME load at HLA-A locus was associated with improved OS primarily because of a reduction in relapse rate posttransplant. HLA-DPB1 nonpermissive mismatch in GVH direction demonstrated also significant relapse protection without increasing the risk of GVHD. Molecular mismatch analysis could be performed routinely in the future in haploidentical donor transplants to assist in risk stratification and optimal donor selection.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Rene Duquesnoy for helpful discussion and preparation of the manuscript and Kevin Harrell and Jar-How Lee from Thermo Fisher Scientific for their help in eplet analysis.
Footnotes
For data sharing requests, e-mails should be sent to the corresponding author, Jun Zou (jzou@mdanderson.org) or Stefan O. Ciurea (sciurea@hs.uci.edu).
Authorship
Contribution: J.Z., S.O.C., and K.C. designed the study, contributed to data collection and interpretation, and manuscript writing; J.Z. wrote the initial draft of the manuscript; P.K. and M.Y. contributed statistical analysis, interpretation of statistical results, and reviewed and approved the manuscript; Y.C. contributed to data collection and data analysis and reviewed and approved the manuscript; G.R. contributed to data collection and reviewed and approved the manuscript; S.S., P.K., and D.P. contributed to data interpretation and reviewed and approved the manuscript; and S.O.C. and R.E.C. contributed to treatment of patients, reviewed, edited, and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.O.C. is Division of Hematology/Oncology, Department of Medicine, Chao Family Comprehensive Cancer Center, University of California, Irvine, Orange, CA.
Correspondence: Jun Zou, Department of Laboratory Medicine, HLA Laboratory, University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030; e-mail: jzou@mdanderson.org; or Stefan O. Ciurea, Division of Hematology/Oncology, Department of Medicine, Chao Family Comprehensive Cancer Center, University of California, Irvine, 101 The City Dr South, ZOT 4061, Orange, CA 92868; e-mail: sciurea@hs.uci.edu.
References
- 1.Luznik L, Jalla S, Engstrom LW, Iannone R, Fuchs EJ. Durable engraftment of major histocompatibility complex-incompatible cells after nonmyeloablative conditioning with fludarabine, low-dose total body irradiation, and posttransplantation cyclophosphamide. Blood. 2001;98(12):3456-3464. [DOI] [PubMed] [Google Scholar]
- 2.Ciurea SO, Bayraktar UD. “No donor”? Consider a haploidentical transplant. Blood Rev. 2015;29(2):63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciurea SO, Zhang MJ, Bacigalupo AA, et al. . Haploidentical transplant with posttransplant cyclophosphamide vs matched unrelated donor transplant for acute myeloid leukemia. Blood. 2015;126(8):1033-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw BE. Related haploidentical donors are a better choice than matched unrelated donors: Counterpoint. Blood Adv. 2017;1(6):401-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCurdy SR, Kasamon YL, Kanakry CG, et al. . Comparable composite endpoints after HLA-matched and HLA-haploidentical transplantation with post-transplantation cyclophosphamide. Haematologica. 2017;102(2):391-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gragert L, Eapen M, Williams E, et al. . HLA match likelihoods for hematopoietic stem-cell grafts in the U.S. registry. N Engl J Med. 2014;371(4):339-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pidala J, Kim J, Schell M, et al. . Race/ethnicity affects the probability of finding an HLA-A, -B, -C and -DRB1 allele-matched unrelated donor and likelihood of subsequent transplant utilization. Bone Marrow Transplant. 2013;48(3):346-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciurea SO, Bittencourt MCB, Milton DR, et al. . Is a matched unrelated donor search needed for all allogeneic transplant candidates? Blood Adv. 2018;2(17):2254-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SJ, Klein J, Haagenson M, et al. . High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576-4583. [DOI] [PubMed] [Google Scholar]
- 10.Beatty PG, Clift RA, Mickelson EM, et al. . Marrow transplantation from related donors other than HLA-identical siblings. N Engl J Med. 1985;313(13):765-771. [DOI] [PubMed] [Google Scholar]
- 11.Szydlo R, Goldman JM, Klein JP, et al. . Results of allogeneic bone marrow transplants for leukemia using donors other than HLA-identical siblings. J Clin Oncol. 1997;15(5):1767-1777. [DOI] [PubMed] [Google Scholar]
- 12.Raiola AM, Risitano A, Sacchi N, et al. . Impact of HLA disparity in haploidentical bone marrow transplantation followed by high-dose cyclophosphamide. Biol Blood Marrow Transplant. 2018;24(1):119-126. [DOI] [PubMed] [Google Scholar]
- 13.Lorentino F, Labopin M, Fleischhauer K, et al. . The impact of HLA matching on outcomes of unmanipulated haploidentical HSCT is modulated by GVHD prophylaxis. Blood Adv. 2017;1(11):669-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazaryan A, Wang T, Spellman SR, et al. . Human leukocyte antigen supertype matching after myeloablative hematopoietic cell transplantation with 7/8 matched unrelated donor allografts: a report from the Center for International Blood and Marrow Transplant Research. Haematologica. 2016;101(10):1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duquesnoy RJ. The eplet load concept in clinical transplantation. Pediatr Transplant. 2016;20(7):884-885. [DOI] [PubMed] [Google Scholar]
- 16.Duquesnoy RJ. Reflections on HLA epitope-based matching for transplantation. Front Immunol. 2016;7:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duquesnoy RJ, Takemoto S, de Lange P, et al. . HLAmatchmaker: a molecularly based algorithm for histocompatibility determination. III. Effect of matching at the HLA-A,B amino acid triplet level on kidney transplant survival. Transplantation. 2003;75(6):884-889. [DOI] [PubMed] [Google Scholar]
- 18.Goodman RS, Taylor CJ, O’Rourke CM, Lynch A, Bradley JA, Key T. Utility of HLAMatchmaker and single-antigen HLA-antibody detection beads for identification of acceptable mismatches in highly sensitized patients awaiting kidney transplantation. Transplantation. 2006;81(9):1331-1336. [DOI] [PubMed] [Google Scholar]
- 19.van Seventer GA, Huis B, Melief CJ, Iványi P. Fine specificity of human HLA-B7-specific cytotoxic T-lymphocyte clones. I. Identification of HLA-B7 subtypes and histotopes of the HLA-B7 cross-reacting group. Hum Immunol. 1986;16(4):375-389. [DOI] [PubMed] [Google Scholar]
- 20.Smith KD, Epperson DF, Lutz CT. Alloreactive cytotoxic T-lymphocyte-defined HLA-B7 subtypes differ in peptide antigen presentation. Immunogenetics. 1996;43(1-2):27-37. [DOI] [PubMed] [Google Scholar]
- 21.Hiraiwa M, Yamamoto J, Matsumoto K, et al. . T cell can recognize the allospecificities formed by the substitution of amino acids associated with HLA-Bw4/Bw6 public epitopes. Hum Immunol. 1991;32(1):41-45. [DOI] [PubMed] [Google Scholar]
- 22.Solomon SR, Aubrey MT, Zhang X, et al. . Selecting the best donor for haploidentical transplant: impact of HLA, killer cell immunoglobulin-like receptor genotyping, and other clinical variables. Biol Blood Marrow Transplant. 2018;24(4):789-798. [DOI] [PubMed] [Google Scholar]
- 23.Oran B, Saliba RM, Carmazzi Y, et al. . Effect of nonpermissive HLA-DPB1 mismatches after unrelated allogeneic transplantation with in vivo T-cell depletion. Blood. 2018;131(11):1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleischhauer K, Shaw BE, Gooley T, et al. ; International Histocompatibility Working Group in Hematopoietic Cell Transplantation . Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13(4):366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pidala J, Lee SJ, Ahn KW, et al. . Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood. 2014;124(16):2596-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaballa S, Ge I, El Fakih R, et al. . Results of a 2-arm, phase 2 clinical trial using post-transplantation cyclophosphamide for the prevention of graft-versus-host disease in haploidentical donor and mismatched unrelated donor hematopoietic stem cell transplantation. Cancer. 2016;122(21):3316-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciurea SO, de Lima M, Cano P, et al. . High risk of graft failure in patients with anti-HLA antibodies undergoing haploidentical stem-cell transplantation. Transplantation. 2009;88(8):1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorror ML, Sandmaier BM, Storer BE, et al. . Comorbidity and disease status based risk stratification of outcomes among patients with acute myeloid leukemia or myelodysplasia receiving allogeneic hematopoietic cell transplantation. J Clin Oncol. 2007;25(27):4246-4254. [DOI] [PubMed] [Google Scholar]
- 29.Armand P, Kim HT, Logan BR, et al. . Validation and refinement of the Disease Risk Index for allogeneic stem cell transplantation. Blood. 2014;123(23):3664-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crivello P, Zito L, Sizzano F, et al. . The impact of amino acid variability on alloreactivity defines a functional distance predictive of permissive HLA-DPB1 mismatches in hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2015;21(2):233-241. [DOI] [PubMed] [Google Scholar]
- 31.Symons HJ, Leffell MS, Rossiter ND, Zahurak M, Jones RJ, Fuchs EJ. Improved survival with inhibitory killer immunoglobulin receptor (KIR) gene mismatches and KIR haplotype B donors after nonmyeloablative, HLA-haploidentical bone marrow transplantation. Biol Blood Marrow Transplant. 2010;16(4):533-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wanquet A, Bramanti S, Harbi S, et al. . Killer cell immunoglobulin-like receptor-ligand mismatch in donor versus recipient direction provides better graft-versus-tumor effect in patients with hematologic malignancies undergoing allogeneic T cell-replete haploidentical transplantation followed by post-transplant cyclophosphamide. Biol Blood Marrow Transplant. 2018;24(3):549-554. [DOI] [PubMed] [Google Scholar]
- 33.Ruggeri L, Capanni M, Urbani E, et al. . Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295(5562):2097-2100. [DOI] [PubMed] [Google Scholar]
- 34.Kramer C, Heidt S, Claas FHJ. Towards the identification of the relative immunogenicity of individual HLA antibody epitopes. Hum Immunol. 2019;80(4):218-220. [DOI] [PubMed] [Google Scholar]
- 35.Rimando J, Slade M, DiPersio JF, et al. . HLA epitope mismatch in haploidentical transplantation is associated with decreased relapse and delayed engraftment. Blood Adv. 2018;2(24):3590-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanda Y, Chiba S, Hirai H, et al. . Allogeneic hematopoietic stem cell transplantation from family members other than HLA-identical siblings over the last decade (1991-2000). Blood. 2003;102(4):1541-1547. [DOI] [PubMed] [Google Scholar]
- 37.Morishima Y, Yabe T, Matsuo K, et al. ; Japan Marrow Donor Program . Effects of HLA allele and killer immunoglobulin-like receptor ligand matching on clinical outcome in leukemia patients undergoing transplantation with T-cell-replete marrow from an unrelated donor. Biol Blood Marrow Transplant. 2007;13(3):315-328. [DOI] [PubMed] [Google Scholar]
- 38.Wiebe C, Rush DN, Nevins TE, et al. . Class II eplet mismatch modulates tacrolimus trough levels required to prevent donor-specific antibody development. J Am Soc Nephrol. 2017;28(11):3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tambur AR. HLA-epitope matching or eplet risk stratification: the devil is in the details. Front Immunol. 2018;9:2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kramer CSM, Roelen DL, Heidt S, Claas FHJ. Defining the immunogenicity and antigenicity of HLA epitopes is crucial for optimal epitope matching in clinical renal transplantation. HLA. 2017;90(1):5-16. [DOI] [PubMed] [Google Scholar]
- 41.Kasamon YL, Luznik L, Leffell MS, et al. . Nonmyeloablative HLA-haploidentical bone marrow transplantation with high-dose posttransplantation cyclophosphamide: effect of HLA disparity on outcome. Biol Blood Marrow Transplant. 2010;16(4):482-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colvin GA, Berz D, Ramanathan M, et al. . Nonengraftment haploidentical cellular immunotherapy for refractory malignancies: tumor responses without chimerism. Biol Blood Marrow Transplant. 2009;15(4):421-431. [DOI] [PubMed] [Google Scholar]
- 43.Guo M, Hu KX, Yu CL, et al. . Infusion of HLA-mismatched peripheral blood stem cells improves the outcome of chemotherapy for acute myeloid leukemia in elderly patients. Blood. 2011;117(3):936-941. [DOI] [PubMed] [Google Scholar]
- 44.Dey BR, McAfee S, Colby C, et al. . Anti-tumour response despite loss of donor chimaerism in patients treated with non-myeloablative conditioning and allogeneic stem cell transplantation. Br J Haematol. 2005;128(3):351-359. [DOI] [PubMed] [Google Scholar]
- 45.Li HW, Sykes M. Emerging concepts in haematopoietic cell transplantation. Nat Rev Immunol. 2012;12(6):403-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubio MT, Kim YM, Sachs T, Mapara M, Zhao G, Sykes M. Antitumor effect of donor marrow graft rejection induced by recipient leukocyte infusions in mixed chimeras prepared with nonmyeloablative conditioning: critical role for recipient-derived IFN-gamma. Blood. 2003;102(6):2300-2307. [DOI] [PubMed] [Google Scholar]
- 47.Rubio MT, Saito TI, Kattleman K, Zhao G, Buchli J, Sykes M. Mechanisms of the antitumor responses and host-versus-graft reactions induced by recipient leukocyte infusions in mixed chimeras prepared with nonmyeloablative conditioning: a critical role for recipient CD4+ T cells and recipient leukocyte infusion-derived IFN-gamma-producing CD8+ T cells. J Immunol. 2005;175(2):665-676. [DOI] [PubMed] [Google Scholar]
- 48.Fleischhauer K, Ahn KW, Wang HL, et al. . Directionality of non-permissive HLA-DPB1 T-cell epitope group mismatches does not improve clinical risk stratification in 8/8 matched unrelated donor hematopoietic cell transplantation. Bone Marrow Transplant. 2017;52(9):1280-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wachsmuth LP, Patterson MT, Eckhaus MA, Venzon DJ, Gress RE, Kanakry CG. Post-transplantation cyclophosphamide prevents graft-versus-host disease by inducing alloreactive T cell dysfunction and suppression. J Clin Invest. 2019;129(6):2357-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruggeri L, Mancusi A, Capanni M, et al. . Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood. 2007;110(1):433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petersdorf EW, Gooley T, Malkki M, et al. . The biological significance of HLA-DP gene variation in haematopoietic cell transplantation. Br J Haematol. 2001;112(4):988-994. [DOI] [PubMed] [Google Scholar]
- 52.Crivello P, Heinold A, Rebmann V, et al. . Functional distance between recipient and donor HLA-DPB1 determines nonpermissive mismatches in unrelated HCT [published correction appears in Blood. 2016:128(23):2744]. Blood. 2016;128(1):120-129. [DOI] [PubMed] [Google Scholar]
- 53.Fleischhauer K, Shaw BE. HLA-DP in unrelated hematopoietic cell transplantation revisited: challenges and opportunities. Blood. 2017;130(9):1089-1096. [DOI] [PubMed] [Google Scholar]
- 54.Fleischhauer K, Beelen DW. HLA mismatching as a strategy to reduce relapse after alternative donor transplantation. Semin Hematol. 2016;53(2):57-64. [DOI] [PubMed] [Google Scholar]
- 55.Zino E, Frumento G, Marktel S, et al. . A T-cell epitope encoded by a subset of HLA-DPB1 alleles determines nonpermissive mismatches for hematologic stem cell transplantation. Blood. 2004;103(4):1417-1424. [DOI] [PubMed] [Google Scholar]
- 56.Petersdorf EW, Malkki M, O’hUigin C, et al. . High HLA-DP expression and graft-versus-host disease. N Engl J Med. 2015;373(7):599-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crocchiolo R, Zino E, Vago L, et al. ; Italian Bone Marrow Donor Registry . Nonpermissive HLA-DPB1 disparity is a significant independent risk factor for mortality after unrelated hematopoietic stem cell transplantation. Blood. 2009;114(7):1437-1444. [DOI] [PubMed] [Google Scholar]
- 58.Shimoni A, Labopin M, Lorentino F, et al. . Killer cell immunoglobulin-like receptor ligand mismatching and outcome after haploidentical transplantation with post-transplant cyclophosphamide. Leukemia. 2019;33(1):230-239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.