Abstract
Increased exposure to estrogen is associated with an elevated risk of breast cancer. Considering estrogen as a possible mutagen, we hypothesized that exposure to estrogen alone or in combination with the DNA-damaging chemotherapy drug, cisplatin, could induce expression of genes encoding enzymes involved in APOBEC-mediated mutagenesis. To test this hypothesis, we measured the expression of APOBEC3A (A3A) and APOBEC3B (A3B) genes in two breast cancer cell lines treated with estradiol, cisplatin or their combination. These cell lines, T-47D (ER+) and MDA-MB-231 (ER−), differed by the status of the estrogen receptor (ER). Expression of A3A was not detectable in any conditions tested, while A3B expression was induced by treatment with cisplatin and estradiol in ER+ cells but was not affected by estradiol in ER− cells. In The Cancer Genome Atlas, expression of A3B was significantly associated with genotypes of a regulatory germline variant rs17000526 upstream of the APOBEC3 cluster in 116 ER− breast tumors (P = 0.006) but not in 387 ER+ tumors (P = 0.48). In conclusion, we show that in breast cancer cell lines, A3B expression was induced by estradiol in ER+ cells and by cisplatin regardless of ER status. In ER+ breast tumors, the effect of estrogen may be masking the association of rs17000526 with A3B expression, which was apparent in ER− tumors. Our results provide new insights into the differential etiology of ER+ and ER− breast cancer and the possible role of A3B in this process through a mitogenic rather than the mutagenic activity of estrogen.
By analyses in cell lines and TCGA tumors, we show that estrogen exposure may increase breast cancer risk by inducing APOBEC3B expression in ER+ breast cancer cells. The effect on APOBEC3B expression supports the mitogenic rather than the mutagenic activity of estrogen in ER+ breast cells.
Introduction
Exposure to sex hormones represents a natural but variable environment for both normal and cancerous breast cells. One of these hormones, estrogen, is endogenously produced by premenopausal ovaries and acts on both reproductive (breast, ovaries, uterus) and non-reproductive (e.g. bone, brain) systems (1). Exposure to estrogen may also be contributed by exogenous sources such as contraception or hormone replacement therapy (HRT) (2–4), also known as menopausal hormone therapy (5). Increased exposure to endogenous and exogenous estrogen has been implicated in an increased risk of breast cancer by epidemiological, molecular and clinical findings (2–7). Two hypotheses have been proposed to explain the role of estrogen in increased cancer risk (8). The first hypothesis suggests that estrogen metabolites are mitogenic, i.e. they can accelerate the cell cycle and fuel the growth of active and dormant cancer cells; the second hypothesis suggests that estrogen metabolites damage DNA and are directly mutagenic. Both hypotheses may partially explain some mechanisms contributing to the development of estrogen-mediated carcinogenesis (8–12).
Extensive resources generated by The Cancer Genome Atlas (TCGA) enabled comprehensive analyses of somatic mutagenesis, including in breast tumors (13). Based on mutational patterns, at least 12 somatic mutational signatures have been annotated in TCGA breast tumors (14). Two of these mutational signatures have been attributed to the activity of APOBEC (apolipoprotein B mRNA editing enzyme, catalytic-polypeptide-like) enzymes that belong to a family of proteins with cytidine deaminase activity (15,16). In particular, APOBEC3A (A3A) and APOBEC3B (A3B) have been considered the main mutagenic enzymes that generate APOBEC-signature mutations in breast and other tumor types (17–20).
Expression of APOBEC3s, including in breast cancer cell lines, is inducible in response to various environmental exposures (20,21). We hypothesized that estrogen, the main environmental exposure for breast cells, could induce A3A and A3B expression in vitro. Breast cancer cells can also be exposed to anticancer chemotherapy drugs, such as DNA-damaging platinum-based drugs cisplatin and its analogs (22). To evaluate the effects of these exposures, we measured A3A and A3B expression in ER+ and ER− breast cancer cell lines treated with estradiol and/or cisplatin. Our analyses in breast cancer cell lines and TCGA breast tumors suggest that A3B expression is induced by estradiol in an estrogen receptor (ER)-dependent way but without significant contribution to APOBEC-mediated mutagenesis. Our results provide indirect support to the mitogenic hypothesis of estrogen action independent of somatic mutagenesis (mutagenic hypothesis) and may improve our understanding of the etiology and clinical outcomes of breast cancer.
Materials and methods
Cell lines
Breast cancer cell lines: triple-negative, claudin-low MDA-MB-231 (ER−/PR−/HER2−/TP53mut) and luminal A cell line T-47D (ER+/PR+/HER2−/TP53mut) were purchased from the American Type Culture Collection (ATCC). MDA-MB-231 and T-47D cells were grown in Dulbecco’s modified Eagle’s medium and RPMI-1640 media, respectively (both from Quality Biological), supplemented with 10% fetal bovine serum (ThermoFisher Scientific) and 1X antibiotic-antimycotic (Quality Biological). Cell lines were regularly tested for mycoplasma contamination using the MycoAlert Mycoplasma Detection kit (Lonza). Cell lines were acquired from ATCC in 2016 and their reauthentication was done by the NCI Cancer Genomics Research Laboratory every 6 months after purchase using the AmpFLSTR Identifiler Plus PCR Amplification Kit (ThermoFisher Scientific).
According to the COSMIC database (https://cancer.sanger.ac.uk/cosmic) (23), both cell lines have functional mutations in the tumor suppressor gene TP53 (24). Specifically, MDA-MB-231 cells carry the TP53-R280K mutation (AGA to AAA) that contributes to spheroid disorganization and mammary architecture disruption (25,26) and T-47D cells carry the TP53-L194F mutation (CTT to TTT) that contributes to improved cell survival (27). According to information from ATCC, the MDA-MB-231 cell line originates from a Caucasian female, but the ancestry of the T-47D cell line is unknown. However, Estimated Cell Line Ancestry analysis (28) annotated both cell lines as of similar and predominantly European ancestry https://web.expasy.org/cellosaurus/CVCL 0553 (or CVCL 0062).
Genotyping
DNA from T-47D and MDA-MB-231 cells was isolated using the DNeasy Blood and Tissue Kit (Qiagen). Samples were genotyped for single nucleotide polymorphisms (SNPs) rs12628403 with a custom TaqMan assay (20) and rs17000526 with a commercial assay (C___2189439_10) from ThermoFisher Scientific, using Type-It Fast SNP Probe Master Mix (Qiagen). HapMap samples with known genotypes and water were used as positive and negative controls, respectively.
Estradiol and cisplatin treatments
Cells were seeded in four six-well plates with 300,000 cells/3 ml media per well. After reaching 80% confluency, one plate was harvested (0 h), one left untreated to grow for 72 h and the remaining two plates were incubated for 72 h in phenol-free media with 5% charcoal-stripped fetal bovine serum and 1X antibiotic-antimycotic. Serum starvation was done to mimic a low-hormone environment and minimize the confounding effects of growth factors and hormones present in the standard media. 17-Beta-estradiol (referred to as estradiol, Sigma–Aldrich), a synthetic analog of estrogen, was reconstituted in 100% ethanol to make a 1 mM stock. Cisplatin (Enzo Life Sciences) was reconstituted in dimethyl sulfoxide to make a 3.33 mM stock. Cells were treated for 8 or 24 h at final concentrations of 10 or 100 nM of estradiol with or without 40 µM of cisplatin, according to an ENCODE protocol (https://www.encodeproject.org/experiments/ENCSR000DMN/) and pilot optimization for the specific experiments. Treatments with ethanol and dimethyl sulfoxide alone were included as negative controls. All experiments were independently performed at least three times.
Analysis of gene expression
After treatment, plated cells were washed once with PBS and lysed in 350 μl of RLT buffer (Qiagen). RNA was extracted from the lysed cells with the RNeasy Mini QIAcube kit with an on-column DNase I treatment (all from Qiagen). The RNA concentration and quality were measured with the NanoDrop 8000 (Thermo Scientific). cDNA was prepared using the RT2 HT First Strand Kit (Qiagen) with an additional DNA removal step and equal amounts of RNA input for all samples. All expression assays and TaqMan Gene Expression Master Mix were from ThermoFisher Scientific. The following predesigned TaqMan expression assays were used—PGR (Hs01556702_m1, FAM/MGB), A3A (Hs00377444_m1, FAM/MGB), A3G (Hs01043989_m1, FAM/MGB), ESR1 (Hs00174860_m1, FAM/MGB), ESR2 (Hs00230957_m1, FAM/MGB), and endogenous controls GAPDH (4333764T, FAM/MGB) and PPIA (4326316E, VIC/MGB). A3B was measured with a custom-designed TaqMan expression assay—Forw: ACCAGCACATGGGCTTTCTATG, Rev: AAAGAAGGAACCAGGTCCAAGAAG, Probe: CGAGGCTAAGAATCT (FAM/MGB), with the probe crossing exon–exon junction.
Expression was measured using QuantStudio 7 Flex (Applied Biosystems) in four technical replicates with standard conditions and 45 PCR cycles. Water and genomic DNA were included as negative controls for all assays. Expression was measured in the cycle at threshold (Ct) values, normalized by geometric means of Ct values for endogenous controls (GAPDH and PPIA); expression differences between experimental conditions were calculated as fold = 2−(dCt control − dCt treatment).
Analysis of ER binding sites within the A3A/A3B genomic region
Genome-wide binding sites for transcription factors, including ESR1 (ERα), were mapped by ENCODE based on ChIP-sequencing and explored using UCSC Genome Browser (https://genome.ucsc.edu/). The ESR1 sites were analyzed within the ~67 kb genomic region (chr22:39,323,512-39,390,946, GRCh37/hg19) that includes both the A3A and A3B genes, as well as the predicted enhancer of A3B with the regulatory germline variants rs1014971 and rs17000526.
Statistical analysis
Results were plotted with GraphPad Prism 7 (GraphPad Software) and analyzed with SPSS 25 (IBM SPSS Statistics). The significance between the group means was calculated with unpaired two-sided Student’s t-tests (for two groups), one-way analysis of variance (for sets with more than two groups) or with analysis of variance with Tukey test (comparing specific groups in sets with multiple groups). TCGA data for breast tumors were acquired as previously described (20) and analyzed with multivariable linear regression models for A3B expression controlling for covariates considered relevant—age at diagnosis (years), tumor stage (1, 2, 3, 4) and race (White, n = 416, Black or African American, n = 54, Asian, n = 32/American Indian or Alaska Native, n = 1) in 503 samples with data available for ER status and all the covariates used. Additionally, we adjusted for the expression of the A3AB deletion transcript (represented by the SNP rs12628403), which involves complete elimination of the A3B gene by a germline deletion. Expression in ER+ versus ER− tumors was analyzed with one-way analysis of covariance, adjusting for age, race and tumor stage.
Results
Expression of A3B is induced by treatment with estradiol and cisplatin but only in an ER+ breast cancer cell line
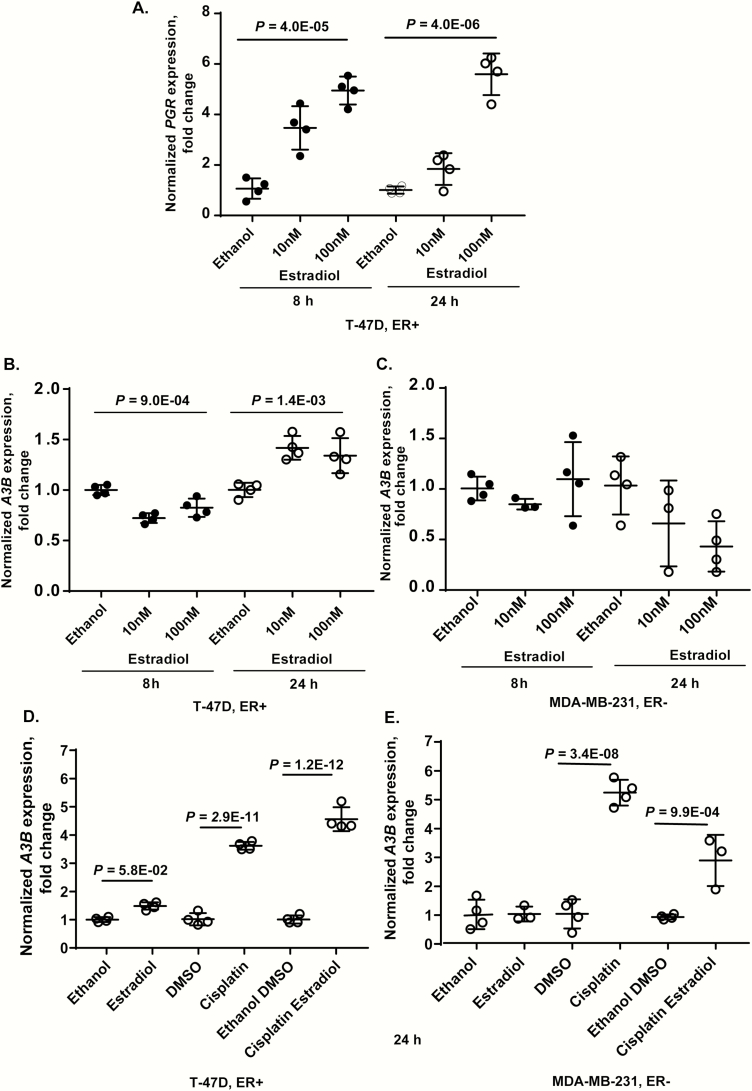
First, we evaluated the expression of the progesterone receptor (PGR), which is known to be induced by estrogen (29,30). As expected, there was significant induction of PGR in an ER+ cell line T-47D after treatment with estradiol for 8 and 24 h (Figure 1A), while PGR was not expressed at baseline or after estradiol treatment in an ER− cell line MDA-MB-231 (data not shown). Thus, our system was sensitive enough to detect the effects of estradiol exposure.
Figure 1.
Expression of PGR and APOBEC3B (A3B) in breast cancer cell lines. (A) Estradiol treatment significantly induced PGR expression in the ER+ cell line T-47D but expression of PGR was not detectable in the ER− cell line MDA-MB-231 at baseline or after treatment (data not shown); (B and C) estradiol treatment reduced A3B expression at 8 h but induced at 24 h in the ER+ cell line, without causing significant effects in the ER− cell line; (D and E) expression of A3B in ER+ and ER− breast cancer cell lines treated for 24 h with 100 nM estradiol, 40 µM cisplatin or a combination of these drugs. In the ER+ cell line, expression of A3B induced by the combination of estradiol and cisplatin was significantly higher than by estradiol alone (P = 2.64E−12) or cisplatin alone (P = 1.25E−04). In the ER− cell line, expression of A3B induced by the combination of estradiol and cisplatin was significantly higher than by estradiol alone (P = 3.0E−3) but lower than by cisplatin alone (P = 1.39E−04). All results are presented as fold difference comparing treatment conditions to vehicle only (ethanol and dimethyl sulfoxide for estradiol and cisplatin, respectively) at corresponding time points. Plots show values for individual replicates, group means and standard deviations. P-values are for one-way analysis of variance, based on biological replicates. For D and E plots—P-values are for one-way analysis of variance between all groups with Tukey test applied for comparisons between the indicated groups.
Next, we evaluated the expression of A3A and A3B in these cell lines treated with estradiol, cisplatin or a combination of these drugs. Expression of A3A was not detectable in either cell line at baseline or after 24 h of treatment (data not shown). This is consistent with our previous observations that A3A was not expressed at baseline in any cell lines tested and was strongly induced by a viral infection but not the DNA-damaging drug bleomycin (20). In the ER+ cell line, A3B expression was reduced by treatment with estradiol for 8 h but was induced at 24 h (Figure 1B), while in the ER− cells, A3B expression was not significantly affected by estradiol treatment (Figure 1C).
Additional analyses in these cell lines treated for 24 h with 100 nM estradiol, 40 µM cisplatin or their combination showed that estradiol and cisplatin had a positive additive effect on A3B expression in the ER+ cells (Figure 1D). However, in the ER− cells, A3B expression was induced by cisplatin alone but was significantly decreased by combination with estradiol (Figure 1E).
Expression of A3B is significantly affected by two germline genetic variants (20). The first variant is a 30 kb deletion that eliminates A3B and creates a chimeric A3AB transcript; this deletion was associated with breast cancer risk and an increased load of APOBEC-signature mutations in breast tumors (31–33). The second variant is a regulatory SNP rs1014971 (and its proxy rs17000526 in TCGA), located upstream of the APOBEC3 gene cluster on chromosome 22q13 (20). The cell lines tested were identical for these genetic variants—homozygous for the rs12628403-A allele, which corresponds to the absence of the A3B deletion, and homozygous for the rs17000526-A allele, which was associated with increased A3B expression in bladder and breast tumors (20). Thus, the observed differences in A3B expression in these cell lines could not be explained by the germline variants tested.
Expression of ESR1 is necessary but not sufficient to explain the differential induction of A3B expression in ER+ and ER− cell lines
The ER is a dimer that is formed by the ERα and ERβ subunits encoded by ESR1 and ESR2 genes, respectively. To further explore the link between A3B expression and ER status, we measured the expression of these transcripts in cells treated with estradiol compared with control. Expression of ESR1 in the ER+ cell line T-47D was high at baseline and unaffected at 8 h but moderately decreased after 24 h of treatment, Supplementary Figure S1A, available at Carcinogenesis Online), while ESR1 was not expressed in the ER− cell line MDA-MB-231 at baseline or after estradiol treatment (data not shown). Expression of ESR2 was very low at baseline but was significantly induced by 3.3 and 4.5 Ct values (~10- and 22-fold) by estradiol treatment in ER+ and ER− cell lines, respectively, although a longer treatment time (24 h) was required for ESR2 induction in ER− cells (Supplementary Figure S1B and C, available at Carcinogenesis Online).
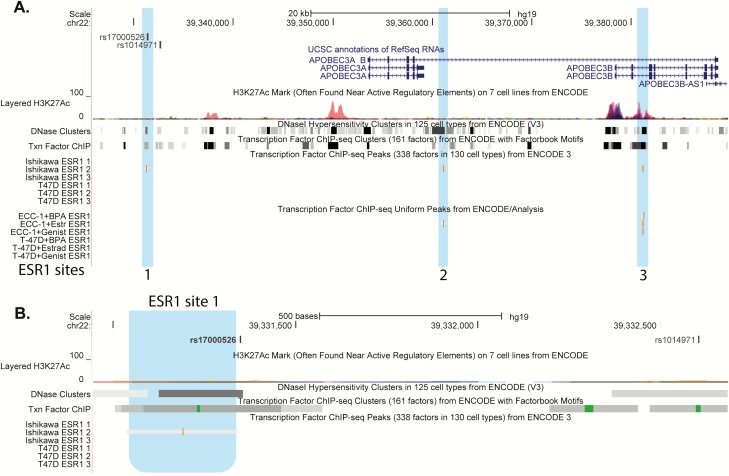
Analysis of the A3B genomic region on chromosome 22 using the ENCODE resources showed three locations of ESR1 (ERα) binding sites in ER+ positive endometrial cancer cell lines Ishikawa and ECC-1 but none in T-47D (Figure 2A). Notably, one of the ESR1 binding sites (detected in Ishikawa cell line) maps just 6 bp upstream of SNP rs17000526 (Figure 2B), for which we previously demonstrated strong but non-allele-specific interaction with nuclear proteins in MCF7, an ER+ breast cancer cell line (20). Thus, ESR1 binding to the A3B enhancer in some ER+ cells might induce A3B expression regardless of the rs17000526 genotype. However, in T-47D cells, A3B expression might be regulated by estrogen through other, indirect mechanisms, explaining the relatively modest induction of A3B by estradiol in this cell line. These results suggest that A3B induction by estradiol likely requires ERα activity, but without significant correlation between ESR1 and A3B on the mRNA level.
Figure 2.
Location of the ESR1 (ERα) binding sites within the APOBEC3B genomic region on chromosome 22. Genome-wide maps of transcription factor binding sites were generated by ENCODE based on ChIP-sequencing and viewed using the UCSC genome browser (GRCh37/hg19). (A) The 67 kb genomic region that includes A3A and A3B genes and SNPs rs1014971 and rs17000526, which are located within the A3B enhancer. (B) Zoom-in view of the 1.7 kb genomic region centered on the ESR1 site 1, which is mapped just 6 bp upstream of rs17000526 in the ER+ Ishikawa endometrial cancer cell line. ESR1 binding sites are highlighted in blue.
Analysis of A3B expression in TCGA breast tumors
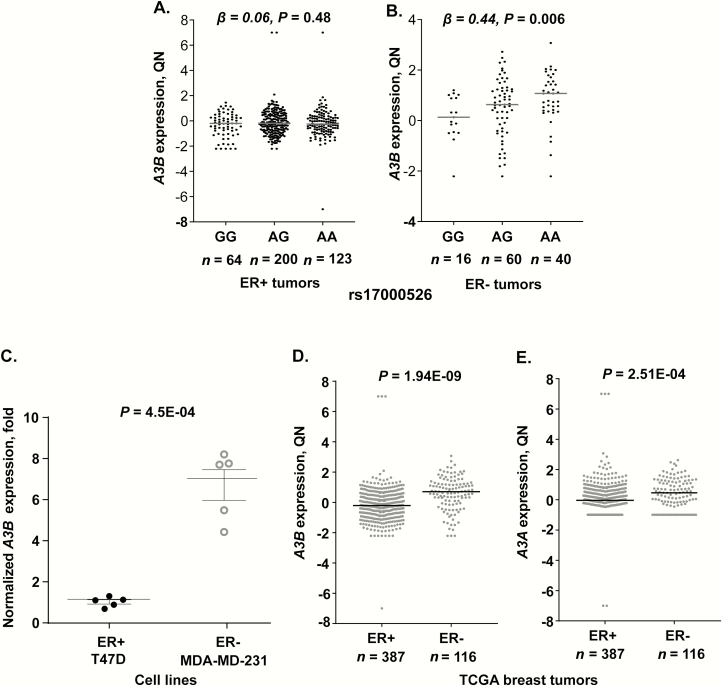
Previously, we reported that A3B expression in 533 breast tumors from TCGA was significantly increased (P = 5.9E−03) in carriers of the rs17000526-A allele (20), but this analysis did not include information on the ER status of breast tumors. In light of our observations on the differential induction of A3B by estradiol in ER+ and ER− cell lines, and the presence of an ERα binding site adjacent to rs17000526 in some ER+ cell lines (Figure 2), we performed stratified analysis of A3B expression in TCGA ER+ and ER− breast tumors. The breast cancer TCGA dataset comprised of 503 tumors with data available for all of these variables. In these 503 tumors, A3B expression was significantly associated with increased counts of rs17000526-A allele (β = 0.17; P = 0.026), controlling for age (P = 0.064), tumor stage (P = 0.12), race (P = 0.13) and expression of the A3AB deletion transcript (P = 0.007) in a multivariable linear regression model. Stratified analysis of these tumors showed that in the 387 TCGA ER+ tumors, none of these factors (except increased tumor stage) were associated with A3B expression—rs17000526 (β = 0.06; P = 0.48), age (P = 0.080), tumor stage (P = 0.046), race (P = 0.60) and A3AB deletion transcript (P = 0.36) (Figure 3A). A similar analysis limited to the 116 TCGA ER− tumors showed that A3B expression was significantly associated with increased counts of rs17000526-A allele (β = 0.44; P = 0.006) (Figure 3B), controlling for age (P = 0.85), tumor stage (P = 0.63), race (P = 0.48) and expression of the A3AB deletion transcript (P = 0.016). Thus, the association of rs17000526 with A3B expression was observed only in TCGA ER− tumors, possibly because of the dominant effect of the ER masking effects of all other factors.
Figure 3.
A3B mRNA expression in 503 breast tumors in TCGA and breast cancer cell lines. (A and B) A3B expression in relation to genotypes of the regulatory germline SNP rs17000526 in 387 ER+ and 116 ER− tumors. Per-allele β (effect size) and P-values are for multivariable linear regression models, controlling for age, race, tumor stage and expression of the A3AB isoform that corresponds to the germline deletion of A3B. (C). Baseline A3B expression normalized to endogenous controls is higher in the ER− cell line, MDA-MB-231, compared with the ER+ cell line, T-47D. P-value is for an unpaired two-sided t-test based on 5 biological replicates. (D and E) Expression of A3B and A3A in 503 breast tumors in TCGA (387 ER+ and 116 ER−); P-values are for one-way analysis of covariance adjusting for age, race and tumor stage. Plots show individual data values, group means and standard deviations. QN-quantile-normalized expression.
Baseline A3B expression in our experiments was higher in an ER− than in an ER+ cell line (P = 4.5E−04, Figure 3C). In TCGA, expression of A3B was also higher in 116 ER− compared with 387 ER+ tumors (P= 1.94E−09, controlling for age, tumor stage and race, Figure 3D). Although A3A expression was not detected in the cell lines at baseline or under experimental conditions, similar to A3B, A3A was expressed higher in ER− compared with ER+ breast tumors (P = 2.51E−04, Figure 3E).
Discussion
Here, we demonstrated that in human breast cancer cell lines, A3B expression was induced by exposure to estradiol in an ER+ but not in an ER− cell line, and this induction likely required ERα activity. A3B expression was also induced by a DNA-damaging chemotherapy drug, cisplatin, regardless of the ER status. Expression of A3A in these cell lines was not detectable in any conditions tested, suggesting that A3B is an estrogen-responsive gene and A3A is not. The expression of A3B was significantly higher in ER− compared with ER+ breast cancer cell line and breast tumors in TCGA. The dominant effect of endogenous estrogen on A3B expression in ER+ breast tumors might be masking the weaker effects of other factors.
Because A3B encodes a mutagenic APOBEC3 enzyme, the observed estrogen-dependent induction of A3B expression in ER+ cells could potentially correspond to APOBEC-mediated mutagenesis reported in many cancer types, including breast cancer (13), which supports the proposed direct mutagenic effect of estrogen metabolites (8). The mutagenic deaminase activity of A3B has been associated with reduced efficacy of tamoxifen in ER+ breast tumors due to the generation of treatment-resistant mutations (34). The contribution of A3B to tamoxifen resistance in a xenograft model for ER+ breast cancer was confirmed by improved sensitivity to tamoxifen after the elimination of A3B expression (34). Cooperation between A3B and the ER was also proposed based on an observed enrichment of A3B binding sites near estrogen-responsive genes (35). Activation and nuclear translocation of the ER recruits A3B to ER-responsive genes where deamination activity of A3B can generate DNA strand breaks, triggering DNA repair and chromatin remodeling within ER-responsive elements of these genes (35). Thus, the mutagenic activity of A3B regulates estrogen signaling. On the other hand, our data suggest a feedback loop in which A3B is induced by estrogen to assists in its action at ER-responsive promoters in ER+ breast cancer cells. We also showed that the induction of A3B expression by estradiol in an ER+ cell line is likely dependent on ERα (and not ERβ), although ESR1 expression was not inducible and thus did not correlate with A3B expression.
Although in TCGA breast tumors A3B mRNA expression was not a good proxy for APOBEC-mediated mutagenesis, which was primarily associated with the expression of A3A and its deletion isoform (A3AB) (20), increased A3B expression was identified as a prognostic biomarker of poor outcomes in breast cancer patients with ER+ tumors (34,36,37). Thus, the importance of A3B in breast cancer might be related to its activity not causing mutagenesis. An alternative, deaminase-independent activity of A3B was recently reported in liver cancer (38). According to this mechanism, increased A3B expression modulates the tumor microenvironment and promotes tumor progression without using the deaminase activity of A3B to generate somatic mutations. This mechanism has not yet been tested in relation to breast cancer, but the promotion of cancer progression by estrogen-induced A3B expression affecting the tumor microenvironment in ER+ cells would be consistent with the proposed mitogenic effect of estrogen (8). Importantly, the effect of A3B expression on tumor microenvironment cannot be evaluated in cell lines because they lack immune and other cell types that constitute a microenvironment.
The mitogenic effect of estrogen is also supported by observations of increased cellular proliferation induced by A3B expression in breast cancer cells (39). Furthermore, elimination of A3B expression by siRNA resulted in a reduction of estrogen-induced growth in ER+, but not ER− breast cancer cell lines (35). The potent effect of estrogen on the activation of transcriptional programming and acceleration of the cell cycle accompanied by DNA damage in cotranscriptional structures (RNA:DNA hybrids) in ER+ breast cancer cells was compared with oncogene exposure (40). Analysis of estrogen metabolites in a large cohort of women also supported the mitogenic rather than the direct mutagenic effect of estrogen metabolites (10).
It is unclear if A3B expression might be induced by the naturally high and variable levels of endogenous estrogen (such as in younger women) or by the levels outside of the normal age-related range (such as in HRT in older women). Increased A3B expression in ER+ normal and dormant breast cancer cells might contribute to the reported association between HRT and cancer risk (2–5). Anticancer chemotherapy drugs, such as cisplatin, represent another common exposure that can induce A3B expression in cell lines. In our experiments, A3B expression was induced by cisplatin above the level already induced by estrogen treatment in ER+ breast cells, but the effect of cisplatin on A3B expression was attenuated by estradiol in ER− breast cells. Thus, treatment with cisplatin in women with high levels of endogenous estrogen or receiving HRT may result in a further increase of A3B expression. However, in women with ER− breast tumors, endogenous estrogen or HRT may not affect A3B expression, even during cisplatin therapy.
It has been previously reported (16,35,41) and reiterated in our current work that A3B is expressed significantly higher in ER− compared with ER+ cell lines (Figure 3C), and in breast tumors in TCGA, even after adjusting for relevant covariates, such as age at diagnosis, race and tumor stage (Figure 3D). A3B expression is controlled by p53 (42,43) and since ER− tumors are enriched for inactivating p53 mutations (13,44), this could contribute to elevated levels of A3B expression in ER− tumors. Both the triple-negative cell line MDA-MB-231 and the ER+ luminal A cell line T-47D used in our experiments carried p53 mutations, but A3B expression was significantly higher in the MDA-MB-231 cells (Figure 3C). This could be because of the different functional properties of these p53 mutations and/or other factors, such as chromosomal instability and replication stress (21) that are more predominant in ER− than ER+ breast cancer cells.
Previously, a germline genetic variant rs17000526-A has been associated with increased A3B expression in bladder and breast tumors (20). A detailed analysis stratified by ER status showed that rs17000526 was associated with increased A3B expression (P = 0.006, Figure 3B) and better survival [hazard ratio = 0.40 (0.18–0.90), P = 0.024] (20) but only in patients with ER− tumors. In patients with ER+ tumors, there was no association for this SNP with A3B expression (Figure 3A) or survival (20). Notably, analysis of ENCODE data showed three ERα binding sites within the A3B genomic region (Figure 2A), including one site mapping just 6 bp upstream of the rs17000526 in one of two ER+ endometrial cancer cell lines tested, while no ERα binding sites were detected in T-47D cells in this genomic region. Previously, we detected strong but non-allele-specific interaction between rs17000526 and nuclear proteins from another ER+ cell line, MCF7 (20). Thus, the non-allele-specific binding of ERα to the A3B enhancer (that includes rs17000526) could contribute to the induction of A3B expression in some ER+ cells, masking more subtle effects, such as of the SNP rs17000526, which is associated with A3B expression in tissues not affected by estrogen, such as bladder tumors (20).
While we found rs17000526 to be associated with A3B expression in ER− breast tumors, this SNP is not significantly associated with breast cancer risk (20). This could be because APOBEC mutagenesis in breast tissue is more strongly associated with A3A than A3B expression (20). Additionally, the strong effect of estrogen on A3B expression in ER+ tumors would mask an effect of rs17000526 and explain why this SNP is not associated with breast cancer risk in ER+ tumors, while this association might be more noticeable in ER− tumors in the absence of the effect of estrogen on A3B expression.
Although our results suggest that estrogen exposure induces A3B expression in ER+ breast cells, A3B expression is unlikely to account for APOBEC-mediated mutagenesis in breast tumors but might contribute to cancer development based on the possible mitogenic effect of A3B. Our findings provide stronger support for the mitogenic rather than mutagenic cancer-promoting effects of estrogen but without a significant impact on APOBEC-mediated mutagenesis detectable in breast tumors in TCGA. We have not addressed the direct causal connection between exposure to endogenous and exogenous estrogen, A3B expression in human samples, and breast cancer risk. Epidemiological studies with large cohorts of breast cancer patients would be necessary to address these relationships in breast tumors of different subtypes with consideration of all relevant covariates such as environmental exposures, germline and somatic variants, and clinical outcomes.
Supplementary Material
Glossary
Abbreviations
- ER
estrogen receptor
- HRT
hormone replacement therapy
- TCGA
The Cancer Genome Atlas
Funding
The work was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics (DCEG) of the National Cancer Institute, NIH. We thank Drs Gretchen Gierach and Xiaohong Rose Yang from the Integrated Tumor Epidemiology Branch (ITEB), DCEG, NCI for critical comments and discussions. The presented results are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Conflict of Interest Statement: None declared.
References
- 1. Cui J., et al. (2013) Estrogen synthesis and signaling pathways during aging: from periphery to brain. Trends Mol. Med., 19, 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ross R.K., et al. (2000) Effect of hormone replacement therapy on breast cancer risk: estrogen versus estrogen plus progestin. J. Natl. Cancer Inst., 92, 328–332. [DOI] [PubMed] [Google Scholar]
- 3. Beral V.; Million Women Study Collaborators (2003) Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet, 362, 419–427. [DOI] [PubMed] [Google Scholar]
- 4. Chlebowski R.T., et al. (2015) Breast cancer after use of estrogen plus progestin and estrogen alone: analyses of data from 2 women’s health initiative randomized clinical trials. JAMA Oncol., 1, 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collaborative Group on Hormonal Factors in Breast, C. (2019) Type and timing of menopausal hormone therapy and breast cancer risk: individual participant meta-analysis of the worldwide epidemiological evidence. Lancet, 394, 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Toniolo P.G., et al. (1995) A prospective study of endogenous estrogens and breast cancer in postmenopausal women. J. Natl. Cancer Inst., 87, 190–197. [DOI] [PubMed] [Google Scholar]
- 7. Rosenberg C.R., et al. (1994) Premenopausal estradiol levels and the risk of breast cancer: a new method of controlling for day of the menstrual cycle. Am. J. Epidemiol., 140, 518–525. [DOI] [PubMed] [Google Scholar]
- 8. Yager J.D., et al. (2006) Estrogen carcinogenesis in breast cancer. N. Engl. J. Med., 354, 270–282. [DOI] [PubMed] [Google Scholar]
- 9. Hankinson S.E., et al. (2004) Towards an integrated model for breast cancer etiology: the lifelong interplay of genes, lifestyle, and hormones. Breast Cancer Res., 6, 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sampson J.N., et al. (2017) Association of estrogen metabolism with breast cancer risk in different cohorts of postmenopausal women. Cancer Res., 77, 918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seeger H., et al. (2006) Comparison of possible carcinogenic estradiol metabolites: effects on proliferation, apoptosis and metastasis of human breast cancer cells. Maturitas, 54, 72–77. [DOI] [PubMed] [Google Scholar]
- 12. Yager J.D. (2000) Endogenous estrogens as carcinogens through metabolic activation. J. Natl. Cancer Inst. Monogr., 27, 67–73. [DOI] [PubMed] [Google Scholar]
- 13. Cancer Genome Atlas, N. (2012) Comprehensive molecular portraits of human breast tumours. Nature, 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nik-Zainal S., et al. (2017) Mutational signatures in breast cancer: the problem at the DNA level. Clin. Cancer Res., 23, 2617–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alexandrov L.B., et al. ; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. (2013) Signatures of mutational processes in human cancer. Nature, 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roberts S.A., et al. (2013) An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet., 45, 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harris R.S. (2015) Molecular mechanism and clinical impact of APOBEC3B-catalyzed mutagenesis in breast cancer. Breast Cancer Res., 17, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burns M.B., et al. (2013) APOBEC3B is an enzymatic source of mutation in breast cancer. Nature, 494, 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nik-Zainal S., et al. ; Breast Cancer Working Group of the International Cancer Genome Consortium. (2012) Mutational processes molding the genomes of 21 breast cancers. Cell, 149, 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Middlebrooks C.D., et al. (2016) Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat. Genet., 48, 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kanu N., et al. (2016) DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer. Genome Biol., 17, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rugo H.S., et al. (2017) Use of neoadjuvant platinum-the ongoing conundrum. JAMA Oncol., 3, 1312–1314. [DOI] [PubMed] [Google Scholar]
- 23. Forbes S.A., et al. (2017) COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res., 45, D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muller P.A., et al. (2014) Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell, 25, 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coffill C.R., et al. (2012) Mutant p53 interactome identifies nardilysin as a p53R273H-specific binding partner that promotes invasion. EMBO Rep., 13, 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Girardini J.E., et al. (2011) A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell, 20, 79–91. [DOI] [PubMed] [Google Scholar]
- 27. Vikhanskaya F., et al. (2007) Cancer-derived p53 mutants suppress p53-target gene expression—potential mechanism for gain of function of mutant p53. Nucleic Acids Res., 35, 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dutil J., et al. (2019) An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines. Cancer Res., 79, 1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horwitz K.B., et al. (1978) Estrogen control of progesterone receptor in human breast cancer: role of estradiol and antiestrogen. Endocrinology, 103, 1742–1751. [DOI] [PubMed] [Google Scholar]
- 30. Mohammed H., et al. (2015) Progesterone receptor modulates ERα action in breast cancer. Nature, 523, 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nik-Zainal S., et al. (2014) Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet., 46, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Caval V., et al. (2014) A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3′UTR enhances chromosomal DNA damage. Nat. Commun., 5, 5129. [DOI] [PubMed] [Google Scholar]
- 33. Xuan D., et al. (2013) APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis, 34, 2240–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Law E.K., et al. (2016) The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv., 2, e1601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Periyasamy M., et al. (2015) APOBEC3B-mediated cytidine deamination is required for estrogen receptor action in breast cancer. Cell Rep., 13, 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sieuwerts A.M., et al. (2014) Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm. Cancer, 5, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tokunaga E., et al. (2016) Expression of APOBEC3B mRNA in primary breast cancer of Japanese women. PLoS One, 11, e0168090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang D., et al. (2019) APOBEC3B interaction with PRC2 modulates microenvironment to promote HCC progression. Gut, 68, 1846–1857. [DOI] [PubMed] [Google Scholar]
- 39. Cescon D.W., et al. (2015) APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc. Natl. Acad. Sci. USA, 112, 2841–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tubbs A., et al. (2017) Endogenous DNA damage as a source of genomic instability in cancer. Cell, 168, 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fujiki Y., et al. (2018) APOBEC3B gene expression as a novel predictive factor for pathological complete response to neoadjuvant chemotherapy in breast cancer. Oncotarget, 9, 30513–30526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Periyasamy M., et al. (2017) p53 controls expression of the DNA deaminase APOBEC3B to limit its potential mutagenic activity in cancer cells. Nucleic Acids Res., 45, 11056–11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Menendez D., et al. (2017) The cytidine deaminase APOBEC3 family is subject to transcriptional regulation by p53. Mol. Cancer Res., 15, 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Curtis C., et al. ; METABRIC Group. (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature, 486, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.