Abstract
The eukaryotic 5’ untranslated region (UTR) is critical for ribosome recruitment to the mRNA and start codon choice, and plays a major role in the control of translation efficiency and shaping the cellular proteome. The ribosomal initiation complex is assembled on the mRNA via a cap-dependent or cap-independent mechanism. We describe various mechanisms controlling ribosome scanning and initiation codon selection by 5’ upstream open reading frames (ORFs), translation initiation factors, and primary and secondary structures of the 5’UTR, including particular sequence motifs. We also discuss translational control via phosphorylation of eIF2, which is implicated in learning and memory, neurodegenerative diseases and cancer.
The scanning mechanism of translation initiation
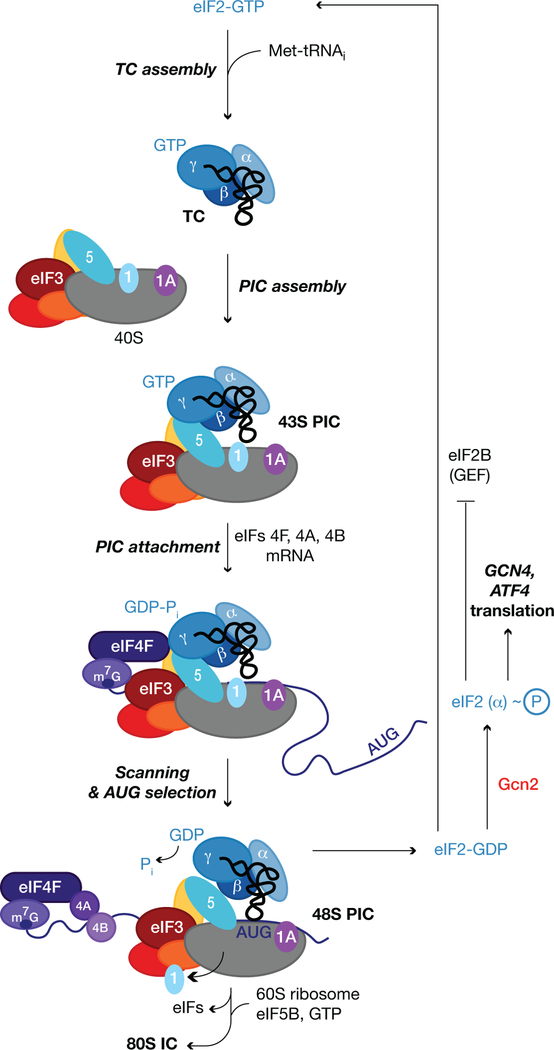
Most eukaryotic mRNAs are translated by the scanning mechanism, which begins with assembly of a 43S preinitiation complex (PIC), containing methionyl-initiator tRNA (Met-tRNAi) in a ternary complex (TC) with GTP-bound eukaryotic initiation factor 2 (eIF2). 43S PIC assembly is stimulated by eIFs 1, 1A, 3, and 5 (Fig. 1). Its subsequent attachment to mRNA at the m7G-capped 5’ end is facilitated by the eIF4F complex, comprised of cap-binding protein eIF4E, eIF4G, and RNA helicase eIF4A, and by poly(A)-binding protein (PABP). The PIC scans the mRNA 5’ untranslated region (UTR) for an AUG start codon using complementarity with the anticodon of Met-tRNAi. AUG recognition evokes hydrolysis of the GTP bound to eIF2 to produce a stable 48S PIC. Release of eIF2-GDP is followed by joining of the large (60S) ribosome subunit, catalyzed by eIF5B, to produce an 80S initiation complex ready to begin protein synthesis (Fig. 1) (1). There are exceptions to the scanning mechanism in which PICs are recruited by specialized sequences in the 5’UTR, called internal ribosome entry sites (IRESes).
Fig. 1. The scanning mechanism of translation initiation.
The simpler 5-subunit version of budding yeast eIF3 is depicted. See text for details.
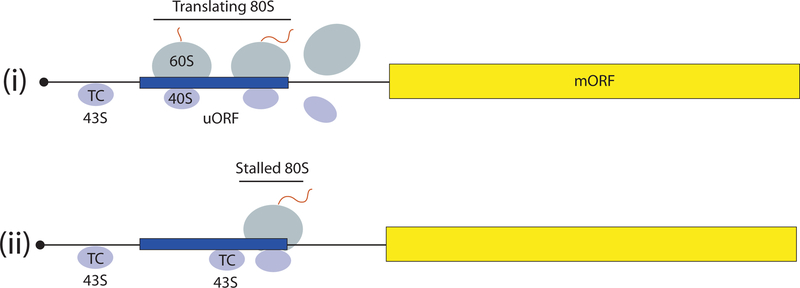
The nature of scanning, its 5’ to 3’ directionality, dictates that the initiation codon is frequently the AUG triplet closest to the 5’end, encountered first by the scanning PIC. The first AUG can be skipped when it is flanked by an unfavorable sequence—a process termed “leaky scanning”—to use a downstream AUG. A favorable sequence context in mammals is the “Kozak consensus”, 5’ (A/G)CCAUGG 3’ (2). While not always the same in plants and fungi, a purine at the −3 position from the AUG is both conserved and functionally predominates over other positions in all organisms (1). When an upstream AUG (uAUG) is in-frame with a downstream AUG without an intervening stop codon, leaky scanning may occur at some frequency to allow production of two protein isomers differing only by an N-terminal extension, with the longer form often targeted to a particular cellular compartment. If the uAUG is followed by a stop codon in the same ORF, then translation of the upstream ORF (uORF) will attenuate translation of the downstream main ORF, because reinitiation is generally inefficient (Fig. 2-i). Some uORFs inhibit downstream translation primarily because ribosomes stall during their translation and create a roadblock to scanning PICs that bypass the uORF start codon (Fig. 2-ii). These principles account for the fact that polycistronic mRNAs, common in bacteria, are rare in eukaryotes (1).
Fig. 2. Mechanisms of translational control by short uORFs.
(i) 43S PICs scanning from the mRNA 5’end translate the uORF (as 80S ribosomes) and free subunits dissociate from the mRNA following termination, preventing translation of the main ORF (mORF). (ii) 80S ribosomes are stalled during elongation or termination by the uORF-encoded attenuator peptide and impose a barrier to scanning 43S PICs that leaky-scan the uORF start codon, preventing translation of the mORF. Stalling is modulated by small molecules (see text).
“Near-cognate” triplets, differing from AUG by a single base can be selected by the scanning PIC, but with lower frequencies, owing to the mismatch with the anticodon of tRNAi and attendant destabilization of the 48S PIC. NUG (N is any nucleotide) triplets generally function the best, while A(A/G)G triplets are the worst; and the use of near-cognates relies more heavily than AUG on optimal context (1). Although CUG codons are usually decoded by Met-tRNAi, leucyl-tRNALeu can be engaged by a scanning PIC in a manner requiring the non-canonical initiation factor eIF2A, but not eIF2. This occurs in the synthesis of antigenic precursors for loading on major histocompatibility complex molecules (3).
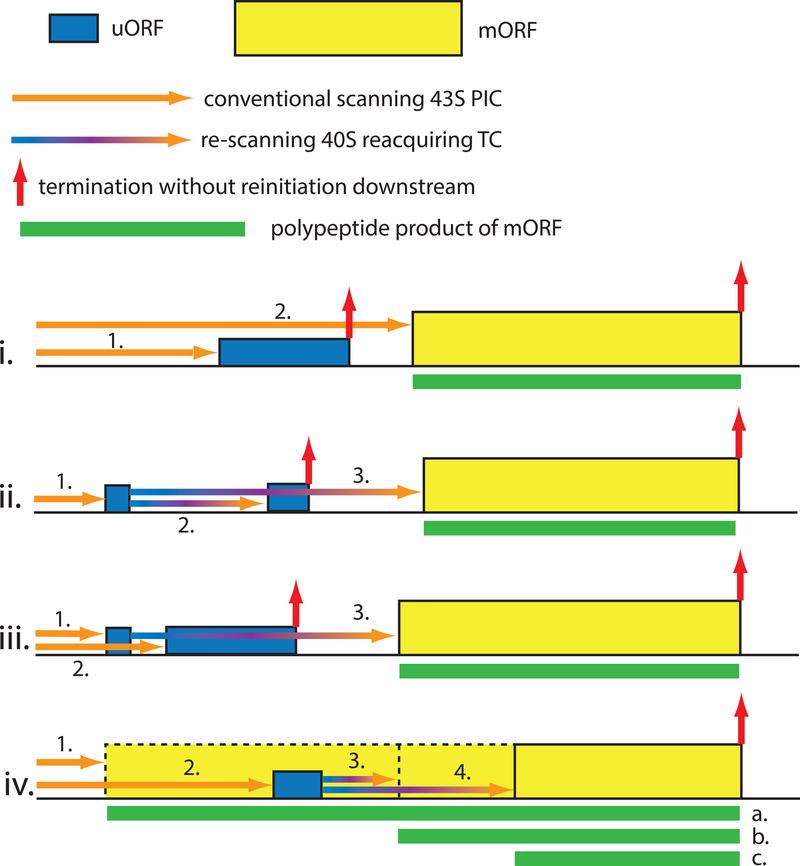
Multiple eIFs, structural elements of tRNAi, and both rRNA and protein components of the 40S subunit participate in discrimination between AUGs and non-AUG triplets as start codons, and good versus poor Kozak context, by the scanning PIC (1). eIF1 and eIF5 have opposing effects, with eIF1 promoting scanning and blocking recognition of non-AUGs and AUGs in poor context, and eIF5 antagonizing eIF1 function. These activities are exploited for autoregulation and cross-regulation of eIF1/eIF5 expression in most eukaryotes (4), as the eIF1 AUG is in weak context and poorly recognized unless eIF1 levels drop below a certain threshold (5). Initiation at the eIF5 start codon is impaired by an uORF whose AUG is in weak context and hence efficiently bypassed by leaky scanning only at low eIF5 (or high eIF1) levels (Fig. 3-i) (4). Yeast mutants of these and other initiation factors (eIF1A, eIF2 subunits, and eIF3c) can increase or decrease discrimination against suboptimal start codons (1, 6). There is a potential for regulating such initiation events through post-translational modifications of these factors, or with small molecules.
Fig 3. Different gene architectures conferring translational control by short uORFs.
i) 1. Scanning PICs that translate the uORF fail to reinitiate at the mORF, as depicted in Fig. 2. 2. A fraction of scanning PICs leaky-scan the uORF start codon, enhanced by its suboptimal context, and initiate at the mORF instead. Leaky-scanning can be induced by elevated eIF5 levels for the eIF5 structural gene (4), by eIF2(αP), eg. for GADD34 (36) and IFRD (35); and by polyamines for AMD1 (encoding SAMDC) and AZIN1 (34). ii) 1. Scanning ribosomes initiate translation of a short uORF whose translation does not preclude reinitiation. 2. Resumed scanning followed by quick reacquisition of TC enables translation of an inhibitory downstream uORF that precludes further reinitiation. 3. Slow reacquisition of TC at reduced TC concentrations evoked by eIF2(αP) allows reinitiation further downstream at the mORF. Examples include GCN4 (37) and ATF4 (38, 39). iii) 1. Scanning ribosomes initiate translation of a short uORF permissive for reinitiation. 2. Ribosomes that leaky-scan the first uORF translate a second inhibitory uORF that precludes reinitiation. 3. Ribosomes that translate the first uORF resume scanning and reacquire TC only after bypassing the second uORF, avoiding its inhibitory effect. Exemplified by polyamine regulation of SAMDC synthesis in plants (34). iv) 1. Ribosomes initiate at an upstream start codon in-frame with the mORF and bypass an inhibitory uORF during elongation while producing protein isoform “a” with specific properties. 2. Scanning subunits bypass the in-frame start site, owing to its suboptimal context, and initiate downstream at the uORF. 3. Re-scanning followed by quick reacquisition of TC leads to reinitiation at a proximal start codon to produce protein isoform “b”. 4. Slow re-acquisition of TC allows initiation further downstream producing the shortest isoform “c”, with activities opposite those of “a” and “b”. Examples include C/EBP-α and C/EBP-β (49).
Based on studies in yeast where dramatically lengthening the 5’UTR conferred no reduction in translational efficiency, it appears that the scanning PIC is highly processive (7). However, an excessively short 5’UTR (< ~20nt) is generally detrimental and can evoke leaky scanning (1, 8). Indeed, genome-wide mapping of yeast 5’UTRs identified many mRNAs with short 5’UTRs exhibiting lower-than-average translational efficiency (TEs_ (9). Leaky scanning induced by short 5’UTRs allows production of different isoforms differing at the N-termini from the same mRNA (1, 10). In contrast, the mammalian 5’UTR element “Translation Initiator of Short 5’UTRs” (TISU) allows cap-dependent, but scanning-independent initiation on mRNAs with 5’UTRs as short as 5 nt. While not requiring eIF4A, TISU’s function paradoxically depends on eIF1 (11), which normally blocks selection of AUGs too close to the cap (8). mRNAs encoding mitochondrial proteins are enriched in TISU, which appears to confer maintenance of translation at low energy levels (11).
Translational control by 5’UTR structure
Secondary structures in the 5’UTR can also influence the initiation efficiency of sub-optimal start codons. A strong stem-loop (SL) structure just downstream of the start codon will stall the scanning 40S subunit, increasing its “dwell time” and thus decreasing the probability of leaky scanning through near-cognates or AUG triplets in poor context (12).
While a precise SL-AUG spacing is required for the SL stimulatory effect, mRNA structures of sufficient stability inhibit all scanning-dependent initiation downstream (1). DEAD-box ATP-dependent RNA helicases can overcome SL structural impediments, and they might be specialized for certain types or locations of mRNA structures. The fact that eIF4A is recruited to the mRNA 5’ end and activated as a component of eIF4F, positions eIF4A to facilitate PIC attachment near the cap to initiate scanning at the mRNA 5’ end (1). eIF4E overexpression preferentially stimulates translation of mRNAs containing excessive secondary structure implying that eIF4F is limiting for translation of mRNAs with structured leaders (13). Mammalian DHX29 and the yeast homolog of DDX3 (Ded1) appear to be crucial for resolving stable structures distal from the cap that impede scanning (14). Indeed, genome-wide analysis of TEs in Ded1 and eIF4A yeast mutants revealed that Ded1-hyperdependent genes tend to have atypically long and structured 5’UTRs, whereas eIF4A contributes more equally to translation of all mRNAs (15). This differs in mammals, where eIF4A-dependence is conferred by long 5’ UTR sequences capable of forming stable secondary structure (16) or G-quadruplex structures (17). Moreover, mammalian mRNAs containing 5’UTR secondary structure are hyper-dependent on eIF4A for translation in vitro (18). Cap-proximal structures can also impede eIF4F binding to the cap (19), and DDX3 was implicated in resolving cap-proximal SLs to enhance eIF4F recruitment (20).
Analogous to the inhibitory effects of cap-proximal SL elements, a paradigm of translational control in mammals involves formation of an mRNA—protein complex comprised of the iron regulatory protein (IRP) and a cap-proximal SL known as the iron response element (IRE), which blocks 43S attachment to mRNAs encoding ferritin or other iron metabolism proteins in iron-deprived cells (21).
Translational control by uORFs
Genome-wide sequencing of 5’UTRs reveals that uORFs are pervasive, occurring in ~50% of mammalian mRNAs, and there is evidence from ribosome footprint profiling that a sizable fraction of uORFs are translated (22–24), although only a small fraction produces peptides sufficiently abundant and stable for detection (25). It is likely that ribosome occupancies of uORFs detected in certain profiling experiments overestimate their TEs in cells (26, 27). This is especially true for uORFs initiated by near-cognates under conditions where bulk protein synthesis is diminished, where their representation is substantially elevated compared with their utilization as start sites for main ORFs. However, the fact that the occurrence of AUG-initiated uORFs is below the frequency predicted by chance, that when present their start codons are in poor initiation context, that their occurrence and translation is associated with below-average TEs for the downstream ORFs genome-wide, and that they show evidence of evolutionary sequence conservation, are good indicators that AUG-initiated uORFs function broadly to throttle-down translation initiation, whereas the same evidence is lacking for most non-AUG initiated uORFs (9, 24, 28, 29). Regulation via uORFs is likely coupled to transcriptional control in yeast meiosis where the transcription start sites of certain genes shift upstream to include one or more AUG-initiated uORFs which is accompanied by diminished TE of the downstream ORFs (29). Termination at an uORF stop codon can elicit the same mRNA destabilization evoked by the nonsense-mediated decay (NMD) pathway at premature termination codons in ORFs, magnifying the inhibitory effects of uORFs (9, 30).
Despite their widespread occurrence, direct evidence that particular uORFs inhibit translation of downstream ORFs exists only for a relatively small number of genes, with two primary control mechanisms at play. For one class of regulatory uORFs, the encoded peptide acts to stall the elongating 80S ribosome engaged in its synthesis at or near the uORF stop codon, creating a “roadblock” to scanning PICs that leaky-scanned the uORF AUG codon (Fig. 2-ii). This roadblock can be modulated by ligands to achieve translational control, e.g. arginine for yeast CPA1 and Neurospora crassa arg-2 attenuator peptides (31, 32), or spermidine for ADM1 (33, 34).
An encoded peptide sequence is irrelevant for a second class of regulatory uORFs which functiononly to waylay scanning PICs from the downstream ORF start codon (Fig. 2-i). That the barrier imposed by such uORFs is generally overcome by leaky scanning is suggested by genome-wide data indicating that uORFs whose AUG codons better conform to the Kozak consensus are more inhibitory (23, 28). Also, upstream start codons tend to be near-cognates or AUGs in poor context, which should favor leaky-scanning (23). Leaky scanning of an inhibitory uORF, through an unknown mechanism, is increased under stress conditions that evoke phosphorylation of eIF2 on serine-51 of its α-subunit; eIF2(αP) (Fig. 3-i) (35). This applies to GADD34, a targeting subunit for protein phosphatase-1 that dephosphorylates eIF2α, enabling autoregulation of eIF2(αP) accumulation (36). Phosphorylation of eIF2α converts eIF2-GDP into a competitive inhibitor of the guanine nucleotide exchange factor eIF2B, and thereby decreases TC assembly (Fig. 1) (37). This might allow a fraction of PICs scanning from the cap to reach the uORF without harboring the TC, bypass the uAUG (owing to the absence of Met-tRNAi), and bind TC while scanning the remainder of the 5’UTR and initiate at the main ORF. Alternatively, phosphorylation might also alter eIF2 function in start codon recognition to allow leaky scanning even with TC bound to the PIC.
The presence of multiple uORFs can greatly amplify the effect of eIF2α phosphorylation on leaky scanning, as demonstrated first for yeast GCN4 (37) and subsequently mammalian ATF4 and ATF5 (38, 39), which encode transcription factors instrumental in responding to stresses that activate eIF2α kinases, such as amino acid deprivation for kinase Gcn2 (Fig. 1), and ER-stress for kinase PERK. The first (uORF1) is translated by most scanning PICs and optimized to allow a fraction of 40S ribosomes to remain attached to the 5’UTR and reinitiate downstream (37). For GCN4 uORF1, sequences/structures upstream of the uORF functionally interact with the a-subunit of eIF3 and AU-rich sequences 3’ of the uORF stop codon to allow scanning to resume (40). With abundant TC in non-stressed cells, “re-scanning” PICs re-bind TC rapidly and efficiently reinitiate at the downstream uORF(s), optimized to evict post-termination 40S subunits from the mRNA and prevent translation of the downstream main ORF. The decreased TC levels evoked by eIF2α phosphorylation allow a fraction of re-scanning PICs to re-bind TC only after leaky-scanning the inhibitory uORFs and initiate downstream at the ORF instead (Fig. 3-ii). Because of minimal leaky-scanning of the inhibitory uORFs in non-stressed cells, owing to their optimum context, only a modest reduction in their recognition engenders large increases in main ORF translation in stressed cells (37). This mechanism enables the rapid, strategic induction of key transcription factors, while the reduced TC levels dampen bulk protein synthesis, for a two-pronged stress response.
The short length of uORF1 is crucial for reinitiation, and might facilitate retention of eIF3 during its translation (37). Reinitiation following longer uORFs requires additional cis-acting sequences, like the “termination upstream ribosomal binding site” (TURBS) element of polycistronic calicivirus mRNA that base-pairs with 18S rRNA sequences of the 40S subunit (41, 42), similar to Shine-Dalgarno sequences in bacterial mRNAs and eukaryotic viral IRESs such as in HCV. Some cellular IRESs might also function in this manner (43, 44). The role of eIF3 subunits in promoting reinitiation appears widespread (45, 46). Accessory factors, including ligatin/eIF2D or related proteins MCT-1 and DENR, can promote reinitiation, possibly through an alternative pathway for recovering Met-tRNAi by re-scanning PICs (42, 47).
The role of uORFs in regulating reinitiation in response to eIF2(αP) was implicated in learning and memory, neurodegenerative diseases, and cancer. For example, eliminating Gcn2 improved memory in mice owing to decreased eIF2(αP), leading to reduced ATF4 translation (48). Alternative outcomes of leaky-scanning versus reinitiation imposed by a uORF in C/EBP mRNAs determine the balance of isoforms differing at their N-termini, one activating and the other inhibiting transcription, important for mouse liver differentiation and regeneration (Fig. 3-iv) (49). A novel role for a uORF in facilitating translational repression of Drosophila msl-2 mRNA in conjunction with 5’UTR binding by Sex-lethal (SXL) protein regulates dosage compensation (50). There is a growing list of mutations associated with human disease that increase or decrease the influence of uAUGs/uORFs on translation of the main ORF (28, 51).
Other 5’UTR regulatory elements
The 5’ terminal oligopyrimidine (5’TOP) motif plays a role in mTOR-dependent stimulation of the expression of proteins of the translation machinery to promote cell growth. mTORC1 activates the La-related protein 1 (LARP1) that binds to the TOP sequence (52). Many less abundant mRNAs lacking 5’TOP exhibit mTOR dependence, encoding mitochondrial and growth/survival-promoting proteins (53). Additionally, the m6-adenine methylation of 5’UTR sequences seems to have stimulatory effects (54).
That uORFs appear to influence translation genome-wide suggests that scanning operates widely in the eukaryotic translatome. However, scanning can be circumvented by specialized elements that enable PICs to enter the 5’UTR internally. mRNAs with a stretch of unstructured nucleotides in the 5’UTR can bypass the requirement for the m7G cap and eIF4F, as shown for poly(A) sequences in poxvirus mRNA 5’UTRs (55) and CAA repeats in the Ω leader of tobacco mosaic virus mRNAs. While dispensable for 48S assembly per se, eIF4A accelerated the process on a Ω reporter (56). eIF4F-independence for 48S PIC assembly was also observed for mRNAs with synthetic unstructured 5’UTRs, which like the poly(A) 5’UTRs, still require eIF1, eIF1A, TC, and (in mammals) eIF3 (8, 57). Unstructured nucleotides might bind directly in the mRNA binding cleft of the 40S subunit, with ATP hydrolysis by eIF4A enabling subsequent 5’-to-3’ directional scanning. A group of mRNAs in yeast are refractory to widespread translational repression in carbon-starved cells and contain a poly(A) stretch in their 5’UTRs that might recruit eIF4F to the 5’UTR via PABP-eIF4G association, augmenting cap-binding by eIF4F (58).
Many viral mRNAs circumvent the scanning process with highly structured IRES elements that interact with the 40S subunit or particular eIFs to recruit the PIC to internal sites in the 5’UTR (59). These mechanisms persist when cap/eIF4F-dependent initiation is impaired, such as in virus infections where eIF4G is cleaved. Whereas viral IRESes have been extensively characterized biochemically and structurally (59), such studies have not been accomplished for potential IRESes in cellular mRNAs (60). A genome-wide search yielded a large number of mammalian cellular IRESes (10% of randomly selected 5’UTRs) (44), which if validated for individual mRNAs would be highly significant for understanding gene regulation in humans.
Important progress has been made in elucidating mechanisms by which the 5’UTR regulates translation initiation. This includes molecular and structural understanding of the assembly and recruitment of the PIC to the 5’UTR, scanning, and start codon selection. Acutely lacking is a precise kinetic analysis of the pathway. Single-molecule approaches can be expected to fill this gap and identify intermediate states too transient for detection by ensemble kinetics. Ribosome profiling (22) can be adapted to analyze the kinetics and regulation of scanning on all 5’UTRs. Advanced cryo-EM will continue to yield high-resolution structures of the PIC in different stages of initiation. The new information is bound to aid efforts to discover new drugs to treat diseases whose etiology is associated with dysregulated translation.
References
- 1.Hinnebusch AG, Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75, 434–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozak M, Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292 (1986). [DOI] [PubMed] [Google Scholar]
- 3.Starck SR et al. , Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336, 1719–1723 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Loughran G, Sachs MS, Atkins JF, Ivanov IP, Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res 40, 2898–2906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanov IP, Loughran G, Sachs MS, Atkins JF, Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci U S A 107, 18056–18060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donahue TF, Cigan AM, Pabich EK, Castilho-Valavicius B, Mutations at a Zn(II) finger motif in the yeast elF-2β gene alter ribosomal start-site selection during the scanning process. Cell 54, 621–632 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Berthelot K, Muldoon M, Rajkowitsch L, Hughes J, McCarthy JE, Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol Microbiol 51, 987–1001 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Pestova TV, Kolupaeva VG, The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16, 2906–2922. (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arribere JA, Gilbert WV, Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res 23, 977–987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sedman SA, Gelembiuk GW, Mertz JE, Translation initiation at a downstream AUG occurs with increased efficiency when the upstream AUG is located very close to the 5’ cap. J Virol 64, 453–457 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinvani H et al. , Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab 21, 479–492 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Kozak M, Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A 87, 8301–8305 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koromilas AE, Lazaris-Karatzas A, Sonenberg N, mRNAs containing extensive secondary structure in their 5’ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 11, 4153–4158 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abaeva IS, Marintchev A, Pisareva VP, Hellen CU, Pestova TV, Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. Embo J 30, 115–129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sen ND, Zhou F, Ingolia NT, Hinnebusch AG, Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res 25, 1196–1205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubio CA et al. , Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol 15, 476 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfe AL et al. , RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 513, 65–70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svitkin Y et al. , The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5’ secondary structure. RNA 7, 382–394 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelletier J, Sonenberg N, Photochemical cross-linking of cap binding proteins to eucaryotic mRNAs: effect of mRNA 5’ secondary structure. Mol Cell Biol 5, 3222–3230 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soto-Rifo R et al. , DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J 31, 3745–3756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hentze M, Gebauer F, Preiss T, cis-Regulatory Sequences and trans-Acting Factors in Translational Control. Translational Control in Biology and Medicine (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2007), pp. 269–295. [Google Scholar]
- 22.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS, Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S et al. , Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A 109, E2424–2432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnstone TG, Bazzini AA, Giraldez AJ, Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J 35, 706–723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanderperre B et al. , Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One 8, e70698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F, Hinnebusch AG, An upstream ORF with non-AUG start codon is translated in vivo but dispensable for translational control of GCN4 mRNA. Nucleic Acids Res 39, 68–86 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerashchenko MV, Gladyshev VN, Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res 42, e134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calvo SE, Pagliarini DJ, Mootha VK, Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A 106, 7507–7512 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brar GA et al. , High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaba A, Jacobson A, Sachs MS, Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol Cell 20, 449–460 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Werner M, Feller A, Messenguy F, Pierard A, The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell 49, 805–813 (1987). [DOI] [PubMed] [Google Scholar]
- 32.Wei J, Wu C, Sachs MS, The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol Cell Biol 32, 2396–2406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raney A, Law GL, Mize GJ, Morris DR, Regulated translation termination at the upstream open reading frame in s-adenosylmethionine decarboxylase mRNA. J Biol Chem 277, 5988–5994 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Ivanov IP, Atkins JF, Michael AJ, A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res 38, 353–359 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andreev DE et al. , Translation of 5’ leaders is pervasive in genes resistant to eIF2 repression. Elife 4, e03971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young SK, Willy JA, Wu C, Sachs MS, Wek RC, Ribosome Reinitiation Directs Gene-specific Translation and Regulates the Integrated Stress Response. J Biol Chem 290, 28257–28271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinnebusch AG, Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59, 407–450 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Lu PD, Harding HP, Ron D, Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167, 27–33 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vattem KM, Wek RC, Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 101, 11269–11274 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szamecz B et al. , eIF3a cooperates with sequences 5’ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev 22, 2414–2425 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luttermann C, Meyers G, The importance of inter- and intramolecular base pairing for translation reinitiation on a eukaryotic bicistronic mRNA. Genes Dev 23, 331–344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zinoviev A, Hellen CU, Pestova TV, Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol Cell 57, 1059–1073 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chappell SA, Dresios J, Edelman GM, Mauro VP, Ribosomal shunting mediated by a translational enhancer element that base pairs to 18S rRNA. Proc Natl Acad Sci U S A 103, 9488–9493 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weingarten-Gabbay S et al. , Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351, (2016). [DOI] [PubMed] [Google Scholar]
- 45.Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA, A plant viral “reinitiation” factor interacts with the host translational machinery. Cell 106, 723–733 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Roy B et al. , The h subunit of eIF3 promotes reinitiation competence during translation of mRNAs harboring upstream open reading frames. RNA 16, 748–761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schleich S et al. , DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature 512, 208–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costa-Mattioli M et al. , Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature 436, 1166–1173 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wethmar K et al. , C/EBPbetaDeltauORF mice--a genetic model for uORF-mediated translational control in mammals. Genes Dev 24, 15–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Medenbach J, Seiler M, Hentze MW, Translational control via protein-regulated upstream open reading frames. Cell 145, 902–913 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Somers J et al. , A common polymorphism in the 5’ UTR of ERCC5 creates an upstream ORF that confers resistance to platinum-based chemotherapy. Genes Dev 29, 1891–1896 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tcherkezian J et al. , Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5’TOP mRNA translation. Genes Dev 28, 357–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gandin V et al. , nanoCAGE reveals 5’ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer KD et al. , 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 163, 999–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shirokikh NE, Spirin AS, Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proc Natl Acad Sci U S A 105, 10738–10743 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agalarov S, Sakharov PA, Fattakhova D, Sogorin EA, Spirin AS, Internal translation initiation and eIF4F/ATP-independent scanning of mRNA by eukaryotic ribosomal particles. Sci Rep 4, 4438 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Algire MA et al. , Development and characterization of a reconstituted yeast translation initiation system. RNA 8, 382–397 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilbert WV, Zhou K, Butler TK, Doudna JA, Cap-independent translation is required for starvation-induced differentiation in yeast. Science 317, 1224–1227 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Kieft JS, Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci 33, 274–283 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson RJ, The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol 5, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]