Abstract
Whole genome sequencing is replacing traditional microbiological typing methods for investigation of outbreaks in clinical settings. Here, we used a clinical microbiology laboratory core genome multilocus sequence typing (cgMLST) workflow to analyze 40 isolates of K. pneumoniae which are part of the Antimicrobial Resistance Leadership Group (ARLG) isolate collection, alongside 10 Mayo Clinic K. pneumoniae isolates, comparing results to those of pulsed-field gel electrophoresis (PFGE). Additionally, we used the whole genome sequencing (WGS) data to predict phenotypic antimicrobial susceptibility (AST). 31/40 ARLG K. pneumoniae isolates belonged to the same PFGE type, all of which, alongside three isolates of different PFGE types, formed a large cluster by cgMLST. PFGE and cgMLST were completely concordant for the 10 Mayo Clinic K. pneumoniae isolates. For AST prediction, the overall agreement between phenotypic AST and genotypic prediction was 95.6%.
Introduction
Klebsiella pneumoniae is a frequent pathogen in nosocomial settings, causing variety of infections, including hospital acquired pneumonia, bacteremia and urinary tract infection (1). An alarming increase in resistance to a broad array of antibiotics in this pathogen is now recognized, with several notable hospital-associated outbreaks reported (2–5). To definitively diagnose an outbreak, isolates need to be typed and shown to be genetically related. Various techniques are employed to type K. pneumoniae isolates to determine genetic relatedness. Now-unavailable methods like PCR electrospray ionization mass spectrometry (PCR/ESI-MS), alongside older methods such as pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST), are being replaced by whole genome sequence (WGS)-based typing methods. WGS data provides high-level differentiation amongst isolates of the same species and can also be used to predict phenotypic antimicrobial susceptibility (AST) (6, 7). WGS-based typing approaches rely on identification of single nucleotide polymorphisms (SNPs) across the entire genome, or genome-wide, gene-by-gene, allelic profiling of core genes or shared genes, known as core or whole genome multilocus sequence typing (cgMLST or wgMLST, respectively). cgMLST has been employed to differentiate outbreak and non-outbreak isolates of Acinetobacter baumannii, Listeria monocytogenes, Staphylococcus aureus, Legionella pneumophilia, Francisella tularensis, K. pneumoniae and Mycobacterium tuberculosis, among others (2, 8–14). Criteria for discrimination of outbreak from non-outbreak isolates (i.e., relatedness thresholds) are incompletely defined for SNP-based WGS analysis, cgMLST and wgMLST, and differ by species (8, 12, 15).
We described the use of cgMLST for clonality testing of S. aureus using an automated analytic system, SeqSphere+ (Ridom, Meunster, DE) (7, 16). Here, we applied the same computerized system to cgMLST analysis of previously characterized collections of K. pneumoniae, including outbreak-associated isolates, in comparison to PFGE. Additionally we used the WGS data to predict phenotypic resistance and susceptibility using the Acuitas® Whole Genome Sequence Analysis pipeline (OpGen, Inc., Gaithersburg, MD).
Materials and Methods
Bacterial isolates
Forty-two isolates from the ARLG and 10 from Mayo Clinic were studied (17). Two ARLG isolates were excluded; one failed to assemble after three sequencing attempts and another was a misidentified species - Escherichia coli - as shown by MALDI-TOF MS and phenotypic testing. ARLG isolates had been recovered from five institutions in the Eastern United States, and were previously described; they all harbored blaKPC, of one of two types, 2 or 3 (Table 1) (17). Most of the isolates were from Mount Sinai Medical Center in New York (MSMCNY) (n=24) or Cleveland Clinic (CC) (n=9), with the rest being from University of Pittsburg Medical Center (UPMC) (n=3), University Hospitals Case Medical Center (UHCMC) (n= 2) and the Louis Stokes Veterans Affairs Medical Center (LSVAMC) (n=2).
Table 1. Antibacterial Resistance Leadership Group (ARLG) Klebsiella pneumoniae isolates showing correlation between core genome multilocus sequence type (cgMLST) and pulsed-field gel electrophoresis type (17).
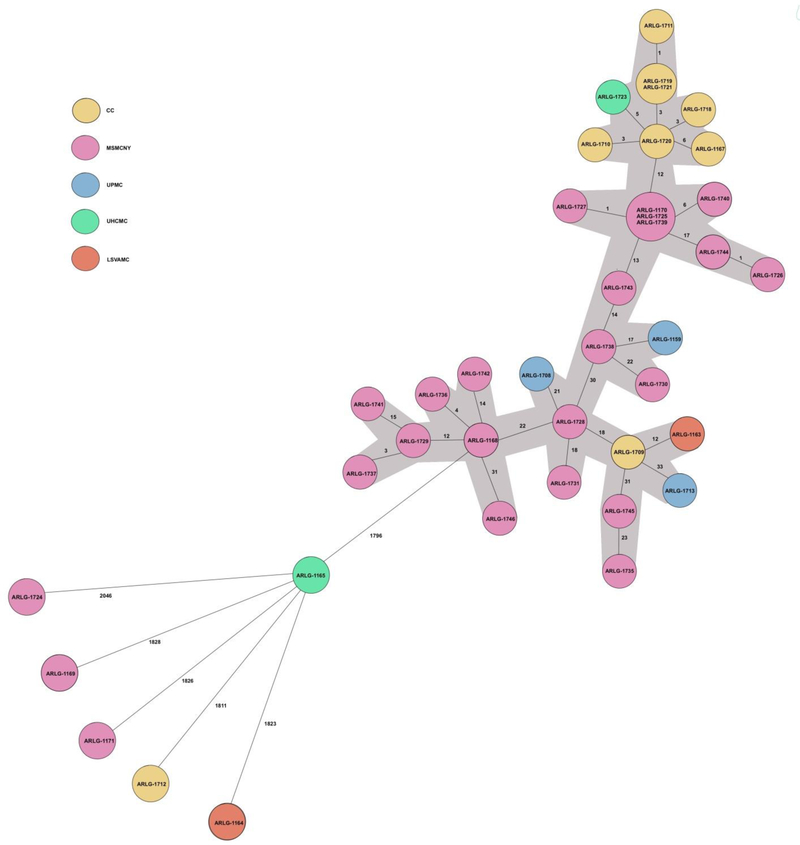
The multilocus sequence type (MLST) shown was based on whole genome sequence assemblies configured from SeqSphere+ independently of cgMLST prediction. A minimum spanning tree illustrating the cgMLST data is shown in Figure 1.
| ARLG Isolate Number | Isolate Designation | Isolate Source | blaKPC type | PFGE Group | cgMLST Group (Number of allelic differences used to assess relatedness shown) | cgMLST Type | MLST Type | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ≤6 | ≤15 | ≤50 | |||||||
| From reference (17) | This study | |||||||||
| ARLG-1168 | VA-397 | MSMCNY | 3 | A1 | None | D | 2 | A | 1089 | ST258 |
| ARLG-1736 | VA-402 | MSMCNY | 3 | A1 | None | D | 2 | A | 1089 | ST258 |
| ARLG-1742 | VA-410 | MSMCNY | 3 | A1 | None | None | 2 | A | 1089 | ST258 |
| ARLG-1741 | VA-409 | MSMCNY | 3 | A2 | None | None | 2 | A | 1089 | ST258 |
| ARLG-1746 | VA-416 | MSMCNY | 3 | A3 | None | None | None | A | 1145 | ST258 |
| ARLG-1728 | VA-392 | MSMCNY | 3 | A4 | None | None | None | A | 1107 | ST258 |
| ARLG-1729 | VA-394 | MSMCNY | 3 | A4 | None | E | 2 | A | 1089 | ST258 |
| ARLG-1735 | VA-401 | MSMCNY | 3 | A4 | None | None | None | A | 1110 | ST512 |
| ARLG-1737 | VA-403 | MSMCNY | 3 | A4 | None | E | 2 | A | 1089 | ST258 |
| ARLG-1745 | VA-414 | MSMCNY | 3 | A4 | None | None | None | A | 305 | ST258 |
| ARLG-1731 | VA-396 | MSMCNY | 2 | A5 | None | None | None | A | 1109 | ST258 |
| ARLG-1730 | VA-395 | MSMCNY | 2 | A7 | None | None | None | A | 1108 | ST258 |
| ARLG-1738 | VA-404 | MSMCNY | 2 | A8 | None | None | 1 | A | 1057 | Indeterminate |
| ARLG-1170 | VA-400 | MSMCNY | 2 | A9 | B | B | 1 | A | 335 | ST258 |
| ARLG-1725 | VA-389 | MSMCNY | 2 | A9 | B | B | 1 | A | 335 | ST258 |
| ARLG-1739 | VA-406 | MSMCNY | 2 | A9 | B | B | 1 | A | 335 | ST258 |
| ARLG-1740 | VA-408 | MSMCNY | 2 | A9 | None | B | 1 | A | 1144 | ST258 |
| ARLG-1727 | VA-391 | MSMCNY | 3 | A11 | None | B | 1 | A | 335 | ST258 |
| ARLG-1726 | VA-390 | MSMCNY | 2 | A12 | None | C | 3 | A | 1106 | ST258 |
| ARLG-1744 | VA-413 | MSMCNY | 2 | A13 | None | C | 3 | A | 1106 | ST258 |
| ARLG-1169 | VA-398 | MSMCNY | 3 | C2 | None | None | None | None | 1090 | ST278 |
| ARLG-1724 | VA-388 | MSMCNY | 3 | G | None | None | None | None | 1103 | ST1697 |
| ARLG-1743 | VA-412 | MSMCNY | 2 | G | None | None | 1 | A | 1057 | ST258 |
| ARLG-1171 | VA-417 | MSMCNY | 3 | H | None | None | None | None | 1099 | ST34 |
| ARLG-1167 | VA-384 | CC | 2 | A13 | None | A | 1 | A | 51 | ST258 |
| ARLG-1710 | VA-360 | CC | 2 | A13 | None | A | 1 | A | 51 | ST258 |
| ARLG-1711 | VA-361 | CC | 2 | A13 | None | A | 1 | A | 51 | ST258 |
| ARLG-1718 | VA-376 | CC | 2 | A13 | None | A | 1 | A | 51 | ST258 |
| ARLG-1719 | VA-378 | CC | 2 | A13 | A | A | 1 | A | 51 | ST258 |
| ARLG-1720 | VA-380 | CC | 2 | A13 | None | A | 1 | A | 51 | ST258 |
| ARLG-1721 | VA-383 | CC | 2 | A13 | A | A | 1 | A | 51 | ST258 |
| ARLG-1709 | VA-357 | CC | 2 | B2 | None | None | 4 | A | 204 | ST258 |
| ARLG-1712 | VA-362 | CC | 2 | D | None | None | None | None | 1101 | ST234 |
| ARLG-1723 | VA-387 | UHCMC | 2 | A13 | None | A | 1 | A | 51 | ST258 |
| ARLG-1165 | VA-375 | UHCMC | 3 | F | None | None | None | None | 1088 | ST133 |
| ARLG-1713 | VA-364 | UPMC | 2 | A6 | None | None | None | A | 1102 | ST258 |
| ARLG-1708 | VA-267 | UPMC | 3 | B1 | None | None | None | A | 1100 | ST258 |
| ARLG-1159 | VA-184 | UPMC | 2 | A10 | None | None | None | A | 237 | ST258 |
| ARLG-1163 | VA-367 | LSVAMC | 3 | A5 | None | None | 4 | A | 204 | ST258 |
| ARLG-1164 | VA-373 | LSVAMC | 2 | C1 | None | None | None | None | 1087 | ST268 |
MSMCNY, Mount Sinai Medical Center in New York City; UPMC, University of Pittsburgh Medical Center; UHCMC, University Hospitals Case Medical Center; CC, Cleveland Clinic; LSVAMC, Louis Stokes Veterans Affairs Medical Center
cgMLST
DNA was extracted and purified using the Quick-DNA™ Fungal/Bacterial Miniprep Kit (Zymo Research Corp.) and normalized to 0.2 ng/μL with a Quantus™ fluorometer (Promega, Madison, WI). Next generation sequencing libraries were prepared from normalized DNA with Nextera® XT (Illumina, San Diego, CA) and dual-indexed. Sequencing was performed on the MiSeq® platform (Illumina) using the V2 2 X 250 bp paired-end chemistry with a maximum pooling capacity of six libraries per flow cell, targeting a minimum coverage of 200X. Reads were processed for adapter and index cleaning utilizing the MiSeq reporter software in real-time. Adapter-clipped read files were imported into SeqSphere+ software version 4.1.9. Velvet de novo assembly and cgMLST were performed within the software suite following default settings. The SeqSphere+ embedded K. pneumoniae cgMLST schema was employed using K. pneumoniae genome NC_012731.1 as a reference genome derived from the analysis of 30 query genome sequences. Minimum spanning trees were generated from the final comparison data table. Grouping clusters obtained with cgMLST typing were compared to data generated by PFGE. In addition, MLST types were generated within the SeqSphere+ tool (18).
PFGE
ARLG isolates had been previously-subjected to PFGE (17). For the remaining 10 isolates, we performed PFGE according to the established method (19). Digestion of whole chromosomal DNA in agarose was performed with Xba I (Sigma-Aldrich St Louis, MO). The resulting restriction fragments were separated in CHEF Mapper® XA system (Bio-Rad Laboratories, Hercules, CA) with 1% agarose gel. The gels were then stained with ethidium bromide, illuminated under UV light and photographed in a Gel Doc™ XR+ Gel Documentation System (Bio-Rad).
Prediction of phenotypic AST
Susceptibility prediction were assessed using the 40 ARLG isolates. WGS assemblies were analyzed with the OpGen’s Acuitas® Whole Genome Sequence Analysis pipeline, a high-resolution tool which identifies antibiotic resistance genotypes using OpGen’s Antibiotic Resistance Genes Database which consists of approximately 2,400 curated antibiotic resistance genes from the following databases: the Antibiotic Resistance Genes Database (ARDB), the Comprehensive Antibiotic Resistance Database (CARD), Resfinder and the Lahey Clinic (20–22). After antibiotic resistance genes were determined by querying assembled whole genome sequences of K. pneumoniae isolates against OpGen’s Antibiotic Resistance Genes Database, a subset of antibiotic resistance genes were used as input for OpGen’s Acuitas Lighthouse® prediction models to predict antibiotic resistance/susceptibility phenotypes (23).
The isolates underwent phenotypic AST by agar dilution, with results interpreted according to CLSI guidelines (24). For the purposes of this study, isolates that tested susceptible by phenotypic AST were considered susceptible, as were those that were susceptible dose dependent (SDD) to cefepime, and those that were either resistant or intermediate were considered non-susceptible.
Results
PFGE types
PFGE had previously classified the ARLG isolates into 21 unique types including A (which was further subgrouped into A1-A13) and non-A (B, C, D, F, G, H, which were further subgrouped into B1, B2, C1 and C2) groups (Table 1) (17). Among Mayo Clinic isolates, 4 were unique, with the rest grouping into one of two groups of three indistinguishable isolates each (Table 2).
Table 2. Mayo Clinic Klebsiella pneumoniae isolates showing complete correlation between core genome multilocus sequence type (cgMLST) and pulsed-field gel electrophoresis type.
The multilocus sequence type (MLST) shown was based on whole genome sequence assemblies configured from SeqSphere+ independently of cgMLST prediction. A minimum spanning tree illustrating the cgMLST data is shown in the Supplemental Figure.
| Isolate Number | PFGE Group (indistinguishable) | cgMLST | MLST Type | |
|---|---|---|---|---|
| Type | Group (≤6 allelic differences) | |||
| BTP113 | Unique | 729 | Unique | ST14 |
| BTP114 | Unique | 1288 | Unique | ST147 |
| BTP115 | Group 2 | 1289 | Group 2 | ST231 |
| BTP116 | Group 1 | 1290 | Group 1 | ST101 |
| BTP117 | Group 1 | 1290 | Group 1 | ST101 |
| BTP118 | Unique | 1293 | Unique | ST307 |
| BTP119 | Group 1 | 1290 | Group 1 | ST101 |
| BTP120 | Unique | 1294 | Unique | ST14 |
| BTP121 | Group 2 | 1289 | Group 2 | ST231 |
| BTP122 | Group 2 | 1289 | Group 2 | ST231 |
MLST
Forty ARLG isolates were clustered into 8 MLST types and one indeterminate type. 7 MLST types (ST512, ST268, ST278, ST234, ST133, ST1697, ST34) were represented by single isolates, with the remaining 32 isolates (80%) belonging to ST258 (Table 1). Among Mayo Clinic isolates, 3 were ST231, 3 were ST101, 2 were ST14 and one each were ST147 and ST307 (Table 2).
cgMLST sequence types
The 40 ARLG isolates encompassed 25 unique cgMLST types with 37 unique allelic profiles. Thirty-four of the ARLG K. pneumoniae isolates formed a large cluster with a maximum cumulative distance (calculated by adding allelic differences between individual isolates) of 147 allelic differences between the most distant isolates by cgMLST. Six isolates were different from all isolates and from each other by ≥1796 allelic differences. For the ARLG isolates, four allelic relatedness thresholds were evaluated, 0, ≤6, ≤15 and ≤50 allelic differences.One pair and one triplet of isolates were indistinguishable. Nineteen isolates were classified into five unique clusters based on a relatedness threshold of ≤6 allelic differences; 25 into 4 unique clusters based on a relatedness threshold of ≤15 allelic differences (with two pairs of clusters collapsing to one and one new cluster appearing compared to groupings with a relatedness threshold of ≤6 allelic differences); and all 44 isolates comprising one large cluster based on a relatedness threshold of ≤50 allelic differences (Figure 1). 21/24 isolates from MSMCNY clustered together in the same group when an allelic threshold of ≤50 was used whereas with an allelic threshold of ≤15, fifteen isolates were grouped into three groups. Similarly all but one of the CC isolates (ARLG-1712) grouped together using an allelic threshold of ≤50. Even after using the allelic threshold of ≤15, 7/9 isolates clustered in same group, suggesting clonal relatedness.
Figure 1. Minimum spanning tree showing core genome multilocus sequence type of the 40 Antibacterial Resistance Leadership Group Klebsiella pneumoniae isolates.
Each isolate is designed with a unique number. Isolates in the same circle having no allelic differences between them. The lines between the circles show the numbers of allelic differences. Thirty-four isolates formed a large cluster (A) with a maximum cumulative distance of 147 allelic differences between the most distant isolates by cgMLST (colored gray), with the remaining isolates differing from the group and from each other by ≥1796 allelic differences.
CC, Cleveland Clinic; MSMCNY, Mount Sinai Medical Center in New York City; UPMC, University of Pittsburgh Medical Center; UHCMC, University Hospitals Case Medical Center; LSVAMC, Louis Stokes Veterans Affairs Medical Center
Of the 10 isolates obtained from Mayo Clinic, 6 grouped into 2 clusters, whereas 4 did not group into any specific cluster and differed from each other and the clustered isolates by ≥78 allelic differences (Supplemental Figure).
Correlation between PFGE, blaKPC-type, and cgMLST
There were four ARLG isolates with different PFGE types (D, F, G and H), that differed from each other and all other isolates by ≥1796 allelic differences, and had unique cgMLST and MLST types. The two PFGE C type isolates (C1 and C2) differed from each other by 3,651 allelic differences; these isolates were from different institutions and of different blaKPC types, suggesting that they were not related to one another. 31/40 ARLG K. pneumoniae isolates belonged to the same PFGE type (A with subtypes A1–13), all of which, alongside three isolates of different PFGE types, formed a large cluster with a maximum cumulative distance of 147 allelic differences (calculated by adding allelic differences between individual isolates) by cgMLST; this cluster (shown as Group 1 in Figure 1) included the four groups of isolates with ≤15 allelic differences. The three isolates in the large cluster of 34 isolates which were not PFGE type A included two which were PFGE type B (ARLG-1708, ARLG-1709) and one which was PFGE type G (ARLG-1743).
The two PFGE type B isolates differed from one another by 39 allelic differences, were from two institutions and of different blaKPC-types suggesting that cgMLST better differentiated them than did PFGE. There were 4 PFGE A9 isolates (ARLG-1170, ARLG1725, ARLG-1739, ARLG-1740), all from the same institution and of the same blaKPC type, of which three were indistinguishable by cgMLST, with the fourth having 6 allelic differences from the other three; the single PFGE A11 type (ARLG-1727) had only one allelic difference from the cluster of three PFGE A9 isolates (ARLG-1170, ARLG-1725 and ARLG-1739) and was from the same institution, but of a different blaKPC type. The single isolate of PFGE A12 (ARLG-1726) and one of the 9 PFGE A13 isolates (ARLG-1744) differed by a single allelic difference, and were from the same institution and of the same blaKPC type. The other 8 PFGE A13 isolates differed from ARLG-1744 by ≥29 allelic differences, and from each other by ≤10 allelic differences; all but one were from the same instution and were all of the same blaKPC type. The three PFGE A1 (ARLG-1168, ARLG-1736, ARLG- 1742) isolates were of the same blaKPC type and from the same instution; they differed from one another by ≤18 allelic differences. The single PFGE A10 isolate (ARLG-1159) had 17 allelic differes from the most closely related isolate, which was the only PFGE A8 isolate (ARLG-1738) and was from a different institution and of the same blaKPC type. There was one PFGE A7 isolate (ARLG-1730), and it differed from the most closely related isolate (ARLG-1738) by 22 allelic differences; the two were from the same instution and of the same blaKPC type. There was a single PFGE A2 isolate (ARLG-1741) which differed by 15 alleles from the most closely related isolate, a PFGE A4 isolate (ARLG-1729) from the same institution and of the same blaKPC type. ARLG-1729 (PFGE A4) was in turn 3 allelic differences away from another PFGE A4 isolate, ARLG-1737. However, the three other PFGE A4 isolates, ARLG-1728, ARLG-1745 and ARLG-1735 were more distantly separated by cgMLST; the last two differening from each other by 23 allelic differences and ARLG-1728 differing from ARLG-1729 by 34 allelic differences. There were two PFGE A5 isolates (ARLG-1163 and ARLG-1731) from two medical centers and of different blaKPC types, that differed from one another by 48 allelic differences. Finally, there was one PFGE A6 isolate (ARLG-1713), which differed from the most closely related isolate (ARLG-1709, which was from a different institution but of the same blaKPC type) by 33 allelic differences.
There was 100% concordance between PFGE and cgMLST groupings for Mayo Clinic isolates.
Correlation between phenotypic AST results and genetic prediction of phenotypic AST
Most isolates were resistant to multiple antibiotic classes. Susceptibility and non-susceptibility predictions were made for ampicillin, cefazolin, ceftriaxone, ceftazidime, cefepime, ertapenem, meropenem, aztreonam, gentamicin, tobramycin, trimethoprim-sulfamethoxazole (TMP-SMX), ciprofloxacin and levofloxacin. Table 3 summarizes the performance of WGS for prediction of phenotypic resistance in terms of the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and agreement of genotypic prediction with phenotypic AST. Agreement represents the correct prediction of phenotypic susceptibility or non-susceptibility. Overall agreement was 95.6% (confidence interval, 93.4– 97.2%). All isolates were resistant to cefazolin, ampicillin, ceftriaxone, ceftazidime, ertapenem, meropenem and aztreonam by phenotypic and genotypic AST. All, except one isolate, were resistant to cefepime. This isolate was dose dependent susceptible however was predicted to be resistant by genotypic testing. Overall agreement of phenotypic AST and genotypic prediction was >87% for all antibiotics, except gentamicin. For gentamicin there was a high false negativity rate (n=22, Table 4) with overall agreement of only 45%. In contrast, overall agreement for tobramycin was 100%.
Table 3:
Performance of genotypic resistance data for prediction of phenotypic antimicrobial resistance in Klebsiella pneumoniae.
| Antibiotic | Sensitivity (%) | Specificity (%) | % agreement (CI) | PPV (100%) | NPV (100%) |
|---|---|---|---|---|---|
| Ampicillin | 100 | N/A | 100 (91.2– 100) | 100 | N/A |
| Ceftriaxone | 100 | N/A | 100 (91.2– 100) | 100 | N/A |
| Ceftazidime | 100 | N/A | 100 (91.2–100) | 100 | N/A |
| TMP/SMX | 91.9 | 33.3 | 87.5 (73.2–95.8) | 94.4 | 25 |
| Ciprofloxacin | 97.3 | 66.6 | 95 (83.1–99.4) | 97.3 | 66.67 |
| Gentamicin | 24.1 | 100 | 45 (29.2–61.5) | 100 | 33.3 |
| Meropenem | 100 | N/A | 100 (91.2–100) | 100 | N/A |
| Levofloxacin | 100 | 100 | 100 (91.19–100) | 100 | 100 |
| Cefepime | 100 | N/A | 97.5 (86.8–99.9) | 97.5 | N/A |
| Tobramycin | 100 | 100 | 100 (91.2– 100) | 100 | 100 |
| Ertapenem | 100 | N/A | 100 (91.2– 100) | 100 | N/A |
| Cefazolin | 100 | N/A | 100 (91.2– 100) | 100 | N/A |
| Aztreonam | 100 | N/A | 100 (91.2– 100) | 100 | N/A |
| Total | 94.7 | 88 | 94.4 (92.0–96.2) | 99.4 | 45.8 |
PPV, positive predictive value; NPV, negative predictive value; CI, confidence intervals; N/A, not available
Table 4: Phenotypic antimicrobial susceptibility (AST) of Klebsiella pneumoniae isolates.
Gray blocks represent interpretative disagreement between phenotypic AST and genotypic susceptibility/non-susceptibility prediction whereas white blocks represent interpretative agreement.
| ARLG Isolate Number | β-lactamase resistance genes | AMP, CFZ, CAZ, CRO, MEM, ETP, ATM | FEP | Fluoroquinolone efflux pump genes, resistance genes and mutations | CIP | LVX | Aminoglycoside resistance genes | GEN | TOB | Trimethoprim resistance genes | Sulfonamide resistance genes | SXT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARLG-1168 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1A | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aph(3′)-Ia, aac(6′)-Ib, strA, strB | R | R | dfrA12, dfrA14 | sul1, sul2 | R |
| ARLG-1736 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aph(3′)-Ia, aac(6′)-Ib, strA, strB | R | R | dfrA12, dfrA14 | sul1, sul2 | R |
| ARLG-1742 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aacA4-8, aph(3′)-Ia, strA, strB | I | R | dfrA12, dfrA14 | sul1, sul2 | R |
| ARLG-1159 | blaKPC-2 blaSHV-11 | R | R | oqxA, aadA, gyrA S83I, parC S80I | R | R | aadA2, aac(6′)-Ib, aph(3′)-Ia | S | R | dfrA12 | sul1, sul3 | R |
| ARLG-1727 | blaKPC3, blaOXA-9, blaSHV-1, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aac(6′)-Ib, aph(3′)-Ia, strA, strB | R | R | dfrA12, dfrA14 | sul1, sul2 | R |
| ARLG-1726 | blaKPC-2, blaOXA-9, blaSHV-11, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aac(6′)-Ib | S | R | dfrA12 | sul1 | R |
| ARLG-1167 | blaKPC-2, blaOXA-9, blaSHV-1, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aacA4-8 | R | R | dfrA12 | sul1 | R |
| ARLG-1710 | blaKPC-2, blaOXA-9, blaSHV167 blaTEM-1 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aacA4-8 | R | R | dfrA12 | sul1 | R |
| ARLG-1711 | blaKPC-2, blaOXA-9, blaSHV-167, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aacA4-8 | R | R | dfrA12 | sul1 | R |
| ARLG-1718 | blaKPC-2, blaOXA-9, blaSHV-167, blaTEM-192, | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aacA4-8 | R | R | dfrA12 | sul1 | R |
| ARLG-1719 | blaKPC-2, blaOXA-9, blaSHV-11, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aacA4-8 | R | R | dfrA12 | sul1 | R |
| ARLG-1720 | blaKP-2, blaOXA-9, blaSHV-167, blaTEM-192, | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aacA4-8 | R | R | dfrA12 | sul1 | R |
| ARLG-1721 | blaKPC-2, blaOXA-9, blaSHV-12, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aacA4-8, aadA2 | R | R | dfrA12 | sul1 | R |
| ARLG-1723 | blaKPC-2, blaOXA-9, blaSHV-11, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aacA4-8 | I | R | dfrA12 | sul1 | R |
| ARLG-1744 | blaKPC-2, blaOXA-9, blaSHV-11, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aac(6′)-Ib | S | R | dfrA12 | sul1 | R |
| ARLG-1741 | blaKPC-3, blaSHV-11, blaOXA-9, blaTEM-1A | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadAl, aacA4-8, strA, strB, | I | R | dfrA14 | sul2 | R |
| ARLG-1746 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA1, aac(6′)-Ib, aph(3′)-Ia, strA, strB | R | R | dfrA14 | sul2 | R |
| ARLG-1728 | blaKPC-3, blaSHV-11, blaTEM-1A | R | R | oqxA, oqxB, gyrA S83I, parCS80I | R | R | aadA1, aadA2, aph(4)-Ia, aph(3′)-Ia, aac(3)-IVa | R | R | dfrA12 | sul1, sul3 | R |
| ARLG-1729 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1A | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA1, aacA4-8, strB, strA | I | R | dfrA14 | sul2 | R |
| ARLG-1735 | blaKPC-3, blaOXA-9, blaSHV-11 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aph(3′)-Ia, aac(6′)-Ib, aadA2 | S | R | dfrA12 | sul1 | R |
| ARLG-1737 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1A | R | R | oqxA, oqxB, gyrAS83I, parC S80I | R | R | aacA4-8, aadA1, strA, strB, | I | R | dfrA14 | sul2 | R |
| ARLG-1745 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aac(6′)-Ib, aph(3′)-Ia, | S | R | dfrA12 | sul1 | R |
| ARLG-1163 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-192 | R | R | oqxA, oqxB, qnrB19, gyrA S83I, parC S80I | R | R | aac(6′)-Ib, aph(3′)-Ia | S | R | dfrA12 | sul1 | R |
| ARLG-1731 | blaKPC-2, blaOXA-9, blaSHV-11, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aph(4)-Ia, aac(6′)-Ib, aac(3)-IVa | R | R | - | sul1, sul3 | S |
| ARLG-1713 | blaKPC-2, blaOXA-9, blaSHV-11, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aph(3′)-Ia, aacA4-8 | R | R | dfrA12 | sul1 | R |
| ARLG-1730 | blaKPC-2, blaOXA-9, blaCARB-2, blaSHV-12, | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aacA4, aadA2, aadB, aph(3′)-Ia | R | R | dfrA12 | sul1, sul2 | R |
| ARLG-1738 | blaKPC-2, blaSHV-11, blaOXA-9, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aph(3′)-Ia, aac(6′)-Ib | R | R | dfrA12 | sul1, sul3 | R |
| ARLG-1170 | blaKPC-2, blaOXA-9, blaSHV-11, blaTEM-192, | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA2, aac(6′)-Ib, aph(3′)-Ia | S | R | dfrA12 | sul1 | R |
| ARLG-1725 | blaKPC-2, blaOXA-9, blaSHV-12, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parCS80I | R | R | aadA2, aph(4)-Ia, aac(6′)-Ib, aph(3′)-Ia, aac(3)-IVa | I | R | dfrA12 | sul1, sul3 | R |
| ARLG-1739 | blaKPC-2, blaOXA-9, blaSHV-167, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parCS80I | R | R | aadA2, aph(4)-Ia, aac(6′)-Ib, aph(3′)-Ia, aac(3)-IVa | I | R | dfrA12 | sul1, sul3 | R |
| ARLG-1740 | blaKPC-2, blaOXA-9, blaSHV-167, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parCS80I | R | R | aadA2, aph(4)-Ia, aac(6′)-Ib, aph(3′)-Ia, aac(3)-IVa, | I | R | dfrA12 | sul1, sul3 | R |
| ARLG-1708 | blaKPC-3, blaOXA-9, blaSHV-12, blaTEM-1A | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aadA1, aac(6′)-Ib, strA, strB | R | R | dfrA14 | sul2 | R |
| ARLG-1709 | blaKPC-2, blaSHV-167, blaTEM-118 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aac(6′)-33 aac(6′)-Ib | I | R | - | sul1, sul2 | R |
| ARLG-1164 | blaKPC-2, blaSHV-167, | R | R | oqxA, oqxB, gyrA S83F | R | R | aadA6, aadB | S | S | dfrA14 | - | S |
| ARLG-1169 | blaKPC-3, blaSHV-5, blaTEM-1A, | R | SDD | oqxA, oqxB | S | S | aadA1 | S | S | dfrA1 | sul1 | R |
| ARLG-1712 | blaKPC-2, blaOXA-9, blaSHV-1, blaTEM-1A, | R | R | oqxA, oqxB, gyrA S83F | R | R | aadA1, aadB, aacA4-8 | R | R | dfrA18 | sul1 | R |
| ARLG-1165 | blaKPC-3, blaOXA-2, blaOXA-9, blaSHV-75, blaTEM-1A, blaCARB-2, blaFOX-5 | R | R | oqxA, oqxB, qnrA1 | S | S | aadA1, aadA2, aacA4-8, aadB, strA, strB | S | R | dfrA14, dfrA18 | sul1, sul2 | R |
| ARLG-1724 | blaKPC-3, blaOXA-9, blaTEM-1A, blaOKP-B | R | R | oqxA, oqxB, | R | S | aadA1, aac(6′)-Ib, strA, strB | S | R | dfrA14 | sul2 | R |
| ARLG-1743 | blaKPC-2, blaOXA-9, blaSHV-12, blaTEM-192 | R | R | oqxA, oqxB, gyrA S83I, parC S80I | R | R | aph(4)-Ia, aph(3′)-Ia, aac(6′)-Ib, aac(3)-IVa | R | R | - | sul1, sul3 | S |
| ARLG-1171 | blaKPC-3, blaOXA-9, blaSHV-26, blaTEM-1A | R | R | oqxA, oqxB, | S | S | aadA1, aacA4-8, strB, strA | I | R | dfrA14 | sul2 | R |
AMP, Ampicillin; CFZ, Cefazolin; CAZ, Ceftazidime; CRO, Ceftriaxone; FEP, Cefepime; MEM, Meropenem; ETP, Ertapenem; ATM, Aztreonam; CIP, Ciprofloxacin; LVX, Levofloxacin; SXT, Trimethoprim-sulfamethoxazole; GEN, Gentamicin; TOB, Tobramycin
β-lactam resistance
All isolates had two or more β-lactamase genes (Table 4). Thirty-three isolates had at least 4; four had 3, two had 2 and one had 7 β-lactamase genes. These genes included blaKPC-2, blaKPC-3, blaOXA-9, blaOXA-2, blaOKP-B, blaSHV-1, blaSHV-5, blaSHV-11, blaSHV-12, blaSHV-26, blaSHV-75, blaSHV-167, blaTEM-1, blaTEM-1A, blaTEM-118, and blaTEM-192. All 40 isolates were correctly found to be blaKPC-positive, with the type correlating with what had been previously determined using PCR and amplified product sequencing (17).
All but five isolates (ARLG-1159, ARLG-1164, ARLG-1169, ARLG-1709 and ARLG-1728) harbored blaOXA-9. The most common blaSHV gene was blaSHV-11, carried by 20 isolates, followed by blaSHV-167 and blaSHV-12 carried by 8 and 6 isolates, respectively. Twenty-two isolates had blaTEM-192.
ARLG-1719 and ARLG-1721, which were indistinguishable by cgMLST, had identical resistance gene profiles, whereas, ARLG-1725, ARLG-1739 and ARLG-1170, which were also indistinguishable by cgMLST, had the same β-lactamase genes with the exception of their blaSHV type.
Fluoroquinolone resistance
Fluoroquinolone resistance-associated efflux pumps, genes and gene mutations identified included oqxA and oqxB; qnrA1 and qnrB19; and gyrA (S83I, and S83F), and parC (S80I), respectively. All isolates harbored oqxA and oqxB with ARLG-1165 and ARLG-1163 additionally harboring qnrA1 and qnrB19, respectively. All isolates except four (ARLG-1165, ARLG-1169, ARLG-1171 and ARLG-1725) harboured a gyrA S83 mutation with gyrA S83I found in 34 isolates and gyrA S83F in two isolates (ARLG-1164 and ARLG-1712). Thirty four isolates carried parC S80I mutation as well.
All four isolates (ARLG-1165, ARLG-1169, ARLG-1171 and ARLG-1724) carrying the combination of oqxA, oqxB, without gyrA or parC mutations were susceptible to levofloxacin. All of these isolates, except ARLG-1724, were also susceptible to ciprofloxacin.
Aminoglycoside resistance
Aminoglycoside resitance genes detected included aac(3)-IVa, aac(6’)-33, aac(6’)-Ib, aacA4–8, aadA1, aadA2, aadA6, aadB, aph(3’)-Ia, and aph(4)-Ia. One isolate had six aminoglycoside resistance conferring elements; 9 had five, 9 had four, 7 had three, 13 had two and 1 had one. There was wide variation in phenotypic AST for gentamicin. Despite having the same resistance genes identified, phenotypic susceptibility was different for ARLG-1168 (gentamicin -resistant) and ARLG-1736 (gentamicin-susceptible) (Table 4). Both isolates were resistant and genotypically predicted to be resistant to tobramycin. All eight isolates (ARLG-1167, ARLG-1710, ARLG-1711, ARLG-1718, ARLG-1719, ARLG-1720, ARLG-1721) carrying aadA2 and aacA4–8 had non-susceptible phenotypes to tobramycin and gentamicin but were genotypically predicted to be susceptible to gentamicin.
Trimethoprim-sulfamethoxazole (TMP-SMX) resistance
Several TMP resitance genes were identified, including dfrA1, dfrA12, dfrA14 and dfrA18; the most common was dfrA12 which was present in 25 isolates, followed by dfrA14, which was present in 13 isolates. Three isolates had dfrA18, with one having dfrA1. All isolates except ARLG-1709, ARLG-1731 and ARLG-1743 carried at least one TMP resistance element. Five isolates had two TMP resistance-associated genes.
Sulfonamide resistance genes identified included sul1, sul2 and sul3 carried by 32, 14 and 8 isolates, respectively. All isolates except ARLG-1169 had at least one sulfonamide resistance gene. Fifteen isolates had two sulfonamide resistance elements identified. All three isolates (ARLG-1731, ARLG-1164, ARLG-1743) that were susceptible to TMP-SMX carried resistance elements to either sulfonamide or trimethoprim but not both.
Discussion
Herein, we compared WGS analysis using cgMLST to PFGE for typing of K. pneumoniae and found a high degree of correlation between the two typing methods. Additionally, we used the same WGS data to predict phenotypic AST and found a high degree of correspondence between the two.
cgMLST can be highly discriminatory, though the ideal allelic threshold used to define relatedness has been incompletely defined. Our data provides information to assist in the interpretation of cgMLST of K. pneumoniae. For ARLG isolates, using an allelic threshold cutoff of ≤15 allelic differences, cgMLST identified four clusters of 2 to 15 isolates and, 15 isolates that differed from all four groups and each other by >15 allelic differences. In contrast, PFGE identified 21 unique subtypes including A (which was further subgrouped into A1-A13 containing 1 to 9 isolates) and non-A (B, C, D, F, G, H, which were further subgrouped into B1, B2, C1 and C2) groups. Using a higher allelic threshold cutoff of ≤50, 34 isolates clustered into one group. At this higher threshold cutoff, results of cgMLST classification broadly matched those of MLST; all but two isolates (ARLG-1735 and ARLG-1745) from this large cluster grouped into MLST type ST258. ARLG-1735 grouped into MLST ST512 and had 23 allelic differences from the closest member of ST258 (ARLG-1745); the MLST type for ARLG-1738 was indeterminate. ARLG-1743 grouped into type G by PFGE which was a non-A PFGE group; by cgMLST (using allelic threshold cutoff of ≤15), it clustered with other isolates of PFGE type A13. The MLST type of this isolate was ST258 which matched the cgMLST grouping. ARLG-1743 had 3,908 allelic differences from ARLG-1724 which was also grouped into G by PFGE. This discrepancy may be secondary to mislabeling at the Mayo Clinic, the ARLG laboratory or the origin laboratory.
Accurate typing is essential for outbreak investigations. Previously used typing methods for K. pneumoniae have included PFGE, PCR/ESI-MS, MLST, and amplified fragment-length polymorphism (AFLP) analysis (25–28). PFGE is one of the most common methods employed for typing of bacteria and is based on digestion of chromosomal DNA with infrequently-cutting restriction endonucleases. The resulting DNA fragments take a specific pattern of discrete bands on the gel. These patterns are then compared with one another to determine relatedness. The procedure itself is standardized; however, the interpretation is not always straightforward. MLST involves sequencing of a small number of housekeeping genes within a bacterial genome. Since it only involves a few loci, it lacks the discriminatory power needed to analyze outbreaks (but is useful for broadly categorizing strains across time and geographical distributions). To distinguish between different isolates, genome-wide analysis should ideally be executed. The resolution capacity of WGS-based methods such as cgMLST is higher than historical methods. And, once the data is generated, it can be shared and utilized by distant laboratories for data assessment, providing unambiguous data comparison. Inter-laboratory standardization of sequencing methods, coverage and typing schemes is evolving.
To date, several studies have investigated cgMLST or wgMLST in outbreak situations, including investigation of K. pneumoniae outbreaks. In a Dutch study, wgMLST was used to identify an outbreak of NDM-1 producing K. pneumoniae (2). A total of 162 K. pneumoniae isolates underwent WGS; MLST and wgMLST were then performed using the WGS data with SeqSphere+. In addition, resistome and plasmid analysis were performed using an online tool ResFinder. Twenty-six of 162 isolates clustered into one group with a maximum allelic difference of 5. Interestingly, the investigators compared these isolates with unrelated isolates originating from five other health care centers and found 7 isolates that clustered with the 26 outbreak isolates with a maximum allelic difference of 5. This cluster had a distance of 2,926 genes from its nearest neighbor. All 33 isolates belonged to same MLST type (ST873), carried the same plasmid and had small differences in their resistomes. In an outbreak of colistin-resistant K. pneumoniae carbapenemase (KPC)-producing K. pneumoniae in the Netherlands that involved two institutions (a hospital and a nursing home), wgMLST was employed (14). During this outbreak, six patients were found to be colonized with KPC-producing K. pneumoniae, and four enviromental samples were positive for KPC-producing K. pneumoniae. Isolates were typed using AFLP and MLST. WGS followed by MLST+ typing was performed using an in-house scheme with 3,102 K. pneumoniae genes as a reference in SeqSphere+. Out of the 3,102 target genes, 3,042 were used in MLST+ typing as these were common to all six isolates. Three isolates of ten (6 patient and 4 environmental isolates), including two isolates from patients and one from the environment, were indistinguishable. Four, two and one isolate had 1, 5 and 2 allelic differences respectively from the three indistinguishable isolates. The maximum allelic difference between any two isolates was 10. MLST+ results in this study were found to be concordant with those of AFLP. All of these isolates belonged to the same MLST type, ST258. WGS data was also used for resistome analysis without making phenotypic predictions. In our study, the most common sequence type of K. pneumoniae was ST258, a hybrid sequence type deriving most of its genome (~80%) from ST11 (29). ST258 represents the majority of KPC-producing K. pneumoniae, has the ability to rapidly disseminate in healthcare settings, and is frequently associated with nosocomial outbreaks (30–32).
WGS can also been used for resistome analysis (2, 14, 33). To take it one step further, and make it clinically-useful, this data can then be utilitized to predict phenotypic AST. Thus far, WGS data has been used to predict phenotypic resistance for A. baumannii, E. coli, K. pneumoniae, S. aureus, and M. tuberculosis, among others (34–39). WGS data has additionally been evaluated for prediction of MIC with reported accuracy up to 92% (37). Overall agreement of phenotypic AST and genetic prediction was 95.6% for our isolates, similar to what has been described in previous studies (23, 35). For gentamicin however, the overall agreement was only 45%. There was a high rate of false negativity (phenotypic resistance, WGS-predicted susceptibility). In contrast, the overall agreement was 100% for tobramycin. The reason for this discrepancy is not clear. In a previous study employing the same platform for genetic prediction of AST for K. pneumoniae, the agreement between phenotypic and genotypic AST for gentamicin was 86% (23). Aminoglycoside resistance is mediated by several mechanisms, including drug modifying enzymes, target site mutations, drug efflux pumps and porin mutations (40). Drug modifying enzymes differentially modify diverse aminoglycosides, based on the presence of target sites on particular aminoglycosides. A limitation of the approach applied is incomplete knowledge of all resistance genes and mutations. In addition, the relationship between genotype and phenotype is not linear. In our study isolates carrying similar genes had different phenotypic resitance profiles. Multiple mutations and/or resistance genes may work in a non-additive manner (antagonist or synergistic) to confer resistance and one mutation may confer resistance to multiple antibiotics. Also, phenotypic AST, which is the current gold standard, is not free from errors and may be operator or instrument dependent, in addition to depending on breakpoints applied. All ARLG isolates were highly resistant to β-lactam antibiotics, an important limitation, as addressed below. A mixture of phenotypes would allow better evaluation of performance of genotypic susceptibiltiy prediction.
We have described cgMLST typing for K. pneumoinae isolates, and have compared this typing method to the traditionally used typing method of PFGE. We have also described relatedness with different cgMLST allelic thresholds for K. pneumoniae; however this is also one of the main limitations of our study as further data are needed to precisely pinpoint allelic threshold criteria for determining relatedness. Previous studies considered isolates with 5–10 allelic differences as being related (2, 14). Our report illustrates the potential of cgMLST as an epidemiological tool to replace PFGE and other traditional typing methods. We have also used the WGS data to predict phenotypic antimicrobial susceptibility with an overall agreement of 95.6%, though high rates of resistance in the study isolates limited evaluation of specificity and negative predictive values.
In conclusion, we evaluated a discriminatory and easy-to-use cgMLST scheme for WGS-based typing for K. pneumoniae. We have been routinely performing cgMLST analyses in our clinical microbiology laboratory for over two and a half years and have found it to be a technologist-friendly methodology. Here, we show that cgMLST correlates with PFGE for K. pneumoniae, appearing to be more discriminatory. cgMLST is a suitable method for investigating the relatedness of K. pneumoniae isolates encountered in outbreak scenarios. Additional benefits of cgMLST are the ability to easily compare nucleotide sequence data between laboratories. We also show that WGS data can predict phenotypic AST.
Supplementary Material
Supplemental Figure. Minimum spanning tree showing core genome multilocus sequence type of the 10 Mayo Clinic Klebsiella pneumoniae isolates. Each isolate is designed with a unique number and shown in a circle, with the lines between the circles showing the numbers of allelic differences between individual isolates. Isolates with ≤6 allelic differences from another isolate are shown as Groups 1 and 2 and colored gray.
Acknowledgements
We thank the Mayo Clinic Center for Individualized Medicine Microbiome Program for supporting this study. We thank Dag Harmsen for his thoughtful review of this manuscript. Dr. Patel is supported, in part, by UM1 AI104681. We thank the ARLG for providing isolates used in this study. We thank OpGen for providing the genotypic antimicrobial susceptibility prediction data. This study was also supported in part by funds the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) to R.A.B. under Award Numbers R01AI100560, R01AI063517, and R01AI072219, and the Cleveland Department of Veterans Affairs Award Number 1I01BX001974 to R.A.B. from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, and the Geriatric Research Education and Clinical Center VISN 10. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Department of Veterans Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch T, Lutgens SPM, Hermans MHA, Wever PC, Schneeberger PM, Renders NHM, Leenders A, Kluytmans J, Schoffelen A, Notermans D, Witteveen S, Bathoorn E, Schouls LM. 2017. Outbreak of NDM-1-producing Klebsiella pneumoniae in a Dutch hospital, with interspecies transfer of the resistance plasmid and unexpected occurrence in unrelated health care centers. J Clin Microbiol 55:2380–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woodford N, Tierno PM Jr., Young K, Tysall L, Palepou MF, Ward E, Painter RE, Suber DF, Shungu D, Silver LL, Inglima K, Kornblum J, Livermore DM. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York Medical Center. Antimicrob Agents Chemother 48:4793–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang T, Mariano N, Urban C, Colon-Urban R, Grenner L, Eng RH, Huang D, Dholakia H, Rahal JJ. 2007. Identification of carbapenem-resistant Klebsiella pneumoniae harboring KPC enzymes in New Jersey. Microb Drug Resist 13:235–39. [DOI] [PubMed] [Google Scholar]
- 5.Samra Z, Ofir O, Lishtzinsky Y, Madar-Shapiro L, Bishara J. 2007. Outbreak of carbapenem-resistant Klebsiella pneumoniae producing KPC-3 in a tertiary medical centre in Israel. Int J Antimicrob Agents 30:525–9. [DOI] [PubMed] [Google Scholar]
- 6.Junemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, von Haeseler A, Stoye J, Harmsen D. 2013. Updating benchtop sequencing performance comparison. Nat Biotechnol 31:294–6. [DOI] [PubMed] [Google Scholar]
- 7.Park KH, Greenwood-Quaintance KE, Uhl JR, Cunningham SA, Chia N, Jeraldo PR, Sampathkumar P, Nelson H, Patel R. 2017. Molecular epidemiology of Staphylococcus aureus bacteremia in a single large Minnesota medical center in 2015 as assessed using MLST, core genome MLST and spa typing. PLoS One 12:e0179003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moran-Gilad J, Prior K, Yakunin E, Harrison TG, Underwood A, Lazarovitch T, Valinsky L, Luck C, Krux F, Agmon V, Grotto I, Harmsen D. 2015. Design and application of a core genome multilocus sequence typing scheme for investigation of Legionnaires’ disease incidents. Euro Surveill 20. [DOI] [PubMed] [Google Scholar]
- 9.Kohl TA, Diel R, Harmsen D, Rothganger J, Walter KM, Merker M, Weniger T, Niemann S. 2014. Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J Clin Microbiol 52:2479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antwerpen MH, Prior K, Mellmann A, Hoppner S, Splettstoesser WD, Harmsen D. 2015. Rapid high resolution genotyping of Francisella tularensis by whole genome sequence comparison of annotated genes (“MLST+”). PLoS One 10:e0123298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins PG, Prior K, Harmsen D, Seifert H. 2017. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS One 12:e0179228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willems S, Kampmeier S, Bletz S, Kossow A, Kock R, Kipp F, Mellmann A. 2016. Whole-Genome Sequencing elucidates epidemiology of nosocomial clusters of Acinetobacter baumannii. J Clin Microbiol 54:2391–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weterings V, Zhou K, Rossen JW, van Stenis D, Thewessen E, Kluytmans J, Veenemans J. 2015. An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur J Clin Microbiol Infect Dis 34:1647–55. [DOI] [PubMed] [Google Scholar]
- 15.Neumann B, Prior K, Bender JK, Harmsen D, Klare I, Fuchs S, Bethe A, Zuhlke D, Gohler A, Schwarz S, Schaffer K, Riedel K, Wieler LH, Werner G. 2019. A Core Genome Multilocus Sequence Typing Scheme for Enterococcus faecalis. J Clin Microbiol 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham SA, Chia N, Jeraldo PR, Quest DJ, Johnson JA, Boxrud DJ, Taylor AJ, Chen J, Jenkins GD, Drucker TM, Nelson H, Patel R. 2017. Comparison of whole-genome sequencing methods for analysis of three methicillin-resistant Staphylococcus aureus outbreaks. J Clin Microbiol 55:1946–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endimiani A, Hujer AM, Perez F, Bethel CR, Hujer KM, Kroeger J, Oethinger M, Paterson DL, Adams MD, Jacobs MR, Diekema DJ, Hall GS, Jenkins SG, Rice LB, Tenover FC, Bonomo RA. 2009. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. J Antimicrob Chemother 63:427–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jolley KA, Maiden MC. 2010. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MA P Chromosomal restriction fragment analysis by pulsed-field gel electrophoresis. Application to molecular epidemiology, p 651–7, Essential Procedures for Clinical Microbiology. ASM Press, Washington, DC. [Google Scholar]
- 20.Liu B, Pop M. 2009. ARDB--Antibiotic Resistance Genes Database. Nucleic Acids Res 37:D443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O’Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker GT, Quan J, Higgins SG, Toraskar N, Chang W, Saeed A, Sapiro V, Pitzer K, Whitfield N, Lopansri BK, Motyl M, Sahm D. 2019. Predicting Antibiotic Resistance in Gram-Negative Bacilli from Resistance Genes. Antimicrob Agents Chemother 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CLSI. 2019. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 25.Mammina C, Palma DM, Bonura C, Anna Plano MR, Monastero R, Sodano C, Cala C, Tetamo R. 2010. Outbreak of infection with Klebsiella pneumoniae sequence type 258 producing Klebsiella pneumoniae carbapenemase 3 in an intensive care unit in Italy. J Clin Microbiol 48:1506–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van ‘t Veen A, van der Zee A, Nelson J, Speelberg B, Kluytmans JA, Buiting AG. 2005. Outbreak of infection with a multiresistant Klebsiella pneumoniae strain associated with contaminated roll boards in operating rooms. J Clin Microbiol 43:4961–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Zou MX, Wang HC, Dou QY, Hu YM, Yan Q, Liu WE. 2016. An outbreak of infections caused by a Klebsiella pneumoniae ST11 clone coproducing Klebsiella pneumoniae carbapenemase-2 and RmtB in a Chinese teaching hospital. Chin Med J (Engl) 129:2033–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su LH, Leu HS, Chiu YP, Chia JH, Kuo AJ, Sun CF, Lin TY, Wu TL. 2000. Molecular investigation of two clusters of hospital-acquired bacteraemia caused by multi-resistant Klebsiella pneumoniae using pulsed-field gel electrophoresis and in frequent restriction site PCR. Infection Control Group. J Hosp Infect 46:110–7. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Mathema B, Pitout JD, DeLeo FR, Kreiswirth BN. 2014. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio 5:e01355–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mezzatesta ML, Gona F, Caio C, Petrolito V, Sciortino D, Sciacca A, Santangelo C, Stefani S. 2011. Outbreak of KPC-3-producing, and colistin-resistant, Klebsiella pneumoniae infections in two Sicilian hospitals. Clin Microbiol Infect 17:1444–7. [DOI] [PubMed] [Google Scholar]
- 31.Tofteland S, Naseer U, Lislevand JH, Sundsfjord A, Samuelsen O. 2013. A long-term low-frequency hospital outbreak of KPC-producing Klebsiella pneumoniae involving Intergenus plasmid diffusion and a persisting environmental reservoir. PLoS One 8:e59015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mammina C, Bonura C, Di Bernardo F, Aleo A, Fasciana T, Sodano C, Saporito MA, Verde MS, Tetamo R, Palma DM. 2012. Ongoing spread of colistin-resistant Klebsiella pneumoniae in different wards of an acute general hospital, Italy, June to December 2011. Euro Surveill 17. [PubMed] [Google Scholar]
- 33.R DS, Pinto NA, Hwang I, Younjee H, Cho Y, Kim H, Yong D, Choi J, Lee K, Chong Y. 2017. Molecular epidemiology and resistome analysis of multidrug-resistant ST11 Klebsiella pneumoniae strain containing multiple copies of extended-spectrum beta-lactamase genes using whole-genome sequencing. New Microbiol 40:38–44. [PubMed] [Google Scholar]
- 34.Kumburu HH, Sonda T, van Zwetselaar M, Leekitcharoenphon P, Lukjancenko O, Mmbaga BT, Alifrangis M, Lund O, Aarestrup FM, Kibiki GS. 2019. Using WGS to identify antibiotic resistance genes and predict antimicrobial resistance phenotypes in MDR Acinetobacter baumannii in Tanzania. J Antimicrob Chemother 74:1484–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TE, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, Earle S, Pankhurst LJ, Anson L, de Cesare M, Piazza P, Votintseva AA, Golubchik T, Wilson DJ, Wyllie DH, Diel R, Niemann S, Feuerriegel S, Kohl TA, Ismail N, Omar SV, Smith EG, Buck D, McVean G, Walker AS, Peto TE, Crook DW, Iqbal Z. 2015. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun 6:10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis JJ, Boisvert S, Brettin T, Kenyon RW, Mao C, Olson R, Overbeek R, Santerre J, Shukla M, Wattam AR, Will R, Xia F, Stevens R. 2016. Antimicrobial Resistance Prediction in PATRIC and RAST. Sci Rep 6:27930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen M, Brettin T, Long SW, Musser JM, Olsen RJ, Olson R, Shukla M, Stevens RL, Xia F, Yoo H, Davis JJ. 2018. Developing an in silico minimum inhibitory concentration panel test for Klebsiella pneumoniae. Sci Rep 8:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long SW, Olsen RJ, Eagar TN, Beres SB, Zhao P, Davis JJ, Brettin T, Xia F, Musser JM. 2017. Population Genomic Analysis of 1,777 Extended-Spectrum Beta-Lactamase-Producing Klebsiella pneumoniae Isolates, Houston, Texas: Unexpected Abundance of Clonal Group 307. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magnet S, Blanchard JS. 2005. Molecular insights into aminoglycoside action and resistance. Chem Rev 105:477–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure. Minimum spanning tree showing core genome multilocus sequence type of the 10 Mayo Clinic Klebsiella pneumoniae isolates. Each isolate is designed with a unique number and shown in a circle, with the lines between the circles showing the numbers of allelic differences between individual isolates. Isolates with ≤6 allelic differences from another isolate are shown as Groups 1 and 2 and colored gray.