Abstract
As exome sequencing expands as a diagnostic tool, patients and providers have voiced concerns about communicating the breadth and scope of potential results when obtaining informed consent. This study aimed to understand how genetic counselors prioritize essential components of the informed consent process and whether counselor factors influence these decisions. Development of a best-worst scaling experiment was informed by a systematic literature review and two focus groups. Eleven choice sets were created using a balanced incomplete block design, where participants selected the most and least important object in each set. Mean best-worst scores were calculated to summarize the relative importance of each object, and mediation analyses assessed whether responses were associated with genetic counselor factors and attitudes. 342 members of the National Society of Genetic Counselors completed the online survey. Ranking of best-worst scores suggests that participants prioritize collaborative decision-making, assessing understanding and managing expectations, with the least emphasis placed on discussing technological complexities. Stratified analyses found that counselors more experienced with obtaining informed consent for exome sequencing and those reporting higher perceptions of patients’ ability to manage information rated discussing variants of uncertain significance as significantly more important (p<0.05). Our results suggest that genetic counselors report intentions to prioritize individual patient needs when obtaining informed consent for exome sequencing. Professional characteristics and attitudes may influence preemptive discussion of uncertain results.
Keywords: Exome sequencing, genetic counseling, informed consent
Introduction
Since its introduction as a research and clinical tool, exome sequencing has been met with both enthusiasm and trepidation. Despite the excitement surrounding its diagnostic potential, there has been much debate about how to manage secondary findings, evolving result interpretation, and variants of uncertain significance (VUS) (Bertier et al. 2017; Green et al. 2013). Uncertainties pervade sequencing as the majority of variants identified are classified as VUS and lack targeted management recommendations (Bertier et al. 2017; Han et al. 2017). The unpredictable nature and broad scope of potential results introduce challenges to traditional informed consent for more targeted genetic testing (Bernhardt et al. 2015).
Effective approaches to consent are essential to ensure that patients and research participants understand the limitations and potential uncertainties of exome sequencing (Bernhardt et al. 2015; Biesecker and Green 2014). Broadly, informed consent should address expected outcomes of testing including the type and likelihood of results that will or will not be returned, and the risks, benefits, limitations, alternatives and potential implications for relatives (ACMG Board of Directors 2013). While these requirements have been supported by professional guidelines and consent documents, there is still considerable variability in the approaches to communicating this information (Henderson et al. 2017).
Most often genetic counselors are the professionals obtaining informed consent for exome sequencing (Machini et al. 2014). They report concerns about the amount and complexity of information included in consent documents and professional guidelines that may negatively impact how they communicate these concepts (Bernhardt et al. 2015; Rigter et al. 2014). Studies of communication about genetic testing have found that professional presentation of biomedical information often dominates the interaction, leading to limited patient participation (Paul et al. 2015; Roter et al. 2006). Despite clear intentions and attempts to engage patients, discussions of exome sequencing have also been shown to be dominated by providers teaching genomics (Walser et al. 2017; Levenseller et al. 2014). This pattern may explain parents’ reports that pretest counseling underemphasizes discussions of the aspects of testing they value, such as how results may impact daily life (Krabbenborg et al. 2016).
The challenges to consent for exome sequencing are particularly salient in the pediatric setting where children undergoing a diagnostic workup have the potential to learn information about risk for unrelated adult onset conditions that could influence developing self-identity (Miller et al. 2017; Werner-Lin et al. 2016). The decision to opt out of receiving medically actionable secondary findings should be made prior to undergoing testing, and parents who have difficulty grasping the potential consequences of the results may report lower confidence in their ability to make an informed choice (Turbitt et al. 2018; O’Daniel et al. 2017). Consent conversations with parents can also be complicated by the involvement of children in providing assent and the possible implications for parents and relatives when trio testing is ordered (Bernhard et al. 2015; Levenseller et al. 2014). Furthermore, families of children affected with developmental delays and/or rare undiagnosed conditions often have high expectations for the diagnostic capability of sequencing that may inhibit their ability to fully accept the limitations of testing (Tomlinson et al. 2016).
Previous research has informed proposed essential elements of consent for exome sequencing, but to our knowledge has not explored how genetic counselors intend to prioritize these factors in consent interactions with patients (Ayuso et al. 2014). Gaining an understanding of genetic counselors’ intentions to obtain informed consent can identify potential discordance between patient and provider priorities and lead to development of more effective patient-centered practices. To address this research gap, we selected a Best-Worst Scaling (BWS) task to assess how genetic counselors prioritize elements of consent when presenting options to parents for their child to undergo exome sequencing. Our analysis aimed to explore the relationship between prioritization decisions and counselor factors, such as experience with exome sequencing and tolerance for ambiguity. We further assessed the role of communication efficacy (one’s perceived ability to communicate information) and target efficacy (belief in the patient’s ability to manage the information), two concepts central to the Theory of Motivated Information Management (Afifi and Weiner 2004). We hypothesized that genetic counselors reporting more experience with exome sequencing and higher communication efficacy would place less emphasis on discussing the technical aspects of sequencing, and prioritize discussions about outcomes of testing and decision-making.
Methods
Best-Worst Scaling Task Design
Best-worst scaling (BWS) is a choice-based preference elicitation measurement that generates a ranking based upon participant report of factors that are most and least valued (Louviere et al. 2013). This approach requires participants to select only the best and worst items in each of a series of choice sets, thereby eliminating the need to make indiscriminate choices about midrange items (Louviere et al. 2103; Peay et al. 2016). This methodology has been utilized extensively in the healthcare field to investigate priorities and preferences among various stakeholders including patients, clinicians and policy makers (Cheung et al. 2016). Previous applications of BWS in the context of genetics research have explored perceived barriers to personalized medicine and prioritized outcomes of genetic testing (Najafzadeh et al. 2012; Severin et al. 2013).
Design of a best-worst scaling (BWS) experiment was conduceted in concordance with current best practices (Bridges et al. 2011). Best-worst scaling Case 1 (object case) was selected based on the exploratory nature of the research question. Objects were proposed using evidence from the published literature and qualitative analysis of focus group data to encompass all relevant aspects of informed consent for exome sequencing (Bridges et al. 2011). Object development was conducted in a two-step process, starting with the conceptual delineation of each item followed by the refinement of meaningful language.
A systematic review of the literature was undertaken to develop a preliminary list of elements essential to informed consent for exome sequencing. PubMed, EMBASE, Scopus, Web of Science and Cochrane databases were queried using the following combinations of key search terms “genome sequencing” OR “exome sequencing” OR “next generation sequencing” AND “informed consent”; AND/OR “genetic counseling” AND “barriers”. This search yielded 294 abstracts, and papers were eligible for inclusion if published from January 2010, to February 2017, written in English, and reporting quantitative or qualitative primary data from patients, research participants or professionals in a research or clinical setting. Eleven papers met our inclusion criteria, and qualitative meta-analysis of data across studies yielded 17 distinct challenges.
Two focus groups were subsequently conducted with six genetic counselors from two institutions, one with a primarily clinical focus and one research institution. All focus group participants reported previous experience with obtaining consent for exome sequencing. Focus group participants were asked to draw upon their experience to provide oral ansd written feedback of the preliminary objects and to suggest additional objects that may not have been captured. Focus group participants generally endorsed the preliminary list and suggested removing select objects due to redundancy and dividing one concept into two distinct items. These results were incorporated to devise the final list of objects, summarized in Table 1.
Table 1.
Finalized list of objects used in the best-worst scaling experiment
| Object Label | Captured Element of Informed Consent |
|---|---|
| Assessing understanding | Assessing patient understanding of key content |
| Complex technology | Describing the technological complexities of sequencing |
| Managing expectations | Managing patient expectations about the results |
| Evolving interpretation | Explaining the evolution of variant interpretation |
| Engaging decision-making | Engaging patients in collaborative decision-making |
| Tailoring to context | Tailoring consent to the indication and context |
| Secondary findings | Deliberating about the implications of secondary findings |
| VUS Results | Explaining variants of uncertain significance |
| Negative results | Clarifying the meaning of a negative result |
| Family implications | Conveying potential implications for relatives |
| Insurance discrimination | Warning for insurance discrimination |
The final list of eleven objects was organized into choice sets using a Balanced Incomplete Block Design (BIBD) so that each item appears the same number of times and co-appears with all other items evenly (Louviere et al. 2013). The BIBD used in this study was generated using the “support.BWS” package for R statistical software (Aizaki and Aizaki 2017). After choice sets were constructed, the objects were randomized within each choice set to control for potential bias from ordering effects. The order in which choice sets were presented was randomized as well.
Study Instrument Design
In addition to the BWS experiment, the survey assessed participant characteristics, professional experience, and attitudes toward informed consent for exome sequencing. Demographic characteristics included age, gender, race and ethnicity, geographic location, years in practice, and practice specialty. Attitude toward uncertainty was assessed using the modified Tolerance for Ambiguity (TFA) scale, a 7-item measure capturing the degree to which an individual is comfortable approaching uncertain situations (Gellar et al. 1993). This scale was previously validated for use in the context of genetic testing. A 6-item knowledge check was adapted from the exome sequencing domain of the UNC Genomic Knowledge Scale to assess understanding of relevant basic concepts (Langer et al. 2017). This scale was originally designed for a patient population; thus, it was expected that most participants would answer correctly allowing responses to be used as a validity check for the sample. Communication efficacy and target efficacy were assessed with four items each rated using a Likert scale. These were modeled on scales previously validated in studies framed by the Theory of Motivated Information Management (Fowler and Afifi, 2011).
A hypothetical pediatric clinical scenario was designed to provide a standardized context for the BWS tasks and a consistent frame of reference for the objects (Bridges et al. 2011). Prior to initiation of the BWS experiment, objects were presented individually, and participants were asked to report the degree to which they perceived each object to be challenging using a sliding scale from 0 to 100. The BWS tasks required participants to select the single ‘most important’ and ‘least important’ elements of informed consent in each five-item choice set. A selection had to be made before advancing to the next choice set. A sample choice set appears in Figure 1.
Figure 1:
Sample choice set.
The online survey was assembled and distributed using Survey Monkey Platinum. Four practicing genetic counselors pilot tested the instrument, and feedback was incorporated prior to survey finalization. Modifications included providing more detailed instructions for the BWS task and presenting explanations of each object below the choice sets in a standardized order for clarity.
Recruitment and Participants
Eligible participants were full members of the National Society of Genetic Counselors (NSGC) practicing in any specialty or setting in the United States or Canada. Previous experience ordering exome sequencing was not required so as to capture a wider range of perspectives and assess potential differences among respondents with varying levels of exposure to exome consent interactions. Study invitations were sent via the NSGC email listserv to over 4,040 genetic counselors, and a reminder email was sent two weeks later. Participants were offered a $10 Amazon gift card for completing the survey. Both the focus group interview study and the anonymous survey were determined to be exempt from IRB approval by the NIH Office of Human Subject Research Protection (OHSRP).
Data Analysis
BWS data was analyzed using a count-based approach. In this method, the relative importance of each object is reported as a Best-Worst (BW) score calculated by subtracting the total number of times that the item was selected as the least important from the number of times that it was chosen as the most important (Louviere et al. 2013; Peay et al. 2016). A mean BW score for each object can then be generated and used for standardized ranking. Individual BW scores were also calculated for every participant, and ranged from −5 to +5 for each object. Standardized mean BW scores were rank ordered and reported with standard errors and 95% confidence intervals. Despite its relative simplicity, the count-based approach has been shown to yield valid object measures comparable to those derived with more complex regression-based models such as conditional logit and linear probability (Louviere et al. 2013). Thus, this method provides valid and reliable results, but is still conceptually accessible to the target population of genetic counselors and clinical policy makers (Peay et al. 2016).
Four stratified analyses were performed among groups that differed by level of previous experience obtaining informed consent for exome sequencing, tolerance for ambiguity, communication efficacy and target efficacy. Mean BW scores were generated for all objects in each group, and two tailed t-tests were conducted to assess whether between-group differences for any of the objects were statistically significant (Peay et al. 2016). Demographic data were reported using descriptive summary statistics for the whole sample and separated based on degree of experience with exome sequencing. The probability test function in Stata v14.0 was used to test whether characteristics varied significantly between the two groups.
Results
Participant Characteristics
Survey responses were collected from 08/17 to 09/17. A total of 375 genetic counselors initiated the survey, calculated as a response rate of 9.3%. Incomplete surveys were excluded, and 342 complete responses were used in the analysis. Ninety-seven percent of participants answered all items on the knowledge scale correctly, supporting validity of the sample. Participants reported a wide range of experience with exome sequencing; 7.6% had ordered 50 or more exome sequencing tests (n=26), 12% (n=41) had ordered 30–49, 19.8% (n=68) had ordered 10–29, the largest group (38.6%, n=132) had ordered sequencing 1–9 times; 22% (n=75) had never ordered it.
Table 2 summarizes respondent characteristics. Some participants declined to answer select demographic questions. The majority of participants (91.5%, n=313) were Caucasian females who reported working primarily in a clinical setting. These demographic findings matched that of the larger membership of NSGC and the target population of genetic counselors (Professional Status Survey 2016). The sample included genetic counselors in a variety of specialties, with the largest proportion working in pediatrics (31.8%, n=109). Most of the respondents had been in practice for less than five years (62.3%, n=213). Tolerance for ambiguity (TFA) varied among the sample, but generally trended towards lower TFA with a mean score of 20.0, ranging from 7 (highest TFA) to 31 (lowest TFA). Communication efficacy and target efficacy were high across the sample, with a mean score of 6 and 5 out of 7, respectively. Scores on these two efficacy scales were significantly correlated with each other (r=0.45, p<0.01).
Table 2.
Demographic characteristics
| Total Sample (n=324) | |
|---|---|
| Gender | n (%) |
| Female | 323 (94.4%) |
| Male | 18 (5.3%) |
| Age | |
| 20–29 years | 191 (55.8%) |
| 30–39 years | 108 (31.6%) |
| 40–49 years | 25 (7.3%) |
| 50–59 years | 14 (4.1%) |
| Ethnicity | |
| Caucasian | 313 (91.5%) |
| African American | 3 (0.9%) |
| Hispanic | 5 (1.5%) |
| Asian | 16 (4.7%) |
| Other | 5 (1.5%) |
| Years in Practice | |
| <5 | 213 (62.3%) |
| 5–9 | 77 (22.5% |
| 10–19 | 34 (9.9%) |
| 20–29 | 16 (4.7%) |
| 30+ | 2 (0.6%) |
| Geographic Region | |
| Northeast | 93 (27.9%) |
| Midwest | 106 (31%) |
| South | 64 (18.7%) |
| West | 58 (17.0%) |
| Primary Role | |
| Clinical Care | 269 (78.7%) |
| Research | 24 (7.0%) |
| Laboratory | 45 (31.2%) |
| Other | 7 (2.0%) |
| Primary Specialty | |
| Pediatric | 109 (31.9%) |
| Prenatal | 56 (16.4%) |
| Cancer | 85 (24.9%) |
| Other | 91 (26.6%) |
The total sample was separated into two groups based on experience completing the informed consent process for exome sequencing. The ‘less experienced’ group (60.8%) included those who had ordered exome sequencing zero to nine times, and the ‘more experienced’ group (39.2%) reported ordering sequencing 10 times or more. This division is consistent with criteria for experience with exome sequencing used in previous studies (Bernhardt et al. 2015).
Table 3 illustrates the results of Fischer’s exact tests that were conducted to determine significance of the association between experience with the consent process for exome sequencing and primary practice role (clinical vs. non-clinical) and specialty (pediatric vs. other specialty). There was a statistically significant association between working in primarily in pediatrics and reporting more experience with exome sequencing. This finding was expected and is consistent with the current clinical use of exome sequencing. The association between primarily clinical versus nonclinical practice and level of experience was not significant.
Table 3.
Participant practice characteristics and attitudes separated by experience with obtaining informed consent for exome sequencing
| Practice Characteristics | Total Sample | Less Experiencea | More Experienceb | P-Valuee |
| Primary Role | ||||
| Clinical Care | 269 | 158 | 111 | |
| Non-clinical Role | 70 | 47 | 23 | 0.22 |
| Specialty | ||||
| Pediatric | 109 | 32 | 77 | |
| Other | 232 | 176 | 56 | <0.01 |
| Attitude Scales | Mean (SD) | Mean (SD) | Mean (SD) | P-Valuef |
| Comm. Efficacyc | 6.09 (1.08) | 5.73 (1.19) | 6.64 (0.52) | <0.01 |
| Target Efficacyc | 4.98 (1.03) | 4.78 (0.97) | 5.32 (1.01) | <0.01 |
| Tol. For Ambiguityd | 20.20 (4.30) | 20.30 (4.46) | 19.60 (4.01) | 0.13 |
‘Less experience’ refers to participants who report having ordered exome sequencing 0–9 times.
‘More experience’ refers to those who have ordered exome sequencing 10 or more times
Efficacy scales are scored on a scale of 1–7 with 7 corresponding to highest perceived efficacy
Scores for Tolerance for Ambiguity range from 7 to 30, with 7 as the highest and 30 the lowest
Calculated via Fischer’s exact test
Calculated via t-test
There were also statistically significant differences in perceptions of both communication and target efficacy. The mean efficacy score reported by counselors in the ‘more experience’ group was significantly higher than mean efficacy scores for those in the ‘less experience’ group according to t-tests completed for both efficacy domains (p<0.01). These analyses are summarized in Table 3.
Prioritized Elements of Informed Consent
Before starting the best-worst scaling tasks, participants rated the degree to which they found each element challenging on a sliding scale from zero to one hundred. The top three challenging objects were: “managing expectations” (mean 41.85), “assessing understanding” (36.39), and “complex technology” (mean 34.98). “Family implications” was rated as least challenging (24.72).
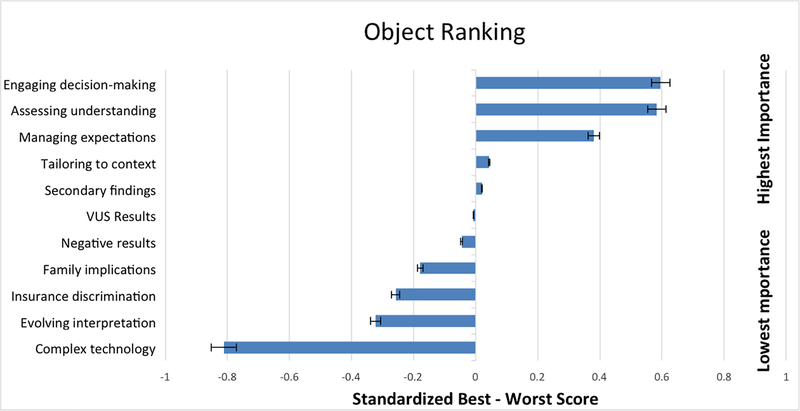
Ranked standardized BW scores with standard errors and 95% confidence intervals are represented graphically in Figure 2. “Engaging decision-making” was endorsed as the most important element of obtaining informed consent (BW=0.596, SE=0.019). Participants also prioritized “assessing understanding” (BW=0.584, SE=0.018) and “managing expectations” (BW=0.380, SE=0.018). The least important objects were “complex technology” (BW=−0.812, SE=0.015) and “evolving interpretation” (BW=−0.322, SE=0.018). “VUS results” (BW=0.250, SE=0.014) was ranked as the midpoint, and this mean BW score was not statically significantly different from zero (95% CI: −0.034, 0.02).
Figure 2:
Objects ranked by relative importance.
Ranking is based on standardized B-W scores. Positive scores indicate higher importance and negative scores represent lower importance.
Error bars represent a 5% error range.
Stratified BWS analyses were conducted by experience with obtaining informed consent for exome sequencing, communication efficacy, target efficacy and tolerance for ambiguity. For these analyses, the efficacy and TFA responses were divided into low and high groups based on the mean scores for each construct. Two-tailed t-tests were conducted to determine whether there were significant differences in standardized BW scores between the low and high groups. For each object comparison we first ran an F test to determine whether variances were equal or unequal, and then ran the appropriate t-test. Significant differences were observed for the following objects after stratifying by experience with consent for exome sequencing: “VUS results” (p=<0.001), “family implications” (p=<s0.001) and “negative results” (p<0.001). Respondents in the high perceived target efficacy group ranked “VUS results” as significantly more important than those in the low target efficacy group (p=0.04). There were no significant differences in standardized BW scores after stratifying by communication efficacy or tolerance for ambiguity. Numerical comparisons across these groups are summarized in Supporting Information Table S1.
Discussion
Our results suggest that genetic counselors most highly value elements of the informed consent process that facilitate patient engagement and shared deliberation. Conversely, they rank discussing complex technical information and variant interpretation as least important. These findings were consistent among counselors with and without direct experience with exome sequencing, suggesting that these intentions are rooted in professional values rather than individual experience with exome sequencing. Because these favored elements focus on understanding preferences of end users for testing, the stated priorities of the counselors are also congruent with the definition of informed choice as a decision that is made and implemented with sufficient knowledge and supported by individual values (Michie et al. 2002). Participants also highly rated the importance of managing expectations for potential results, an essential aspect of pretest counseling that may be challenging to convey (Bernhardt et al. 2015).
This study provides evidence for a unified view of intentions for approaching informed consent for exome sequencing, although additional research is needed to understand how these intentions translate into practice. Observations from a recent study of exome result return clearly demonstrated providers’ attempts to assess patient understanding, but found misunderstandings were not fully addressed and many of these sessions defaulted to information-heavy communication (Walser et al. 2017). This work may be valuable to future research as a framework for assessing concordance between the stated priorities and content of actual consent sessions. Identified gaps between intentions and practice can then be used to guide educational interventions. Genetic counselors have a clear desire and intention to facilitate understanding and collaborative decision-making, and may benefit from training in specific communication strategies, such as teach back methods, to further implement their counseling goals. These tools may be particularly beneficial when involving children or adolescents who may not be accustomed to participating in healthcare decisions (Werner-Lin et al. 2016).
Another major finding of the BWS study was that explaining variants of uncertain significance (VUS) was rated as significantly more important among respondents with more experience ordering exome sequencing and those with higher perceived target efficacy. Questions about the interpretation and communication of VUS results have been central to debates surrounding the use of exome sequencing, particularly given that there is currently a higher yield of VUS results than pathogenic variants. Counselors with more experience ordering exome sequencing may be more acutely aware of the high probability of obtaining a VUS result, and this may motivate them to prioritize this discussion during the consent interaction. More experience with exome sequencing may also lead to increased comfort with discussions of uncertainty and provision of anticipatory guidance prior to results disclosure. As such, counselors can help patients to frame VUS results in a productive manner as they may represent possibilities for clarity and an opportunity for partnering with the care team (Walser et al. 2017).
In the cancer genetics setting, VUS results have been associated with a moderate but significant increase in patient distress (Lumish et al. 2017). There is also evidence that some parents have difficulty conceptualizing the meaning of a VUS in the context of microarray testing (Reiff et al. 2012; Kiedrowski et al. 2016). Knowledge that it can be challenging for parents to understand and adapt to a VUS could explain why participants with lower perceptions of target efficacy ranked this object significantly lower than those in the high target efficacy group. Genetic counselors with more experience ordering exome sequencing reported significantly higher perceptions of target efficacy, suggesting that it may mediate the relationship between experience and value placed on discussing VUS results. While uncertainty cannot be eliminated from exome sequencing, providers have an opportunity to help patients understand differing sources of ambiguity and identify areas where they can attempt to find control in the face of uncertainty (Han et al. 2017). These ideas may be particularly helpful if explored during pretest counseling so that patients can begin to build a concept of uncertainty prior to receiving such a result.
Limitations
The findings of this study are limited by the contributions of participants who did not have direct experience with exome sequencing. More research is needed to understand how intentions as reported by this group may be implemented in practice. Despite this limitation, responses from these participants helped to identify relevant differences between groups with differing levels of experience. Though the sample was representative of the target population of genetic counselors, it may not be generalizable to the experiences of other healthcare providers or researchers who have the responsibility of obtaining consent for exome sequencing.
There could also be limitations in the ability of utilized scales to fully capture the constructs of interest. Particularly, more variance in perceptions of communication and target efficacy may have been identified if more items were included in these scales. Furthermore, tolerance for ambiguity is a complex construct encompassing a variety of cognitive and emotional reactions. A number of scales have been designed, though there is an observed lack of consistency and coherence across these measures (Hillen eti al. 2017). The scale used in this study was selected because it has been previously validated for use with genetics professionals (Gellar et al. 1993). However, we recognize that it may not have addressed the full spectrum of factors that contribute to an understanding of tolerance for ambiguity.
Despite the rigorous object development process, it is possible that important concepts were not captured by the included items. Inclusion of additional objects could have provided more nuanced results. Additionally, participants could have perceived conceptual overlap or clear superiority among some of the objects leading to difficulty in making direct comparisons. Meaning of each object could have been interpreted differently by individual participants, which would also have influenced responses. The study may have failed to capture perceived difference between the consent process for genome sequencing and exome sequencing, as we did not specifically address experience with genome sequencing in our survey. Future studies using a similar method to survey a patient population will make a valuable contribution to the literature.
Practice implications
As exome sequencing becomes more widely used, patients may benefit from an increased understanding of sources of uncertainty as well as clarification of their attitudes towards uncertainty. Acknowledgement of uncertainty should be included in the informed consent process, as uncertainty can be raised with any outcome of sequencing. Among respondents who had never ordered exome sequencing, the BWS task may have acted as a values clarification exercise to help participants begin thinking about what they find most challenging about exome sequencing and how they hope to approach these discussions in practice. While this was not the primary aim of the study, secondary gains of this nature could increase overall awareness of the challenges to consent introduced by exome sequencing. Our results suggest that genetic counselors strongly value collaborative decision-making and ensuring patient understanding. These priorities may be used as a framework to assess current consent proceses and should inform future training initiatives that prepare healthcare providers across all disciplines to incorporate exome sequencing into their clinical and research practices.
Supplementary Material
Acknowledgements
We would like to thank Holly Peay for her guidance and advice about best-worst scaling design and analyses.
The research presented in this paper was conducted while the first author was in training and fulfilled a degree requirement in 2018.
This research study was funded by the National Human Genome Research Institute Intramural Research Program, National Institutes of Health.
Footnotes
Conflict of Interest
Julie Cohen is a consultant for Invitae. Rachel Gore, John Bridges and Barbara Biesecker declare that they have no conflict of interest.
Human Studies and Informed Consent
This study was reviewed by the NIH Office of Human Subject Research Protection (OHSRP) and was determined to be exempt from IRB review in accordance with the US Federal Policy for the Protection of Human Subjects. All participants gave their informed consent prior to initiating the study survey.
Animal Studies
No non-human animal studies were carried out by the authors for this article.
References
- ACMG Board of Directors. (2013). Points to consider for informed consent for genome/exome sequencing. Genetics in Medicine, 15, 748. doi: 10.1038/gim.2013.94 [DOI] [PubMed] [Google Scholar]
- Afifi WA, & Weiner JL (2004). Toward a theory of motivated information management. Communication Theory, 14, 167–190. doi: 10.1111/j.1468-2885.2004.tb00310.x [DOI] [Google Scholar]
- Aizaki H, & Aizaki MH (2017). Package ‘support. BWS’. [Google Scholar]
- Ayuso C, Millán JM, Mancheno M, & Dal-Ré R (2013). Informed consent for whole-genome sequencing studies in the clinical setting. Proposed recommendations on essential content and process. European Journal of Human Genetics, 21, 1054. doi: 10.1038/ejhg.2012.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt B, Roche MI, Perry DL, Scollon SR, Tomlinson AN, & Skinner D (2015). Experiences with obtaining informed consent for genomic sequencing. American Journal of Medical Genetics, 167, 2634–2646. doi: 10.1002/ajmg.a.37256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertier G, Hétu M, & Joly Y (2016). Unsolved challenges of clinical whole-exome sequencing: a systematic literature review of end-users’ views. BMC medical genomics, 9, 52. doi: 10.1186/s12920-016-0213-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, & Green RC (2014). Diagnostic clinical genome and exome sequencing. New England Journal of Medicine, 370, 2418–2425. doi: 10.1056/NEJMra1312543 [DOI] [PubMed] [Google Scholar]
- Bridges JF, Hauber AB, Marshall D, Lloyd A, Prosser LA, Regier DA, … Mauskopf J (2011). Conjoint analysis applications in health—a checklist: a report of the ISPOR Good Research Practices for Conjoint Analysis Task Force. Value in Health, 14, 403–413. doi: 10.1016/j.jval.2010.11.013 [DOI] [PubMed] [Google Scholar]
- Cheung K, Wijnen B, Hollin I, Janssen E, Bridges J, Evers S, & Hiligsmann M (2016). Using best-worst scaling to investigate preferences in health care. Pharmacoeconomics, 34(12):1195–1209. doi: 10.1007/s40273-016-0429-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler C, & Afifi WA (2011). Applying the theory of motivated information management to adult children’s discussions of caregiving with aging parents. Journal of Social and Personal Relationships, 28, 507–535. doi: 10.1177/0265407510384896 [DOI] [Google Scholar]
- Geller G, Tambor ES, Chase GA, & Holtzman NA (1993). Measuring physicians’ tolerance for ambiguity and its relationship to their reported practices regarding genetic testing. Medical Care, 989–1001. [DOI] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, … Rehm HL (2013). ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genetics in Medicine, 15, 565. doi: 10.1038/gim.2013.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han PK, Umstead KL, Bernhardt BA, Green RC, Joffe S, Koenig B, … Biesecker BB (2017). A taxonomy of medical uncertainties in clinical genome sequencing. Genetics in Medicine, 19, 918. doi: 10.1038/gim.2016.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson GE, Wolf SM, Kuczynski KJ, Joffe S, Sharp RR, Parsons DW, … Appelbaum PS (2014). The challenge of informed consent and return of results in translational genomics: empirical analysis and recommendations. The Journal of Law, Medicine & Ethics, 42, 344–355. doi: 10.1111/jlme.12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen MA, Gutheil CM, Strout TD, Smets EM, & Han PK (2017). Tolerance of uncertainty: conceptual analysis, integrative model, and implications for healthcare. Social Science & Medicine,180, 62–75. doi: 10.1016/j.socscimed.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Kiedrowski LA, Owens KM, Yashar BM, & Schuette JL (2016). Parents’ perspectives on variants of uncertain significance from chromosome microarray analysis. Journal of Genetic Counseling, 25, 101–111. doi: 10.1007/s10897-015-9847-3 [DOI] [PubMed] [Google Scholar]
- Krabbenborg L, Schieving J, Kleefstra T, Vissers LELM, Willemsen MA, Veltman JA, & Van Der Burg S (2016). Evaluating a counselling strategy for diagnostic WES in paediatric neurology: an exploration of parents’ information and communication needs. Clinical Genetics, 89, 244–250. doi: 10.1111/cge.12601 [DOI] [PubMed] [Google Scholar]
- Langer MM, Roche MI, Brewer NT, Berg JS, Khan CM, Leos C, … Rini C (2017). Development and validation of a genomic knowledge scale to advance informed decision-making research in genomic sequencing. MDM Policy & Practice, 2. doi: 10.1177/2381468317692582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenseller BL, Soucier DJ, Miller VA, Harris D, Conway L, & Bernhardt BA (2014). Stakeholders’ opinions on the implementation of pediatric whole exome sequencing: implications for informed consent. Journal of Genetic Counseling, 23, 552–565. doi: 10.1007/s10897-013-9626-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louviere J, Lings I, Islam T, Gudergan S, & Flynn T (2013). An introduction to the application of (case 1) best–worst scaling in marketing research. International Journal of Research in Marketing, 30, 292–303. doi: 10.1016/j.ijresmar.2012.10.002 [DOI] [Google Scholar]
- Lumish HS, Steinfeld H, Koval C, Russo D, Levinson E, Wynn J, … Chung WK (2017). Impact of panel gene testing for hereditary breast and ovarian cancer on patients. Journal of Genetic Counseling, 26, 1116–1129. doi: 10.1007/s10897-017-0090-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machini K, Douglas J, Braxton A, Tsipis J, & Kramer K (2014). Genetic counselors’ views and experiences with the clinical integration of genome sequencing. Journal of Genetic Counseling, 23, 496–505. doi: 10.1007/s10897-014-9709-4 [DOI] [PubMed] [Google Scholar]
- Michie S, Dormandy E, & Marteau TM (2002). The multi-dimensional measure of informed choice: a validation study. Patient Education and Counseling, 48, 87–91. doi: 10.1016/S0738-3991(02)00089-7 [DOI] [PubMed] [Google Scholar]
- Miller VA, Werner-Lin A, Walser SA, Biswas S, & Bernhardt BA (2017). An observational study of children’s involvement in informed consent for exome sequencing research. Journal of Empirical Research on Human Research Ethics, 12, 6–13. doi: 10.1177/1556264616674096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafzadeh M, Lynd LD, Davis JC, Bryan S, Anis A, Marra M, & Marra CA (2012). Barriers to integrating personalized medicine into clinical practice: a best–worst scaling choice experiment. Genetics in Medicine, 14, 520. doi: 10.1002/ajmg.a.35811 [DOI] [PubMed] [Google Scholar]
- National Society of Genetic Counselors. (2016). NSGC Professional Status Survey 2016. Chicago: National Society of Genetic Counselors. [Google Scholar]
- O’Daniel JM, McLaughlin HM, Amendola LM, Bale SJ, Berg JS, Bick D, … Cooper GM (2017). A survey of current practices for genomic sequencing test interpretation and reporting processes in US laboratories. Genetics in Medicine, 19, 575. doi: 10.1038/gim.2016.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J, Metcalfe S, Stirling L, Wilson B, & Hodgson J (2015). Analyzing communication in genetic consultations—a systematic review. Patient Education and Counseling, 98, 15–33. doi: 10.1016/j.pec.2014.09.017 [DOI] [PubMed] [Google Scholar]
- Peay HL, Hollin IL, & Bridges JFP (2016). Prioritizing parental worry associated with Duchenne muscular dystrophy using best-worst scaling. Journal of Genetic Counseling, 25, 305–313. doi: 10.1007/s10897-015-9872-2 [DOI] [PubMed] [Google Scholar]
- Reiff M, Bernhardt BA, Mulchandani S, Soucier D, Cornell D, Pyeritz RE, & Spinner NB (2012). “What does it mean?”: Uncertainties in understanding results of chromosomal microarray testing. Genetics in Medicine, 14, 250. doi: 10.1038/gim.2011.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigter T, Van Aart CJA, Elting MW, Waisfisz Q, Cornel MC, & Henneman L (2014). Informed consent for exome sequencing in diagnostics: exploring first experiences and views of professionals and patients. Clinical Genetics, 85, 417–422. doi: 10.1111/cge.12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roter D, Ellington L, Erby LH, Larson S, & Dudley W (2006). The genetic counseling video project (GCVP): models of practice. American Journal of Medical Genetics, 142, 209–220. doi: 10.1002/ajmg.c.30094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F, Schmidtke J, Mühlbacher A, & Rogowski WH (2013). Eliciting preferences for priority setting in genetic testing: a pilot study comparing best-worst scaling and discrete-choice experiments. European Journal of Human Genetics, 21, 1202. doi: 10.1038/ejhg.2013.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson AN, Skinner D, Perry DL, Scollon SR, Roche MI, & Bernhardt BA (2016). “Not tied up neatly with a bow”: professionals’ challenging cases in informed consent for genomic sequencing. Journal of Genetic Counseling, 25, 62–72. doi: 10.1007/s10897-015-9842-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turbitt E, Chrysostomou PP, Peay HL, Heidlebaugh AR, Nelson LM, & Biesecker BB (2018). A randomized controlled study of a consent intervention for participating in an NIH genome sequencing study. European Journal of Human Genetics, 26, 622. doi: 10.1038/s41431-018-0105-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser SA, Werner-Lin A, Mueller R, Miller VA, Biswas S, & Bernhardt BA (2017). How do providers discuss the results of pediatric exome sequencing with families?. Personalized Medicine, 14, 409–422. doi: 10.2217/pme-2017-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Lin A, Tomlinson A, Miller V, & Bernhardt BA (2016). Adolescent engagement during assent for exome sequencing. AJOB Empirical Bioethics, 7, 275–284. doi: 10.1080/23294515.2016.1197983 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.