Abstract
Background
This study investigated changes in rumen protozoal and methanogenic communities, along with the correlations among microbial taxa and methane (CH4) production of six Belmont Red Composite beef steers fed tea seed saponins (TSS). Animals were fed in three consecutive feeding periods, a high-grain basal diet for 14 d (BD period) then a period of progressive addition of TSS to the basal diet up to 30 g/d for 20 d (TSS period), followed by the basal diet for 13 d without TSS (BDP post-control period).
Results
The study found that TSS supplementation decreased the amount of the protozoal genus Entodinium and increased Polyplastron and Eudiplodinium genera. During BDP period, the protozoa community of steers did not return to the protozoal profiles observed in BD period, with higher proportions of Metadinium and Eudiplodinium and lower Isotricha. The addition of TSS was found to change the structure of methanogen community at the sub-genus level by decreasing the abundance of methanogens in the SGMT clade and increasing the abundance of methanogens in the RO clade. The correlation analysis indicated that the abundance of SGMT clade methanogens were positively correlated with Isotricha, and Isotricha genus and SGMT clade methanogens were positively correlated with CH4 production. While RO clade were positively correlated with the proportion of Metadinium genus, which was negatively correlated with CH4 emission.
Conclusions
These results suggest that different genera of rumen protozoa ciliates appear to be selectively inhibited by TSS, and the change in methanogen community at the subgenus level may be due to the mutualistic relationships between methanogens and rumen ciliates.
Keywords: Beef steers, Methane, Methanogen, Protozoa, Tea seed saponin
Background
Specific ciliate protozoa colonize the rumen ecosystem of ruminant animals and contribute 40% ~ 50% of the rumen microbial biomass, which is almost equal to that of rumen bacteria [1]. Rumen protozoa play an important role in feed digestion and homeostasis of the rumen ecosystem due to the predator-prey relationship and the symbiotic relationship between rumen ciliates and prokaryotic cells [2]. Studies using electron microscopy and sequencing revealed the presence of methanogens inside and outside of rumen ciliates [3–5]. And the higher methanogens/bacteria ratio observed within the protozoa fractions suggested that anaerobic ciliates contain symbiotic methanogens, which strengthened the association between rumen ciliates and methanogens [6]. The strong functional association between rumen protozoa and methanogens via interspecies hydrogen transfer contributes to overall methane (CH4) production (9% ~ 37%) in the rumen as demonstrated by defaunation or elimination of certain protozoa [7]. Thus, there is increasing evidence about the relationship between protozoa and methanogenic archaea, and their important role in rumen (CH4) formation [7–9].
Many rumen protozoa contain an organelle (hydrogenosome) which produces hydrogen that attracts methanogens as endosymbionts [10, 11]. Hydrogenosomes have been identified in species of Epidinium, Isotricha and Dasytricha, but they were not found in Entodinium caudatum and Diploplastron affine [12]. Tymensen et al. [4] reported that Methanobrevibacter and Methanomicrobium genera are the major groups of methanogen in the free-living (FL) and protozoa-associated methanogen (PAM) fractions, but their population structure differed between FL and PAM pools. In addition, the populations of PAM were more variable than FL methanogens [5]. It has been found that rumen methanogens closely related to M. smithii were primarily associated with the vestibuliferid protozoa (Isotricha and Dasytricha) [13]. While Polyplastron multivesiculatum harbored a large number of intracellular bacteria, no methanogen was observed [3]. Methanogens also associate with protozoa as ectosymbionts which may be a more stable population compared with the internal dwelling methanogens [11]. Due to the apparent variability in association between methanogens and different protozoal taxa, it is hypothesized that some protozoa species may have a greater effect on rumen methanogenesis and CH4 production than others.
Some studies have shown that variation in CH4 emissions are not necessarily due to the changes in methanogen cell density, but rather the community structure of the methanogens [14, 15]. Phytochemicals such as saponins, tannins and essential oils, have been used to manipulate rumen microbial populations and reduce CH4 emissions from ruminants [16]. Tea seed (Camellia sinensis L.) saponins (TSS) were reported to reduce methane production by an inhibitory effect on ciliate protozoa [17–19]. Tannins, essential oils, and linseed oil are also inhibitory to protozoa, but species are impacted differently [20–22]. Our previous study in beef cattle supplemented with TSS showed reduction in CH4 emissions were associated with changes in methanogen population structure, while total protozoal numbers remained unchanged [23]. It remains to be determined if there was an alteration of rumen ciliate composition in response to supplemental TSS, which further modified the methanogens composition and CH4 emissions. The recent development of sequencing primers that can discriminate members of the rumen protozoal populations may provide an opportunity to characterize changes in the relative abundance of different protozoal taxa that previously could only be determined by microscopy [24].
Therefore to gain greater insight into the community structure of ruminal ciliate protozoa and their relationship to methanogen diversity and CH4 production, we employed next-generation sequencing techniques to investigate these populations in the rumen of beef steers fed a high grain-based diet supplemented with TSS.
Methods
Animals, diet and experimental design
Care of animals and experimental procedures were approved by the CSIRO Animal Ethic Committee protocol No A12/2012. This research is part of a larger study that has been previously reported and see Ramirez-Restrepo et al. [23] for more details. Briefly, six Belmont Red Composite [Africander (African Sanga) × Brahman (Bos indicus) × Hereford-Shorthorn (3/4 B. taurus)] rumen-cannulated steers [327 ± 17.1 kg, initial body weight] were used in a repeated measures design.
After free grazing, the steers were individually allowed to adjust to a conventional-finishing feedlot basal diet (BD) fed ad libitum in two equal parts at 09:00 and 16:30 for 14 d (control BD period). This was followed by supplementing the BD in the morning feeding with 6, 10, 15, 20, 25 and 30 g/d of TSS light-yellow color powder dissolved in ~ 250 mL cold water during 5, 2, 4, 3, 2 and 4 d, respectively (TSS period). After that, using only the BD, steers were fed for other 13 days (post-control period; BDP). The basal diet consisted of Rhodes grass (Chloris gayana; 0.15) and a sorghum high-grain mixture (0.85; Coleman Stock Feeds Pty Ltd., Charters Towers, Australia; Additional file 1: Table S1). The TSS per kg of commercial product (Zhejiang Oriental Tea Technology Co., Ltd., Changshan, China) included 570 g triterpenoid saponin, 330 g crude fiber, 54 g crude protein, 50 g ash, and 1 g water-insoluble matter [25]. Water was available ad libitum and steers were allowed to exercise daily in the cattle yard before the morning feeding to reduce stress, record experimental data and facilitate pen cleaning.
Methane emissions from all steers were measured in open-circuit respiration chambers for each period, and the results of the six cattle in our previous paper [23] showed that the average CH4 production in the BDP period (14.2 g/kg DMI) was significantly lower than the CH4 production in the BD period (18.0 g/kg DMI) and TSS period (18.7 g/kg DMI) respectively (P < 0.05). The lower CH4 emissions in BDP period could be due to the reestablishment of a microbial community that had lower CH4 producing potential than the controls.
Analysing protozoal and methanogenic communities
Rumen samples from each rumen-cannulated steer at the end of each feeding period were collected 2 h after the morning feeding. Raft and liquid components of the rumen content obtained from the dorsal and ventral sacks and were squeezed through two layers of cheese cloth. The fluid was immediately transferred to sterile containers placed on dry ice, and stored at − 80 °C for later DNA extraction.
Genomic DNA was extracted from 2 mL rumen fluid using the standard cetyl-trimethyl ammonium bromide (CTAB) method with some modifications to obtain higher concentrations and better purity of gDNA extracts from the rumen fluid [26]. The procedure used for DNA extraction is described in detail in our previous study [23] using bead beating in CTAB lysis buffer followed by phenol–chloroform separation. DNA extracts were dissolved in a 200 μL buffer EB, and the quality and quantity of the extracted DNA were determined by UV spectrophotometric analysis, using a NanoDrop ND-1000 Spectrophotometer (Nyxor Biotech, Paris, France).
The V3–V4 region of the protozoan 18S rRNA gene was PCR amplified using PSSU316F-GIC758R [24] barcoded primers set, and the V3–V4 region of the methanogen 16S rRNA gene was amplified using 915F-1386R [27] barcoded primers set. The two-step PCR amplification procedure with the barcoded primers (Additional file 2: Table S2) were used to obtain the PCR amplicons for paired-end sequencing on an Illumina MiSeq platform. The first-step PCR amplification setup consisted of 10 μL 5× Phusion HF buffer (including MgCl2), 1 μL dNTP (10 mmol/L), 1 μL inner primer (10 μmol/L), 20–50 ng of DNA template 1 U of Phusion High Fidelity DNA Polymerase (New England Biolabs, Beijing, China) and ddH2O up to 50 μL. Amplification was initiated with denaturation for 2 min at 94 °C, followed by 30 cycles of 94 °C for 30 s, annealing at 55 °C for protozoa or 56 °C for methanogen amplifying for 1 min and 30 s and extension at 72 °C for 30 s, with a final elongation for 2 min. After purification of PCR product, the cleaned PCR fragments were used in the second step PCR amplification. The 2nd-step PCR setup consisted of 8 μL HF buffer, 1 μL dNTP, 1 μL outer primer, 5 μL cleaned PCR product, 0.8 U of DNA Polymerase and ddH2O up to 40 μL. PCR cycles were as follows: 1 cycle at 94 °C for 2 min; 8 cycles at 94 °C for 30 s, 56 °C for 1.5 min and 72 °C for 30 s; and a final extension step at 72 °C for 2 min.
The purified barcoded amplicons were pooled in equimolar quantity to generate the Illumina library and quantified on Qubit 3.0 Fluorometer (Invitrogen, Waltham, USA). Sequencing was conducted on the Illumina multiplex 2 × 300-bp paired-end MiSeq-platform in TINY gene biotechnology Co. Ltd. (Shanghai, China). The sequencing procedure was followed as per Kozich et al. [28]. The raw sequencing data were submitted to the NCBI Sequence Read Archive (SRA), which are available from NCBI under BioProject accession number PRJNA574788 (https://www.ncbi.nlm.nih.gov/bioproject/ PRJNA574788) for both protozoal (n = 18) and methanogenic (n = 18) community datasets.
The paired-end reads were assembled by using Mothur software package [29], and then trimmed and quality-filtered in QIIME using default settings [30]. Briefly, Paired-end reads with ambiguous bases (n > 0), homologous regions (n ≥ 8), mismatches in the primer (n > 0), nontarget areas and errors in barcodes (n > 0) were culled, as well as sequences less than 200 bp, longer than 580 bp. Subsequently, chimeric sequences were removed by UCHIME [31]. And the quality-filtered sequences were clustered into operational taxonomic units (OTUs) for species classification with 96% similarity for ciliate protozoa [24]. An 18S rRNA gene sequence reference, providing the alignment for candidate sequences of ciliate protozoa, contained 131 sequences (≥1,500 bp in length) belonging to the gastrointestinal tract trichostome (subclass Trichostomatia) ciliates and downloaded from NCBI. The sequences of methanogen dataset were assigned to OTUs at a 3% species-level cutoff [32], with the RDP (Ribosomal Database Project) database as the reference. For OTUs grouping, only one or two reads were removed before taxonomic assignment. A good depth of coverage, community diversity indicators, the Shannon-Wiener index, Simpson’s index and community richness estimates, such as the Chao 1 index, were also calculated using Mothur and self-compiled employing program in BioLinker. Phylogenetic tree was constructed to visualize the phylogeny of dominant methanogen OTUs, representing sequences across all samples, using the neighbor-joining method in the MEGA6 [33].
Statistical analyses
All statistical analysis was performed using the R Package (version 3.5.1). Normality test of all the data was analyzed first. The normally distributed data, included OTU richness and diversity indices, were analyzed by using one-way analysis of variance (ANOVA) followed by the application of Tukey test for multiple comparison. The non-normally distributed data, taxonomic richness, were analyzed by using Kruskal-Wallis test to assess overall significant differences among the groups, followed by the post hoc Wilcoxon rank sum test to determine which paired groups differed from each other. Unless otherwise noted, the mean and pooled standard error of the mean (SEM) are reported. Spearman correlation analysis was used to assess microbial community interactions between protozoa and methanogen, as well as the relationship between microbial taxa abundance and CH4. Correlation matrix and graphics were generated using the online resource (https://www.omicshare.com/). P-values of < 0.05 for the overall significant differences and P-values of < 0.05 after FDR correction using the Benjamini–Hochberg procedure for the multiple comparison were considered significant.
Results
Effect on rumen protozoa communities
A total of 457,733 high-quality sequences mapped to protozoan 18S rRNA references generated 1,473 representative OTUs at 96% similarity, for an average of 870 OTUs per sample (range 593–1208). TSS supplementation reduced the number of observed protozoa OTUs and the species richness metrics, Chao 1 and ACE (FDR-P < 0.05; Table 1). This indicates a reduction in protozoal species richness for beef steers fed on a diet containing TSS. However, the alpha diversity indices of rumen protozoa, Shannon-Wiener and Simpson’s indices, were not influenced by TSS supplementation (P > 0.05, Table 1), so the reduction of species richness and similarity of alpha diversity indices with TSS indicated a shift in the evenness of protozoa taxa due to TSS. Good’s Coverage index showed good overall sampling and sequencing, ranging between 96.9% and 98.5% for rumen protozoa.
Table 1.
Effect of tea seed saponins (TSS) supplementation on OTU richness and alpha diversity of ciliate protozoa at 96% similarity
| Treatment | BD | TSS | BDP | SEM | ANOVA_P |
|---|---|---|---|---|---|
| Observed OTUs | 999a | 771b | 840ab | 37 | 0.354 |
| Shannon-Wiener | 3.04 | 3.32 | 2.98 | 0.08 | 0.173 |
| Simpson’s | 0.13 | 0.10 | 0.14 | 0.01 | 0.458 |
| Chao 1 | 2,699a | 1,805b | 2,162ab | 125 | 0.005 |
| Ace | 4,223a | 2,599b | 3,186ab | 226 | 0.004 |
| Good’s coverage, % | 97.78 | 98.02 | 97.68 | 0.13 | 0.613 |
Within the same row, values with a different letter are significantly different (FDR-P < 0.05)
ANOVA_P values were the overall significant differences analyzed using one-way analysis of variance for the normally distributed data
BD Basal Diet, BDP Post-control period, SEM Standard error of the mean
Across all samples, taxonomic classification based on the observed OTUs at the family and genus level resulted in the identification of 3 families and 10 genera of protozoa. The families Ophryoscolecidae and Isotrichidae accounted for 85.9% and 8.7% of the sequences, respectively (Additional file 3: Figure S1A). Entodinium (44.1%), Metadinium (10.5%), Eudiplodinium (5.9%) and Polyplastron (4.7%) were the dominant genera identified in the family Ophryoscolecidae. Another two genera in the family Isotrichidae, Isotricha (8.0%) and Dasytricha (0.7%) were also identified in this study (Additional file 3: Figure S1B). And the community composition of rumen protozoa at the order, family and genus level among three periods are shown in Additional file 3: Figure S2. At the order and family level, less relative abundance of Entodiniomorphida and Ophryoscolecidae were observed in TSS supplementation steers (FDR-P < 0.05) compared with BDP period, while the abundance of Vestibuliferida order and Isotrichidae family in BDP period were much lower than that of BD and TSS periods (FDR-P < 0.05, Table 2). At the genus level, TSS supplementation significantly reduced the abundance of Entodinium (FDR-P < 0.05), while significantly increased the abundance of the Polyplastron genus (FDR-P < 0.05, Table 2). Meanwhile, the abundance of Eudiplodinium in BDP period was higher than the BD period (FDR-P < 0.05). As for the Metadinium genus, its abundance in BDP period was higher than that in other two periods (FDR-P < 0.05), but the abundance of Isotricha in BDP period was lower (FDR-P < 0.05, Table 2).
Table 2.
Effect of tea seed saponins (TSS) on the protozoal communities (% of total sequences)
| Assigned taxa | BD | TSS | BDP | SEM | KW_P |
|---|---|---|---|---|---|
| O-Entodiniomorphida | 83.94ab | 70.50b | 94.96a | 3.51 | 0.007 |
| F-Ophryoscolecidae | 83.73ab | 70.25b | 94.76a | 3.54 | 0.007 |
| G-Entodinium | 52.44a | 29.45b | 39.72ab | 4.11 | 0.009 |
| G-Metadinium | 8.32b | 6.67b | 17.43a | 1.97 | 0.048 |
| G-Polyplastron | 1.72b | 9.10a | 6.22ab | 1.13 | 0.017 |
| G-Eudiplodinium | 2.46b | 7.62ab | 9.71a | 1.14 | 0.030 |
| G-Ostracodinium | 1.00 | 2.81 | 2.08 | 0.60 | 0.806 |
| G-Ophryoscolex | 0.66 | 4.18 | 0.98 | 0.85 | 0.463 |
| O-Vestibuliferida | 12.78a | 13.74a | 2.99b | 2.03 | 0.007 |
| F-Isotrichidae | 12.78a | 13.73a | 2.99b | 2.03 | 0.008 |
| G-Isotricha | 11.82a | 12.74a | 2.83b | 1.91 | 0.008 |
| G-Dasytricha | 0.95 | 0.97 | 0.41 | 0.16 | 0.211 |
| O-Unclassified | 3.32b | 15.86a | 2.07b | 2.25 | 0.011 |
Values within the same row with different letters are significant different (FDR-P < 0.05)
KW_P values were the overall significant differences analyzed using Kruskal-Wallis test for the non-normally distributed data
BD Basal Diet, BDP Post-control period, SEM Standard error of the mean. O Order, F Family, G Genus
Effect on rumen methanogenic communities
A total of 370,065 high-quality sequences were mapped to methanogen 16S rRNA references and generated 1,441 representative OTUs at 97% similarity, for an average of 459 OTUs per sample. The number of observed methanogen OTUs in the rumen of beef steers fed on a basal diet was not affected by TSS supplementation, while higher ACE index for rumen methanogens was observed in TSS supplementation steers compared with BDP period (FDR-P = 0.08, Table 3). However, the alpha diversity indices of rumen methanogen, Shannon-Wiener and Simpson’s indices, were not affected by TSS supplementation (Table 3).
Table 3.
Effect of tea seed saponins (TSS) supplementation on OTU richness and alpha diversity of methanogen at 97% similarity
| Treatment | BD | TSS | BDP | SEM | ANOVA_P |
|---|---|---|---|---|---|
| Observed OTUs | 447 | 490 | 443 | 12 | 0.167 |
| Shannon-Wiener | 2.13 | 2.40 | 2.45 | 0.10 | 0.391 |
| Simpson’s | 0.36 | 0.31 | 0.31 | 0.02 | 0.691 |
| Chao 1 | 1251 | 1335 | 1175 | 48 | 0.421 |
| Ace | 1761 | 2073 | 1672 | 78 | 0.083 |
| Good’s coverage, % | 98.28 | 97.05 | 96.81 | 0.23 | 0.211 |
Within the same row, values with different letters were significantly different (FDR-P < 0.05)
ANOVA_P values were the overall significant differences analyzed using one-way analysis of variance for the normally distributed data
BD Basal Diet, BDP Post-control period, SEM Standard error of the mean
Across all samples, the methanogen OTUs annotated to 2 families and 5 genera (Table 4). Methanobacteriaceae and Methanobrevibacter spp. were the predominant family and genera of methanogen in the rumen of steers during the three periods, accounting for more than 98% of the sequences. The abundance of methanogens at the family and genus levels were not significantly affected by TSS supplementation, while the abundance of Methanosphaera in BDP steers tended to be higher than in the BD period (FDR-P = 0.08; Table 4).
Table 4.
Effect of tea seed saponins (TSS) on the methanogen communities (% of total sequences)
| Assigned taxa | BD | TSS | BDP | SEM | KW_P |
|---|---|---|---|---|---|
| O-Methanobacteriales | 99.42 | 98.55 | 99.55 | 0.22 | 0.440 |
| F-Methanobacteriaceae | 99.42 | 98.55 | 99.55 | 0.22 | 0.440 |
| G-Methanobrevibacter | 99.06 | 98.09 | 98.94 | 0.22 | 0.338 |
| G-Methanosphaera | 0.04b | 0.10ab | 0.14a | 0.02 | 0.010 |
| G-Methanosarcina | 0.01 | 0.01 | 0.01 | 0.00 | 0.892 |
| O-Thermoplasmatales | 0.51 | 1.29 | 0.37 | 0.20 | 0.446 |
| G-Candidatus Methanomethylophilus | 0.51 | 1.29 | 0.37 | 0.20 | 0.446 |
| O-Methanomicrobiales | 0.03 | 0.06 | 0.03 | 0.01 | 0.854 |
| F-Methanomicrobiaceae | 0.03 | 0.06 | 0.03 | 0.01 | 0.854 |
| G-Methanomicrobium | 0.03 | 0.05 | 0.03 | 0.01 | 0.720 |
Within the same row, values with a different letter were significantly different (FDR-P < 0.05)
KW_P values were the overall significant differences analyzed using Kruskal-Wallis test for the non-normally distributed data
BD Basal Diet, BDP Post-control period, SEM Standard error of the mean. O Order, F Family, G Genus
Effect on the dominant methanogen OTUs
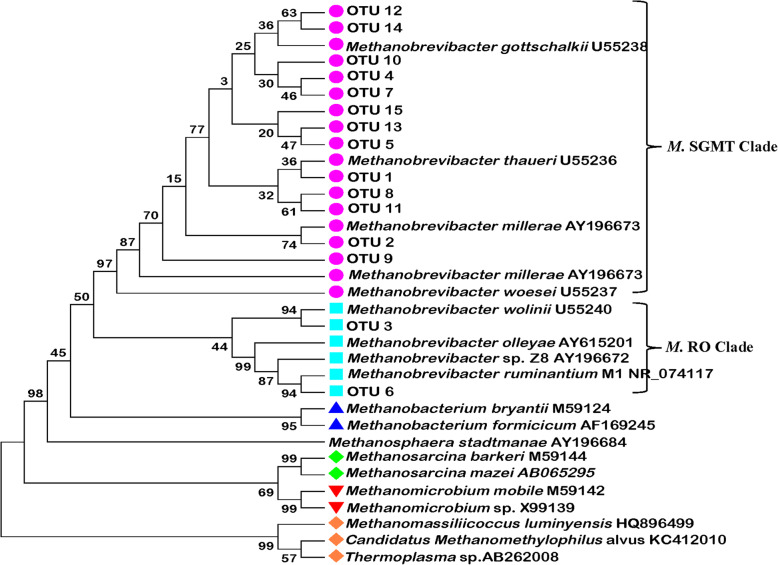
There were 15 dominant methanogen OTUs, averaging greater than 1% of total methanogen sequences across all samples, were used to construct the phylogenetic tree (Fig. 1). The results showed that 86.7% (13 OTUs) of the methanogen OTUs were located in the SGMT clade of the genus Methanobrevibacter (M. smithii, M. gottschalkii, M. millerae and M. thauerii), only 13.3% (2 OTUs) were located in the RO cluster of the genus Methanobrevibacter (M. ruminentium and M. olleyae).
Fig. 1.
Phylogenetic relationships between the 15 dominant methanogenic OTUs of all steers and Methanobrevibacter spp. The sequences of 15 dominant methanogenic OTUs obtained from 16S rRNA illumina library prepared from the community DNA of rumen of Belmont Red Composite steers, and the reference sequences of the Methanobrevibacter genus obtained from GenBank. The phylogenetic tree was constructed using the Neighbor-Joining method with the Kimura two-parameter model
The two predominant OTUs, OTU 1 and OTU 2, both representing approximately 30% of all sequences, were located in the SGMT clade of the Methanobrevibacter genus. The relative abundances of the 15 dominant methanogen OTUs were compared among the three feeding periods (Table 5). Compared with BD period, TSS supplementation significantly reduced the relative abundance of OTU 4 and OTU 7 (FDR-P < 0.05), while increased the proportion of OTU 5 and OTU 6 (FDR-P < 0.05). However, the abundance of SGMT clade and OTU 1 in BDP period was much lower than that in the BD period (FDR-P < 0.05), while the proportion of RO clade of the Methanobrevibacter genus and OTU 6 in BDP period was higher than BD period (FDR-P < 0.05).
Table 5.
Effect of tea seed saponins (TSS) on the abundance of dominant methanogen clade/OTUs
| Methanogen clade/OTUs | BD | TSS | BDP | SEM | KW_P |
|---|---|---|---|---|---|
| SGMT clade | 74.95a | 62.15ab | 42.14b | 4.16 | 0.005 |
| OTU 1 | 20.97a | 32.23a | 9.14b | 2.78 | 0.009 |
| OTU 2 | 14.75 | 9.25 | 8.05 | 1.09 | 0.172 |
| OTU 4 | 10.78a | 0.88b | 4.68ab | 1.21 | 0.017 |
| OTU 5 | 2.06b | 13.99a | 4.07b | 1.45 | 0.002 |
| OTU 7 | 10.91a | 0.12b | 2.18ab | 1.40 | 0.001 |
| Other 8 OTUs | 15.49a | 5.68b | 14.01ab | 1.80 | 0.027 |
| RO clade | 11.72b | 17.99ab | 35.52a | 3.07 | 0.009 |
| OTU 3 | 7.63 | 9.70 | 20.87 | 1.77 | 0.164 |
| OTU 6 | 4.09b | 8.29a | 14.65a | 1.66 | 0.002 |
SGMT clade: M. smithii, M. gottschalkii, M. millerae and M. thauerii, RO clade: M. ruminentium and M. olleyae
Other 8 OTUs are the OTU 8, OTU 9, OTU 10, OTU 11, OTU 12, OTU 13, OTU 14, OTU 15
In the same row, values with different letter mean significant difference (FDR-P < 0.05)
BD Basal Diet, BDP Post-control period, SEM Standard error of the mean
KW_P values were the overall significant differences analyzed using Kruskal-Wallis test for the non-normally distributed data
Within- and between-domain associations among protozoa and methanogen
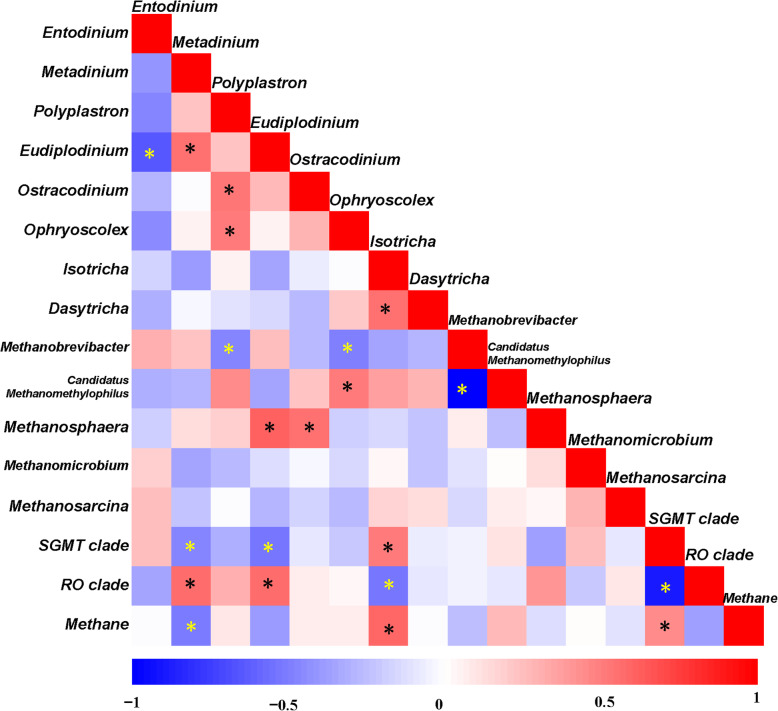
Pearson’s correlations indicated significant associations among protozoa genera, and between protozoa genera and methanogen taxa. Within ciliate protozoa, positive correlations were observed between the abundance of Metadinium and Eudiplodinium (r = 0.55; P = 0.02), between Isotricha and Dasytricha (r = 0.55; P = 0.02), as well as between Polyplastron genus with Ostracodinium and Ophryoscolex (r = 0.54 and 0.51, respectively; P < 0.05), and strong negative correlation was noticed between Entodinium and Eudiplodinium (r = − 0.66; P = 0.01; Fig. 2). In relation to protozoa-methanogen interactions, negative correlations were observed between the Methanobrevibacter genus with Polyplastron and Ophryoscolex (r = − 0.48 and − 0.49, respectively; P < 0.05). While positive correlations occurred between the Methanosphaera genus with Eudiplodinium and Ostracodinium (r = 0.62 and 0.56, respectively; P < 0.05), as well as between the Methanomethylovorans genus with Polyplastron and Ophryoscolex (r = 0.48 and 0.54, respectively; P < 0.05; Fig. 2). More significant correlations between the abundance of protozoal genera and the two clades of the genus Methanobrevibacter. SGMT clade was positively correlated with Isotricha (r = 0.51; P = 0.03), and negatively with Eudiplodinium and Eudiplodinium (r = − 0.49 and 0.51, respectively; P = 0.03), the RO clade was positively correlated with Metadinium and Eudiplodinium (r = 0.57 and 0.56, respectively; P < 0.05), and negatively with Isotricha (r = − 0.51; P = 0.01; Fig. 2).
Fig. 2.
Correlation matrix between protozoal taxa with methanogens and methane. Positive correlations are shown in red and negative correlations in blue. Color intensity represents r-values of correlation, ∗indicates P < 0.05. The scale of the colors is denoted as follows: the more positive the correlation (closer to 1), the darker the shade of red; the more negative the correlation (closer to − 1), the darker the shade of blue. ∗indicates P < 0.05
Correlations between microbial taxa with rumen CH4 emission
The yield of CH4 per unit of DMI (dry matter intake) was positively correlated with the relative abundance of Isotricha genus (r = 0.59, respectively; P < 0.05), and negatively correlated with the genus Metadinium (r = − 0.50; P < 0.05). As for methanogens, only SGMT clade of the genus Methanobrevibacter was positively correlated with CH4 production (r = 0.46; P = 0.05), while no significant correlation between CH4 emissions and methanogen abundance at the genus and family level were observed (Fig. 2).
Discussion
Rumen microbial community is shaped by complex relationships between bacteria, ciliate protozoa, fungi and archaea. Previous studies have shown that TTS as a feed additive can potentially mitigate CH4 emissions from ruminants through their inhibitory effect on protozoa, which subsequently may alter the abundance/diversity of methanogens [18, 34]. In the present study, TSS supplementation reduced the protozoal diversity metrics, but diversity still remained high throughout the study. We speculate that the high value diversity metrics is probably linked with the high-concentrate diet (concentrate to forage, 75:25) used, which can promote the growth of rumen ciliates [35, 36]. Furthermore, the high 18S rRNA gene copies number for large entodiniomorphids protozoa can partially explain the high diversity of rumen protozoa in this study [37]. In this study, the rumen ciliates were taxonomically assigned to 3 protozoan families and 10 genera. The genus Entodinium spp. was the predominant genus in the most dominant family, Ophryoscolecidae, comprising up to 85% of the total ciliate population, which is in agreement with other studies with cattle and sheep [27, 38]. In the present study, the TSS supplementation in a grain-based diet modified the community composition of rumen ciliate protozoa. The relative abundance of the family Ophryoscolecidae and the Entodinium genus in particular were reduced by TSS, however, concurrently there was an increase in the relative proportion of Polyplastron and Eudiplodinium genera, which demonstrated that there was a significant selective inhibitory effect of TSS on the ciliate protozoa community at the family and genus level. This selective inhibitory effect of TSS on rumen protozoa could be due to the physical or chemical differences in the cilia, the pellicle or metabolic activity of rumen ciliates. The concentration and composition of ciliate protozoa in the rumen can be influenced by diet type, pH, supplements, concentrate and roughage level [35, 39]. The most consistent effect of tea saponin supplementation is a decrease in the abundance of rumen protozoa in vitro [40], as well as in the rumen of goats and sheep [17, 18].
Most Ophryoscolecidae ciliates seem to be able to harbor both intracellular and extracellular methanogens, whereas only endosymbiont methanogens have been observed with the Isotrichidae ciliates [41]. In addition, previous studies have reported that Entodinium spp. and Isotricha spp. contain intracellular methanogens, whereas Polyplastron multivesiculatum harbored mainly intracellular bacteria [42]. In the present study, after the withdrawal of TSS supplementation, the ciliate community structure was also dissimilar with a higher proportion of Metadinium, Polyplastron and Eudiplodinium and lower Isotricha, which may also affect other rumen microbial communities due to the predator-prey relationship and the symbiotic relationship between rumen ciliates and prokaryotes (rumen bacteria and methanogens) [43]. The CH4 production in the rumen is significantly influenced by the protozoa population due to inter-species transfer of hydrogen from the protozoa to methanogens [44]. It has also been suggested that the number and activity of endosymbiotic population of methanogens are greater than the ectosymbionts attached to the surface of protozoa [45]. In most cases, the variation in CH4 emissions, caused by the addition of plant secondary compounds or microbial modifiers, could not be attributed to differences in methanogen cell density, but rather the composition of the methanogen community [46]. These results suggest that the shift of the ciliate community composition may result in dissimilarity in methanogen community that could vary in their potential for CH4 production.
In this study, the Methanobacteriaceae and Methanobrevibacter spp. were the predominant family and genus of methanogen in the rumen of steers throughout the trial, which is consistent with the previous findings [47, 48]. We suggest that this may be related to the high-concentrate diet used in our study. As Hook et al. [35] reported, approximately 99.3% of methanogen clones were assigned to the genus Methanobrevibacter in the rumen of cattles fed 65% concentrate diet. Furthermore, Hook et al. [35] and Franzolin [36] found that a high-concentrate diet promoted the abundance of protozoa. And other studies have found that the Methanobrevibacter was the predominant genus associated with protozoa [4, 5]. Therefore, the high density of rumen protozoa may contribute to the dominance of Methanobrevibacter in the rumen of catle fed the high-concentrate diet. St-Pierre et al. [46] observed that Methanobrevibacter related methanogen 16S rRNA gene sequences are mainly distributed between two large clades: SGMT clade, closely related to M. smithii, M. gottschalkii, M. millerae and M. thaueri, and RO clade, closely related to M. ruminantium and M. olleyae. The phylogenetic tree of the 15 dominant methanogen OTUs in this study showed that 86.7% methanogen OTUs, accounted for more than 70% of the sequences, and were located in the SGMT clade of the genus Methanobrevibacter with only 13.3% OTUs located in the RO clade. A re-examining report compared the sequence distribution between the SGMT clade and the RO clade and found that methanogens in the SGMT clade were distinctively more highly represented than the RO clade in impalas, wallabies, Holstein dairy cows and water buffaloes [44], which is consistent with the results of the present study. In the current study, the lower abundance of SGMT clade and higher proportion of RO clade were observed after the withdrawal of TSS supplementation, which was also associated with the lower proportion of Isotricha and higher Metadinium. What’s more, a positive correlation was observed between SGMT clade methanogens and Isotricha, as well as between the RO clade for Metadinium. Therefore, changes in the structure and composition of protozoan community should alter the structure of the methanogen community at the subgenus level.
The correlations between microbial taxa with CH4 emissions in the present study showed that CH4 production was positively correlated with Isotricha, but negatively correlated with Metadinium. This finding is supported by a previous study which reported that CH4 production and methanogen population in sheep inoculated with Isotrichidae ciliates were similar to the total protozoa of inoculated sheep [49]. The correlation analysis also showed that CH4 production was positively correlated with the abundance of SGMT clade methanogens. The results indicated that the shift in the methanogen community profile at the subgenus level were more effective in producing CH4, which could help explain the lower amount of CH4 emissions in the steers after withdrawing TTS from the diet, with a reduction in predominant methanogen species (SGMT clade methanogens) and increase in minor species (RO clade methanogens).
Conclusions
Adding TSS in the basal diet of beef steers caused significant alterations in the rumen protozoa community structure at the family and genus level, as well as rumen methanogen community structure. After withdrawing TSS, the ciliate community of steers did not return to the same protozoa community prior to TSS supplementation, and the new structure of the rumen microbial community maybe shaped by complex relationship between ciliate protozoa and methanogenc archaea. A number of protozoa genera and methanogen species were found to be correlated with CH4 emissions. Therefore, we speculated that decreasing the abundance of protozoa belonging to the Isotricha genus, and increasing members of the Metadinium genus, are likely to reduce CH4 emissions.
Supplementary information
Additional file 1 Table S1. Table S1 Composition and nutrient concentration of the basal diet.
Additional file 2 Table S2. The primers used in the construction of Illumina sequencing library.
Additional file 3 Figure S1. Relative abundance of ruminal protozoa genera of steers at the family level (A) and the genus level (B).
Additional file 3 Figure S2. The community composition of rumen protozoa at the order (A), family (B) and genus (C) level among three periods.
Acknowledgements
We would like to thank scholarships from the Chinese Scholarship Council (CSC) awarded to Cui Tan for 12 months of cooperative research at CSIRO. We also thank Jagadish. Padmanabha for assistance on sample analyzing.
Abbreviations
- TSS
Tea seed saponins
- BD
Basal diet
- BDP
Basal diet post
- OTU
Operational taxonomic unit
- SGMT
M.smithii, M.gottschalkii, M.millerae and M.thauerii
- RO
M.ruminentium and M.olleyae
Authors’ contributions
CT, ZW and CM designed the experiments. CT and CR performed the experiments. CT, AMS and RH analyzed the experimental data. CT, CR and CM wrote the draft manuscript, ZW, MB, CR and CM reviewed and edited the manuscript. The author(s) read and approved the final manuscript.
Funding
This study was supported by grants from the Natural Science Foundation of China (31272472), and project grants from Meat & Livestock Australia, Fisheries and Forestry in Australia and Commonwealth Scientific and Industrial Research Organization (CSIRO).
Availability of data and materials
All sequencing data are available in the NCBI Sequence Read Archive (SRA), under the bioproject number PRJNA574788.
Ethics approval and consent to participate
Care of animals and experimental procedures were carried out following the approved CSIRO Animal Ethic Committee protocol No A12/2012.
Consent for publication
Not applicable.
Competing interests
All the authors declare they have no competing interest.
Contributor Information
Cui Tan, Email: tancuiwbl@126.com.
Carlos A. Ramírez-Restrepo, Email: c.ramirez@creac.com.au
Ali Mujtaba Shah, Email: alimujtabashah@sbbuvas.edu.pk.
Rui Hu, Email: 876377758@qq.com.
Matt Bell, Email: matt.bell@nottingham.ac.uk.
Zhisheng Wang, Email: wangzs1968@gmail.com.
Chris McSweeney, Email: Chris.Mcsweeney@csiro.au.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40104-020-00491-w.
References
- 1.Sylvester JT, Karnati SKR, Yu Z, Morrison M, Firkins JL. Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J Nutr. 2004;134:3378–3384. doi: 10.1093/jn/134.12.3378. [DOI] [PubMed] [Google Scholar]
- 2.Newbold CJ, de la Fuente G, Belanche A, Ramos-Morales E, McEwan NR. The role of ciliate protozoa in the rumen. Front Microbiol. 2015;6:1313. doi: 10.3389/fmicb.2015.01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lloyd D, Williams AG, Amann R, Hayes AJ, Durrant L, Ralphs JR. Intracellular prokaryotes in rumen ciliate protozoa: detection by confocal laser scanning microscopy after in situ hybridization with fluorescent 16S rRNA probes. Eur J Protistol. 1996;32:523–531. [Google Scholar]
- 4.Tymensen LD, Beauchemin KA, McAllister TA. Structures of free-living and protozoa-associated methanogen communities in the bovine rumen differ according to comparative analysis of 16S rRNA and mcrA genes. Microbiology. 2012;158:1808–1817. doi: 10.1099/mic.0.057984-0. [DOI] [PubMed] [Google Scholar]
- 5.Belanche A, de la Fuente G, Newbold CJ. Study of methanogen communities associated with different rumen protozoal populations. FEMS Microbiol Ecol. 2014;90:663–677. doi: 10.1111/1574-6941.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ushida K, Newbold CJ, Jouany JP. Interspecies hydrogen transfer between the rumen ciliate Polyplastron multivesiculatum and Methanosarcina barkeri. J Gen Appl Microbiol. 1997;43:129–131. doi: 10.2323/jgam.43.129. [DOI] [PubMed] [Google Scholar]
- 7.Hook SE, Wright A-DG, McBride BW. Methanogens: methane producers of the rumen and mitigation strategies. Archaea. 2010;2010:945785. doi: 10.1155/2010/945785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malmuthuge N, Guan LL. Understanding host-microbial interactions in rumen: searching the best opportunity for microbiota manipulation. J Anim Sci Biotechnol. 2017;8:8. doi: 10.1186/s40104-016-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace RJ, Snelling TJ, McCartney CA, Tapio I, Strozzi F. Application of meta-omics techniques to understand greenhouse gas emissions originating from ruminal metabolism. Genet Sel Evol. 2017;49:9. doi: 10.1186/s12711-017-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finlay BJ, Esteban G, Clarke KJ, Williams AG, Embley TM, Hirt RP. Some rumen ciliates have endosymbiotic methanogens. FEMS Microbiol Lett. 1994;117:157–161. doi: 10.1111/j.1574-6968.1994.tb06758.x. [DOI] [PubMed] [Google Scholar]
- 11.Patra A, Park T, Kim M, Yu Z. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J Anim Sci Biotechnol. 2017;8:13. doi: 10.1186/s40104-017-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis JE, Lindmark DG, Williams AG, Lloyd D. Polypeptides of hydrogenosome-enriched fractions from rumen ciliate protozoa and trichomonads: immunological studies. FEMS Microbiol Lett. 1994;117:211–216. doi: 10.1111/j.1574-6968.1994.tb06767.x. [DOI] [PubMed] [Google Scholar]
- 13.Ohene-Adjei S, Teather RM, Ivan M, Forster RJ. Postinoculation protozoan establishment and association patterns of methanogenic archaea in the ovine rumen. Appl Environ Microbiol. 2007;73:4609–4618. doi: 10.1128/AEM.02687-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi W, Moon CD, Leahy SC, Kang D, Froula J, Kittelmann S, et al. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 2014;24:1517–1525. doi: 10.1101/gr.168245.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramos-Morales E, Rossi G, Cattin M, Jones E, Braganca R, Newbold CJ. The effect of an isoflavonid-rich liquorice extract on fermentation, methanogenesis and the microbiome in the rumen simulation technique. FEMS Microbiol Ecol. 2018;94:fiy009. doi: 10.1093/femsec/fiy009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamra DN, Pawar M, Singh B. Effect of plant secondary metabolites on rumen methanogens and methane emissions by ruminants. In: Patra KA, editor. Dietary phytochemicals and microbes. Dordrecht: Springer Netherlands; 2012. pp. 351–370. [Google Scholar]
- 17.Mao HL, Wang JK, Zhou YY, Liu JX. Effects of addition of tea saponins and soybean oil on methane production, fermentation and microbial population in the rumen of growing lambs. Livest Sci. 2010;129:56–62. [Google Scholar]
- 18.Zhou YY, Mao HL, Jiang F, Wang JK, Liu JX, McSweeney CS. Inhibition of rumen methanogenesis by tea saponins with reference to fermentation pattern and microbial communities in Hu sheep. Anim Feed Sci Technol. 2011;166:93–100. [Google Scholar]
- 19.Wang JK, Ye JA, Liu JX. Effects of tea saponins on rumen microbiota, rumen fermentation, methane production and growth performance: a review. Trop Anim Health Prod. 2012;44:697–706. doi: 10.1007/s11250-011-9960-8. [DOI] [PubMed] [Google Scholar]
- 20.Makkar HPS, Blümmel M, Becker K. In vitro effects of and interactions between tannins and saponins and fate of tannins in the rumen. J Sci Food Agric. 1995;69:481–493. [Google Scholar]
- 21.Patra AK, Kamra DN, Agarwal N. Effect of spices on rumen fermentation, methanogenesis and protozoa counts in in vitro gas production test. Int Congr Ser. 2006;1293:176–179. [Google Scholar]
- 22.Benchaar C, Romero-Perez GA, Chouinard PY, Hassanat F, Eugene M, Petit HV, Cortes C. Supplementation of increasing amounts of linseed oil to dairy cows fed total mixed rations: effects on digestion, ruminal fermentation characteristics, protozoal populations, and milk fatty acid composition. J Dairy Sci. 2012;95:4578–4590. doi: 10.3168/jds.2012-5455. [DOI] [PubMed] [Google Scholar]
- 23.Ramirez-Restrepo CA, Tan C, O'Neill CJ, Lopez-Villalobos N, Padmanabha J, Wang J, et al. Methane production, fermentation characteristics, and microbial profiles in the rumen of tropical cattle fed tea seed saponin supplementation. Anim Feed Sci Technol. 2016;216:58–67. [Google Scholar]
- 24.Ishaq SL, Wright A-DG. Design and validation of four new primers for next-generation sequencing to target the 18s rRNA genes of gastrointestinal ciliate protozoa. Appl Environ Microbiol. 2014;80:5515–5521. doi: 10.1128/AEM.01644-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez-Restrepo CA, O'Neill CJ, Lopez-Villalobos N, Padmanabha J, Wang J, McSweeney CS. Effects of tea seed saponin supplementation on physiological changes associated with blood methane concentration in tropical Brahman cattle. Anim Prod Sci. 2016;56:457–465. [Google Scholar]
- 26.Minas K, McEwan NR, Newbold CJ, Scott KP. Optimization of a high-throughput CTAB-based protocol for the extraction of qPCR-grade DNA from rumen fluid, plant and bacterial pure cultures. FEMS Microbiol Lett. 2011;325:162–169. doi: 10.1111/j.1574-6968.2011.02424.x. [DOI] [PubMed] [Google Scholar]
- 27.Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, Gordon JI, et al. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One. 2013;8:e47879. doi: 10.1371/journal.pone.0047879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin W, Meng Z, Wang J, Cheng Y, Zhu W. Effect of nitrooxy compounds with different molecular structures on the rumen methanogenesis, metabolic profile, and methanogenic community. Curr Microbiol. 2017;74:891–898. doi: 10.1007/s00284-017-1261-7. [DOI] [PubMed] [Google Scholar]
- 33.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu WL, Wu YM, Liu JX, Guo YQ, Ye JA. Tea saponins affect in vitro fermentation and methanogenesis in faunated and defaunated rumen fluid. J Zhejiang Univ Sci B. 2005;6:787–792. doi: 10.1631/jzus.2005.B0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hook SE, Steele MA, Northwood KS, Wright ADG, McBride BW. Impact of high-concentrate feeding and low ruminal pH on methanogens and protozoa in the rumen of dairy cows. Microb Ecol. 2011;62:94–105. doi: 10.1007/s00248-011-9881-0. [DOI] [PubMed] [Google Scholar]
- 36.Franzolin R, Dehority BA. Effect of prolonged high-concentrate feeding on ruminal protozoa concentrations. J Anim Sci. 1996;74:2803–2809. doi: 10.2527/1996.74112803x. [DOI] [PubMed] [Google Scholar]
- 37.Firkins JL, Yu Z, Park T, Plank JE. Extending Burk Dehority's perspectives on the role of ciliate protozoa in the rumen. Front Microbiol. 2020;11:123. doi: 10.3389/fmicb.2020.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kittelmann S, Devente SR, Kirk MR, Seedorf H, Dehority BA, Janssen PH. Phylogeny of intestinal ciliates, including Charonina ventriculi, and comparison of microscopy and 18s rRNA gene pyrosequencing for rumen ciliate community structure analysis. Appl Environ Microbiol. 2015;81:2433–2444. doi: 10.1128/AEM.03697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goel G, Makkar HPS, Becker K. Effects of Sesbania sesban and Carduus pycnocephalus leaves and fenugreek (Trigonella foenum-graecum L.) seeds and their extracts on partitioning of nutrients from roughage- and concentrate-based feeds to methane. Anim Feed Sci Technol. 2008;147:72–89. [Google Scholar]
- 40.Guo YQ, Liu JX, Lu Y, Zhu WY, Denman SE, McSweeney CS. Effect of tea saponin on methanogenesis, microbial community structure and expression of mcrA gene, in cultures of rumen micro-organisms. Lett Appl Microbiol. 2008;47:421–426. doi: 10.1111/j.1472-765X.2008.02459.x. [DOI] [PubMed] [Google Scholar]
- 41.Ushida K. Symbiotic methanogens and rumen ciliates. In: JHP H, editor. (Endo) symbiotic methanogenic archaea. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010. pp. 25–34. [Google Scholar]
- 42.Irbis C, Ushida K. Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J Gen Appl Microbiol. 2004;50:203–212. doi: 10.2323/jgam.50.203. [DOI] [PubMed] [Google Scholar]
- 43.Park T, Yu Z. Do ruminal ciliates select their preys and prokaryotic symbionts? Front Microbiol. 2018;9:1710. doi: 10.3389/fmicb.2018.01710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Deng Q, Liu Y, Yan T, Li F, Cao Y, et al. Dynamics of methanogenesis, ruminal fermentation and fiber digestibility in ruminants following elimination of protozoa: a meta-analysis. J Anim Sci Biotechnol. 2018;9:89. [DOI] [PMC free article] [PubMed]
- 45.Morgavi DP, Forano E, Martin C, Newbold CJ. Microbial ecosystem and methanogenesis in ruminants. Animal. 2010;4:1024–1036. doi: 10.1017/S1751731110000546. [DOI] [PubMed] [Google Scholar]
- 46.St-Pierre B, Cersosimo LM, Ishaq SL, Wright A-DG. Toward the identification of methanogenic archaeal groups as targets of methane mitigation in livestock animals. Front Microbiol. 2015;6:776. doi: 10.3389/fmicb.2015.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Z, Meng Q, Li S, Jiang L, Wu H. Effect of urea-supplemented diets on the ruminal bacterial and archaeal community composition of finishing bulls. Appl Environ Microbiol. 2017;101:6205–6216. doi: 10.1007/s00253-017-8323-4. [DOI] [PubMed] [Google Scholar]
- 48.Carberry CA, Waters SM, Kenny DA, Creevey CJ. Rumen methanogenic genotypes differ in abundance according to host residual feed intake phenotype and diet type. Appl Environ Microbiol. 2014;80:586–594. doi: 10.1128/AEM.03131-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belanche A, de la Fuente G, Newbold CJ. Effect of progressive inoculation of fauna-free sheep with holotrich protozoa and total-fauna on rumen fermentation, microbial diversity and methane emissions. FEMS Microbiol Ecol. 2015;91:fiu026. doi: 10.1093/femsec/fiu026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Table S1. Table S1 Composition and nutrient concentration of the basal diet.
Additional file 2 Table S2. The primers used in the construction of Illumina sequencing library.
Additional file 3 Figure S1. Relative abundance of ruminal protozoa genera of steers at the family level (A) and the genus level (B).
Additional file 3 Figure S2. The community composition of rumen protozoa at the order (A), family (B) and genus (C) level among three periods.
Data Availability Statement
All sequencing data are available in the NCBI Sequence Read Archive (SRA), under the bioproject number PRJNA574788.