Summary
Analysis of the updated reference tomato genome found 34 full‐length TPS genes and 18 TPS pseudogenes.
Biochemical analysis has now identified the catalytic activities of all enzymes encoded by the 34 TPS genes: one isoprene synthase, 10 exclusively or predominantly monoterpene synthases, 17 sesquiterpene synthases and six diterpene synthases. Among the monoterpene and sesquiterpene and diterpene synthases, some use trans‐prenyl diphosphates, some use cis‐prenyl diphosphates and some use both. The isoprene synthase is cytosolic; six monoterpene synthases are plastidic, and four are cytosolic; the sesquiterpene synthases are almost all cytosolic, with the exception of one found in the mitochondria; and three diterpene synthases are found in the plastids, one in the cytosol and two in the mitochondria.
New trans‐prenyltransferases (TPTs) were characterised; together with previously characterised TPTs and cis‐prenyltransferases (CPTs), tomato plants can make all cis and trans C10, C15 and C20 prenyl diphosphates. Every type of plant tissue examined expresses some TPS genes and some TPTs and CPTs.
Phylogenetic comparison of the TPS genes from tomato and Arabidopsis shows expansions in each clade of the TPS gene family in each lineage (and inferred losses), accompanied by changes in subcellular localisations and substrate specificities.
Keywords: functional characterisation, prenyltransferase, specialised metabolism, subcellular localisation, terpene synthase, terpenes, tomato
Introduction
Terpenoids are a class of compounds, made of isoprene building blocks, that are found in all living organisms (Gershenzon & Dudareva, 2007). In plants, terpenoids play essential roles in myriad general physiological and biochemical processes such as photosynthesis, electron transport, developmental regulation and membrane architecture, with many of these roles shared with other organisms (Pichersky & Raguso, 2018). However, the majority of terpenoids in plants identified by far are those that evolved in different lineages as adaptations for specific ecological niches and are therefore defined as specialised terpenoid metabolites. Such metabolites serve in attracting pollinators and seed dispersers, in defence against pathogens and herbivores, and in attracting useful soil microorganism (Gershenzon & Dudareva, 2007; Pichersky & Raguso, 2018). Because of their functions, biosynthesis of specialised terpenes in plants is usually restricted to specific tissues or cell types, such as flowers (Dudareva et al., 1998, 2003; Chen et al., 2003; Tholl et al., 2004), roots (Chen et al., 2004; Vaughan et al., 2013) or glandular trichomes found at the surface of leaves, stems and fruits (Iijima et al., 2004; Schilmiller et al., 2009; Booth et al., 2017).
Regular terpenoids are synthesised by condensing one dimethylallyl diphosphate (DMAPP) molecule with one or more molecules of its isomer, isopentenyl diphosphate (IPP) (McGarvey & Croteau, 1995). In plants, these C5 precursors are the products of two separate pathways, the cytosolic mevalonate (MVA) pathway and the plastidic 2‐C‐methyl‐d‐erythritol 4‐phosphate (MEP) pathway (Vranová et al., 2013). IPP and DMAPP are further converted by trans/cis‐prenyltransferases (TPTs/CPTs) to polyprenyl diphosphates such as geranyl/neryl diphosphate (GPP/NPP, C10; neryl is the cis‐isomer of geranyl), trans/cis‐farnesyl diphosphate (E,E‐FPP/Z,Z‐FPP, C15), geranylgeranyl/nerylneryl diphosphate (GGPP/NNPP, C20) or longer‐chain prenyl diphosphates. The C5–C25 prenyl diphosphate precursors are subsequently utilised by a large family of structurally related enzymes known as terpene synthase/cyclases (TPSs) to form the basic skeletons of isoprene (C5), monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20) and sesterterpenes (C25) (Bohlmann et al., 1998; Chen et al., 2011; Sharkey et al., 2013; Huang et al., 2017). The terpene skeletons can be further modified by various enzymes, such as the cytochrome P450 oxygenases (CYPs), dehydrogenases, methyltransferases, acyltransferases, and glycosyltransferases to form more diverse compounds (Pichersky et al., 2006; Boutanaev et al., 2015).
TPS enzymes fall into two classes, type I and type II, based on structure and catalytic mechanisms. Type I TPS enzymes contain the aspartate‐rich DDxx(D,E) motif at their C‐terminal domain, called the ‘α domain’, that binds the metal cofactor (Mg2+ or Mn2+) that interacts with the prenyl diphosphate substrates and facilitates the substrate cation formation (Christianson, 2017). Type II terpene synthases contain a DxDD motif in the ‘β domain’ near the N‐terminus, with the second aspartate essential for the protonation‐initiated cyclisation of GGPP to form copalyl diphosphate (CPP) or other cyclic diterpene diphosphates (Zerbe & Bohlmann, 2015). The single functional TPS gene in Physcomitrella patens is bifunctional, encoding an enzyme with both type I and type II active domains. This enzyme first catalyses the conversion of GGPP to CPP with its β domain, then of CPP to ent‐kaurene (a precursor of gibberellins) with its α domain (Hayashi et al., 2006). However, angiosperms have a separate gene for CPP synthase (CPS) in which the α domain is nonfunctional, and a second gene for kaurene synthase (KS), in which the β domain is nonfunctional (Köksal et al., 2011). These two genes appear to have evolved by gene duplication of the original CPS/KS bifunctional gene, followed by divergence (Gao et al., 2012). The TPS gene family in plants continued to evolve by gene duplication and divergence, with some diterpene synthases still containing both functional domains, but many diterpene synthases and all monoterpene, sesquiterpene, and sesterterpene synthases containing only the functional α domain and some diterpene synthases containing only the functional β domain (Gao et al., 2012). Phylogenetic analysis of plant TPS genes/proteins has divided these into seven subfamilies: type I TPS sequences form clades TPS‐a, TPS‐b, TPS‐d (gymnosperm‐specific), TPS‐e/f, TPS‐g and TPS‐h (specific to Selaginella spp.); type II TPSs form clade TPS‐c (Chen et al., 2011).
It has generally been observed that monoterpenes and diterpenes are produced in the plastids by the respective terpene synthases, where GPP and GGPP are produced as well, while sesquiterpenes are produced by sesquiterpene synthases in the cytosol, where FPP is synthesised (Vranová et al., 2013). However, over the last decade, multiple researchers have reported the TPS‐catalysed formation of diterpenes and monoterpenes in the cytosol (Aharoni et al., 2004; Herde et al., 2008; Dong et al., 2013; Falara et al., 2014) and sesquiterpenes in the plastids (Sallaud et al., 2009).
The TPS gene family has been studied most extensively in the model plant Arabidopsis (Arabidopsis thaliana). In total, 32 TPS genes encode potentially functional TPSs in at least one accession (Aubourg et al., 2002), but still the functions of seven of these have not been determined in any accession (Tholl & Lee, 2011; Q. Wang et al., 2016; Huang et al., 2017). Of the TPS genes characterised so far in Arabidopsis, six encode monoterpene synthases, six encode sesquiterpene synthases, eight encode diterpene synthases, and five encode sesterterpene synthases (Chen et al., 2011; Huang et al., 2017). In addition to Arabidopsis, genome‐wide analyses of the TPS gene family have been conducted in other plant and algal species, such as red algae (Wei et al., 2019), the nonvascular moss Physcomitrella patens (Hayashi et al., 2006) and liverwort Marchantia polymorpha (Kumar et al., 2016a), the nonseed vascular plant Selaginella moellendorffii (Li et al., 2012), gymnosperms Picea spp. (Keeling et al., 2011), and multiple angiosperm species (Martin et al., 2010; Falara et al., 2011; Zhuang et al., 2012; Alquézar et al., 2017; Booth et al., 2017). These analyses have revealed that the size of the plant TPS gene family varies from one functional gene in Physcomitrella patens (Chen et al., 2011) to 69 putatively functional genes in Vitis vinifera (Martin et al., 2010). However, to date, in no species has the complete set of the TPS genes been functionally characterised.
Previous studies have already indicated that tomato (Solanum lycopersicum) plants synthesise an unusual number and types of specialised terpenes, particularly in their trichomes (Falara et al., 2011; McDowell et al., 2011). A unique feature of terpenoid metabolism in tomato and other Solanum species is the use by TPS enzymes of both trans‐ and cis‐prenyl diphosphates (Sallaud et al., 2009; Schilmiller et al., 2009). The release of the first high‐quality genome sequence of tomato (Tomato Genome Consortium, 2012) made possible a thorough examination of its TPS gene family, albeit 10 of the identified 30 functional genes (out of a total of 45 loci) could not be functionally characterised (Colby et al., 1998; van Schie et al., 2007; Schilmiller et al., 2009, 2010; Bleeker et al., 2011; Falara et al., 2011, 2014; Matsuba et al., 2013, 2015). Based on the analysis of the latest tomato genome release (2017 release of v. SL3.0), we now report that the tomato genome contains 34 functional TPS genes, and we have successfully determined the catalytic activities, gene expression patterns and subcellular localisations of this entire group of enzymes. These results make the tomato TPS family the only one to be fully characterised so far.
Materials and Methods
Plant materials and chemicals
Tomato (Solanum lycopersicum L. cv MP1) seeds were obtained from the Tomato Genetic Resource Center (https://tgrc.ucdavis.edu). Throughout the text, when not specifically indicated, the tomato plants used were of cv MP1. Nicotiana benthamiana and Arabidopsis thaliana (Col‐0 ecotype) seeds were obtained from our laboratory and Arabidopsis Biological Resource Center, respectively. Details of plant growth conditions can be found in Supporting Information Methods S1.
IPP, DMAPP, GPP, NPP, E,E‐FPP, Z,Z‐FPP and GGPP were obtained from Echelon Biosciences (Salt Lake City, UT, USA). Terpene standards were purchased from Sigma Aldrich (St Louis, MO, USA). All other chemicals and solvents were obtained from Fisher Scientific (Hampton, NH, USA).
Identification of new TPS gene models in the tomato genome
The most recent tomato genome (https://solgenomics.net) was used for blastn searches with known tomato TPS genomic sequences. The resulting candidates were manually annotated to accomplish a more accurate result. To verify and fill the gaps of the newly discovered TPS sequences (TPS47 through TPS53), genomic DNA was extracted from tomato leaves and used as template for PCR with gene‐specific primers (Table S1). PCR amplification was performed with KOD Hot Start DNA polymerase (EMD Millipore, Burlington, MA, USA). DNA fragments obtained were extracted with EZNA Gel Extraction Kit (Omega Bio‐tek, Norcross, GA, USA) and then verified by the Sanger sequencing method.
RNA isolation, cDNA synthesis and quantitative real‐time PCR
Different tissues of S. lycopersicum plants were collected and immediately frozen in liquid nitrogen and stored at −80°C until use. Trichomes were collected by gently shaking the corresponding tissues in liquid nitrogen. Details of RNA isolation, cDNA synthesis and quantitative real‐time PCR (RT‐qPCR) can be found in Methods S1.
Isolation of full‐length TPS and prenyltransferase cDNAs
The full‐length cDNAs of TPS10, TPS16, TPS27, TPS28, TPS33, TPS35 and TPS36 were obtained by PCR amplification with gene‐specific primers (Table S1) based on a previous publication (Falara et al., 2011). The full‐length cDNAs of TPS25, TPS47, TPS48, TPS51, TPS52, GGPPS3, SSU I, SSU II, TPT1 and TPT2 were obtained by PCR amplification with gene‐specific primers (Table S1) based on annotated results from the genome database and our manual annotation (Fig. S1). The cDNA used as template was synthesised from MP1 leaf, flower, mature red fruit, stem trichome and root RNA. The other TPS and prenyltransferase genes were obtained as described by Falara et al. (2011) (TPS24 and TPS40), Matsuba et al. (2013) (TPS18 and TPS41), Akhtar et al. (2013) (CPT1, CPT2 and CPT6), Gaffe et al. (2000) (FPPS1) and Ament et al. (2006) (GGPPS1 and GGPPS2). Amino acid sequences of all cloned TPS and prenyltransferase genes from MP1 are identical to the tomato reference genome obtained from Heinz 1706.
Synthesis of codon‐optimised genes
Escherichia coli codon‐optimised versions of TPS10, TPS16, TPS47, TPS51 and TPS52 were synthesised by Gene Universal (Newark, DE, USA).
Multiple sequence alignment and phylogenetic analysis
The full‐length amino acid sequences were aligned using the clustalw program (Thompson et al., 1994) with default parameters. A maximum‐likelihood phylogenetic tree was constructed using the Mega7 tool (Kumar et al., 2016b) with default settings, bootstrap values were performed with 1000 repetitions. The scale bar corresponds to 20% amino acid sequence divergence.
Protoplast transformation and confocal microscopy
Regions corresponding to the first c. 120 or full‐length amino acids of the tested proteins were amplified (detailed in Table S1), digested accordingly and ligated into pEZS‐NL vector (Zhou et al., 2017), creating an in‐frame C‐terminal fusion protein with EGFP. The resulting constructs were applied for PEG‐mediated transient expression in Arabidopsis mesophyll protoplasts as described before (Yoo et al., 2007). MitoTracker Red (Invitrogen, Carlsbad, CA, USA) was used as a mitochondrial marker. Fluorescence signals were observed using a Leica SP5 laser scanning confocal microscope as described previously (Falara et al., 2011). All the transient expression experiments were repeated independently at least three times.
Expression in Escherichia coli and TPS enzyme assays
TPS genes without their fragments corresponding to putative transit peptides were cloned in pET32b or pET28a to express TPS‐His recombinant proteins (detailed in Table S1). Protein expression and purification are detailed in Methods S1.
TPS enzyme assays were performed as described previously (Falara et al., 2011) using GPP, NPP, E,E‐FPP, Z,Z‐FPP and GGPP as substrates. After enzyme assay incubation, products were extracted with methyl tert‐butyl ether (MTBE) and analysed using gas chromatography–mass spectrometry (GC–MS).
Isoprene synthase enzyme assays
Enzymatic assays for isoprene synthase were performed as previously described (Sasaki et al., 2005) using DMAPP as the substrate. After enzyme assay incubation, headspace products were collected with a 100 μm solid‐phase micro extraction (SPME) fibre (Supelco, St Louis, MO, USA) at 42°C for 15 min and analysed by GC–MS.
Prenyltransferase assays
For enzymatic assays, truncated trans‐prenyltransferases (TPTs) without stop codons were cloned into pET32b upstream and in‐frame of the (His)6‐tag sequence to express TPT‐His recombinant proteins. Truncated SSU I and SSU II were cloned into pET28a downstream and in‐frame of the (His)6‐tag sequence to express His‐SSU recombinant proteins. For co‐expression with His‐SSU, truncated GGPPSs with stop codons were subcloned into pET32b to express nontagged GGPPS proteins (detailed in Table S1).
The prenyltransferase assays were performed using IPP and DMAPP as substrates as described previously (Zhou et al., 2017). After enzyme assay incubation, products were hydrolysed to their corresponding alcohols, extracted with MTBE and analysed by GC–MS. To calculate the molar ratio of formed products, standard curves were constructed using commercial geraniol, E,E‐farnesol and geranylgeraniol.
Agrobacterium‐mediated transient expression of heterologous proteins in tobacco
For constructing of plant transformation vectors, full‐length or truncated ORFs of the tested genes (detailed in Table S1) were amplified by PCR and cloned into the binary vector pEAQ‐HT (Sainsbury et al., 2009) that contains the cauliflower mosaic virus 35S promoter and nopaline synthase terminator. In planta transient expression in tobacco (Nicotiana benthamiana) leaves was performed as previously described (Sainsbury et al., 2009). A pEAQ‐HT construct carrying GFP alone was used as a control. Tobacco leaves were harvested 3 d after infiltration and immediately dipped in MTBE for 30 min to extract terpene products. The MTBE extracts were concentrated by evaporating the solvent to a final volume of c. 100 μl for GC–MS analysis.
Profiling of tomato terpene volatiles and correlation analysis
All the tested tissues were freshly harvested and submerged in MTBE containing 10 ng μl−1 of the tetradecane internal standard. Metabolites were extracted for 15 min with gentle shaking and the resulting extracts were concentrated to a final volume of c. 100 μl for GC–MS analysis. The correlation analyses of tomato terpene synthase genes and volatile terpenes are detailed in Methods S1.
GC–MS analysis of terpenes
The analysis of terpene products (C10–C25) was performed as described previously (Falara et al., 2011). Briefly, 1 μl of sample was autoinjected into a Shimadzu QP‐2010 SE GC‐MS system equipped with a Rxi‐5Sil column (30 m length, 0.25 mm i.d., and 0.25 μm film thickness; Restek, Bellefonte, PA, USA). Analysis of isoprene was performed by directly injecting the SPME fibre into the Shimadzu QP‐2010 SE GC‐MS system equipped with the Rxi‐5Sil column. The analysis of prenyltransferase assay products was performed using an EC‐WAX column (30 m length, 0.32 mm i.d., and 0.25 μm film thickness; Grace, Columbia, MD, USA). The identification methods for each terpene compound are listed in Table S2. Details of GC–MS parameters can be found in Methods S1.
Results
Identification of new TPS genes in the tomato genome
A search for TPS genes in the most recently updated tomato genome (https://solgenomics.net/organism/Solanum_lycopersicum/genome; the 2017 release of v. SL3.0) identified 52 gene models, of which 45 had previously been reported (TPS1–TPS46, not including TPS45, which is present in S. habroichaites but not in S. lycopersicum; Tables S3, S4). The seven additional loci were designated as TPS47–TPS53. TPS47 and TPS48 appeared to have uncompromised open reading frames (Table S3), whereas TPS49, TPS50 and TPS53 appeared to have mutations and deletions (Table S4). Genomic sequences of TPS51 and TPS52 had c. 1 kb gaps between exons 2 and 5 (Fig. S1). To verify the mutations and fill the gaps in the new gene sequences, genomic DNA of these seven genes was amplified by PCR and fully sequenced. The results indicated that TPS47, TPS48, TPS51 and TPS52 encoded potentially functional TPSs, with protein lengths of 562, 560, 550 and 551 amino acids, respectively (Table S3). By contrast, TPS49, TPS50 and TPS53 were pseudogenes (Table S4).
TPS47 and TPS49 are located on chromosome 3, TPS48 and TPS50 are located on chromosome 4, TPS51 and TPS52 are close to each other and in the same orientation on chromosome 7, and TPS53 resides on chromosome 10 (Fig. 1). Phylogenetic analysis indicates that TPS47 belongs to the TPS‐b clade, together with five previously characterised monoterpene synthases, one previously characterised sesquiterpene synthase and two uncharacterised enzymes (TPS25 and TPS27) (Fig. 2). TPS48, TPS51, TPS52 and six other uncharacterised enzymes (TPS10, TPS16, TPS28, TPS33, TPS35 and TPS36, which was previously shown to have activity with Z,Z‐FPP, but the structure of the sesquiterpene product could not be identified) (Falara et al., 2011), belong to the TPS‐a clade, a clade that also contains seven previously characterised enzymes, all of which are sesquiterpene synthases. Thus, the tomato genome contains at least 52 TPS genes including 34 putative functional TPSs, among these 14 (the 12 genes listed above, plus TPS18 and TPS41, which belong to the e/f clade) have not been previously functionally characterised. To determine the terpene synthase activity of the 14 uncharacterised TPS enzymes, their full‐length cDNAs were obtained from different tissues and functionally tested by expression in Escherichia coli or in Nicotiana benthamiana.
Figure 1.
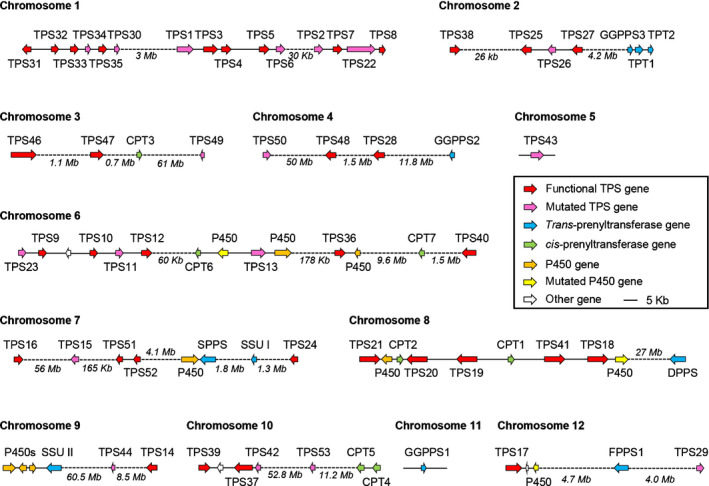
The organisation of tomato TPS genes and other related genes on the chromosomes. CPT, cis‐prenyltransferase; DPPS, decaprenyl diphosphate synthase; FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; P450, putative cytochrome P450 oxidoreductase; SPPS, solanesyl diphosphate synthase; SSU, small subunit of geranyl diphosphate synthase; TPS, terpene synthase; TPT, trans‐prenyltransferase.
Figure 2.
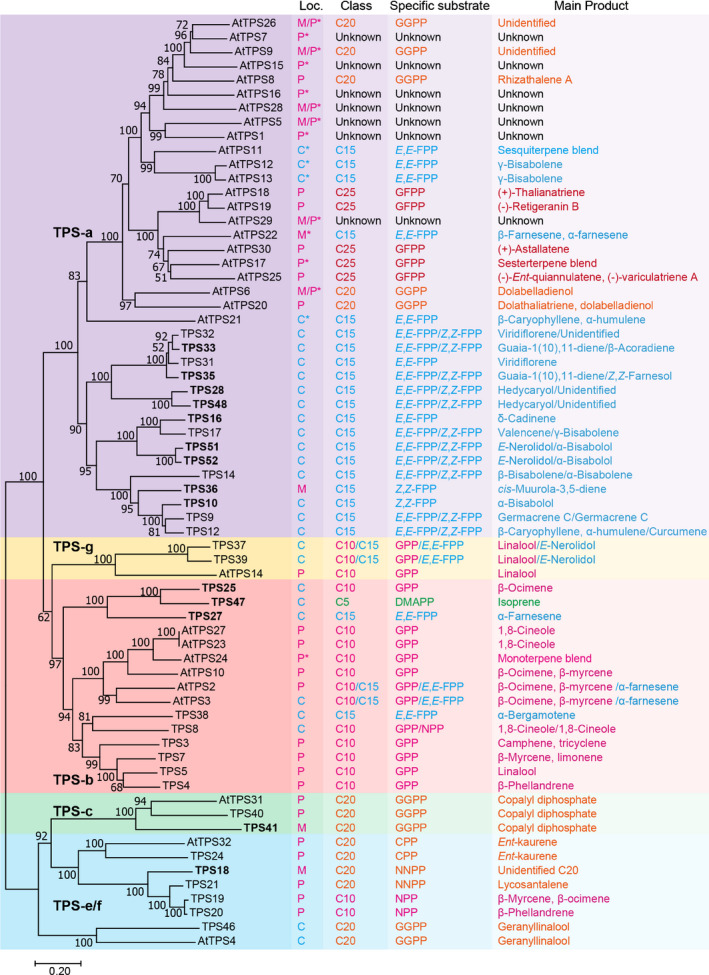
Phylogenetic analysis of tomato and Arabidopsis TPS gene family. A maximum‐likelihood phylogenetic tree of the TPS proteins is shown on the left panel, while subcellular localisation (Loc.), substrate class, specific substrate and main product of the corresponding enzymes are shown on the right four panels. The TPS‐a, TPS‐g, TPS‐b, TPS‐c and TPS‐e/f clades are shading in purple, yellow, pink, green and blue, respectively. Cytosolic (C) localisation is shown in blue, plastidic (P) and mitochondrial (M) localisations are shown in magenta. Localisations predicted by TargetP and ChloroP are denoted by asterisks. C5, C10, C15, C20 and C25 compounds are shown in green, magenta, blue, orange and red, respectively. Terpene synthases characterised in this work are shown in bold. At, Arabidopsis thaliana; CPP, ent‐copalyl diphosphate; DMAPP, dimethylallyl diphosphate; FPP, farnesyl diphosphate; GFPP, geranylfarnesyl diphosphate; GGPP, geranylgeranyl diphosphate; GPP, geranyl diphosphate; NNPP, nerylneryl diphosphate; NPP, neryl diphosphate; TPS, terpene synthase.
Characterisation of the enzymatic activities of 14 previously uncharacterised TPS genes
TPS enzymes of the TPS‐a clade – TPS48, TPS51, TPS52, TPS10, TPS16, TPS28, TPS33, TPS35 and TPS36
All soluble TPS proteins (Fig. S2) described in this section (and subsequent ones) were assayed in vitro using GPP, NPP, E,E‐FPP, Z,Z‐FPP and GGPP. Results indicated that both TPS28 and TPS48 (which show 85% amino acid sequence identity) used E,E‐FPP to produce a product identified as elemol (Figs 3a, S3; Table S2); elemol is reported to be the thermal breakdown product of (+)‐hedycaryol (Fig. S4; Hattan et al., 2016). TPS28 and TPS48 also catalysed the formation of a major sesquiterpene from Z,Z‐FPP that is currently unidentified (Figs 3b, S3; Table S2). TPS33 and TPS35 (91.7% identical) both catalysed the formation of guaia‐1(10),11‐diene from E,E‐FPP (Figs 3a, S3; Table S2). When assayed with Z,Z‐FPP, TPS33 made several sesquiterpenes including β‐acoradiene, β‐curcumene, α‐cedrene and cis‐α‐bergamotene, while TPS35 made predominantly Z,Z‐farnesol (Figs 3b, S3; Table S2). TPS36 only had activity using Z,Z‐FPP and produced mostly cis‐muurola‐3,5‐diene (Figs 3b, S3; Table S2). TPS51 and TPS52 (94.7% sequence identity) catalysed the formation of mostly E‐nerolidol from E,E‐FPP (Figs 3a, S3; Table S2) and several sesquiterpenes (α‐bisabolol, Z‐nerolidol, α‐bisabolene, β‐bisabolene and Z‐β‐farnesene) from Z,Z‐FPP (Figs 3b, S3; Table S2). No activity was observed for either of these proteins with any of the other tested substrates.
Figure 3.
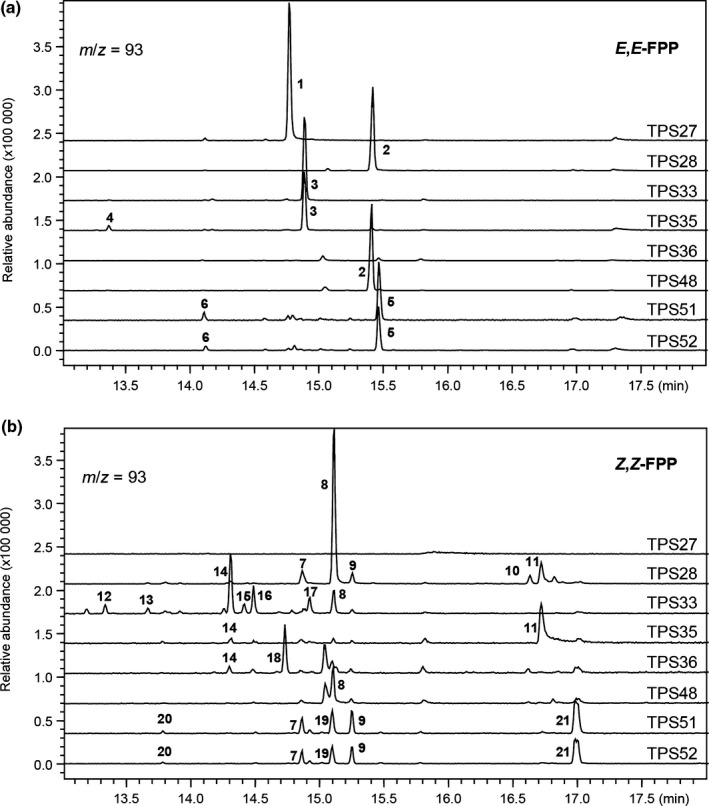
GC–MS analysis of the products formed in vitro by the enzymatic activities of tomato TPS proteins. Enzymes were incubated with E,E‐FPP (a) and Z,Z‐FPP (b) and products were analysed as described in the ‘Materials and Methods’ section. Reaction products were identified by comparison of their mass spectra and retention indices with authentic standards and NIST libraries: 1, α‐farnesene; 2, elemol; 3, guaia‐1(10),11‐diene; 4, β‐elemene; 5, E‐nerolidol; 6, E‐β‐farnesene; 7, E‐β‐bisabolene; 8, unknown 1; 9, α‐bisabolene; 10, α‐eudesmol; 11, (Z,Z)‐farnesol; 12, α‐cedrene; 13, cis‐α‐bergamotene; 14, β‐acoradiene; 15, unknown 2; 16, unknown 3; 17, β‐curcumene; 18, cis‐muurola‐3,5‐diene; 19, Z‐nerolidol; 20, Z‐β‐farnesene; 21, α‐bisabolol. Mass spectra and retention indices of the terpene products can be found in Supporting Information Fig. S3 and Table S2, respectively. FPP, farnesyl diphosphate; TPS, terpene synthase.
We could not obtain soluble proteins from TPS10 and TPS16 when expressed in E. coli, and we therefore transiently expressed their full‐length cDNAs in N. benthamiana. Using this approach, we determined that TPS16 catalysed the formation of δ‐cadinene from the endogenous E,E‐FPP pool (Figs 4a, S3; Table S2). However, no product was detected when TPS10 was expressed alone (Fig. 4a). TPS10 is located on chromosome 6 c. 60 kb away from CPT6, the gene encoding the cis‐prenyltransferase responsible for the biosynthesis of Z,Z‐FPP (Fig. 1), and when TPS10 was co‐expressed with CPT6 in N. benthamiana, the formation of α‐bisabolol was detected (Figs 4a, S3; Table S2). TPS10 and TPS16 showed no activity when their genes were co‐expressed with either CPT1 or CPT2 (Fig. S5).
Figure 4.
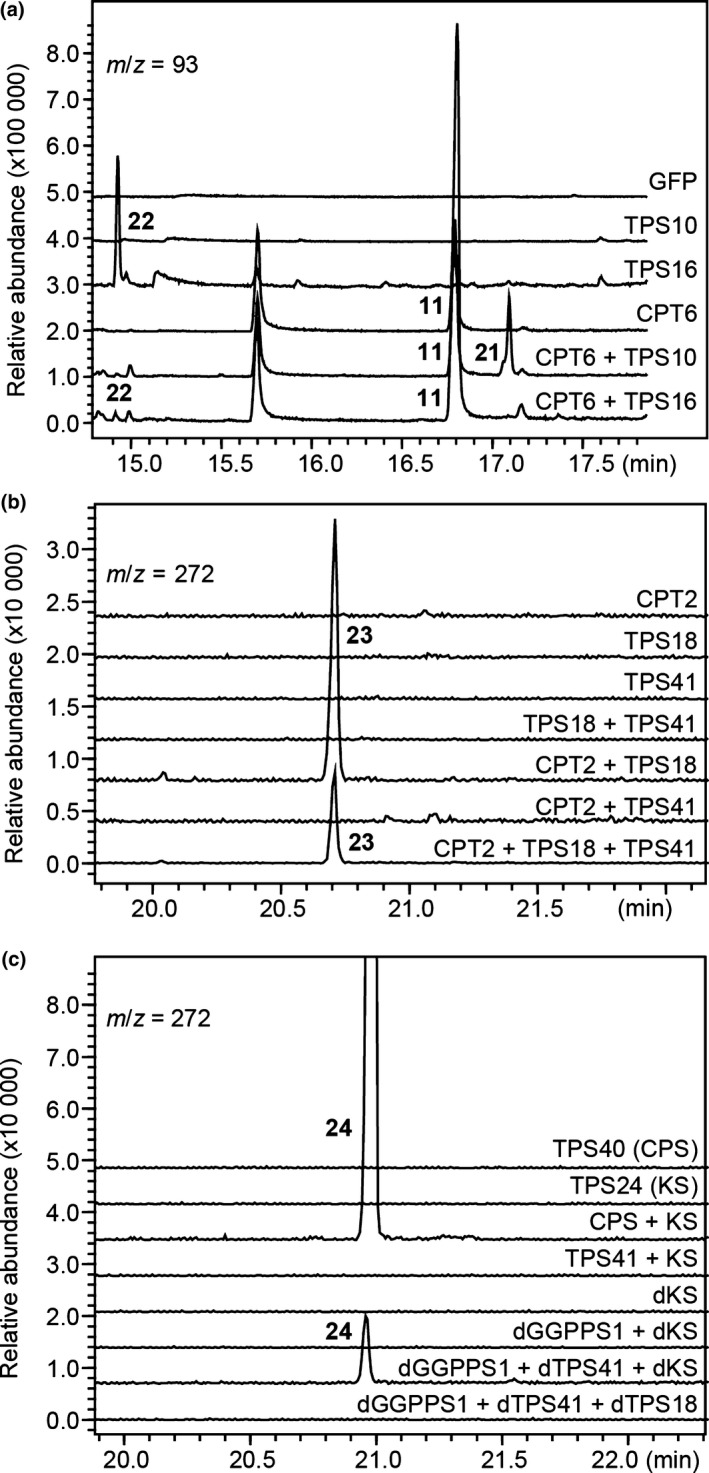
GC–MS analysis of the products formed in planta by transiently expressing TPS genes in Nicotiana benthamiana leaves. (a) Analysis of sesquiterpenes formed by TPS10 and TPS16. Nicotiana benthamiana leaves expressing GFP alone was used as a negative control. (b) Analysis of the formation of an unknown diterpene by co‐expressing TPS18 with CPT2. (c) Analysis of the formation of ent‐kaurene by expressing the full‐length and transit peptide deleted version (‘d’) of TPS genes. Products were extracted with methyl tert‐butyl ether (MTBE) and analysed as described in the ‘Materials and Methods’ section. Reaction products were identified by comparison of their mass spectra and retention indices with authentic standards and NIST libraries: 11, (Z,Z)‐farnesol; 21, α‐bisabolol; 22, δ‐cadinene; 23, unknown 4; 24, ent‐kaurene. Mass spectra and retention indices of the terpene products can be found in Supporting Information Fig. S3 and Table S2, respectively. CPS, ent‐copalyl diphosphate synthase; CPT, cis‐prenyltransferase; GGPPS, geranylgeranyl diphosphate synthase; KS, ent‐kaurene synthase; TPS, terpene synthase.
TPS enzymes of the TPS‐b clade – TPS25, TPS27 and TPS47
These three enzymes fall into a separate subclade within the TPS‐b clade (Fig. 2). TPS25 had activity only with GPP as the substrate, producing mostly β‐ocimene (Figs 5a, S3; Table S2). TPS27 had activity only with E,E‐FPP to form α‐farnesene (Figs 3a, S3; Table S2). TPS47 showed no activity towards any of the initially tested substrates (GPP, NPP, E,E‐FPP, Z,Z‐FPP and GGPP). Further phylogenetic analysis with terpene synthases from other plants indicated that isoprene synthases (IspSs), which catalyse the enzymatic conversion of DMAPP to isoprene, from various species are closely related to these three enzymes (Fig. S6a). Structural and functional analysis have established four ‘isoprene score’ amino acids that are specific to isoprene synthases (Köksal et al., 2010; Sharkey et al., 2013; Ilmén et al., 2015). These four amino acids are F338, S445, F485 and N505 (numbering refers to IspS from Populus alba) (Fig. S6b), of which only F338 is strictly conserved in all functional IspSs presently known, and when both F338 and F485 are present, the enzyme is always isoprene synthase (Ilmén et al., 2015). Among the tomato TPS enzymes, TPS36 has the first Phe residue, while TPS47 contains both Phe residues (Fig. S6b). When tested in vitro, TPS47 catalysed the formation of isoprene from DMAPP (Figs 5b, S3). By contrast, no isoprene synthase activity was observed for TPS25, TPS27 or TPS36 (Fig. 5b).
Figure 5.
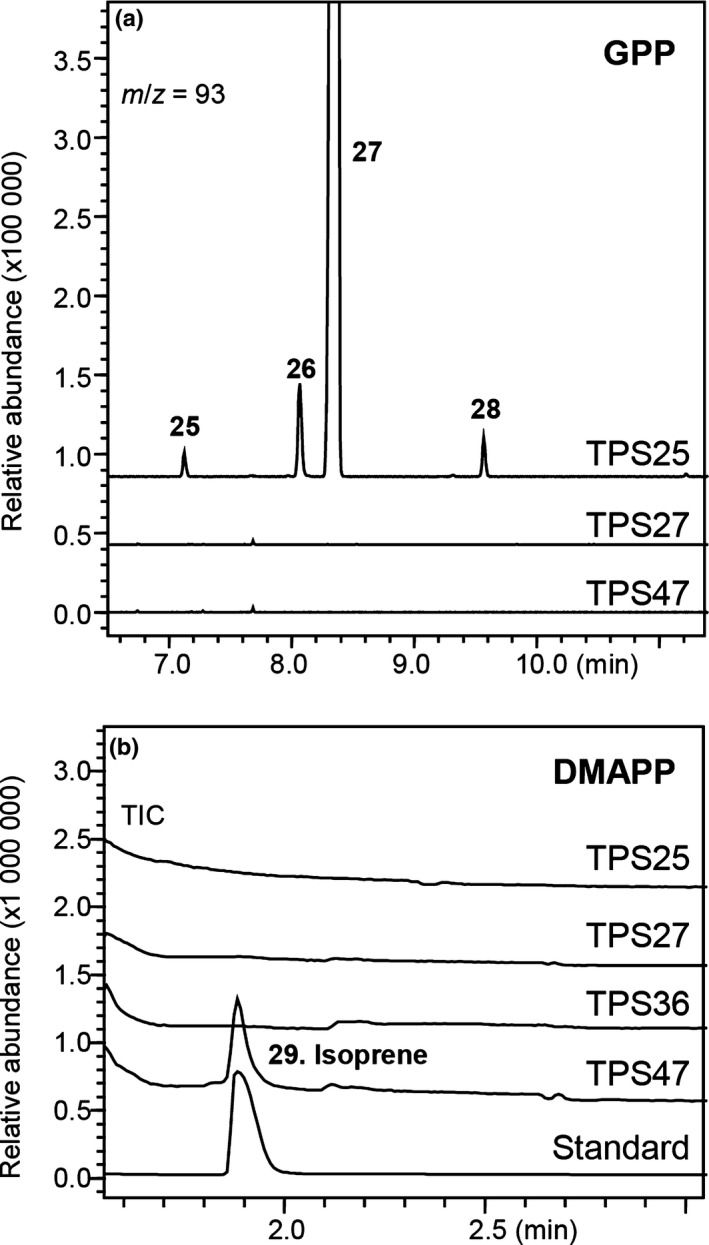
GC–MS analysis of the products formed in vitro by the enzymatic activities of tomato terpene synthase (TPS) proteins using geranyl diphosphate (GPP) (a) and dimethylallyl diphosphate (DMAPP) (b) as substrates. Reaction products were collected and analysed as described in the ‘Materials and Methods’ section, commercial isoprene was used as a standard. Reaction products were identified by comparison of their mass spectra and retention indices with authentic standards and NIST libraries: 25, β‐myrcene; 26, β‐cis‐ocimene; 27, β‐trans‐ocimene; 28, linalool; 29, isoprene. Mass spectra and retention indices of the terpene products can be found in Supporting Information Fig. S3; Table S2, respectively.
TPS enzymes of the TPS‐e/f and TPS‐c clades – TPS18 and TPS41
TPS18 and TPS41, which were not previously biochemically characterised, belong to the e/f and c clades of the TPS family, respectively (Fig. 2). Proteins in these two clades are typically longer (c. 800 amino acids) than other TPS proteins, and past attempts to express such proteins, including TPS18 and TPS41, in E. coli have met with meagre success in obtaining soluble, enzymatically functional proteins (Falara et al., 2011; Matsuba et al., 2013). We transiently expressed these two genes in N. benthamiana, but no terpene products were observed (Fig. 4b). TPS18 and TPS41 are present in a cluster on chromosome eight where CPT1 (encoding neryl diphosphate synthase) and CPT2 (encoding nerylneryl diphosphate synthase) are also present (Fig. 1), suggesting that they may be functionally associated. To test this hypothesis, TPS18 and TPS41 were each co‐expressed with either CPT1 or CPT2 in N. benthamiana leaves. When TPS18 was co‐expressed with CPT2, an unidentified diterpene was produced (Figs 4b, S3; Table S2), while no monoterpene was detected when co‐expressed with CPT1 (Fig. S5). By contrast, TPS41 showed no activity when co‐expressed with either CPT1 (Fig. S5) or CPT2 (Fig. 4b). Neither TPS18 nor TPS41 showed activity when their genes were co‐expressed with CPT6 (Fig. S5).
TPS41 is most similar to TPS40, another TPS‐c member with a proven ent‐copalyl diphosphate synthase (CPS) activity (Bensen & Zeevaart, 1990; Falara et al., 2011). To test whether TPS41 has CPS activity, we co‐expressed TPS41 with the previously characterised ent‐kaurene synthase (TPS24, KS; Falara et al., 2011). No terpene product was detected when the full‐length cDNAs of TPS41 and KS were co‐expressed. By contrast, co‐expression of TPS40 and TPS24 led to the production of ent‐kaurene (Fig. 4c). TPS41 has a long N‐terminal extension (Fig. S7) and was predicted to be localised to the mitochondria by TargetP, which is different from the predicted plastidic localisation of KS (Fig. S8 and see below subcellular localisation section). We then co‐expressed the truncated versions of TPS41, KS, as well as GGPPS1 (Ament et al., 2006) to provide GGPP in the cytosol, and ent‐kaurene was detected, indicating that TPS41 does have CPS activity (Figs 4c, S3; Table S2). Since the TPS18 protein was also shown to be in the mitochondria (see below subcellular localisation section), we expressed a TPS18 construct lacking a transit peptide‐encoding region together with a similarly truncated construct for TPS41 and GGPPS1 in N. benthamiana, but no new product was detected in those plants (Fig. 4c).
Characterisation of the enzymatic activities of previously uncharacterised trans‐prenyltransferases
The cis‐prenyltransferase (CPT) gene family in tomato has been fully characterised (Akhtar et al., 2013), but the composition and enzymatic functions of the trans‐prenyltransferase (TPT) family members have not been thoroughly investigated. Our analysis shows that the tomato genome contains 10 putative full‐length TPTs (Fig. 1; Table S5), including one enzymatically characterised FPP synthase (FPPS1; Gaffe et al., 2000), two GGPP synthases (GGPPS1 and GGPPS2; Ament et al., 2006) and two long‐chain (C45 and C50) prenyl diphosphate synthases (SPPS and DPPS; Jones et al., 2013). Our analysis of the genome sequence also identified one homologue each of type I and type II small subunit genes of GPP synthases (GPPSs), called respectively SSU I and SSU II (Wang & Dixon, 2009) (Figs 1, S9; Table S5). SSU I and SSU II proteins are catalytically inactive by themselves but each is able to bind a subunit of GGPPS to form a heterodimer that can produce GPP (Orlova et al., 2009; Zhou et al., 2017). To examine the prenyltransferase activities of the newly identified TPT enzymes (Table S5), affinity‐purified His‐tagged recombinant proteins were incubated with IPP and DMAPP, and the hydrolysed products were analysed by GC–MS. Consistent with previous reports, GGPPS1 and GGPPS2 exhibited GGPPS activity, producing GGPP as the main product (66.3% and 86.3%, respectively) with small amounts of GPP (28.1% and 9.6%, respectively) and FPP (5.6% and 4.1%, respectively) (Fig. 6). Solyc02g085700 also showed predominantly GGPPS activity, producing a mixture of GGPP (61.2%), FPP (2.8%) and GPP (36.0%) (Fig. 6), and was thus designated as GGPPS3. No activity was observed for TPT1 and TPT2 (Fig. 6). SSU I and SSU II displayed no catalytic activity alone, but SSU I was able to change the product specificity of GGPPS1, GGPPS2 as well as GGPPS3 from GGPP to predominantly GPP (95.3%, 93.3% and 87.1%, respectively), while the heterodimers formed between SSU II and GGPPS1, GGPPS2 and GGPPS3 catalysed the formation of mainly GGPP (92.0%, 97.4% and 79.0%, respectively) (Fig. 6). Taken together, these results indicated that GPP and GGPP pools in tomato cells are provided by heteromeric GGPPSs/SSU I and three GGPPSs, respectively.
Figure 6.
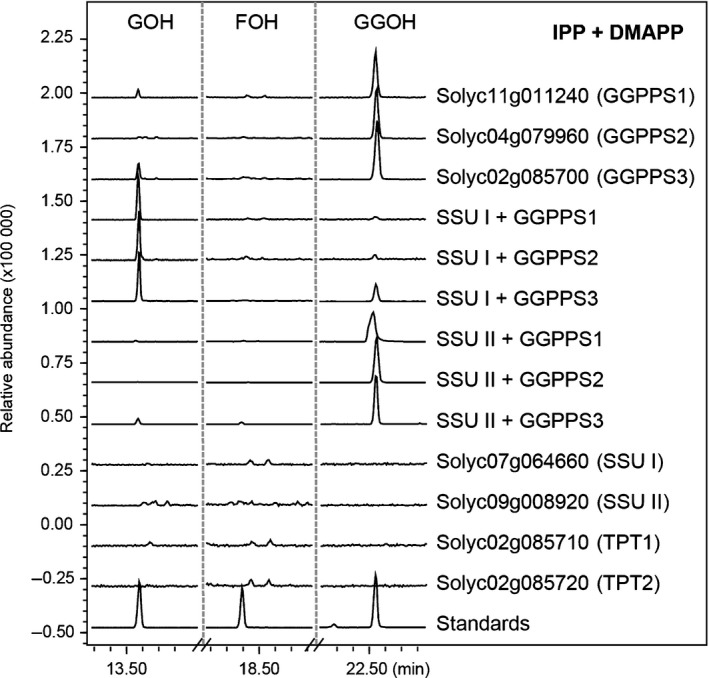
Characterization of the tomato trans‐prenyltransferase proteins. In vitro enzyme assays of recombinant transprenyltransferases (TPTs) and the geranylgeranyl diphosphate synthase (GGPPS)/SSU heterodimers with isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). Products were hydrolysed to corresponding alcohols (GOH, geraniol; FOH, farnesol; GGOH, geranylgeraniol) and analysed with GC–MS. m/z = 93 was monitored for terpene products, only compounds corresponding to the prenyl alcohols were shown. SSU, small subunit of geranyl diphosphate synthase.
The current tomato genome does not contain sesterterpene synthase genes
TPT1 and TPT2 are the only two remaining unidentified TPTs (Fig. 6). Phylogenetic analysis indicates that TPT1 and TPT2 are clustered with Arabidopsis GFPP synthases (Fig. S9), however, neither enzyme is predicted to have GFPP synthase activity by the well established ‘three floors’ model (Table S5; C. Y. Wang et al., 2016). Since no activity was detected when TPT1 and TPT2 were assayed in vitro, we co‐expressed both genes with a sesterterpene synthase (sesterTPS) gene (AtTPS19; Shao et al., 2017) in N. benthamiana cytosol to test whether they have GFPP synthase activity. As a positive control (−)‐retigeranin B was produced when AtGFPPS2 was co‐expressed with AtTPS19, while no sesterterpenes were formed when either TPT1 or TPT2 was co‐expressed with AtTPS19 (Fig. S10), indicating that TPT1 and TPT2 do not have GFPP synthase activity. Arabidopsis sesterTPSs belong to the TPS‐a clade (Fig. 2), and a recent study revealed that a single amino acid with a small side chain – Gly or Pro – near the active centre determines the substrate specificity of sesterTPSs (Chen et al., 2019), providing a large active‐site cavity for the larger GFPP substrates (Fig. S11a,c). However, TPS enzymes from tomato all possess a large‐side‐chain amino acid at the same site (Fig. S11a,b). These results suggest that the current tomato genome does not contain GFPPS or sesterTPS genes.
Subcellular localisation of tomato TPS and TPT proteins
The subcellular localisation of only a handful of TPS enzymes in tomato has been reported. TPS14 was previously shown to be localised in the cytosol and TPS36 was targeted to the mitochondria (Falara et al., 2011). CPT1, CPT2 and CPT6 were shown to be localised to the plastids (Akhtar et al., 2013), but no short‐chain TPT has yet been localised to a subcellular compartment. We therefore endeavoured to determine the subcellular localisation of the all TPS and relevant TPT proteins. Sequence alignment showed that proteins in the TPS‐a clade have a short N‐terminal extension (9–18 residues) upstream the conserved R(R,P)(x)8W motif, except for TPS36 (48 residues) and TPS14 (27 residues) (Fig. S12). Most proteins in the TPS‐b clade and TPS‐g clade have a relatively long N‐terminal extension (28–56 residues), except for TPS38 (11 residues) (Fig. S13). Proteins in TPS‐e/f clade appear to have a transit peptide except for TPS46 (Fig. S14). TPS40 and TPS41 in TPS‐c clade both have a long N‐terminal extension (Fig. S7). All TPT proteins seem to have a transit peptide (Fig. S15).
To experimentally determine their subcellular localisations, the part of each gene encoding the c. 120 amino acids of the N‐terminus was fused to the N‐terminus of GFP and transiently expressed in Arabidopsis mesophyll protoplasts. Consistent with predictions made by TargetP and ChloroP (Fig. S8), GFP fusion proteins of TPS3, TPS4, TPS5, TPS7, TPS19, TPS20, TPS21, TPS24, TPS40, GGPPS1, GGPPS2, GGPPS3 and SSU II were localised in the plastids, while GFP‐fusions of TPS8, TPS9, TPS10, TPS12, TPS16, TPS17, TPS25, TPS27, TPS28, TPS31, TPS32, TPS33, TPS35, TPS38, TPS39, TPS46, TPS47, TPS48, TPS51, TPS52 and FPPS1 were localised in the cytosol (Fig. 7). TPS18, TPS41, TPT1 and TPT2 were targeted to mitochondria as predicted by TargetP (Figs 8, S8a). TPS37 was predicted to be localised in the plastids by both TargetP and ChloroP (Fig. S8), however fluorescence signal of TPS37‐GFP fusion protein was only observed in the nonorganellar area of the cell, presumably the cytosol (Fig. 7). SSU I was targeted to plastids instead of the predicted cytosol localisation (Fig. 7). Taken together, three of the tomato TPS proteins are localised in the mitochondria, nine of them are targeted to the plastids and all the rest reside in the cytosol (Figs 7, 8). Analysis of the TPTs relevant to the production of TPS substrates indicates that FPPS1 is localised in the cytosol, while GGPPS1, GGPPS2, GGPPS3, SSU I and SSU II are localised in the plastids (Fig. 7). The speckled fluorescence pattern of GGPPS3, SSU I and SSU II‐GFP fusion proteins inside the plastid also suggests a suborganelle localisation, presumably the thylakoid, as reported in other plants (Zhou et al., 2017; Wang et al., 2018). It must be noted that these results, obtained in Arabidopsis protoplasts, will need to be confirmed in the future using tomato cells.
Figure 7.
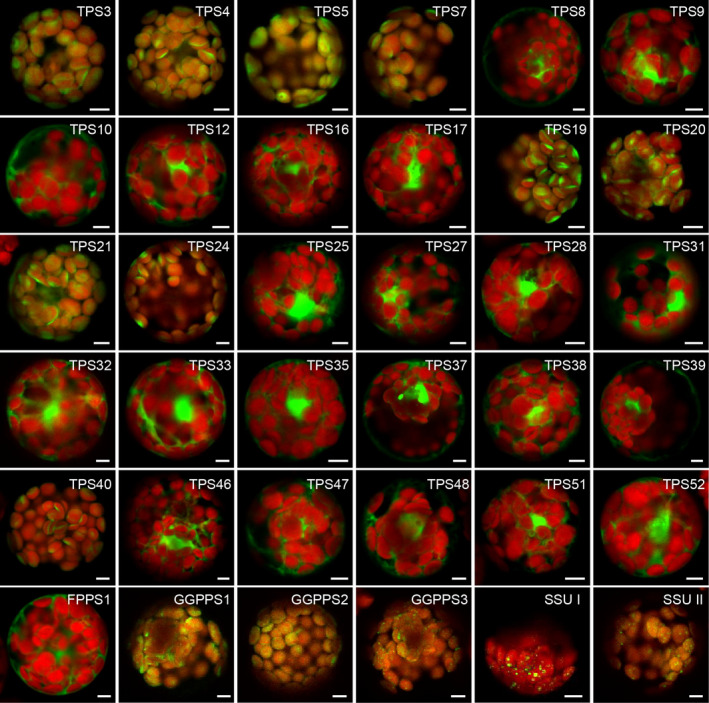
Subcellular localisation of the tomato terpene synthase (TPS) and trans‐prenyltransferase (TPT) proteins. The complete open reading frames of TPTs (bottom line) and the first c. 120 codons of TPSs were fused to a downstream GFP and transiently expressed in Arabidopsis leaf protoplasts. GFP fluorescence indicates the location of each fusion protein (shown in green) and the location of chloroplasts was determined by chlorophyll autofluorescence (shown in red), pictures shown are the merged channel. Bars, 5 μm. FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; SSU, small subunit of geranyl diphosphate synthase.
Figure 8.
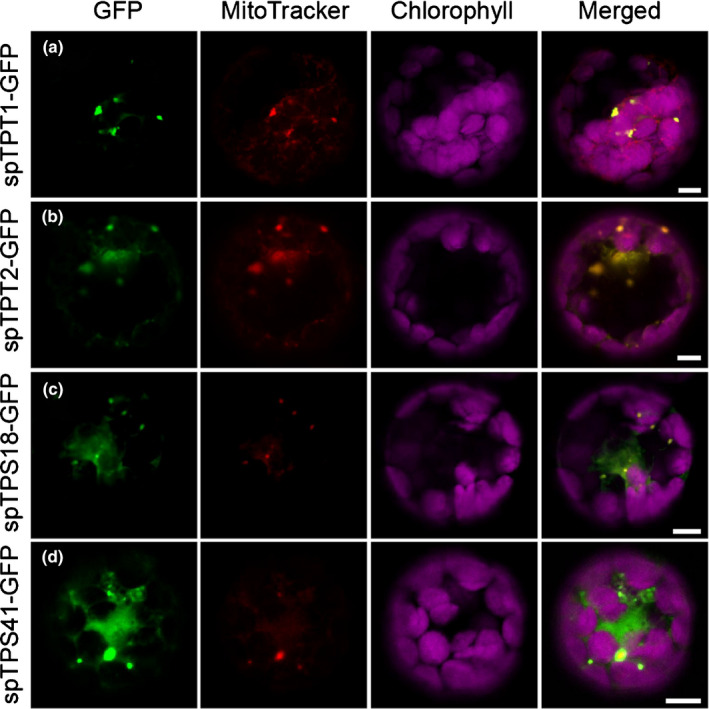
Subcellular localisation of the four mitochondrial proteins. (a, b) Localisation of trans‐prenyltransferase (TPT) 1 and 2, respectively. (c, d) Localisation of terpene synthase (TPS) 18 and 41, respectively. Sequences corresponding to the putative mitochondria transit peptide (sp) were fused to a downstream GFP and transiently expressed in Arabidopsis leaf protoplasts. GFP fluorescence indicates the location of each fusion protein (shown in green), the location of chloroplasts was determined by chlorophyll autofluorescence (shown in magenta), and the localisation of mitochondria was determined by the fluorescence of MitoTracker (shown in red). Column labelled ‘Merged’ represents all the combined fluorescent signals. Bars, 5 μm.
Expression of TPS and prenyltransferase genes and metabolic profiling of terpenes
To determine the relative amount and tissue distribution of all the TPS transcripts as well as TPT and cis‐prenyltransferase transcripts encoding the enzymes that form the substrates of TPS enzymes, the expression of each gene was measured in a total of 17 tomato tissues using quantitative real‐time PCR (RT‐qPCR) with gene‐specific primers. Expression was quantified and is presented as a heat map (Fig. 9) to compare the relative expression levels for each gene in different tissues. To identify any correlation between TPS and prenyltransferase gene expression and terpene profiles, terpene volatiles in these tissues were analysed as well (Figs 10, S16). Transcripts of the four new TPS genes (TPS47, TPS48, TPS51 and TPS52) are present at low levels in multiple tissues, with slightly higher amounts observed in young leaf, flower bud, petal and mature leaf, respectively (Fig. 9). Among the other previously functionally uncharacterised TPS genes, transcripts of TPS16 and TPS41 are almost exclusively found in various trichomes. TPS10 transcripts are enriched in immature green fruit in addition to trichomes, while TPS25 is expressed in multiple tissues primarily in petiole and flower bud, TPS27 is maximally expressed in young leaf trichomes, and TPS28 is mainly expressed in flower bud at low levels. TPS18 is mostly expressed in roots and flower buds, TPS36 is mainly expressed in mature leaf trichomes and immature green and yellow fruits. TPS33 is primarily expressed in immature green fruit and flower bud, while TPS35 is mainly expressed in stem trichomes, flower bud and immature green fruit (Fig. 9).
Figure 9.
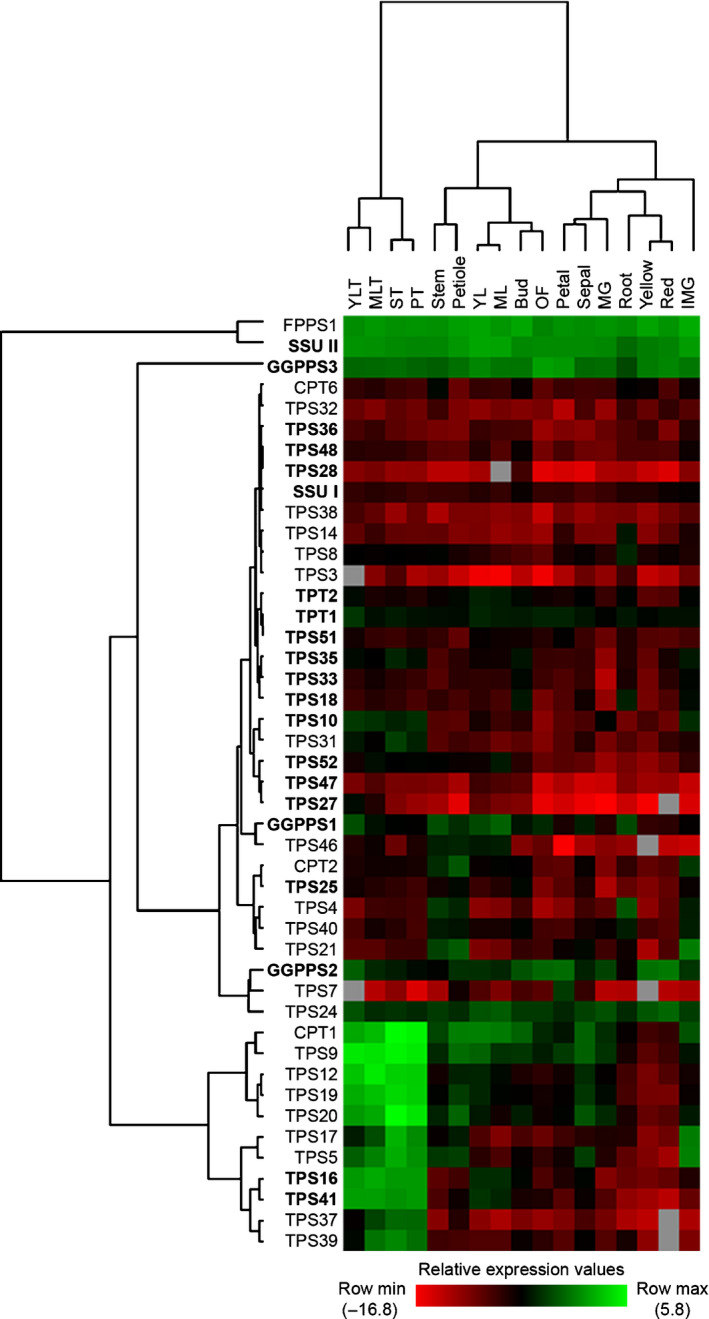
Gene expression analysis of terpene metabolism‐related genes. Heat map showing the relative transcript abundance of terpene synthase and prenyltransferase genes from tomato plant tissues taken at different developmental stages. Genes functionally characterised in this work are shown in bold. The values in the colour bar are log‐transformed values of relative transcript levels (2−∆Ct) determined by quantitative real‐time PCR (RT‐qPCR), grey colour indicates missing of transcript reads. The hierarchical clustering was performed using average linkage method by cluster 3.0. Abbreviations for tissues: IMG, immature green fruit; MG, mature green fruit; ML, mature leaf; MLT, mature leaf trichome; OF, fully opened flower; PT, petiole trichome; ST, stem trichome; YL, young leaf; YLT, young leaf trichome. Abbreviations for genes: CPT, cis‐prenyltransferase; FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; SSU, small subunit of geranyl diphosphate synthase; TPS, terpene synthase; TPT, trans‐prenyltransferase.
Figure 10.
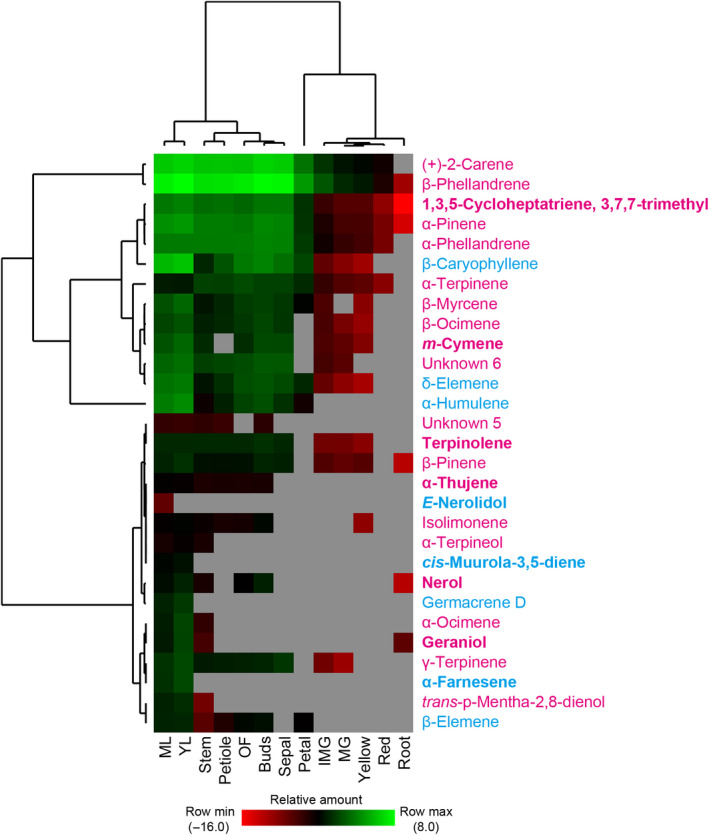
Terpene profiles of different tomato tissues. The peak areas normalised to the internal standard and tissue wet weight were calculated for each of the 29 terpene signals in 13 tomato tissues. Mass spectra and retention indices of the terpene products can be found in Supporting Information Fig. S3; Table S2, respectively. The average from two to three replicates was calculated and log‐transformed for the construction of heatmap. Grey colour indicates undetectable. The hierarchical clustering was performed using average linkage method by cluster 3.0. Monoterpenes are shown in magenta and sesquiterpenes are shown in blue. New terpenes identified in this work are shown in bold. IMG, immature green fruit; MG, mature green fruit; ML, mature leaf; OF, fully opened flower; YL, young leaf.
The TPT genes FPPS1, SSU II and GGPPS3 are highly expressed in all 17 tissues, GGPPS1 is mostly expressed in leaf tissues, GGPPS2 is mainly expressed in fruits and flower parts, and SSU I is mainly expressed in flower bud and fruits (Fig. 9). Our results for CPTs were similar to those previously reported (Akhtar et al., 2013; Matsuba et al., 2013), with the highest expression of CPT1 found in trichomes, lower expression of CPT2 mostly in petiole and stem, and lowest expression of CPT6 mainly in stem and root tissues (Fig. 9).
Our metabolic profiling of multiple tomato tissues detected a total of 21 monoterpenes and eight sesquiterpenes (Figs 10, S3; Table S2). In addition to the abundant and previously reported monoterpenes (+)‐2‐carene and β‐phellandrene and the sesquiterpenes β‐caryophyllene and α‐humulene (Schilmiller et al., 2009; Falara et al., 2011), we also identified several new terpenes (Fig. 10). Particularly noteworthy are α‐farnesene and cis‐muurola‐3,5‐diene, detected only from leaf tissues (Fig. 10), positively correlated with the expression patterns of TPS27 (α‐farnesene synthase) and TPS36 (cis‐muurola‐3,5‐diene synthase) (Fig. S17), which are responsible for their synthesis respectively (Figs 3, 11; Table S6).
Figure 11.
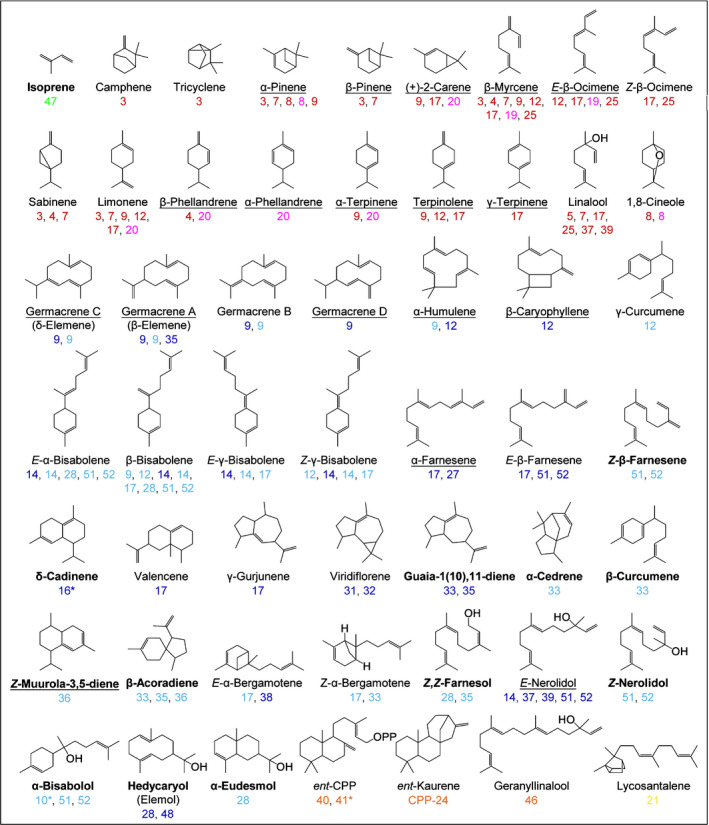
Terpene skeletons formed by the 34 functional tomato terpene synthases. New enzymatic terpene products reported in this study are shown in bold, the corresponding terpene synthase (TPS) enzymes are showing beneath each compound with different colours: light green, dimethylallyl diphosphate (DMAPP) as substrate; dark red, geranyl diphosphate (GPP) as substrate; magenta, neryl diphosphate (NPP) as substrate; dark blue, trans‐farnesyl diphosphate (E,E‐FPP) as substrate; light blue, cis‐farnesyl diphosphate (Z,Z‐FPP) as substrate; orange, geranylgeranyl diphosphate (GGPP) as substrate; yellow, nerylneryl diphosphate (NNPP) as substrate. Enzymes characterised by transiently expressing in tobacco are indicated with asterisks, all other enzymes are characterised by in vitro enzyme assays (detailed in Supporting Information Table S6). Elemol is the thermal breakdown product of (+)‐hedycaryol; β‐elemene and δ‐elemene are the thermal rearrangements of germacrene A and germacrene C, respectively. Terpenes detected from tomato tissues are underlined.
Discussion
The arrangement of TPS genes in the tomato genome
The majority of the 34 functional tomato TPS genes are located in clusters on chromosomes 1, 2, 6, 7, 8 and 10, while the other TPS genes are located on chromosomes 3, 4, 9 and 12 without another TPS genes nearby (Fig. 1). Examination of previously performed plantiSMASH analysis of the tomato genome (Kautsar et al., 2017) identified eight gene clusters related to terpene metabolism (Fig. S18). The clusters on chromosomes 1 and 2 consist of only TPS genes, while the other clusters also contain putative cytochrome P450s, methyltransferases, acyltransferases and glycosyltransferases (Fig. S18). Potential modifications of the direct terpene products (Fig. 11) by these enzymes may lead to the formation of new terpene skeletons with new biological functions, and will need further investigation.
Evolution of the TPS family
While there are still seven remaining enzymes in the Arabidopsis TPS family that have not yet been biochemically characterised (a quarter of the members) and subcellular localisations from many of the Arabidopsis TPS proteins have not yet been examined experimentally (Table S7), a comparison of the TPS families from tomato and Arabidopsis affords unprecedented and illuminating view of the dynamic evolution of the terpene synthase family. However, the fact that these two angiospermous species are both eudicots means that the generality of any conclusion drawn from such a comparison must await confirmation from additional comparisons taken among more distantly related land plants. With this caveat, we note that while the number of functional TPS genes in both dicot species is similar (Tables S3, S7), this similarity is misleading, masking major changes that have occurred in the TPS family in the two lineages since they last shared a common ancestor.
There are similar numbers of members in the TPS‐a clade in tomato and Arabidopsis – 15 and 22, respectively – but the phylogenetic analysis makes clear that the two species shared only one common TPS‐a ancestor from which each species evolved, by gene duplications, its extant group of TPS‐a genes (Fig. 2). This observation almost surely means that each lineage must have also lost TPS‐a genes along the way, and the presence of many TPS pseudogenes in both genomes (Table S4; Aubourg et al., 2002) lends strong support to this supposition. More interesting, while multiple gene duplications occurred in the TPS‐a clade in both lineages, the process of functional divergence of the duplicated genes appears to be quite different in the two lineages. In tomato, all 15 TPS‐a genes encode sesquiterpene synthases, and all but one of these enzymes (TPS36) are localised to the cytosol (Figs 2, 7, 12). However, they have clearly diverged in the type of products they produce, and also in the type of substrate they use; some use only E,E‐FPP, some can also use Z,Z‐FPP, and some use Z,Z‐FPP exclusively (Fig. 2). By contrast, only four of the 22 Arabidopsis TPS‐a genes are cytosolic sesquiterpene synthases. The rest are plastidic and/or mitochondrial diterpene synthases, plastidic sesterterpene synthases and one is a mitochondrial sesquiterpene synthase, the same as tomato TPS36 (seven of the Arabidopsis TPS‐a enzymes are still not functionally characterised; Fig. 2).
Figure 12.
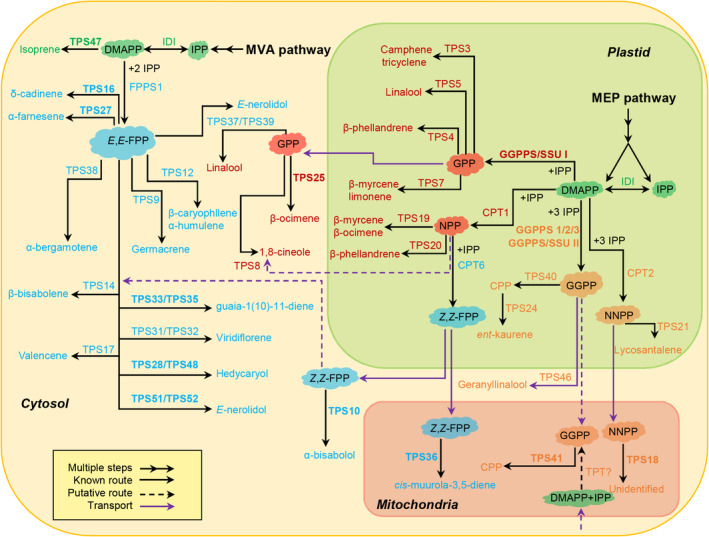
Terpene biosynthesis pathway in tomato. Terpene precursors are synthesised by the cytosolic mevalonic acid (MVA) pathway and the plastidic methylerythritol phosphate (MEP) pathway. The prenyl diphosphate substrates are labelled with clouds. C5, C10, C15, C20 compounds and their related enzymes are shown in green, red, light blue and orange, respectively. Enzymes characterised in this work are shown in bold. CPP, copalyl diphosphate; CPT, cis‐prenyltransferase; DMAPP, dimethylallyl diphosphate; E,E‐FPP, trans‐farnesyl diphosphate; FPPS, E,E‐FPP synthase; GPP, geranyl diphosphate; GGPP, geranylgeranyl diphosphate; GGPPS, GGPP synthase; IDI, isopentenyl diphosphate isomerase; IPP, isopentenyl diphosphate; NPP, neryl diphosphate; NNPP, nerylneryl diphosphate; SSU, small subunit of GGPPS; TPS, terpene synthase; TPT, trans‐prenyltransferase; Z,Z‐FPP, cis‐farnesyl diphosphate.
Regarding the TPS‐b clade, the Arabidopsis genome is missing (Fig. 2) representatives from a deep branch called the ‘β‐ocimene synthase branch’ to which all known isoprene synthases belong (Sharkey et al., 2013). There are three members in this branch in the tomato genome, one of which, TPS47, encodes a cytosolic isoprene synthase (all other known isoprene synthases are plastidic; Sharkey et al., 2013). The other two members, TPS25 and TPS27, encode cytosolic monoterpene and sesquiterpene synthases, respectively (Figs 3, 5, 7). The second branch of the TPS‐b clade contains six genes in both tomato and Arabidopsis (Fig. 2), and again the phylogenetic analysis indicates that both species had only a single common TPS gene ancestor of this branch, and that the six genes of this branch in each species are the result of more recent gene duplications (and loss). While most of these genes in each species encode plastidic monoterpene synthases, a cytosolic TPS‐b C10/C15 synthase evolved in this group in the Arabidopsis lineage, while in the tomato lineage this group also contains one cytosolic monoterpene synthase and one cytosolic sesquiterpene synthase (Fig. 2).
The TPS‐g clade is present in both genomes, with one representative – a plastidic monoterpene synthase – in Arabidopsis, and two representatives – both cytosolic C10/C15 synthases – in tomato (Fig. 2). Similarly, there is one Arabidopsis gene in the TPS‐c clade, a bona fide plastidic CPS, while tomato has two CPS genes, TPS40 and TPS41 (Fig. 2). TPS40 is plastidic (Fig. 7) and was shown to be involved in gibberellin biosynthesis (Rebers et al., 1999; Falara et al., 2011). TPS41 is localised to the mitochondria (Fig. 8d) and its physiological role is presently unknown. It should be pointed out that N. tabacum has at least one additional member in the TPS‐c clade besides its bona fide CPS, but this additional enzyme has been shown to produce 8‐hydroxy copalyl diphosphate (rather than CPP) that is the substrate of abienol synthase (Sallaud et al., 2012).
The TPS‐e/f clade in general contains two deep branches that diverged before the split of the gymnosperm/angiosperm lineages (Fig. 2), one encoding a plastidic KS (Falara et al., 2011) and one encoding a cytosolic geranyllinalool synthase (GLS) (Falara et al., 2014). The enzymes encoded by both genes use GGPP as the substrate (Fig. 2). There seems to have been little evolutionary novelty in this clade in the Arabidopsis genome. It contains a single GLS gene and a single KS gene (Fig. 2; Table S7). Tomato also contains a single GLS gene and a single KS gene (Fig. 2; Table S3). However, as previously shown (Matsuba et al., 2013), the tomato genome also contains four TPS‐e/f genes in a clade that diverged from KS before the split of the Arabidopsis and tomato lineages, two of which encode plastidic monoterpene synthases using NPP as the substrate, and a third one encoding a plastidic diterpene synthase whose substrate is NNPP (Fig. 2). Here we show that the fourth gene in this group, TPS18, encodes a mitochondrial diterpene synthase that uses NNPP (Figs 4b, 8c). While the functions of these four genes are likely to have evolved recently, the orthologue of these genes must have been lost in the lineage leading to Arabidopsis.
Biosynthesis of different classes of short‐chain terpenes in different subcellular compartments
Early analysis of the subcellular distribution of short‐chain (C10–C25) trans‐prenyltransferases indicated that GPPS and GGPPS are localised in the plastid, while E,E‐FPPS is localised mostly to the cytosol and perhaps to the mitochondria as well (Sun et al., 2016). This led to the general assumption that monoterpenes and diterpenes are synthesised in the plastids, while sesquiterpenes are synthesised in the cytosol (McGarvey & Croteau, 1995). Short‐chain CPTs, which provide substrates to some TPSs in Solanum, were shown to be all localised to the plastids (Akhtar et al., 2013), and the mono‐ and diterpene synthases that were previously shown to use these substrates were also localised to the plastids, thus providing no exception to the rule. However, Z,Z‐FPPS was also localised to the plastid, and a plastidic TPS‐e/f enzyme capable of using Z,Z‐FPP to produce sesquiterpenes was discovered in S. harbochaites (Sallaud et al., 2009).
In the last decade, there have been additional examples of rule‐breaking TPSs. GLS is a cytosolic diterpene synthase in both Arabidopsis (Herde et al., 2008) and tomato (Fig. 7). A cytosolic monoterpene synthase, geraniol synthase, from Lippia dulcis was shown to produce geraniol from GPP in planta (Dong et al., 2013), a cytosolic C10/C15 synthase was shown to use GPP to produce the monoterpene linalool in strawberry (Aharoni et al., 2004), and a basil cytosolic C10/C15 synthase was shown to use GPP to produce monoterpenes in a tomato heterologous system (Gutensohn et al., 2013). Although a cytosolic GGPPS has now been molecularly identified from several species (Coman et al., 2014), a cytosolic GPPS has not, although there are reports of cytosolic localisation of GPPS activity in the roots of Lithospermum erythrorhizon (Sommer et al., 1995). Additionally, accumulating results have led to the realisation that plant cells have mechanisms for transport of various prenyl diphosphates between compartments. The observations presented here indicate that more than a few tomato TPS enzymes are present in subcellular compartments in which their substrate may not be synthesised (Fig. 12). To highlight one example, mitochondrial TPS36 used Z,Z‐FPP to catalyse the formation of cis‐muurola‐3,5‐diene (Fig. 3b), a sesquiterpene that is not produced by any other tomato TPS enzyme (Fig. 11) but can be detected from tomato leaves (Fig. 10), suggesting that Z,Z‐FPP can also be imported into mitochondria from plastid (Fig. 12). Taken together, it appears that any class of short‐chain terpenes can no longer be assumed to be limited to a specific cellular compartment (Fig. 12).
Conclusions
The analysis of the biochemical activities and subcellular localisations of all the enzymes encoded by the functional TPS genes in tomato and of the genes encoding the enzymes that synthesise the substrates of terpene synthases provides an unprecedented view of the complexity of the terpenoid metabolic network. The terpenes directly produced by tomato TPS genes, and those that are obtained after modification of the direct products by additional enzymes, some of which may be encoded by genes clustered together with TPS genes, participate in multiple processes in the plants. Some have well established physiological roles such as hormones (e.g. gibberellins). The roles of others may be hypothesised to be ecological in nature, such as defence against insects or microorganisms, attraction of beneficial organisms, or even interaction with other plants (Pichersky & Raguso, 2018). The complete functional elucidation of the tomato TPS family now allows such roles to be investigated in detail.
Author contributions
EP and FZ designed the research. FZ performed all the experiments. EP and FZ interpreted the results and wrote the paper.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Annotated sequences of TPS51 and TPS52.
Fig. S2 SDS‐PAGE analysis of the purified recombinant His‐tagged TPS proteins.
Fig. S3 Mass spectra of identified terpenes from Figs 3–5 & 10.
Fig. S4 GC–MS analysis of elemol and (+)‐hedycaryol.
Fig. S5 GC–MS analysis of the products formed in planta by transiently co‐expressing TPS with CPT genes in Nicotiana benthamiana leaves.
Fig. S6 Analysis of tomato terpene synthases with isoprene synthases from other plants.
Fig. S7 Sequence alignment of the proteins encoded by the functional tomato TPS‐c genes and CPS genes from other plants.
Fig. S8 Predictions of subcellular localisation for TPS and prenyltransferase proteins made by TargetP 1.1 and ChloroP 1.1.
Fig. S9 Phylogenetic analysis of tomato TPT homologues and TPTs identified in other plants.
Fig. S10 GC–MS analysis of the products formed in planta by transiently co‐expressing TPS with TPT genes in Nicotiana benthamiana leaves.
Fig. S11 Sequence alignment of five Arabidopsis sesterterpene synthases and tomato TPSs from TPS‐a, TPS‐b and TPS‐g clades.
Fig. S12 Sequence alignment of the proteins encoded by the functional tomato TPS‐a genes.
Fig. S13 Sequence alignment of the proteins encoded by the functional tomato TPS‐b and TPS‐g genes.
Fig. S14 Sequence alignment of the proteins encoded by the functional tomato TPS‐e/f genes.
Fig. S15 Sequence analysis of TPT homologues from tomato and other plants.
Fig. S16 GC–MS analysis of terpenes extracted from tomato leaves.
Fig. S17 Correlation analysis of terpene biosynthesis genes and volatile terpenes from tomato.
Fig. S18 Terpene biosynthesis gene clusters identified from tomato genome by plantiSMASH.
Methods S1 Details on experimental procedures.
Table S1 Primers used in this study.
Table S2 A list of terpene compounds identified from Figs 3–5 & 10.
Table S3 A list of tomato terpene synthase genes, their characteristics and the enzymes they encode.
Table S4 A list of tomato terpene synthase pseudogenes.
Table S5 A list of the trans‐prenyltransferase genes in tomato.
Table S6 A list of terpene compounds produced by all the functional tomato terpene synthases.
Table S7 A list of Arabidopsis terpene synthase genes, their characteristics and the enzymes they encode.
Acknowledgements
This work was supported by National Science Foundation (USA) grant 1546617.
References
- Aharoni A, Giri AP, Verstappen FWA, Bertea CM, Sevenier R, Sun ZK, Jongsma MA, Schwab W, Bouwmeester HJ. 2004. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16: 3110–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar TA, Matsuba Y, Schauvinhold I, Yu G, Lees HA, Klein SE, Pichersky E. 2013. The tomato cis‐prenyltransferase gene family. The Plant Journal 73: 640–652. [DOI] [PubMed] [Google Scholar]
- Alquézar B, Rodríguez A, de la Peña M, Peña L. 2017. Genomic analysis of terpene synthase family and functional characterization of seven sesquiterpene synthases from Citrus sinensis . Frontiers in Plant Science 8: 1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament K, Van Schie CC, Bouwmeester HJ, Haring MA, Schuurink RC. 2006. Induction of a leaf specific geranylgeranyl pyrophosphate synthase and emission of (E, E)‐4,8,12‐trimethyltrideca‐1,3,7,11‐tetraene in tomato are dependent on both jasmonic acid and salicylic acid signaling pathways. Planta 224: 1197–1208. [DOI] [PubMed] [Google Scholar]
- Aubourg S, Lecharny A, Bohlmann J. 2002. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana . Molecular Genetics and Genomics 267: 730–745. [DOI] [PubMed] [Google Scholar]
- Bensen RJ, Zeevaart JAD. 1990. Comparison of ent‐kaurene synthetase A‐activity and B‐activity in cell‐free‐extracts from young tomato fruits of wild‐type and gib‐1, gib‐2, and gib‐3 tomato plants. Journal of Plant Growth Regulation 9: 237–242. [Google Scholar]
- Bleeker PM, Spyropoulou EA, Diergaarde PJ, Volpin H, De Both MT, Zerbe P, Bohlmann J, Falara V, Matsuba Y, Pichersky E et al 2011. RNA‐seq discovery, functional characterization, and comparison of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites trichomes. Plant Molecular Biology 77: 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann J, Meyer‐Gauen G, Croteau R. 1998. Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proceedings of the National Academy of Sciences, USA 95: 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth JK, Page JE, Bohlmann J. 2017. Terpene synthases from Cannabis sativa . PLoS ONE 12: e0173911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev AM, Moses T, Zi JC, Nelson DR, Mugford ST, Peters RJ, Osbourn A. 2015. Investigation of terpene diversification across multiple sequenced plant genomes. Proceedings of the National Academy of Sciences, USA 112: E81–E88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Ro DK, Petri J, Gershenzon J, Bohlmann J, Pichersky E, Tholl D. 2004. Characterization of a root‐specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8‐cineole. Plant Physiology 135: 1956–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E. 2011. The family of terpene synthases in plants: a mid‐size family of genes for specialized metabolism that is highly diversified throughout the kingdom. The Plant Journal 66: 212–229. [DOI] [PubMed] [Google Scholar]
- Chen F, Tholl D, D'Auria JC, Farooq A, Pichersky E, Gershenzon J. 2003. Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QW, Jiang T, Liu YX, Liu HL, Zhao T, Liu ZX, Gan XC, Hallab A, Wang XM, He J et al 2019. Recently duplicated sesterterpene (C25) gene clusters in Arabidopsis thaliana modulate root microbiota. Science China Life Sciences 62: 947–958. [DOI] [PubMed] [Google Scholar]
- Christianson DW. 2017. Structural and chemical biology of terpenoid cyclases. Chemical Reviews 117: 11570–11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby SM, Crock J, Dowdle‐Rizzo B, Lemaux PG, Croteau R. 1998. Germacrene C synthase from Lycopersicon esculentum cv. VFNT cherry tomato: cDNA isolation, characterization, and bacterial expression of the multiple product sesquiterpene cyclase. Proceedings of the National Academy of Sciences, USA 95: 2216–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coman D, Altenhoff A, Zoller S, Gruissem W, Vranová E. 2014. Distinct evolutionary strategies in the GGPPS family from plants. Frontiers in Plant Science 5: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong LM, Miettinen K, Goedbloed M, Verstappen FWA, Voster A, Jongsma MA, Memelink J, van der Krol S, Bouwmeester HJ. 2013. Characterization of two geraniol synthases from Valeriana officinalis and Lippia dulcis: Similar activity but difference in subcellular localization. Metabolic Engineering 20: 198–211. [DOI] [PubMed] [Google Scholar]
- Dudareva N, Martin D, Kish CM, Kolosova N, Gorenstein N, Fäldt J, Miller B, Bohlmann J. 2003. (E)‐beta‐ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 15: 1227–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Raguso RA, Wang J, Ross JR, Pichersky E. 1998. Floral scent production in Clarkia breweri. III. Enzymatic synthesis and emission of benzenoid esters. Plant Physiology 116: 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falara V, Akhtar TA, Nguyen TT, Spyropoulou EA, Bleeker PM, Schauvinhold I, Matsuba Y, Bonini ME, Schilmiller AL, Last RL et al 2011. The tomato terpene synthase gene family. Plant Physiology 157: 770–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falara V, Alba JM, Kant MR, Schuurink RC, Pichersky E. 2014. Geranyllinalool synthases in Solanaceae and other angiosperms constitute an ancient branch of diterpene synthases involved in the synthesis of defensive compounds. Plant Physiology 166: 428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffe J, Bru JP, Causse M, Vidal A, Stamitti‐Bert L, Carde JP, Gallusci P. 2000. LeFPS1, a tomato farnesyl pyrophosphate gene highly expressed during early fruit development. Plant Physiology 123: 1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Honzatko RB, Peters RJ. 2012. Terpenoid synthase structures: a so far incomplete view of complex catalysis. Natural Product Reports 29: 1153–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J, Dudareva N. 2007. The function of terpene natural products in the natural world. Nature Chemical Biology 3: 408–414. [DOI] [PubMed] [Google Scholar]
- Gutensohn M, Orlova I, Nguyen TT, Davidovich‐Rikanati R, Ferruzzi MG, Sitrit Y, Lewinsohn E, Pichersky E, Dudareva N. 2013. Cytosolic monoterpene biosynthesis is supported by plastid‐generated geranyl diphosphate substrate in transgenic tomato fruits. The Plant Journal 75: 351–363. [DOI] [PubMed] [Google Scholar]
- Hattan J, Shindo K, Ito T, Shibuya Y, Watanabe A, Tagaki C, Ohno F, Sasaki T, Ishii J, Kondo A et al 2016. Identification of a novel hedycaryol synthase gene isolated from Camellia brevistyla flowers and floral scent of Camellia cultivars. Planta 243: 959–972. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Kawaide H, Notomi M, Sakigi Y, Matsuo A, Nozaki H. 2006. Identification and functional analysis of bifunctional ent‐kaurene synthase from the moss Physcomitrella patens . FEBS Letters 580: 6175–6181. [DOI] [PubMed] [Google Scholar]
- Herde M, Gärtner K, Köllner TG, Fode B, Boland W, Gershenzon J, Gatz C, Tholl D. 2008. Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect‐induced volatile C16‐homoterpene TMTT. Plant Cell 20: 1152–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Kautsar SA, Hong YJ, Medema MH, Bond AD, Tantillo DJ, Osbourn A. 2017. Unearthing a sesterterpene biosynthetic repertoire in the Brassicaceae through genome mining reveals convergent evolution. Proceedings of the National Academy of Sciences, USA 114: E6005–E6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y, Davidovich‐Rikanati R, Fridman E, Gang DR, Bar E, Lewinsohn E, Pichersky E. 2004. The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant Physiology 136: 3724–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilmén M, Oja M, Huuskonen A, Lee S, Ruohonen L, Jung S. 2015. Identification of novel isoprene synthases through genome mining and expression in Escherichia coli . Metabolic Engineering 31: 153–162. [DOI] [PubMed] [Google Scholar]
- Jones MO, Perez‐Fons L, Robertson FP, Bramley PM, Fraser PD. 2013. Functional characterization of long‐chain prenyl diphosphate synthases from tomato. Biochemical Journal 449: 729–740. [DOI] [PubMed] [Google Scholar]
- Kautsar SA, Suarez Duran HG, Blin K, Osbourn A, Medema MH. 2017. plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Research 45: W55–W63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling CI, Weisshaar S, Ralph SG, Jancsik S, Hamberger B, Dullat HK, Bohlmann J. 2011. Transcriptome mining, functional characterization, and phylogeny of a large terpene synthase gene family in spruce (Picea spp.). BMC Plant Biology 11: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köksal M, Hu H, Coates RM, Peters RJ, Christianson DW. 2011. Structure and mechanism of the diterpene cyclase ent‐copalyl diphosphate synthase. Nature Chemical Biology 7: 431–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köksal M, Zimmer I, Schnitzler JP, Christianson DW. 2010. Structure of isoprene synthase illuminates the chemical mechanism of teragram atmospheric carbon emission. Journal of Molecular Biology 402: 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Kempinski C, Zhuang X, Norris A, Mafu S, Zi J, Bell SA, Nybo SE, Kinison SE, Jiang Z et al 2016a. Molecular diversity of terpene synthases in the liverwort Marchantia polymorpha . Plant Cell 28: 2632–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016b. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Kollner TG, Yin Y, Jiang Y, Chen H, Xu Y, Gershenzon J, Pichersky E, Chen F. 2012. Nonseed plant Selaginella moellendorffii has both seed plant and microbial types of terpene synthases. Proceedings of the National Academy of Sciences, USA 109: 14711–14715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J. 2010. Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biology 10: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba Y, Nguyen TT, Wiegert K, Falara V, Gonzales‐Vigil E, Leong B, Schäfer P, Kudrna D, Wing RA, Bolger AM et al 2013. Evolution of a complex locus for terpene biosynthesis in Solanum . Plant Cell 25: 2022–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba Y, Zi J, Jones AD, Peters RJ, Pichersky E. 2015. Biosynthesis of the diterpenoid lycosantalonol via nerylneryl diphosphate in Solanum lycopersicum . PLoS ONE 10: e0119302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell ET, Kapteyn J, Schmidt A, Li C, Kang JH, Descour A, Shi F, Larson M, Schilmiller A, An LL et al 2011. Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiology 155: 524–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey DJ, Croteau R. 1995. Terpenoid metabolism. Plant Cell 7: 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova I, Nagegowda DA, Kish CM, Gutensohn M, Maeda H, Varbanova M, Fridman E, Yamaguchi S, Hanada A, Kamiya Y et al 2009. The small subunit of snapdragon geranyl diphosphate synthase modifies the chain length specificity of tobacco geranylgeranyl diphosphate synthase in planta. Plant Cell 21: 4002–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Noel JP, Dudareva N. 2006. Biosynthesis of plant volatiles: nature's diversity and ingenuity. Science 311: 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Raguso RA. 2018. Why do plants produce so many terpenoid compounds? New Phytologist 220: 692–702. [DOI] [PubMed] [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang YY, Imai R, Sekimoto H, Kamiya Y. 1999. Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. The Plant Journal 17: 241–250. [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP. 2009. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnology Journal 7: 682–693. [DOI] [PubMed] [Google Scholar]
- Sallaud C, Giacalone C, Töpfer R, Goepfert S, Bakaher N, Rösti S, Tissier A. 2012. Characterization of two genes for the biosynthesis of the labdane diterpene Z‐abienol in tobacco (Nicotiana tabacum) glandular trichomes. The Plant Journal 72: 1–17. [DOI] [PubMed] [Google Scholar]
- Sallaud C, Rontein D, Onillon S, Jabès F, Duffe P, Giacalone C, Thoraval S, Escoffier C, Herbette G, Leonhardt N et al 2009. A novel pathway for sesquiterpene biosynthesis from Z, Z‐farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21: 301–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Ohara K, Yazaki K. 2005. Gene expression and characterization of isoprene synthase from Populus alba . FEBS Letters 579: 2514–2518. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Miner DP, Larson M, McDowell E, Gang DR, Wilkerson C, Last RL. 2010. Studies of a biochemical factory: tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiology 153: 1212–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E. 2009. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proceedings of the National Academy of Sciences, USA 106: 10865–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Chen QW, Lv HJ, He J, Liu ZF, Lu YN, Liu HL, Wang GD, Wang Y. 2017. (+)‐Thalianatriene and (−)‐retigeranin B catalyzed by sesterterpene synthases from Arabidopsis thaliana . Organic Letters 19: 1816–1819. [DOI] [PubMed] [Google Scholar]
- Sharkey TD, Gray DW, Pell HK, Breneman SR, Topper L. 2013. Isoprene synthase genes form a monophyletic clade of acyclic terpene synthases in the TPS‐b terpene synthase family. Evolution 67: 1026–1040. [DOI] [PubMed] [Google Scholar]
- Sommer S, Severin K, Camara B, Heide L. 1995. Intracellular‐localization of geranylpyrophosphate synthase from cell‐cultures of Lithospermum Erythrorhizon . Phytochemistry 38: 623–627. [Google Scholar]
- Sun P, Schuurink RC, Caissard JC, Hugueney P, Baudino S. 2016. My way: noncanonical biosynthesis pathways for plant volatiles. Trends in Plant Science 21: 884–894. [DOI] [PubMed] [Google Scholar]
- Tholl D, Kish CM, Orlova I, Sherman D, Gershenzon J, Pichersky E, Dudareva N. 2004. Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell 16: 977–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D, Lee S. 2011. Terpene specialized metabolism in Arabidopsis thaliana . The Arabidopsis Book 9: e0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium . 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie CC, Haring MA, Schuurink RC. 2007. Tomato linalool synthase is induced in trichomes by jasmonic acid. Plant Molecular Biology 64: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan MM, Wang Q, Webster FX, Kiemle D, Hong YJ, Tantillo DJ, Coates RM, Wray AT, Askew W, O'Donnell C et al 2013. Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 25: 1108–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W. 2013. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annual Review of Plant Biology 64: 665–700. [DOI] [PubMed] [Google Scholar]
- Wang CY, Chen QW, Fan DJ, Li JX, Wang GD, Zhang P. 2016. Structural analyses of short‐chain prenyltransferases identify an evolutionarily conserved GFPPS clade in Brassicaceae plants. Molecular Plant 9: 195–204. [DOI] [PubMed] [Google Scholar]
- Wang G, Dixon RA. 2009. Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proceedings of the National Academy of Sciences, USA 106: 9914–9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Huang XQ, Cao TJ, Zhuang Z, Wang R, Lu S. 2018. Heteromeric geranylgeranyl diphosphate synthase contributes to carotenoid biosynthesis in ripening fruits of red pepper (Capsicum annuum var. conoides). Journal of Agricultural and Food Chemistry 66: 11691–11700. [DOI] [PubMed] [Google Scholar]
- Wang Q, Jia MR, Huh JH, Muchlinski A, Peters RJ, Tholl D. 2016. Identification of a dolabellane type diterpene synthase and other root‐expressed diterpene synthases in Arabidopsis. Frontiers in Plant Science 7: 1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Jia Q, Chen X, Köllner TG, Bhattacharya D, Wong GK, Gershenzon J, Chen F. 2019. Terpene biosynthesis in red algae is catalyzed by microbial type but not typical plant terpene synthases. Plant Physiology 179: 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocol 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zerbe P, Bohlmann J. 2015. Plant diterpene synthases: exploring modularity and metabolic diversity for bioengineering. Trends in Biotechnology 33: 419–428. [DOI] [PubMed] [Google Scholar]
- Zhou F, Wang CY, Gutensohn M, Jiang L, Zhang P, Zhang D, Dudareva N, Lu S. 2017. A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice. Proceedings of the National Academy of Sciences, USA 114: 6866–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Köllner TG, Zhao N, Li G, Jiang Y, Zhu L, Ma J, Degenhardt J, Chen F. 2012. Dynamic evolution of herbivore‐induced sesquiterpene biosynthesis in sorghum and related grass crops. The Plant Journal 69: 70–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Annotated sequences of TPS51 and TPS52.
Fig. S2 SDS‐PAGE analysis of the purified recombinant His‐tagged TPS proteins.
Fig. S3 Mass spectra of identified terpenes from Figs 3–5 & 10.
Fig. S4 GC–MS analysis of elemol and (+)‐hedycaryol.
Fig. S5 GC–MS analysis of the products formed in planta by transiently co‐expressing TPS with CPT genes in Nicotiana benthamiana leaves.
Fig. S6 Analysis of tomato terpene synthases with isoprene synthases from other plants.
Fig. S7 Sequence alignment of the proteins encoded by the functional tomato TPS‐c genes and CPS genes from other plants.
Fig. S8 Predictions of subcellular localisation for TPS and prenyltransferase proteins made by TargetP 1.1 and ChloroP 1.1.
Fig. S9 Phylogenetic analysis of tomato TPT homologues and TPTs identified in other plants.
Fig. S10 GC–MS analysis of the products formed in planta by transiently co‐expressing TPS with TPT genes in Nicotiana benthamiana leaves.
Fig. S11 Sequence alignment of five Arabidopsis sesterterpene synthases and tomato TPSs from TPS‐a, TPS‐b and TPS‐g clades.
Fig. S12 Sequence alignment of the proteins encoded by the functional tomato TPS‐a genes.
Fig. S13 Sequence alignment of the proteins encoded by the functional tomato TPS‐b and TPS‐g genes.
Fig. S14 Sequence alignment of the proteins encoded by the functional tomato TPS‐e/f genes.
Fig. S15 Sequence analysis of TPT homologues from tomato and other plants.
Fig. S16 GC–MS analysis of terpenes extracted from tomato leaves.
Fig. S17 Correlation analysis of terpene biosynthesis genes and volatile terpenes from tomato.
Fig. S18 Terpene biosynthesis gene clusters identified from tomato genome by plantiSMASH.
Methods S1 Details on experimental procedures.
Table S1 Primers used in this study.
Table S2 A list of terpene compounds identified from Figs 3–5 & 10.
Table S3 A list of tomato terpene synthase genes, their characteristics and the enzymes they encode.
Table S4 A list of tomato terpene synthase pseudogenes.
Table S5 A list of the trans‐prenyltransferase genes in tomato.
Table S6 A list of terpene compounds produced by all the functional tomato terpene synthases.
Table S7 A list of Arabidopsis terpene synthase genes, their characteristics and the enzymes they encode.


