Abstract
A NADH‐dependent engineered amine dehydrogenase from Geobacillus stearothermophilus (LE‐AmDH‐v1) was applied together with a NADH‐oxidase from Streptococcus mutans (NOx) for the kinetic resolution of pharmaceutically relevant racemic α‐chiral primary amines. The reaction conditions (e. g., pH, temperature, type of buffer) were optimised to yield S‐configured amines with up to >99 % ee.
Keywords: biocatalysis, kinetic resolution, amine dehydrogenases, α-chiral amines, oxidative deamination
Oxidative kinetic resolution catalysed by LE‐AmDH‐v1: LE‐AmDH‐v1 previously engineered from the ϵ‐deaminating L‐lysine dehydrogenase from Geobacillus s. catalyses the oxidative kinetic resolution of α‐methyl‐ and α‐ethyl‐benzylamines, 1‐aminotetralin, 4‐aminochromane, and 1‐aminoindan. The reaction yields S‐configured amines with up to >99 % ee in aqueous buffer at pH 7.4, at 30–50 °C within 8–16 h.

α‐Chiral amines are fundamental building blocks for the manufacturing of many active pharmaceutical ingredients (APIs), fine chemicals, and agrochemicals.1 An increasing number of newly approved drugs contain an α‐chiral amine core, and the legislative regulations for their commercialisation are stringent in terms of chemical and enantiomerical purity (i. e., total impurity amount <0.15 %). In this context, several biocatalytic methods to obtain enantiopure α‐chiral amines have been developed, including asymmetric synthesis from prochiral ketones using either ω‐transaminases (ωTA),2 or dehydrogenases (i. e., reductive aminases (RedAm), imine reductases (IRed), amine dehydrogenases (AmDH);3 from alkenes using either ammonia lyases4 or engineered cytochrome c;5 and from alkanes using engineered cytochrome P411 monooxygenases.3c, 6 Enantiomerically pure amines can also be obtained from a racemic mixture by either kinetic resolution (KR) or applying one among several available deracemisation strategies.7 KR is based on the use of an enantioselective catalyst that acts exclusively on one enantiomer while leaving the other untouched. Additionally, KR is a practical approach to assess the efficiency of a biocatalyst that can possibly then be implemented in a deracemisation method. For instance, KR can be combined with a racemisation catalyst (i. e., dynamic KR using e. g., Pd/C, Pd/AIO(OH), VOSO4, Ru or Ir complexes) or a hydride transfer reagent (e. g., NaCNBH3, NaBH4).7, 8 The applicability of KR and DKR for chiral amine synthesis has been demonstrated using hydrolases,8a ωTAs,2a, 2b, 2c, 2e, 2f, 9 and monoamine oxidases (MAOs),10 as well as with AmDHs or RedAms in combination with either a NADH oxidase or an alanine dehydrogenase.11
Complementary strategies include deracemisation cascades in which stereocomplementary AmDH and ωTA, or vice versa, perform stereoselective oxidative deamination followed by stereoselective reductive amination.12
The KR of α‐chiral amines can be performed using AmDHs in combination with a H2O‐forming NADH oxidase (NOx), thereby maximising the atom‐economy, as dioxygen acts as an oxidant and water is the by‐product. However, the restricted substrate scope of AmDHs imposes a limitation on the applicability of the AmDH‐NOx system, as most of the currently available AmDHs were engineered starting from structurally related wild‐type enzymes and using a similar engineering approach. However, we recently reported a highly stereoselective AmDH (LE‐AmDH‐v1), which was engineered starting from a wild‐type enzyme that does not catalyse any apparent asymmetric transformation in its natural reaction, namely the ϵ‐deaminating L‐lysine dehydrogenase (LysEDH) from Geobacillus stearothermophilus.[3°] LE‐AmDH‐v1 (i. e., LysEDH F173A variant) exhibited excellent activity and stereoselectivity in the asymmetric synthesis of pharmaceutically relevant R‐configured α‐chiral amines, including (R)‐α‐methylbenzylamines, (R)‐1‐aminotetraline, and (R)‐4‐aminochromane. Notably, the previously discovered AmDHs do not possess this property.3a, 3c, 3k, 3l, 3m, 3n, 3o, 3p, 3q, 3r, 3s, 3t, 3u Herein, we report the harnessing of the catalytic and stereoselective properties of LE‐AmDH‐v1 to carry out the KR (i. e., enantioselective oxidative deamination) of pharmaceutically relevant α‐chiral amines starting from a racemic mixture of α‐methyl‐ and α‐ethyl‐benzylamines (1–4 a), 1‐aminotetralin (5 a), 4‐aminochromane (6 a), and 1‐aminoindan (7 a).
Initially, we determined the optimum pH for the biocatalytic KR of 1 a as a substrate. The rate for KR of 1 a (10 mM) was investigated in a pH range from 2.5 to 9.8 using Britton‐Robinson's universal buffer and using LE‐AmDH‐v1 (45 μM), NAD+ (1 mM) and NOx from Streptococcus mutans 13 for cofactor recycling (3 μM) (Figure 1, Table S2). The study was performed at 40 °C due to the reported thermal stability of NOx, which is limited to 52 °C.13 We previously demonstrated that LE‐AmDH‐v1 is a thermostable enzyme (T m: 69 °C). Figure 1 shows that the reaction rate was highest at pH 7.4 (56.2 μM/min; for details see SI, Table S2). Although we previously showed that LE‐AmDH‐v1 better performed the reductive amination of ketones at pH 9–9.5, such basic pH values reduced the reaction's rate in the oxidative deamination direction. For instance, the reaction rates were 34.6 μM min−1 and 18.34 μM min−1 at pH 9.2 and 9.7, respectively. Notably, the reported maximum catalytic activity for the oxidative deamination of L‐lysine catalysed by the wild‐type LysEDH was also at pH 10.14 In the case of the applied NOx, the maximum catalytic activity was reported at pH 7–7.5, but the enzyme retained approximately 85 % of its activity at pH 10.13b Therefore, we conclude that the optimal pH of 7.4 for the KR of rac‐1 a is not due to the inherent catalytic activity and stability profile of the enzymes vs. pH. In contrast, we assume that the varied protonation state of the selected racemic amine substrates between pH 7 and 10 may influence the overall kinetics of the KR.
Figure 1.

Reaction rates for the oxidative KR of rac‐1 a (10 mM) at different pH values using Britton‐Robinson's buffer. Reaction conditions: LE‐AmDH‐v1 (45 μM), NAD+ (1 mM), NOx (3 μM), final volume 0.5 mL, at 40 °C. Time range varied from 5 to 40 min.
Next, we investigated the influence of different types of buffers (50 mM Tris‐HCl, 100 mM KPi, 100 mM MOPS, and 100 mM HEPES) at the optimum pH of 7.4 for the KR of 1 a (10 mM) catalysed by LE‐AmDH‐v1 (45 μM) and NOx (3 μM). The reaction time was set for 24 h and the temperature was reduced to 30 °C to avoid any evaporation of the product at longer reaction times such as was partially observed in the previous set of experiments. Figure 2 and Table S3 show that the KRs proceeded equally well in 50 mM Tris/HCl and 100 mM KPi buffers and reached the highest conversions of 48 % and 49 %, respectively. In both cases, the reaction reached completion after 24 h, as the enantiomeric excess (ee) of the remaining amine enantiomer (S) was >99 %.
Figure 2.
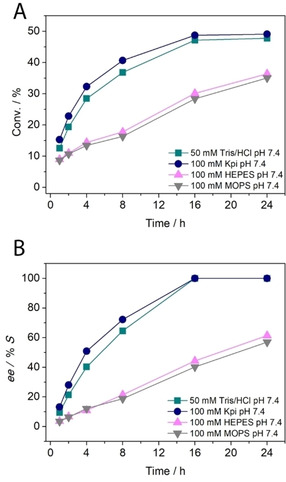
KR of rac‐1 a (10 mM) catalysed by LE‐AmDH‐v1 (45 μM) and using different types of buffers with NAD+ (1 mM) and NOx (3 μM) at 30 °C. (A) Progress of the KR over the time. (B) Variation of the ee of the unreacted enantiomer over the time.
A more detailed study showed that enantiopure (S)‐1 a was obtained after 16 h using both buffers, thus indicating that prolonging the reaction time is unnecessary (Figure 2B). In contrast, the LE‐AmDH‐v1 performed less efficiently in 100 mM MOPS buffer; a maximum conversion of only 35 % was obtained after 24 h (ee=57 % S). A similar scenario was observed when using a HEPES buffer (100 mM, pH 7.4), which provided a maximum conversion of 36 % (ee=61 % S) after 24 h.
With the aim of investigating the influence of temperature on the oxidative KR of 1 a, we monitored the progress of the reactions and variations in the ee of the remaining enantiomer at different temperatures (30 °C, 40 °C and 50 °C). Reactions were performed in Tris‐HCl (50 mM, pH 7.4) containing LE‐AmDH‐v1 (90 μM), NOx (10 μM), and analysed after 2, 4, 8, 16, and 24 h. Figure 3 and Table S4 show that complete KR of 1 a (ee>99 % S) was obtained after 16 h at 30 °C and 40 °C. However, the KR proceeded at higher rate at 40 °C; the ee reached 99 % after only 8 h, whereas it was only 95 % at 30 °C after the same amount of time. At 50 °C, the KR proceed even more rapidly, and only (S)‐1 a was detected (ee>99 %) in the reaction mixture after 8 h. However, due to the reported mesophilic thermal stability of NOx (Tm 52 °C), we decided to continue the study of KR on rac‐2–7 a at 30 °C (Table 1) and to eventually increase the temperature only in those cases in which resolution would not reach completion at 30 °C within 24 h.
Figure 3.

Progress of the KR of rac‐1 a (10 mM) at different temperatures (30 °C–50 °C) catalysed by LE‐AmDH‐v1 (90 μM) with NAD+ (1 mM) and NOx (10 μΜ).
Table 1.
KR of racemic amines 1–7 a employing LE‐AmDH‐v1.
|
| |||
|---|---|---|---|
|
Substrate |
Time [h] |
Conv. [%][a] |
ee [c] |
|
1 a |
24 |
49.8 |
>99.3 % (S) |
|
2 a |
48 |
49.7[b] |
>99.2 % (S) |
|
3 a |
48 |
46.1[b] |
95 % (S) |
|
4 a |
24 |
50.4 |
>99.4 % (S) |
|
5 a |
24 |
49.4 |
>99.5 % (S) |
|
6 a |
24 |
49.9 |
>99.3 % (S) |
|
7 a |
24 |
50.4 |
>99.6 % (S) |
[a] Reported conversions are normalised based on the response factors of the obtained amines/ketones as described in the SI, Table S1; reaction conditions: [substrate] 10 mM, [NAD+] 1 mM, [LE‐AmDH‐v1] 90 μM, [NOx] 10 μM, T 30 °C, time 24 h, [b] reaction time: 48 h, [c] level of accuracy based on GC‐detection limit and concentration of product.
The conversions and ee of the obtained amines are reported in Table 1. The production of enantiopure amines was generally achieved using LE‐AmDH‐v1 under the reported conditions, with the exception being fluoro‐substituted α‐methylbenzylamines. Starting from these racemic amines, the ee values of 2 a and 3 a were respectively 94 % and 77 % (S), thus indicating that the reaction did not reach completion at 30 °C after 24 h. Longer reaction times (48 h) allowed the KR to reach completion in the case of 2 a (ee=>99 %), whereas 3 a was obtained in higher enantioenriched form (ee=95 %). The same results were observed when rac‐2 a and rac‐3 a were resolved at 50 °C for 24 h, thereby affording ee values of >99 % and 95 %, respectively. These results indicate that higher temperatures can indeed accelerate the kinetics of the reaction, as previously reported for reductive amination of ketones catalysed by LE‐AmDH‐v1.[3°] Finally, LE‐AmDH‐v1 was able to resolve 5–7 a at 30 °C after 24 h; these bicyclic aromatic enantiopure amines are motives in many important pharmaceuticals, such as Rotigotine (a dopamine agonist), Norsertraline (a selective serotonin reuptake inhibitor, SSRI), and Resagiline (an irreversible inhibitor of monoamine oxidase‐B). Interestingly, LE‐AmDH‐v1 was unable to resolve racemic aliphatic amines such as 4‐methyl‐2‐pentylamine, 2‐pentylamine, 2‐hexylamine and 2‐heptylamine, although the related ketones were converted in the reductive amination reaction's direction.[3°]
In a previous work by Yun's group,11a the KR of rac‐1 a was attempted using the cFL1‐AmDH (also abbreviated as Ch1‐AmDH) that was previously engineered by Bommarius’ group.3m However, due to the significant inhibitory effect of 1 a on the activity of cFL1‐AmDH‐which was demonstrated in a recent publication,[3°] the authors had to use a large amount of whole cell biocatalysts (100 mgDCW mL−1) co‐expressing cFL1‐AmDH and a NADH oxidase (NOx) from Lactobacillus brevis 15 in order to resolve 20 mM of 1 a. We previously determined that LE‐AmDH‐v1 possesses 20‐fold lower IC50 and 38‐fold lower K I values for (R)‐1 a as an inhibitor compared with cFL1‐AmDH; therefore, we investigated the KR of 1 a catalysed by LE‐AmDH‐v1 (90 μM) at varied substrate concentrations. Figure 4 and Table S5 show that 90 μM of LE‐AmDH‐v1 was sufficient for resolving 50 mM of rac‐1 a after 24 h, thereby producing enantiopure (S)‐1 a (conv. 50 %) at 30 °C. Elevated ee (95 % S) and conversion into 1 b (48 %) were also obtained at 75 mM of 1 a, whereas KR was not effective at 100 mM of 1 a (ee=11 % S, conv. 14 %). Increasing the temperature to 40 °C resulted in complete resolution of 75 mM of rac‐1 a after 24 h using the same amount of LE‐AmDH‐v1. The resolution of 100 mM of rac‐1 a was also improved by increasing the temperature from 30 °C to 40 °C as the conversion increased from 14 % to 57 %.
Figure 4.

Influence of the substrate concentration in the KR of rac‐1 a (10–100 mM) catalysed by LE‐AmDH‐v1 (90 μM) with NAD+ (1 mM), NOx (10 μM) and at 30 °C, for 24 h.
Finally the synthetic applicability of the LE‐AmDH‐v1/NOx system was evaluated in a hundred‐milligram scale KR of rac‐ 1 a (50 mM). After 24 h, the KR reached completion (49.8 % conversion into 1 b) and 1 b was removed by extraction with MTBE under acidic conditions; after basification, (S)‐1 a was obtained in 43.6 % isolated yield (theoretical maximum is 50 %) and with 99.2 % ee.
In summary, we have applied the recently engineered LE‐AmDH‐v1 to the kinetic resolution of a selection of substrates that have thus far proved challenging for the other available AmDHs. For instance, only low activities were observed for the reductive amination of the respective ketone 5 b using Bb‐PhAmDH, cFL1‐AmDH and Cal‐AmDH,3l, 3m, 3p and no activity towards 6 b or 7 b was reported with any of the AmDHs developed to date. Similarly, cFL1‐AmDH performed the reductive amination of respective ketones 1–4 b yielding poor‐to‐moderate conversions after 48 h,3n and low activity was observed with Es‐LeuDH (AmDH) for the reductive amination of 1 b and 4 b.3s In the oxidative deamination direction (KR), only a few α‐methylbenzylamines could be resolved, but at the expense of a large amount of whole cells biocatalysts,11a and using a less atom‐efficient pyruvate/AlaDH system for NAD+ recycling.11b Herein, we used the LE‐AmDH‐v1/NOx system to access pharmaceutically relevant, optically active amines in S‐configuration starting from the respective racemic mixtures. The oxidative kinetic resolution proceeded efficiently at the optimum pH of 7.4 in 50 mM Tris‐HCl or 100 mM KPi. At 30 °C, the KR reached completion after 16 h and increasing the reaction temperature from 30 °C to 50 °C resulted in the resolution of 1 a within 8 h. Using these optimal conditions, the kinetic resolution of α‐methylbenzylamines, 1‐aminotetralin, 1‐aminoindane, and 4‐aminochromane was complete in all cases, with the exception of 3 a, which reached 95 % ee after 48 h. This work highlights the potential of AmDHs for the highly selective manufacture of API intermediates and the need to discover and engineer new AmDH variants in order to extend substrate scope and synthetic applicability.
Experimental Section
For general information, material, details and analytics, see SI.
Procedure for oxidative kinetic resolution in 100 mg scale.
KR of 1 a (50 mM, 100 mg, 0.825 mmol) was performed in a total volume of 16.5 mL of Tris‐HCl buffer (50 mM, pH 7.4), at 30 °C. The reaction mixture contained LE‐AmDH‐v1 (90 μM, 4 mg mL−1), NOx (10 μM, 0.5 mg mL−1), NAD+ (1 mM). After 24 h, a conversion of 49.8 % to 1 b was obtained. The reaction was acidified with concentrated HCl (1 mL) and ketone 1 b was extracted with MTBE (2×20 mL). After the addition of KOH (10 M, 1 mL), the amine (S)‐1 a was extracted with MTBE (3×20 mL). The combined organic layers were dried over MgSO4, filtered and concentrated under reduced pressure. Without any further purification step, (S)‐1 b was obtained with 43.6 % isolated yield (colourless liquid, 43.1 mg, 0.356 mmol) and with 99.2 % ee.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 Research and Innovation programme (ERC‐StG, grant agreement No 638271, BioSusAmin). Dutch funding from the NWO Sector Plan for Physics and Chemistry is also acknowledged.
V. Tseliou, T. Knaus, J. Vilím, M. F. Masman, F. G. Mutti, ChemCatChem 2020, 12, 2184.
References
- 1.
- 1a. Wittcoff H. A., Rueben B. G., Plotkin J. S., Industrial Organic Chemicals, 2nd ed., Wiley-Interscience, New York, 2004; [Google Scholar]
- 1b. Ghislieri D., Turner N. J., Top. Catal. 2013, 57, 284–300; [Google Scholar]
- 1c. Constable D. J. C., Dunn P. J., Hayler J. D., Humphrey G. R., Leazer J. J. L., Linderman R. J., Lorenz K., Manley J., Pearlman B. A., Wells A., Zaks A., Zhang T. Y., Green Chem. 2007, 9, 411–420; [Google Scholar]
- 1d. Nugent T. C., Chiral amine synthesis: Methods, Developments and Applications, Wiley-VCH, 2010. [Google Scholar]
- 2.
- 2a. Slabu I., Galman J. L., Lloyd R. C., Turner N. J., ACS Catal. 2017, 7, 8263–8284; [Google Scholar]
- 2b. Guo F., Berglund P., Green Chem. 2017, 19, 333–360; [Google Scholar]
- 2c. Kelly S. A., Pohle S., Wharry S., Mix S., Allen C. C. R., Moody T. S., Gilmore B. F., Chem. Rev. 2018, 118, 349–367; [DOI] [PubMed] [Google Scholar]
- 2d. Gomm A., O‘Reilly E., Curr. Opin. Chem. Biol. 2018, 43, 106–112; [DOI] [PubMed] [Google Scholar]
- 2e. Patil M. D., Grogan G., Bommarius A., Yun H., Catalysts 2018, 8, 254; [Google Scholar]
- 2f. Fuchs M., Farnberger J. E., Kroutil W., Eur. J. Org. Chem. 2015, 6965–6982; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2g. Savile C. K., Janey J. M., Mundorff E. C., Moore J. C., Tam S., Jarvis W. R., Colbeck J. C., Krebber A., Fleitz F. J., Brands J., Devine P. N., Huisman G. W., Hughes G. J., Science 2010, 329, 305–309; [DOI] [PubMed] [Google Scholar]
- 2h. Pavlidis I. V., Weiss M. S., Genz M., Spurr P., Hanlon S. P., Wirz B., Iding H., Bornscheuer U. T., Nat. Chem. 2016, 8, 1076–1082. [DOI] [PubMed] [Google Scholar]
- 3.
- 3a. Sharma M., Mangas-Sanchez J., Turner N. J., Grogan G., Adv. Synth. Catal. 2017, 359, 2011–2025; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3b. Mangas-Sanchez J., France S. P., Montgomery S. L., Aleku G. A., Man H., Sharma M., Ramsden J. I., Grogan G., Turner N. J., Curr. Opin. Chem. Biol. 2017, 37, 19–25; [DOI] [PubMed] [Google Scholar]
- 3c. Patil M. D., Grogan G., Bommarius A., Yun H., ACS Catal. 2018, 10985–11015; [Google Scholar]
- 3d. Dong J., Fernandez-Fueyo E., Hollmann F., Paul C. E., Pesic M., Schmidt S., Wang Y., Younes S., Zhang W., Angew. Chem. Int. Ed. 2018, 57, 9238–9261; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3e. Aleku G. A., France S. P., Man H., Mangas-Sanchez J., Montgomery S. L., Sharma M., Leipold F., Hussain S., Grogan G., Turner N. J., Nat. Chem. 2017; [DOI] [PubMed] [Google Scholar]
- 3f. Wetzl D., Gand M., Ross A., Müller H., Matzel P., Hanlon S. P., Müller M., Wirz B., Höhne M., Iding H., ChemCatChem 2016, 8, 2023–2026; [Google Scholar]
- 3g. Scheller P. N., Lenz M., Hammer S. C., Hauer B., Nestl B. M., ChemCatChem 2015, 7, 3239–3242; [Google Scholar]
- 3h. Roiban G.-D., Kern M., Liu Z., Hyslop J., Tey P. L., Levine M. S., Jordan L. S., Brown K. K., Hadi T., Ihnken L. A. F., Brown M. J. B., ChemCatChem 2017, 9, 4475–4479; [Google Scholar]
- 3i. Schober M., MacDermaid C., Ollis A. A., Chang S., Khan D., Hosford J., Latham J., Ihnken L. A. F., Brown M. J. B., Fuerst D., Sanganee M. J., Roiban G.-D., Nat. Can. 2019, 2, 909–915; [Google Scholar]
- 3j. Matzel P., Gand M., Höhne M., Green Chem. 2017, 19, 385–389; [Google Scholar]
- 3k. Abrahamson M. J., Vazquez-Figueroa E., Woodall N. B., Moore J. C., Bommarius A. S., Angew. Chem. Int. Ed. 2012, 51, 3969–3972; [DOI] [PubMed] [Google Scholar]
- 3l. Abrahamson M. J., Wong J. W., Bommarius A. S., Adv. Synth. Catal. 2013, 355, 1780–1786; [Google Scholar]
- 3m. Bommarius B. R., Schurmann M., Bommarius A. S., Chem. Commun. 2014, 50, 14953–14955; [DOI] [PubMed] [Google Scholar]
- 3n. Knaus T., Böhmer W., Mutti F. G., Green Chem. 2017, 19, 453–463; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3o. Tseliou V., Knaus T., Masman M. F., Corrado M. L., Mutti F. G., Nat. Commun. 2019, 10, 3717; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3p. Pushpanath A., Siirola E., Bornadel A., Woodlock D., Schell U., ACS Catal. 2017, 7, 3204–3209; [Google Scholar]
- 3q. Chen F.-F., Zheng G.-W., Liu L., Li H., Chen Q., Li F.-L., Li C.-X., Xu J.-H., ACS Catal. 2018, 8, 2622–2628; [Google Scholar]
- 3r. Ye L. J., Toh H. H., Yang Y., Adams J. P., Snajdrova R., Li Z., ACS Catal. 2015, 5, 1119–1122; [Google Scholar]
- 3s. Lowe J., Ingram A. A., Gröger H., Bioorg. Med. Chem. 2018, 26, 1387–1392; [DOI] [PubMed] [Google Scholar]
- 3t. Mayol O., Bastard K., Beloti L., Frese A., Turkenburg J. P., Petit J.-L., Mariage A., Debard A., Pellouin V., Perret A., de Berardinis V., Zaparucha A., Grogan G., Vergne-Vaxelaire C., Nat. Can. 2019, 2, 324–333; [Google Scholar]
- 3u. Mayol O., David S., Darii E., Debard A., Mariage A., Pellouin V., Petit J.-L., Salanoubat M., de Berardinis V., Zaparucha A., Vergne-Vaxelaire C., Catal. Sci. Technol. 2016, 6, 7421–7428. [Google Scholar]
- 4.
- 4a. Bartsch S., Vogel A., in Science of Synthesis, Biocatalysis in Organic Synthesis 2, Chapter 2.3.3 Addition of Ammonia and Amines to C=C Bonds (Eds.: K. Faber, W.-D. Fessner, N. J. Turner), Georg Thieme Verlag KG, Stuttgart (Germany), 2015, pp. 291–311; [Google Scholar]
- 4b. Li R., Wijma H. J., Song L., Cui Y., Otzen M., e Tian Y., Du J., Li T., Niu D., Chen Y., Feng J., Han J., Chen H., Tao Y., Janssen D. B., Wu B., Nat. Chem. Biol. 2018, 14, 664–670. [DOI] [PubMed] [Google Scholar]
- 5. Cho I., Prier C. K., Jia Z. J., Zhang R. K., Gorbe T., Arnold F. H., Angew. Chem. Int. Ed. 2019, 58, 3138–3142. [DOI] [PubMed] [Google Scholar]
- 6. Prier C. K., Zhang R. K., Buller A. R., Brinkmann-Chen S., Arnold F. H., Nat. Chem. 2017, 9, 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Musa M. M., Hollmann F., Mutti F. G., Catal. Sci. Technol. 2019, 9, 5487–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.
- 8a. Verho O., Backvall J. E., J. Am. Chem. Soc. 2015, 137, 3996–4009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b. Schrittwieser J. H., Velikogne S., Hall M., Kroutil W., Chem. Rev. 2018, 118, 270–348. [DOI] [PubMed] [Google Scholar]
- 9. Shin J.-S., Kim B.-G., Biotechnol. Bioeng. 1998, 60, 534–540. [DOI] [PubMed] [Google Scholar]
- 10.
- 10a. Turner N. J., Curr. Opin. Chem. Biol. 2010, 14, 115–121; [DOI] [PubMed] [Google Scholar]
- 10b. Dunsmore C. J., Carr R., Fleming T., Turner N. J., J. Am. Chem. Soc. 2006, 128, 2224–2225; [DOI] [PubMed] [Google Scholar]
- 10c. Ghislieri D., Green A. P., Pontini M., Willies S. C., Rowles I., Frank A., Grogan G., Turner N. J., J. Am. Chem. Soc. 2013, 135, 10863–10869; [DOI] [PubMed] [Google Scholar]
- 10d. Yasukawa K., Nakano S., Asano Y., Angew. Chem. Int. Ed. 2014, 53, 4428–4431; [DOI] [PubMed] [Google Scholar]
- 10e. Heath R. S., Pontini M., Bechi B., Turner N. J., ChemCatChem 2014, 6, 996–1002; [Google Scholar]
- 10f. Li G., Ren J., Yao P., Duan Y., Zhang H., Wu Q., Feng J., Lau P. C. K., Zhu D., ACS Catal. 2014, 4, 903–908; [Google Scholar]
- 10g. Leisch H., Grosse S., Iwaki H., Hasegawa Y., Lau P. C. K., Can. J. Chem. 2012, 90, 39–45; [Google Scholar]
- 10h. Herter S., Medina F., Wagschal S., Benhaim C., Leipold F., Turner N. J., Bioorg. Med. Chem. 2018, 26, 1338–1346; [DOI] [PubMed] [Google Scholar]
- 10i. Batista V. F., Galman J. L., Pinto D. C. G. A., Silva A. M. S., Turner N. J., ACS Catal. 2018, 8, 11889–11907. [Google Scholar]
- 11.
- 11a. Jeon H., Yoon S., Ahsan M., Sung S., Kim G.-H., Sundaramoorthy U., Rhee S.-K., Yun H., Catalysts 2017, 7, 251; [Google Scholar]
- 11b. Patil M. D., Yoon S., Jeon H., Khobragade T. P., Sarak S., Pagar A. D., Won Y., Yun H., Catalysts 2019, 9, 600; [Google Scholar]
- 11c. Aleku G. A., Mangas-Sanchez J., Citoler J., France S. P., Montgomery S. L., Heath R. S., Thompson M. P., Turner N. J., ChemCatChem 2018, 10, 515–519. [Google Scholar]
- 12. Yoon S., Patil M. D., Sarak S., Jeon H., Kim G.-H., Khobragade T. P., Sung S., Yun H., ChemCatChem 2019. [Google Scholar]
- 13.
- 13a. Matsumoto J., Higuchi M., Shimada M., Yamamoto Y., Kamio Y., Biosci. Biotechnol. Biochem. 1996, 60, 39–43; [DOI] [PubMed] [Google Scholar]
- 13b. Higuchi M., Shimada M., Yamamoto Y., Hayashi T., Koga T., Kamio Y., J. Gen. Microbiol. 1993, 139, 2343–2351. [DOI] [PubMed] [Google Scholar]
- 14. Heydari M., Ohshima T., Nunoura-Kominato N., Sakuraba H., Appl. Environ. Microbiol. 2004, 2004, 937–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geueke B., Riebel B., Hummel W., Enzyme Microb. Technol. 2003, 32, 205–211. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary



