Graphical abstract
Abstract
Current treatment of patients with coronavirus 2019 (COVID-19) involves repurposed drugs that inhibit viral infection by either binding to their respective targets or via modulating cellular signal transduction. However, there is still a great deal of efficacy enhancement through combination therapy and derivatization. Combination therapy should involve agents with significant activity and different mechanisms of action. The structural map of the interaction between a drug and its target protein will help guide drug discovery for devising safe and effective ways to treat COVID-19. Herein, we report numerous synthetic designs based on enhanced affinity to the viral carbohydrate-rich protein spikes and protein-binding sites of COVID-19.
Teaser
We report the structural design of several analogs of current COVID-19 repurposed drugs. These analogs are designed to enhance the drug affinity to either the protein-binding sites or to the viral protein spikes.
Introduction
In December 2019, the city of Wuhan located in the Hubei Province of China detected a mysterious case of pneumonia and, shortly after, the symptoms were evident across a larger population within the region [1]. Deep sequence analysis revealed a novel strain of coronaviruses that was termed Severe Acute Syndrome Coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy and Viruses and on February 11 2020, the WHO named the disease caused by SARS-CoV-2 as COVID-19 [2].
Coronaviruses (CoV) are single-stranded RNA viruses that can cause animal or human diseases and are divided into four subfamilies (alpha, beta, gamma-, and delta-CoV), with SARS-CoV-2 belonging to the beta-CoV subfamily. While the alpha- and beta-CoV viruses infect mammals, the gamma- and delta-CoV viruses appear to infect birds [3]. Historically, six CoV have been previously identified as human-infecting viruses: HCoV-229E, HCoV-0C43, SARS-CoV, HCoV-NL63, HCoV-HKU1 and MERS-CoV [4].
SARS-CoV-2 has its name from the crown-like carbohydrate-covered protein spikes sticking out from a relatively large viral centre with a diameter of 60–140 nm [5]. As of April 2020, >1000 genomic sequences of COVID-19 have been globally reported (https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/), while the RNA strand of COVID-19 was shown to encompass ∼30 000 nucleotides [6]. So far, 27 proteins have been identified within the viral genome, some of which are RNA-dependent RNA polymerase (RdRp) [6]. In addition, four structural proteins were shown to be encoded in the corona-viral genome: the nucleocapsid protein (N), a small envelop protein (E), the matrix proteins (M), and the spike surface glycoprotein (S) [7]. The genetic code of COVID-19 was shown to be similar to that of SARS-CoV and MERS-CoV, despite differences in protein structures 7. For instance, amino acid substitutions were observed only in the nonstructural proteins nsp2, nsp3, and spike protein levels, whereas varying substitutions were also observed in nsp7, nsp13, envelope, matrix, and other proteins [7].
Mechanism of action
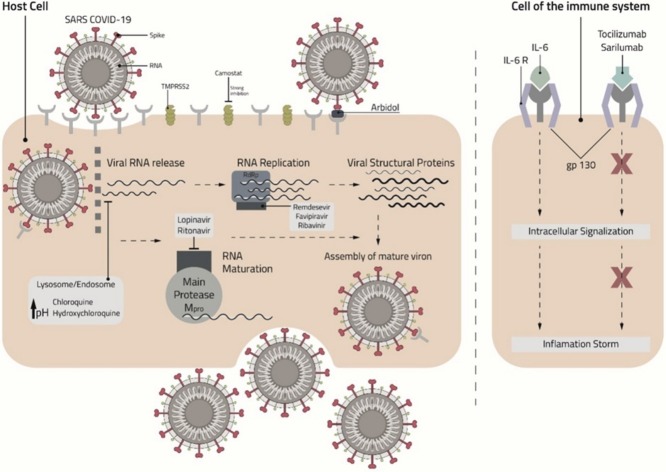
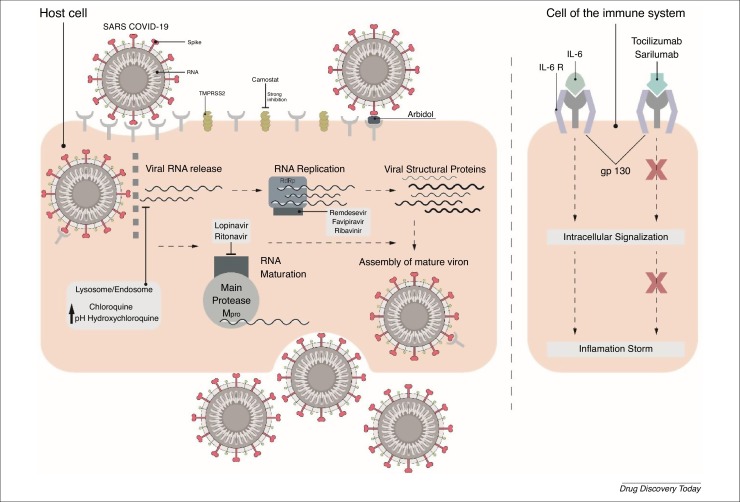
The viral mechanism of action is not yet fully understood, although studies suggest that the S genetic code of the receptor-binding spike protein mediates the viral entry into the host cell [8] while the N, E and M proteins hold general functions, such as encasing the RNA, protein assembly, budding, envelope generation, and pathogenesis [7]. The human angiotensin-converting enzyme 2 (ACE2), which is expressed in abundance in the lung and small intestines, is the main cell receptor for COVID-19 [6]. The fusion of the viral envelope through the ACE2 receptor, which is 10–20-fold higher compared with SARS-CoV, is facilitated upon its priming by Transmembrane Protease Serine 2 (TMPRSS2) [9]. Inside the cell, viral RNA is liberated into the cytoplasm, causing the translation of two polyproteins (pp1a and pp1ab) and leading to the formation of subgenomic RNAs because of replicate–transcription complex replication. At this stage, the N and E proteins congregate and generate viral material, which, having been contained in vesicles, then mingles with the plasma membrane and performs exocellular migration, releasing the virus [10].
Transmission, symptoms, and diagnosis of COVID-19
In March 2020, the WHO confirmed that human to human transmission of COVID-19 occurs through either the respiratory system or physical contact [11]. COVID-19 also appeared to be highly contagious and age related. Lai et al. reported that the reproduction rate of the virus follows an exponential growth pattern, and R0 (i.e., number of individuals who could be infected by a patient or carrier of COVID-19) was estimated to vary between 2.24 and 3.58 (SARS-CoV; R0 = 1.77), with 6.4 days being the epidemic doubling time [2].
The relatively high transmission rate (which follows a Lévy flight behavior) of COVID-19 is tightly connected to its relatively long incubation time (from 2 to 14 days), thereby rendering COVID-19 a disease that is hard to control, hence resulting in a pandemic [12]. Individuals who contract COVID-19 are divided into asymptomatic, pre-symptomatic, and symptomatic cases [13]. In general, symptomatic patients with COVID-19 show dominant nonspecific symptoms (similar to SARS and MERS), such as fever and cough. Clinical data from 278 adult patients (aged >18 years; 62% males) in Wuhan in February 2020 showed that the most common symptoms were fever (92.8%), cough (69.8%), dyspnea (34.5%), myalgia (27.7%), headache (7.2%), diarrhea (6.1%) rhinorrhea (4.0%), sore throat (5.1%), and pharyngalgia (17.4%) [2].
Medical identification of COVID-19 was first performed following the classical Koch’s postulates and viral morphology was determined via electron microscopy [14]. Later, infectious cases were identified in one of two ways, either symptomatic diagnosis or molecular laboratory techniques [15,16]. Currently, and in addition to syndromic diagnosis, the identification of new cases is based on COVID-19 nucleic acid detection in samples from nasal and throat swabs or other respiratory tract samples by real-time PCR methods and confirmed via next-generation sequencing [15,16].
Mortality rate, treatment, and prevention
COVID-19 appears to be less virulent than SARS-CoV (∼40 %) and MERS-CoV (∼10 %) with a lower mortality rate (∼2 %) [17,18]. However, despite the higher case fatality ratio (CFR) for SARS and MERS, COVID-19 has led to more fatalities because of its long incubation time, quick transmission, and the difficulties in detecting pre- and asymptomatic cases. Needless to say, the death rate of COVID-19 is higher among older patients and those with pre-existing chronic disease, such as cancer (5.6%) and cardiovascular diseases (10.5%) [19,20]. In the absence of a specific treatment, the most common therapy provided for patients with severe clinical COVID-19 symptoms is oxygen therapy (high-flow nasal oxygen; HFNO) and non-invasive ventilation. However, HFNO therapy appears to be insufficient in some cases and, thus, patients are sustained with protective mechanical ventilators, which are necessary for managing septic shock. Therefore, the scientific community has been working relentlessly to find anti-COVID-19 drugs and, thus far, a multitude of approaches have been proposed and applied, most notably, repurposing of already existing drugs.
On January 25 2020 a research team at the Shanghai institute of Materia Medica and Shanghai Tech University listed 30 drugs along with Chinese herbal medicines that could be effective against COVID-19 [21]. The list was then shortened and now includes the most promising antiviral agents: remdesivir, interferon-α (IFN-α), ribavirin, lopinavir/ritonavir, favipiravir, chloroquine phosphate (CQ), hydroxychloroquine sulfate (HCQ), umifenovir, tocilizumab, darunavir, and imatinib [18,21]. In addition, glucocorticoids were trialed over short periods of time (3–5 days) in severe cases where patients had a progressive deterioration of oxygenation indicators; however, there is a lack of scientific evidence to either support or prohibit their usage [22].
By contrast, the use of antibiotics against COVID-19 was quickly refuted by the WHO because they are not effective against the virus itself and might even reduce the ability of the body to mount an immunological response [13]. However, antibiotics could be administered to patients with COVID-19 to treat bacterial infections [13]. In addition to antivirals and immunosuppressive agents, trials on repurposing some anti-inflammatory medicines have been conducted [15,18,19,23], such as the kinase inhibitor baricitinib [24] and leronlimab, which is a humanized monoclonal antibody [25]. Plasma therapy is another therapeutic mode that has shown promising results in the fight against COVID-19; clinical trials noted symptomatic reduction within 3 days of therapy, while lung recovery was initiated after 7 days of treatment [26].
As is the case with any pandemic outbreak where R0 is >1, COVID-19 will continue to spread if no vaccine or treatment is available. Until then, prevention appears to be the only course of action to contain the outbreak. Worldwide, containment strategies underlying four basic actions (isolation, quarantine, social distancing, and community containment) have been adopted [27]. However, these measures will have a detrimental impact on national and global economies and, hence, might not be sustainable. This was evident through the ‘herd immunity approach’, which was adopted by some countries with the hope of overcoming the outbreak sooner rather than later so that they can resume their economic dealings at the earliest possible time.
Despite the multitude of approaches for prevention techniques, studies showed that current antiviral agents and various therapeutic modes have not displayed effective healing potential, although they might help to mitigate any viral effects. Although combination therapy involving two or more of the current and potential COVID-19 agents could provide a more effective approach towards patient safety, the need for derivatization of the most promising agents could prove to be the best route in the fight against COVID-19. The rationale behind derivatization should be based on increasing the activity of the drug against COVID-19 through enhanced affinity towards protein-binding sites as well as the carbohydrate-rich viral spikes of the virus.
Repurposed drugs
Remdesivir
Remdesivir (GS-5734) (Fig. 1 a) is a monophosphoramidate prodrug and a nucleotide analog [28] with antiviral activity against several RNA viruses belonging to the Coronaviridae (SARS-CoV and MERS-CoV), Paramyxoviridae (Nipah virus and Hendra virus), and Filoviridae (Ebola virus; EBOV) [29].
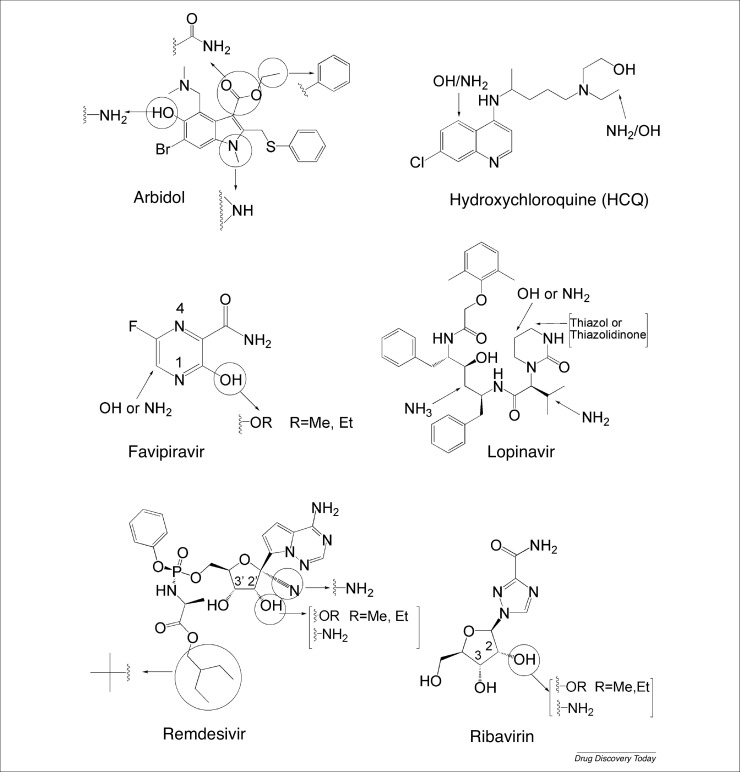
Figure 1.
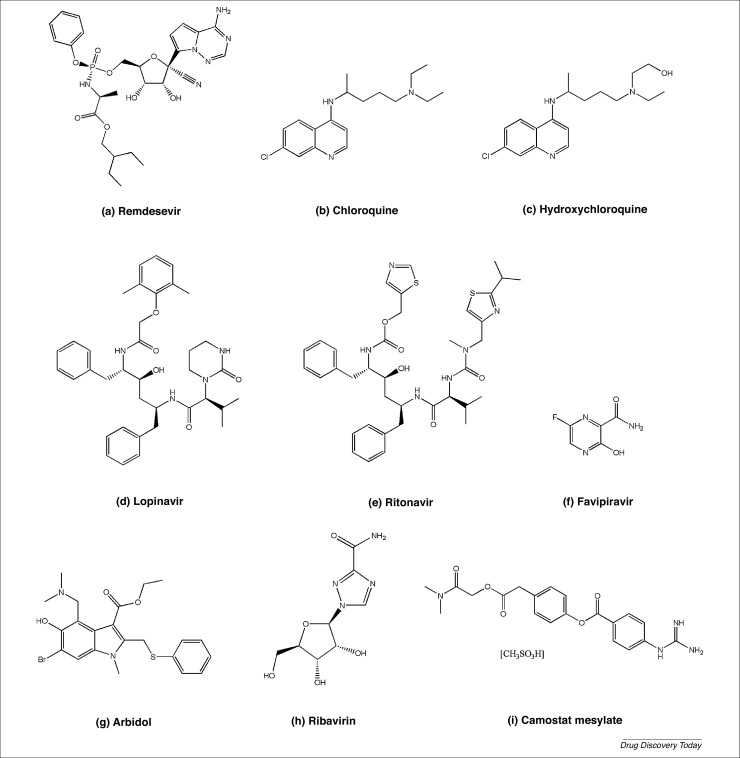
Chemical structures of (a) remdesivir, (b) chloroquine, (c) hydroxychloroquine, (d) lopinavir, (e) ritonavir, (f) favipiravir, (g) umifenovir, (h) ribavirin, and (i) camostat.
EC50 values of remdesivir varied between 0.074 μM in primary human airway epithelial cells (HAE) for MERS-CoV, 0.069 μM in HAE for SARS-CoV, and 0.03 μM in DBT cells for mouse hepatitis virus (MHV) [28,30]. The maximal inhibition of MHV in infected cells was obtained when remdesivir was added into cells between 2 h pre-infection and 2 h post infection. However, less inhibition was observed after 4 h post infection [28]. This showed that remdesivir blocks the virus at the early stage when RNA replication occurs (Fig. 2 ). Moreover, the decrease in viral titer (i.e., quantity of virus per volume of body fluid) was 100-fold greater in nsp14 exoribonuclease ExoN (-) mutant MHV compared with wild type (WT) because ExoN has a proofreading activity and might be responsible for the virus resistance against remdesivir [28].
Figure 2.
Schematic presentation of the mechanism of action of chloroquine, hydroxychloroquine, umifenovir, lopinavir, ritonavir, remdesivir, favipiravir, ribavirin, tocilizumab, and sarilumab against Coronavirus 2019 (COVID-19). Abbreviations: IL, interleukin; SARS-CoV-2, Severe Acute Syndrome Coronavirus 2; TMPRSS2, Transmembrane Protease Serine 2.
Upon stimulation by two cofactors, the nsp7 and nsp8 proteins, RdRp (also called nsp12) catalyzes the replication of RNA from an RNA strand [31]. Remdesivir then competes with nucleotides for RdRp active sites and is incorporated into the growing RNA strand. Sequence alignment of COVID-19 RdRp showed 90.18% and 56.76% sequence identity with SARS RdRp and MERS RdRp, respectively, [32] and high conservation in the active site that contains two aspartate residues (D255 and D256) and mediates nucleotide interaction during RNA replication [32].
The mechanism of action of remdesivir on MERS-CoV RdRp was studied after co-expressing the protein with the cofactors nsp7 and nsp8 and the protease nsp5 in insect cells [33]. RNA template, ATP, and remdesivir-triphosphate were then added for RNA synthesis and nucleotide incorporation. The insertion of remdesivir-triphosphate at position i inhibited RNA synthesis at position i+3 through its hydroxyl group present in position 3′, which allowed the incorporation of three more nucleotides until the RNA synthesis stopped at position i+3. Thus, the active form of remdesivir caused RNA chain termination and the additional three nucleotides protected remdesivir from ExoN proofreading activity [33]. Moreover, the increase in ATP concentrations was accompanied by increased IC50 values of remdesivir-triphosphate, thus confirming competition between ATP and the drug during incorporation in the RNA strand [33]. Similar effects were visible when remdesivir-triphosphate was incorporated at position i during EBOV-RdRp co-expression, and RNA synthesis inhibition occurred at position i+5. These results confirm the role of remdesivir in the delayed inhibition of RNA prolongation and highlights possible mechanisms of action, such as unfavorable interactions between elements of the polymerase and the adenosine analog, and/or structural modifications of the newly produced RNA [34]. As support for these findings, the interaction between remdesivir and COVID-19 RdRp was predicted using molecular docking, where remdesivir was found to bind to the active site with a lower binding energy (–7.5 Kcal/mol) than that observed with SARS-CoV [32]. In vitro studies on Vero E6 cells infected with COVID-19 showed that remdesivir inhibited the infection at a low concentration (EC50 = 0.77 u M) and revealed a high selectivity index (SI > 129.87) [35].
The first successful use of remdesivir for treating COVID-19 was for a patient in the USA, who was found to have COVID-19 and was treated with remdesivir on the evening of the 7th day of admission [36]. After 2 days of intravenous injection of the drug, the patient’s clinical symptoms were ameliorated, and their blood oxygen increased from 90% to 94%. In a recent study, patients found to have COVID-19 received a 10-day treatment with remdesivir, where 200 mg of the drug was administered intravenously on day 1, followed by 100 mg for 9 days. Improvement in clinical symptoms and oxygen support was observed in 68% of patients; among these, 57% were receiving mechanical ventilation; and 13% died. Adverse events, such as diarrhea, hypotension, and rash, were observed in patients receiving mechanical ventilation [37]. Thus, although remdesivir might be a promising drug in treating COVID-19, more randomized, and placebo-regulated trials are required to confirm its efficacy.
Chloroquine and hydroxychloroquine
CQ (Fig. 1b), an analog of 4-aminoquinoline, is among the most important antimalarial drugs and it therapeutic effect is most pronounced against Plasmodium falciparum [38]. One of the many derivatives of CQ is HCQ (Fig. 1c), which also has a main role in prophylactic measures against malaria [39]. Unlike the N-diethyl side chain of CQ, HCQ has an N-hydroxyethyl group that renders it more water soluble and less toxic than CQ [40].
Given the similarities in their chemical structures, CQ and HCQ are thought to have similar mechanisms of action against several viral strains [40]. CQ is a weak alkaline molecule that increases the pH of intracellular organelles upon its entry into the cell (Fig. 2). This pathway decreases the rate of viral infections, such as the Zika virus, which invades the cell via the endosome [41]. Moreover, some viruses, such as EBOV and Marburg virus (MARV), depend mainly on the low pH of lysosomes and endosomes to infect the cell by fusing their surface glycoproteins through cellular membranes. Both CQ and HCQ increase the pH of intracellular organelles, consequently preventing the invasion of the aforementioned viruses, among others [42]. Thus, CQ and HCQ inhibit the protease that is necessary for cleaving the spikes of the virus, a step essential for the successful fusion of the virus into the cell [43]. The most prominent mechanistic pathway associated with CQ and HCQ revolves around ACE2. According to Jia and Li, SARS-CoV-1 enters the cell by binding its spike glycoprotein to ACE2 receptors, which are mainly present on the cells of apical human airway epithelia [44,45]. The spikes of the virus bind to several residues of ACE2, mainly the first αβ-helix, Lys353, and the N-terminal of its fifth β-sheet. In addition, an essential factor hypothesized for the successful binding of the SARS spike is glycosylation of the 90th residue of ACE2 [46]. CQ has been shown to inhibit in vitro infection by SARS-CoV-1 [47] and, in the process, weakens the glycosylation of ACE2, which leads to a lower affinity between the virus and the receptor, causing a decrease in SARS-CoV-1 infection [48].
Consequently, and based on the mechanistic insight proposed earlier, many scientists have hypothesized that CQ and HCQ could decrease the prevalence of SARS-CoV-2 [35]. SARS-CoV-2, similar to SARS-CoV-1, binds the same ACE2 receptor for entry and infection of cells [9]; thus, researchers speculated that CQ and HCQ would have antiviral effects against SARS-CoV-2 just as they do against SARS-CoV-1. According to Wrapp et al., SARS-CoV-2 binds with a 10- to 20-fold higher affinity to the ACE2 receptor than does SARS-CoV-1, which might explain the high human-to-human transmission [8]. Given that CQ impairs the glycosylation of ACE2, which effectively decreases the rate of infection by other viruses, it is hypothesized that it has the same mechanism with SARS-CoV-2 [48]. Additionally, because CQ and HCQ are immunomodulators, researchers have highlighted that both drugs can reduce some of the immune responses caused by the virus [49]. Specifically, HCQ was shown to inhibit the activities of lysosomes in antigen-presenting cells (APCs), such as B cells, by rendering the cellular medium more alkaline [49]. This suggests that both CQ and HCQ can act as antiviral factors and immunomodulators, making them among the leading candidates for the treatment of COVID-19. A plethora of clinical trials has already been initiated, and several of these clinical trials with >100 patients have shown that CQ significantly decreases the severe symptoms of pneumonia with nonsignificant adverse effects [50]. Moreover, a clinical trial testing the effect of HCQ on patients showed promising results attributed to its anti-inflammatory and immunomodulatory role, whereby the symptoms were relieved after 5 days of treatment with 400 mg/day of HCQ oral tablets [51].
Although these drugs have shown potential in treating the COVID-19 viral strain, adverse effects, such as gastrointestinal reactions, indicate that novel or different agents are required [52]. One possible strategy would be to synthesize analogs of these agents with the hope of retaining their activity against COVID-19 while reducing their adverse effects.
Lopinavir and ritonavir
Lopinavir (Fig. 1d) is a peptidomimetic molecule that inhibits protease activity and has high specificity for HIV-1 protease, which has an essential role in the formation of functional polypeptides. The protease is a homodimer that contains 99 amino acids in each monomer and its active site includes three amino acids (Asp25–Thr26–Gly27) forming a loop structure that is stabilized by hydrogen bonds [53]. The two aspartate residues (Asp25 and Asp25′) from both chains are involved in drug binding. Lopinavir binds to the active site of the protease (Fig. 2) via a strong hydrogen bond with the oxygen of Asp25 and a weak hydrogen bond with the carboxylic acid of Asp25′ [54]. However, Lopinavir is rapidly metabolized in the liver by CYP3A4 and CYP3A5, thus lowering its bioavailability [55].
Ritonavir (Fig. 1e) was found to inhibit CYP450-3A4 (Fig. 2) in human liver microsomes, which results in a general increase in drug plasma concentrations [55]. CYP450-3A4 is an enzyme responsible for metabolizing drugs, such as antibiotics and protease inhibitors [56]; ritonavir binds to the heme iron of the active site of CYP450-3A4 via its thiazole nitrogen and reduces the enzyme redox potential. Moreover, the crystal structure of the complex CYP450-3A4/ritonavir shows that the drug binds well to the active site of the enzyme via hydrophobic interactions [57]. Hence, ritonavir increases the bioavailability of certain drugs by deactivating CYP450-3A4, which is responsible for the metabolic breakdown of xenobiotic substances.
Administration of ritonavir with lopinavir inhibits the metabolism of lopinavir by CYP450-3A4, which increases the lopinavir plasma concentration [58]. Therefore, a formulation of lopinavir/ritonavir was approved for clinical use by the US Food and Drug Administration (FDA) in September 2000 for the treatment of HIV [59]. Studies have shown that lopinavir has antiviral activity against SARS-CoV and MERS-CoV with EC50 values of 17.1 μM and 8.0 μM, respectively [60]. The main protease of CoVs, Mpro, has an important role in the formation of new active viral proteins [61]. Results from molecular dynamic simulations showed that lopinavir and ritonavir bind to different residues of the active site of the SARS-CoV Mpro, forming six and seven hydrogen bonds, respectively [62]. The Mpro of COVID-19 shows a higher identity with SARS Mpro (96.08%) than with MERS Mpro (51.61%) [63]. The structure of COVID-19 Mpro was predicted from the structure of SARS-CoV Mpro using modeler algorithms, and virtual docking of lopinavir/ritonavir to COVID-19 was elaborated. Results showed that lopinavir and ritonavir bind to Thr24, Thr26, and Asn119, with two and three hydrogen bonds, respectively [61].
In vitro, lopinavir has antiviral effects against SARS-CoV-2 with an EC50 of 26.1 μM [64]. However, results from the treatment of patients with COVID-19 with lopinavir/ritonavir remain unclear. In China, patients with SARS-CoV-2 were divided into two groups: the first (99 patients) received 400 mg of lopinavir and 100 mg of ritonavir twice daily for 14 days, whereas the second group (100 patients) acted as the positive control and received standard care. The time required for patients to exhibit clinical improvement along with the rate of mortality were shown to be similar for both groups; however, adverse events (nausea and diarrhea) were detected more in the group treated with lopinavir/ritonavir. Moreover, the treatment did not reduce viral RNA loads, as was initially hoped for, compared the standard care group. The combination therapy of lopinavir/ritonavir against those patients from within the Chinese population was shown not only to be ineffective, but also to cause adverse effects [65]. In a separate clinical trial, five patients in Taiwan received two pills of lopinavir (200 mg)/ritonavir (50 mg) twice daily. Viral shedding in patients with mild pneumonia was not reduced upon treatment [66]; instead, several adverse effects, such as gastrointestinal problems, nausea, vomiting, and diarrhea, were observed [65]. Thus, based on the results of these clinical trials, lopinavir/ritonavir might not be recommended for the treatment of patients with COVID-19. However, more data and further research are needed to investigate the effect of the lopinavir/ritonavir combination on COVID-19.
Favipiravir
Favipiravir (Fig. 1f) (T-705 or 6-fluoro-3-hydroxy-2-pyrazinecarboxamide) is a synthetic pyrazinecarboxamide and an antiviral agent designed to treat influenza [67]. Favipiravir showed effective antiviral activity by inhibiting the genomic replication of viruses that use RdRp (Fig. 2), meaning that it has a broad spectrum of activity as an antiviral agent [68].
Favipiravir showed anti-influenza effects both in vitro and in vivo [67]. Once in proximity of an infected cell, the prodrug crosses the cellular membrane and undergoes phosphoribosylation followed by phosphorylation to produce the active form favipiravir ribofuranosyl 5’-triphosphate (T-705RTP), which has a role in inhibiting the RdRp, leading to the inhibition of viral replication [69]. T-705RTP is a purine nucleic acid analog and, hence, a competitive inhibitor because it prevents the binding of ATP or GTP to the RdRp. The incorporation of T-705RTP prevents the continuous extension of the RNA strand being synthesized, also known as chain termination [70]. In addition to influenza, favipiravir has shown to have antiviral activity in vitro and in vivo against EBOV. According to Oestereich et al., T-705 prevented the spreading of the virus in Vero E6 cells in vitro and improved the symptoms of the disease while decreasing significantly the degree of lethality in mice if treatment was started 6 days post infection. In addition, all surviving mice developed specific anti-EBOV antibodies [71].
Currently, numerous clinical trials are under way to test the ability of favipiravir to treat SARS-CoV-2. The results of a clinical trial performed between February 20 2020 and March 12 2020 in three hospitals in China (Trial Registration Number: ChiCTR2000030254) showed that the drug considerably shortened the relief time against most symptoms (fever and cough) of COVID-19 with only minor and easily treated adverse effects still observed [72]. Another open-label non-randomized trial in China one (Trial Registration Number: ChiCTR2000029600) showed that the viral clearance period was shorter when infected patients were treated with favipiravir than with lopinavir/ritonavir, with a significantly more improved chest image. An improvement of 91.23% in the chest image of the patients treated with favipiravir revealed the efficiency of the drug in treating the symptoms of COVID-19 [73].
As a result of the mode of action on RdRp, Favipiravir might yet have a significant role in treating SARS-CoV-2. However, more clinical studies are needed to show whether favipiravir can have effective antiviral activity against SARS-CoV-2 infection with minimal adverse effects. Some adverse effects appeared in the aforementioned clinical trials and mainly include a higher concentration of uric acid in the serum along with digestive tract complications [72].
Umifenovir
Ethyl-6-bromo-4-[(dimethylamino)methyl]-5-hydroxy-1-methyl-2[(phenylsulfanyl)-methyl]-1H-indole-3-carboxylate, umifenovir (Fig. 1g) is an indole compound [74]. It is an antiviral drug that was licensed in Russia in 1993 and in China in 2006 for the treatment of influenza A and B [75]. Umifenovir has also been shown to have antiviral activity against other pH-dependent/independent viruses, such as respiratory syncytial virus [76] and corona viruses [75]. It inhibits Zika and West Nile viruses with an EC50 ranging between 10.57 and 19.16 mM [75]. It has also proved to be effective against EBOV in addition to foot and mouth disease [77].
The mechanism of action of umifenovir (Fig. 2) against the influenza virus involves inhibiting the fusion of the virus with target cell membranes [78,79]. Hemagglutinin (HA) is a glycoprotein on the surface of the influenza virus and has a role in the infection of host cells. At low pH, HA rearranges itself to uncover the fusion peptide, thus permitting the fusion of the viral membrane with the endosome [80] Umifenovir attaches to the hydrophobic cavity of HA and creates an extensive network of noncovalent interactions that stabilizes the conformation of HA preventing viral membrane fusion with the endosomes [78,80]. Moreover, umifenovir boosts the host immune response by the production of endogenous interferon, which acts against virus replication, and improves macrophage phagocytic activity and the activation of natural killer cells [79]. Umifenovir has been also shown to possibly prevent the interaction of the S protein (the viral surface binding protein) with the ACE2 host cell receptor, thus preventing viral envelope membrane fusion [81]. In vitro studies have shown that umifenovir inhibits SARS-CoV-2 at a concentration of 10–30 mM [21].
A nonrandomized study in China on 69 patients with COVID-19, showed a higher discharge (33%) and 0% mortality for 36 patients treated with umifenovir (0.4 g for 9 days) versus 19% discharge and 16% mortality for untreated patients [82]. In a separate study conducted in China, 59.2% patients had a virological conversion from positive to negative after receiving 200 mg of umifenovir three times daily compared with 40.3% for the patients who did not receive umifenovir. Bradycardia was observed in one patient who received umifenovir, and the other usual adverse effects of umifenovir (nausea, diarrhea, and dizziness) were minimal in this study [77]. In a third study, among 120 patients who received umifenovir (200 mg/three times daily for 10 days), 51.67% recovered [72]. Two clinical trials are ongoing to test the effectiveness of umifenovir against SARS-CoV-2 (NCT04260594 and NCT04255017) [83]. Thus, because of its minimal adverse effects, umifenovir should be further investigated with a larger sample to confirm its potential inhibition of SARS-CoV-2.
Ribavirin
Ribavirin (1-b-D-ribofuranosyl-1H-1,2,4-triazole- 3-carboxamide) (Fig. 1h) is a purine analog (guanosine, adenosine, and inosine) that shows antiviral activity against DNA and RNA viruses via different mechanisms of action [84]. One of the mechanisms involves the inhibition of inosine monophosphate dehydrogenase (IMPDH), which catalyzes the conversion of inosine monophosphate (IMP) into guanosine monophosphate via 2 steps (Fig. 2). First, IMPDH oxidizes IMP to form xanthosine monophosphate in the presence of NAD+. Second, guanosine monophosphate synthase (GMPS) transfers an amino group to xanthosine monophosphate to obtain GMP. Given that ribavirin mimics inosine, it binds to the active site of IMPDH, thus reducing the production of GMP, which is essential for RNA replication [85]. This significant reduction in GTP concentration (by 90%) might improve the incorporation of ribavirin triphosphate by RdRp, which might either cause an increase in random mutations in the RNA strand or result in RNA chain termination and blocking of viral RNA synthesis [86]. Moreover, given that ribavirin is a guanosine analog, it might also interact with enzymes implicated in the synthesis of the 7-methylguanosine RNA cap structure that prevents RNA degradation and is essential for RNA translation [87].
Oral ribavirin has been FDA authorized for the treatment of hepatitis C virus (HCV) [88] in addition to being also approved for the treatment of SARS-COV [89] and MERS-COV [90]. However, ribavirin monotherapy is associated with severe adverse effects, including hemolysis, elevation of transaminases, and bradycardia [91]. Currently, ribavirin is one of many agents being tested for the treatment of COVID-19. Wang et al. determined the EC50 of ribavirin in vitro on Vero E6 cells infected with COVID-19 to be 109.50 μM, indicating that high concentrations of ribavirin are required to cause a significant reduction in the in vitro viral infection [35]. The interaction between ribavirin triphosphate and the active site of COVID-19 RdRp was estimated using AutoDock Vina software and highlighted that ribavirin binds tightly to the active site of COVID-19 RdRp with 13 hydrogen bonds and a binding energy of –7.8 Kcal/mol [32].
Tocilizumab and sarilumab
Tocilizumab and sarilumab are monoclonal antibodies [92] used to control cytokine release syndrome (CRS) (Fig. 2), which is an inflammatory response caused by viral infections as well as certain drugs [93]. CRS is characterized by a significant increase in proinflammatory cytokines, including interleukin-6 (IL-6) [93]. IL-6 is a small glycosylated protein (21–28 kDa) that has four alpha helices [94] and is produced by immune system cells, such as lymphocytes B and C, macrophages, and others [95]. IL-6 binds to its receptor IL-6R (membrane bound or soluble) and the complex IL-6/IL-6R attaches to the gp130 membrane protein to trigger intracellular signaling [96] and to activate various cell populations with a role in host defence [95]. Despite the protective function and activation of the immune system, the high release of IL-6 can maintain chronic inflammation, as seen in arthritis or autoimmune diseases [97].
SARS-CoV-2 binds the ACE2 receptor and infects alveolar epithelial cells, which results in activation of immune system cells, such as macrophages, and the liberation of a significant number of cytokines, including IL-6 [95]. The main symptoms of CRS occur in patients with SARS and MERS [95]. Multiorgan dysfunction, such as acute respiratory distress syndrome, kidney or cardiac injuries, and liver disfunction can occur in patients with severe COVID-19. Moreover, an increase in cytokines and chemokines is observed more in patients with severe COVID-19 compared with patients with mild symptoms [98]. The high level of IL-6 has been noted in many studies concerned with COVID-19 and, thus, might act as a biomarker for the determination of the severity of COVID-19 infection [99].
Tocilizumab and sarilumab bind to both soluble and membrane-bound IL-6R, inhibiting the formation of the IL-6/IL-6R complex and, thus, activation of intracellular signaling via gp130 [92]. Tocilizumab is an IL-6 receptor antagonist that inhibits CRS and has been authorized for the treatment of rheumatoid arthritis [100]. Similarly, sarilumab is an anti-IL-6R antibody used to treat rheumatoid arthritis by improving the symptoms of patients [92].
The effect of tocilizumab on patients with COVID-19 was investigated in China among 21 patients, whereby a total recovery for 20 patients was observed within 2 weeks after the administration of tocilizumab. Treated patients had a significant drop in body temperatures along with the disappearance of all COVID-19 symptoms while displaying no adverse drug reactions [101]. Hence, in a separate study also conducted in China, five patients with COVID-19 (with a level of IL-6 from 16.4 to 627.1 pg/ml) received tocilizumab twice or more (80–600 mg each time) [102]. The results suggested that a repeated dose of tocilizumab should be administered for critically ill patients with a high plasma concentration of IL-6 [102]. Thus, tocilizumab would be mainly restricted for the treatment of patients with severe COVID-19 with the hope of reducing the mortality rate. More clinical trials associated with tocilizumab are ongoing on 500 patients with severe COVID-19 in China (ChiCTR2000029765) (http://www.chictr.org.cn/enindex.aspx (accessed April 30, 2020)).
In terms of sarilumab, an ongoing randomized clinical trial in the USA (Trial Registration ID: NCT04315298) is recruiting ∼400 patients who are 18 years old or more, have tested positive for SARS-CoV-2, and have from severe symptoms, pneumonia, and/or multiorgan dysfunction (https://clinicaltrials.gov/ct2/home (accessed April 30, 2020)).
Camostat mesylate
Camostat mesylate [N,N-dimethylcarbamoylmethyl 4-(4- guanidinobenzoyloxy)-phenylacetate; Fig. 1i] is a serine protease inhibitor developed and approved in Japan for reflux esophagitis and chronic pancreatitis and can also act as an antifibrinolytic agent [103,104]. Its main mechanism of action is through the inhibition of TMPRSS2 [104]. SARS-CoV-2 uses TMPRSS2 for spike protein priming and activation, a process that is essential for the viral cell diffusion through the ACE2 receptor. Thus, the inhibition of TMPRSS2 blocks SARS-CoV-2 entry into the cell. In vitro, camostat has been shown to significantly decrease the entry of MERS-CoV, SARS-CoV, and SARS-CoV-2 into human lung cells at concentrations of 10 and 50 μM. This was accompanied with minimal cytotoxic effects, thus rendering camostat a potential and promising agent for the treatment of COVID-19 [105]. Docking analysis further revealed that camostat interacts strongly with the active site residues of TMPRSS2, such as His296 and Ser441, via hydrogen bonding [106].
Currently, there are many ongoing trials investigating the ability of camostat to act against SARS-CoV-2, among other diseases. According to a current clinical trial on patients with dyspepsia, camostat treatment displayed no severe adverse effects in treated patients [107]. In other trials, adverse effects, such as edema and urticaria, were observed when camostat administrated daily at a dose of 900 mg, yet none were observed when the daily dose was <600 mg [104]. Such findings render camostat a well-tolerated drug. A randomized trial is currently being conducted on 580 participants at the Aarhus University in Denmark using tolerable doses of camostat (2 pills/100 mg each/three times daily/5 days) (NCT04321096 - https://clinicaltrials.gov/ct2/home (accessed April 30, 2020)). Another ongoing trial at Yale University in USA is testing the use of 200 mg of camostat administered orally three times daily over 7 days in 114 participants (NCT04353284 - https://clinicaltrials.gov/ct2/home (accessed April 30, 2020)). Camostat is also being tested in combination with HCQ, whereby a dose of 400 mg of HCQ is administered twice on day 1 followed by the administration of a 200 mg dose (twice daily) on days 2–5. In combination, patients are also being treated with tolerable concentrations of camostat (200 mg/three times daily/10 days) (NCT04355052 - https://clinicaltrials.gov/ct2/home (accessed April 30, 2020)). At the University of Kentucky, another trial is being run in which patients are also administered a tolerable camostat dose (2 pills/three times daily/14 days) (NCT04374019 - https://clinicaltrials.gov/ct2/home (accessed April 30, 2020)). However, the results of these ongoing clinical trials are not yet published and, thus, it is not possible to either confirm or deny the potential effect of camostat in treating SARS-CoV-2.
Combination therapy
One way in which therapeutic remedies can achieve enhanced activity while also minimizing adverse effects and overcoming drug resistance is through combination therapy, in which promising drugs with varying mechanisms of action are administered together. Clinical trials using a combination of HCQ and azithromycin (9-deoxo-9a-aza-9a-methyl-9a-homoerythromycin) in patients with COVID-19 was carried in France (EU Clinical Trials Register: 2020-000890-25) [108]. Azithromycin is a broad-spectrum macrolide antibiotic that has mainly immunomodulatory properties. It is mainly used to treat respiratory tract infections, skin infections, and even early stages of sexually transmitted infections (STIs) [109]. Gautret et al. reported that 20 patients with SARS-CoV-2 were given oral HCQ (200 mg, three times daily, for 10 days) and six of these patients were also treated with azithromycin (500 mg on the first day and 250 mg, once daily for the next 4 days) [108]. The results showed that viral clearance in all patients occurred within 6 days after treatment for both groups, with the combination therapy proving to be only slightly more efficient [108]. By contrast, the combination effect of HCQ and azithromycin was tested in parallel on 11 patients in France, whereby one patient died, whereas eight others continued to test positive for SARS-CoV-2 after 6 days of treatment. It is noteworthy that one of the patients also developed adverse effects of QT prolongation, leading to discontinuation of treatment [110]. In a separate study, 21 patients were treated with ribavirin, lopinavir/ritonavir and interferon-α, whereas 46 other patients were treated with the same combination but without ribavirin. All patients were discharged and results in both groups showed a positive correlation between hospital stay and mRNA clearance time [111].
At the University of Hong Kong, the combination of ribavirin with lopinavir/ritonavir and interferon beta-1B is currently being tested on 127 participants in which ribavirin (400 mg twice daily for 14 days) is being administered alongside lopinavir/ritonavir (400 mg/100 mg twice daily for 14 days) and interferon beta-1B (0.25 mg for 3 alternating days) (NCT04276688 - https://clinicaltrials.gov/ct2/home (accessed April 30, 2020)). Another combination currently on trial in China in 36 patients (ChiCTR2000029387) is the combination of ribavirin, lopinavir/ritonavir and interferon α-1B 112. Other clinical trials with combination therapy include favipiravir with CQ (NCT04319900) and favipiravir with lopinavir (800 mg once daily)/ritonavir (200 mg once daily) (NCT04303299). Also, a combination between favipiravir and the antibiotic tocilizumab is being studied in China in 150 patients (NCT04310228 - https://clinicaltrials.gov/ct2/home (accessed April 30, 2020)).
Ligand–protein interaction
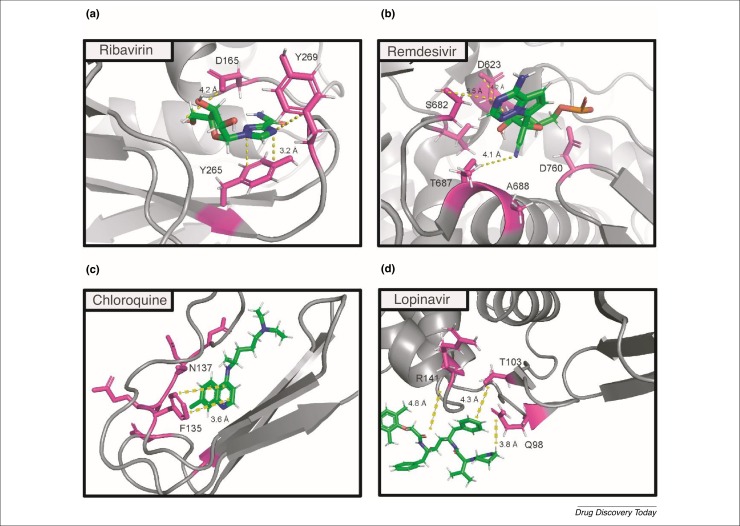
Using a combination of structural and molecular modeling approaches, antiviral drugs have been investigated for binding to proteins encoded by SARS-CoV-2 genes, including the spike protein, RdRp, which comprises nonstructural proteins (nsp7, 8, 12), and Papain-like protease (PLpro). According to Wu et al., ribavirin binds to PLpro in the active site of the enzyme by hydrophobic and hydrogen bonding to G164, D165, and Y269. In addition, π-π stacking strengthens the interaction between Y265 and the triazole ring in the compound [113] (Fig. 3 a). By contrast, remdesivir binds to the nsp12–nsp7–nsp8 complex through hydrogen bonding to the residues D623, S682, T687, A688, and D760 [114] (Fig. 3b). Given that the S protein uses the ACE2 receptor for entry and is linked to host cell surface gangliosides; the binding of CQ to the N-terminal ganglioside-binding domain (GM1) is a suitable approach to targeting a well-established COVID-19 infection [115]. The aromatic ring of F135 stacks onto the glucose cycle of GM1 via a CH-π stacking mechanism and is strengthened by additional interaction with the residue N137 (Fig. 3c). Accumulating evidence shows that the molecular interactions of lopinavir with the protease model of COVID-19 involve Q98, T103, R141, E175, and H176 as the potential drug binding sites [116] (Fig. 3d).
Figure 3.
Title. (a) Ribavirin bound to Coronavirus 2019 (COVID-19) PLpro via aromatic interaction with residue Y265 and hydrogen bonding with residues G164, D165, and Y269 [113]. (B) Binding of remdesivir to the COVID-19 nsp12–nsp7–nsp8 complex [Protein Data Bank (PDB) 7bv2], where the binding pocket is formed by residues D623, S682, T687, A688, and D760 through hydrogen bonding [114]. (C) Binding of chloroquine to the N-terminal domain of SARS-Cov-2 Spike protein (PDB 6vsb) [115]. (D) Binding of lopinavir to the protease model through Q98, T103, and R141 [116].
Derivatization
As previously mentioned, and despite promising therapeutic effects against COVID-19, the repurposed drugs display several adverse effects. In addition, viral strains, including COVID-19, mutate to achieve drug resistance [117] and Tang et al. revealed the evolution of COVID-19 genetic material into two main types, the L-type (70%) and the S-type (30%) [118]. Furthermore, Tang et al. noted that the L-type, derived from the S-type, is more dangerous on both the epidemiological and pathogenetic levels. As a result, current agents might become ineffective once the mutations exceed a given threshold. Therefore, predicting derivatives of currently used drugs, while aiming to enhance their draggability and reduce possible adverse effects, is vital to accelerate the hunt for a specific and efficient therapeutic agent against COVID-19.
Artificial Intelligence (AI) is a powerful tool for the de novo design of promising drugs. Machine learning helps predict quantitative structure–property relationships (QSPR) and quantitative structure–activity relationships (QSAR) for the generation of new biologically active molecules [119]. A precise prediction of the specific derivatization of a drug requires at first a full understanding of the mechanism of interaction between the drug and the virus. Data have been accumulating for such interactions with the Spike glycoproteins, nucleoproteins, and replicase polyproteins 1a/1ab of COVID-19 [15]. However, COVID-19 has only recently emerged and further investigations are needed to unravel the interaction pathways between the active therapeutic agents and the virus. Once this information is known, the de novo design and synthesis of potent, and less toxic derivatives will accelerate.
Herein, we propose short-step and facile derivatization schemes to modify current agents used against COVID-19, whereby functional group transformations are based on structural and mechanistic features of the selected drugs. For instance, the virus spike proteins are coated with carbohydrates, which, in turn, mask the spikes from the immune system and prevent an attack on the virus [120,121]. One potential strategy is to introduce functional groups with high affinity to carbohydrates (such as amino groups) to enhance drug binding to the sugar-coated virus spikes and, in the process, achieve enhanced activity. Also, derivatization of current drugs could be optimized via SARs in which newly synthesized drugs are designed to enhance binding to viral RNA to inhibit RNA replication.
Currently, the drugs showing the most potential in the fight against COVID-19 can be classified into three groups. Group 1 comprise site-directed drugs that target the interactions between the ACE2 receptor and the spike glycoprotein, thus inhibiting viral fusion into the cell. Group 2 comprises agents that disrupt COVID-19 RNA replication through incorporation into nascent viral RNA and inhibiting viral polymerases and proteases, and Group 3 comprises drugs that inhibit TMPRSS2.
Group 1: umifenovir, CQ/HCQ
The antiviral effect of umifenovir on influenza is attributed to its inhibition of viral fusion with cellular membranes by disrupting the pH that is essential for the efficient interaction between the virus and cellular receptors. Against COVID-19, umifenovir have been shown to alter the interaction of the S viral protein with the ACE2 host cell receptor, thus preventing fusion into the host cell. In addition, noncovalent hydrophobic interactions with the virus membrane were shown to be another antiviral mechanism [78,80,81]. Influenza and COVID-19 share similar pathogenic features, which suggests that increasing the basicity of the active molecule could enhance its antiviral activity. Such a modification could be accomplished by replacing the hydroxyl group on the aromatic ring with an amine group or by replacing the ester group attached to the five-membered ring with an amide group, whereby either transformation would increase the basicity of the molecule (Fig. 4 ). Alternatively, the methylated nitrogen within the five-membered ring can be reduced to a secondary NH group (Fig. 4). Incorporating an NH group into the umifenovir framework would increase the ability for noncovalent H-bonding with the viral membranes in addition to rendering the molecule more water soluble. To enhance the capacity for noncovalent hydrophobic bonding, the ethyl ester can be converted into a benzylic ester. Another option might lie in replacing the bromine with a chlorine because both are weak deactivating groups and ortho and para directors, yet the smaller chlorine might provide less hinderance for the aromatic system to engage in noncovalent hydrophobic bonding with the viral membranes.
Figure 4.
Derivatization of (a) umifenovir, (b) hydroxychloroquine, (c) favipiravir, (d) lopinavir, (e) remdesivir, and (f) ribavirin.
CQ and HCQ display the same mechanism of action against COVID-19 in that they both increase the pH of intracellular fluid and alter glycosylation on the 90th residue on ACE2 [46], thus lowering the affinity of the viral spikes to the cellular receptors. Being more water soluble and less toxic than CQ [40], HCQ makes a better candidate for derivatization–reactivity enhancement plots. To increase their basicity, an additional amine group could be added to the benzene ring at the ortho position, because both OH and NH2 are activating and ortho/para orienting groups (Fig. 4). Also, an additional OH (or NH2) group on the N(ethyl) substituent would render HCQ more basic and more water soluble.
Group 2: remdesivir, favipiravir, lopinavir, ritonavir, and ribavirin
Favipiravir is a prodrug and a purine nucleic acid analog that undergoes a phosphoribosylation step followed by phosphorylation to produce the active form of the drug [69]. Having a similar structure to ATP and GTP, the mechanism of action of the active form of favipiravir is based on its ability to compete with these molecules. Consequently, it prevents the binding between RdRp of the virus to ATP or GTP [70]. The N1 and N4 positions in the pyrazine ring are activated and deactivated by both the hydroxyl and amide functional groups (Fig. 4). An additional OH or NH2 group at the remaining ortho position to N1 would increase its affinity towards ribose. Such a modification should be accompanied with a slight decrease in the degree of ring activation that can be achieved by transforming the hydroxy group into an ether moiety, whereby a methyl ether derivative could also tune the activity of the N1 position and maintain the polarity of the molecule. Alternatively, the amide group should not be modified, otherwise the molecule would lose its nucleotide similarity and the electronic push–pull system would be altered.
Lopinavir and ritonavir are antiviral drugs the mechanism of action of which against COVID-19 is based on inhibiting Mpro protease activities of the virus by establishing several hydrogen bonds with two threonine and one asparagine residue in the protease [62]. However, lopinavir is quickly metabolized by the liver via redox reaction [55]. By contrast ritonavir has a redox active group, the thiazole substituent, which binds the heme iron through its nitrogen and, thus, inhibits the redox capacity of CYP450-3A4 [57]. Used in combination therapy, ritonavir helps to maintain an effective dose of lopinavir in the body. Therefore, it would be interesting to design a lopinavir derivative with higher bioavailability by offering the novel derivative some redox resistance. This could be acquired by attaching a thiazole group or any other group with similar effect, such as thiazolidinone, on lopinavir (Fig. 4). In addition, both drugs could use more functional groups that are capable of hydrogen bonding to the oxygen on the amino acid residues. Such a modification would strengthen the binding between the drugs and the protease, allowing them to further destabilize the Mpro–cell membrane interaction. Thus, an additional OH or NH2 group on the piperazine substituent of lopinavir, or on the tertiary carbon of the isopropyl substituent in both structures, could augment the hydrogen-bonding capacity of lopinavir and ritonavir.
Remdesivir is a prodrug and nucleotide (adenosine) analog that undergoes phosphorylation to generate the active triphosphate derivative. The antiviral mechanism resides in the ability of the triphosphate derivative to compete with the required nucleotide for the replication of the viral RNA. This results from the hydroxyl group at the 3′ position that enables the drug to replace ATP and incorporate into the growing RNA strands [33]. To enhance the activity of remdesivir, we propose to functionalize the OH group at the 3′ position, making it more reactive with the RdRp of COVID-19 and to introduce functional groups that would increase the drug affinity towards the carbohydrate coat of the S protein (Fig. 4). First, we suggest the reduction of the nitrile group on the ribose structure to yield an amine that has higher nucleophilic affinity towards the carbohydrate-rich spikes. Also, the OH group (2′ position) could be transformed into either an amine or an ether via the Williamson reaction. Such a modification would halt any possible competition between both hydroxyl groups vis-à-vis the phosphorylation step. Moreover, the 2-ethylbutyl chain of the ester linkage could be replaced with a less lipophilic alkyl chain (isobutyl), thus enhancing the water solubility of the remdesivir analog.
Ribavirin is an antiviral and purine analog with a similar structure to inosine, adenosine, and guanosine. It is hypothesized to affect viral replication by binding to the active site of RdRp of COVID-19 via hydrogen bonding [32]. Similarly, to remdesivir, the OH group on the 3′ position could be boosted by either replacing the OH group on the 2′ position with an amine or by transforming it into an ether (Fig. 4).
Group 3: camostat
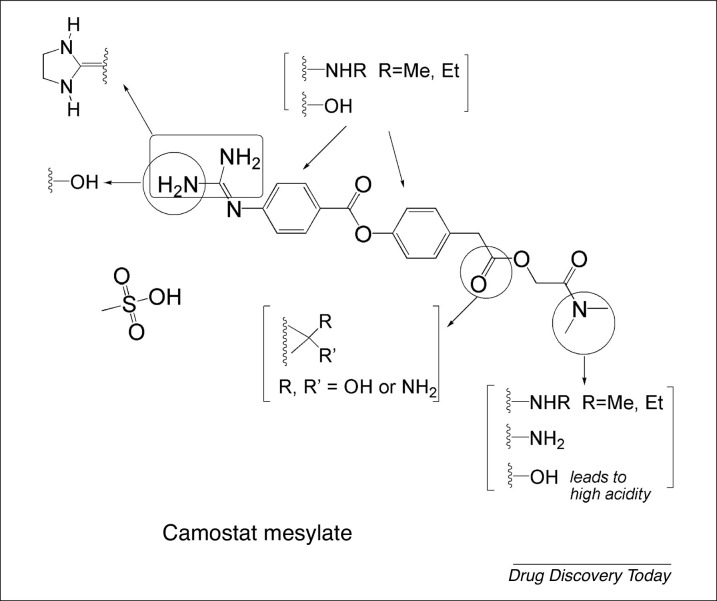
Camostat alters the spread and the pathogenesis of SARS-CoV-2 through strong interactions with different active site residues of TMPRSS2 via hydrogen bonding [106]. Despite the presence of several nitrogen and oxygen atoms in the camostat structure, only the guanidine moiety can engage in hydrogen bonding. Furthermore, the two NH2 groups are close enough to possibly establish intramolecular hydrogen bonds, which could weaken the interaction between the camostat structure and any potential active site on TMPRSS2. Consequently (Fig. 5 ), the replacement of guanidine by imidazolidine might strengthen any possible intermolecular hydrogen bonding with the protease residues, such as His296 and Ser441, because the cyclic rigid structure will hold both NH groups apart, preventing them from intramolecular hydrogen bonding. On a parallel note, it might also be worthwhile replacing one of the guanidine amino groups with a hydroxyl substituent, which in turn would alter the polarity of the molecule. Another approach would entail the addition of either a hydroxyl group or an amino group on the aromatic ring and/or reducing the ester functional group into an either an alcohol or an amine. Such structural modification would increase the ability of the molecule to interact via noncovalent hydrogen bonding, in addition to rendering it more water soluble. In another approach, the tertiary dimethylamine group could be reduced into a secondary or even primary amine (or a hydroxyl group), in turn enhancing the hydrogen-bonding ability of the molecule with TMPRSS2. Incorporating a carboxyl group in the camostat framework would increase the acidity of the molecule, which might result in some additional adverse effects, such as stomach-lining irritation. However, only a few studies/trials have been carried out and finalized with regards to the mechanism of action of camostat on COVID19 and in patients with COVID-19. As more mechanistic data are published, more derivatization schemes could be proposed.
Figure 5.
Derivatization of camostat.
Concluding remarks
In conclusion, COVID-19 is a serious infectious disease that is highly transmitted through physical contact. It infects host cells via the spike protein, which binds to the ACE2 receptor and its main symptoms are fever and cough. Currently, there are no specific treatments available and researchers are actively searching for effective drugs to combat COVID-19. Here, we have provided a framework to better understand the mechanism of action of 11 repurposed drugs and their potential role in treating COVID-19. The repurposed drugs inhibit viral infection by either binding to enzyme active sites and receptors or modulating intracellular signaling. For instance, remdesivir, favipiravir, and ribavirin are believed to bind to the RdRp active site, which inhibits the viral RNA replication. CQ and HCQ bind to the ACE2 receptor to block virus entry into the cells, and can decrease viral infection by modifying the pH of intracellular organelles. The ACE2 receptor is also a target for umifenovir, which appears to prevent the interaction between the spike protein of the virus and its receptor. Lopinavir binds to the active site of the SARS-CoV main protease and inhibits the production of new active viral proteins. Ritonavir is always used with lopinavir to inhibit its metabolic degradation by the cytochrome P450 enzyme and increase its plasma concentration. In contrast to these eight drugs, tocilizumab and sarilumab are two antibodies that control the CRS observed in patients with severe COVID-19. They bind to IL-6R and decrease the intracellular signaling responsible for acute inflammation. Camostat inhibits TMPRSS2, which is needed by SARS-CoV-2 to prime the spike protein and activate the ACE2 receptor for viral entry. Some clinical trials have shown the potential of these drugs in the treatment of COVID-19, and several clinical trials are ongoing to confirm their efficacy and safety.
Many of the current drugs used for the treatment of patients with COVID-19 include agents that are originally designed and on the market for treating other viral infections, such as EBOV and Malaria. On the one hand, the strategy to use repurposed drugs could be commercially viable and a time-saving route; on the other hand, this suggests that these agents are not optimized in terms of SARs against COVID-19 and, hence, there is still a great deal of efficacy enhancement through derivatization. The main problem associated with derivatization is the length of time required (at least 12–18 months) to design, synthesize, test, and acquire approval for any given new drug. As a consequence of the economic impact as well as the significant death rate of COVID-19, time is of the essence, and there is an imminent need to halt the spread of this virus as soon as possible to avoid devastating global health and economic consequences. Thus, we must make do with currently available agents to contain the virus until a treatment is on the market. Until then, combination therapy using current agents offers the greatest potential for overcoming the pandemic and should involve agents with significant enough activity against COVID-19 but with different mechanisms of action, because this would provide the best path towards overcoming viral resistance. For this purpose, we have divided the drugs reviewed herein into six categories based on their mechanism of action (Table 1 ). Formulations of significant potential could be developed by combining two or more drugs from different categories, such as remdesivir/HCQ/lopinavir/umifenovir/tocilizumab/camostat. This technique leads to multiple formulations (>100) in which no fewer than two drugs and no more than six drugs can be combined.
Table 1.
Drug categorization based on mechanism of action
| Category I: inhibit RNA-dependent RNA Pol | Category II: bind ACE2 receptor and increase endosomal pH | Category III: inhibit SARS-COV-2 main protease | Category IV: prevent interaction of S protein with ACE2 | Category V: bind IL-6R | Category VI: inhibit TMPRSS2 |
|---|---|---|---|---|---|
| Remdesivir, favipiravir, ribavirin | HCQ, CQ | Lopinavir, ritonavir (inhibits CYP450-3A4) | Umifenovir | Tocilizumab, sarilumab | Camostat |
In Table 2 , we present a comparative analysis of clinical trial results for the repurposed drugs described herein. Among the listed clinical trials, only two trials were statistically insignificant (population <20). Taking into consideration efficacy, low death rate and varying modes of action, we believe that pharmaceutical formulations based on the information in Table 1 would provide the best chance of overcoming the viral spread. These formulations, while potentially posing a low fatality risk, would ensure that administered drug doses are lowered (for each drug compared with its effective dose), thereby reducing the risk of chromic toxicity and adverse effects. With formulations based on drugs from different mechanistic categories, the issue of viral resistance could also be overcome, lowering, in the process, both the survival and transmission rates of COVID-19.
Table 2.
Comparative analysis of clinical trials results for drugs repurposed for the treatment of COVID-19
| Drug | Mechanism of action | Clinical trial | Community | No. of treated Patients | % Patients recovered/ improved | % Patients unaffected | % Patients deceased | Adverse effects in clinical trial | Notes | Refs |
|---|---|---|---|---|---|---|---|---|---|---|
| Remdesivir | Adenosine analog: inhibits RNA-dependent RNA Pol | Remdesivir administration (unspecified dose) on day 7 of hospitalization | USA | 1 | 100 | – | – | No obvious adverse effects | – | [37] |
| 10-day treatment: Day 1: 200 mg i.v.; Days 2–10: 100 mg i.v. | USA, Japan, Italy, Austria, France, Germany, The Netherlands, Spain, Canada | USA: 22; Japan: 9; Italy: 12; Austria: 1; France: 4; Germany: 2; The Netherlands: 1; Spain: 1; Canada: 1 (total: 53) | 47 | 68 | 13 | Common: increased hepatic enzymes, diarrhea, rash, renal impairment, hypotension. Serious: organ-dysfunction syndrome, septic shock, acute kidney injury, hypotension | Initial number of patients was 61 but data from 8 patients not analyzed | [38] | ||
| HCQ | Increases pH of endosomes, inhibiting viral entry; binds ACE2 receptor preventing binding to SARS-CoV-2 spike | Trial no: ChiCTR2000029559; 200 mg HCQ twice daily orally | Wuhan, China | 31 | 80.7 | 12.9 | No severe adverse effects; mild adverse effects included rash and headache | 6.5% of patients reported exacerbated symptoms | [51] | |
| HCQ with azithromycin | Azithromycin (antibiotic) | Trial no: 2020-000890-25; all patients received 200 mg HCQ, three times daily for 10 days; 6 of these patients received azithromycin 500 mg on Day 1 and 250 mg, once daily for next 4 days | France | 20 (14 patients treated with HCQ only, whereas 6 treated with both drugs) | 100 | 0 | 0 | No severe adverse effects | One patient died and 3 were transferred to ICU; however, these patients were not included in analysis | [112] |
| Favipiravir | Adenosine analog: Inhibits RNA-dependent RNA Pol | Trial no: ChiCTR2000029600; 1600 mg favipiravir twice daily on Day 1; 600 mg Favipiravir twice daily from Day 2 to Day 14 | Shenzhen, China | 35 | 91.43 | 6.45 | No severe adverse effects; mild adverse effects included diarrhea, liver injury, poor diet | Condition of 3.23% of patients worsened | [74] | |
| Trial no: ChiCTR2000030254; 1600 mg favipiravir twice daily on Day 1; 600 mg favipiravir twice daily until end of study | Wuhan, China | 116 | 61.21 | No severe adverse effects; mild adverse effects included raised serum uric acid, digestive tract reactions, irregular liver function tests, and psychiatric symptoms | ||||||
| Lopinavir/ritonavir | Both inhibit SARS-CoV main protease; ritonavir also inhibits CYP450-3A4, responsible for metabolism of lopinavir | Trial no: ChiCTR2000029308; 14 days: lopinavir: 400 mg twice daily; ritonavir: 100 mg, twice daily | Wuhan, China | 99 | 45.5 | – | 19.2 | Gastrointestinal adverse events : nausea, vomiting, diarrhea | Lopinavir–ritonavir treatment stopped early in 13.8% of cases because of adverse events; results with lopinavir–ritonavir similar to standard care | [68] |
| lopinavir (200 mg)/ritonavir (50 mg) twice daily | Taiwan | 5 | 100 | 0 | 0 | Vomiting, diarrhea | Results with lopinavir–ritonavir similar to standard care | [67] | ||
| Umifenovir | Prevents interaction of S protein with ACE2 host cell receptor | 9 days: 400 mg/three times daily | Wuhan, China | 36 | 33 | 67 | 0 | – | – | [82] |
| 200 mg/three times daily | Wuhan, China | 49 | 59.2 | – | – | Usual adverse effects (nausea, diarrhea) minimal in this study; bradycardia observed in 1 patient | – | [77] | ||
| Trial no: ChiCTR2000030254; 200 mg/three times daily for 10 days | Wuhan, China | 120 | 51.67 | – | – | Increased serum uric acid, digestive tract reactions, psychiatric reactions | – | [72] | ||
| Tocilizumab | Binds to IL-6R, inhibiting CRS | 13 days: 400 mg once i.v. | Wuhan, China | 21 | 100 | – | 0 | No adverse drug reactions | – | [101] |
For the long term, drug derivatization of current agents along with novel synthetic designs based on viral protein-binding sites provide the best route towards mitigating and stopping the pandemic. For this purpose, we have reported numerous synthetic designs based on enhanced affinity to the carbohydrate-rich protein spikes, given that the spikes provide the easiest route for targeting the virus. We have also designed and reported the synthesis of multiple derivatives of current COVID-19 agents with enhanced basicity (to increase lysosomal pH) and/or enhanced hydrophobic interactions/H-bonding (enabling agents to bind to viral RdRp or other viral binding sites). Such synthetic designs would provide a new generation of drugs with more specificity, enhanced immunity against virus mutation, and fewer adverse effects. Notably, derivatization through minor modifications of already existing agents is in coherence with the repurposing strategy adopted by international research centers. The repurposing strategy utilizes agents the skeletal structure of which is commercially available, ensuring that the derivatization scope becomes an economic and time-saving route to prepare new and more efficient anti-COVID19 remedies.
Finally, the structural map of the interaction between a drug and its target protein will help guide drug discovery to devise safe and effective ways to treat COVID-19. Understanding the drug-binding sites at the molecular level will shed light on basic mechanisms of action underlying this rapidly spreading virus.
Acknowledgments
We would like to thank Hadi Mroueh for his contribution to Fig. 2, which pertains to the mechanism of action of all the reviewed drugs.
Biographies

Elias Akoury holds a doctorate in physical chemistry from Max Planck Institute for Biophysical Chemistry (Germany) with a focus on the field of NMR and drug discovery. After postdoctoral studies at the University of Munich, he joined the Lebanese American University in 2019 as an assistant professor of chemistry. His research includes structural elucidation of protein complexes, and spectroscopic and computational mechanisms in drug design. Dr Akoury has broad technical experience in scientific instrumentation, including NMR, MS, X-ray crystallography, small angle X-ray scattering (SAXS), and cryo-electron microscopy (EM).

Robin Taleb holds a doctorate in medicinal chemistry from Western Sydney University, Australia, with a focus on drug design and synthesis. In 2011, he joined the Lebanese American University, at which he is now an associate professor of chemistry and the Assistant Dean of the School of Arts and Sciences. His current research focuses on the synthesis of anticancer platinum complexes as well as the biological investigation of synthetic and natural products. Dr Taleb has technical experience in several different types of scientific equipment, including nuclear magnetic resonance (NMR), liquid chromatography (LC), gas chromatography (GC), mass spectrometry (MS), inductively coupled plasma (ICP)-MS, and protein synthesizers.
References
- 1.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C.C., et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miłek J., Blicharz-Domańska K. Coronaviruses in avian species-review with focus on epidemiology and diagnosis in wild birds. J. Vet. Res. 2018;62:249–255. doi: 10.2478/jvetres-2018-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu N., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu A., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song W., et al. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Wilde A.H., et al. Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO Modes of transmission of virus causing COVID-19, implications for IPC precaution recommendations. Sci. Br. 2020;29:10–12. [Google Scholar]
- 12.Lauer S.A., et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO . WHO; 2020. Coronavirus Disease 2019 (COVID-19) Situation Report. [Google Scholar]
- 14.Lu H., et al. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J. Med. Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y.R., et al. The origin, transmission and clinical therapies on coronavirus disease (2019 (COVID-19) outbreak - an update on the status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Udugama B., et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 17.De Wit E., et al. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang F., et al. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19) J. Gen. Intern. Med. 2020;35:1545–1549. doi: 10.1007/s11606-020-05762-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adhikari S.P., et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect. Dis. Poverty. 2020;9:29. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Zhang L. Risk of COVID-19 for patients with cancer. Lancet Oncol. 2020;21:e181. doi: 10.1016/S1470-2045(20)30149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong L., et al. Discovering drugs to treat coronavirus disease (2019 (COVID-19) Drug Discov. Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 22.Russell B., et al. Associations between immune-suppressive and stimulating drugs and novel COVID-19—a systematic review of current evidence. Ecancermedicalscience. 2020;14:1022. doi: 10.3332/ecancer.2020.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cascella M., et al. StatPearls; 2020. Features, Evaluation and Treatment Coronavirus (COVID-19) [PubMed] [Google Scholar]
- 24.Richardson P., et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30304-4. e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duddu P. Clinical Trials Arena; 2020. Coronavirus Treatment: Vaccines/Drugs in the Pipeline for COVID-19. [Google Scholar]
- 26.Duan K., et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 2020;117 doi: 10.1073/pnas.2004168117. 202004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du Z., et al. Risk for transportation of 2019 novel coronavirus disease from Wuhan to other cities in China. Emerg. Infect. Dis. 2020;26:1049–1052. doi: 10.3201/eid2605.200146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agostini M.L., et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9:e00221–00218. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amirian E.S., Levy J.K. Current knowledge about the antivirals remdesivir (GS-5734) and GS-441524 as therapeutic options for coronaviruses. One Heal. 2020;9:100128. doi: 10.1016/j.onehlt.2020.100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheahan T.P., et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;XX:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon C.J., et al. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tchesnokov E.P., et al. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao Y., et al. Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19, An evaluation of the evidence. Travel Med. Infect. Dis. 2020;35:101647. doi: 10.1016/j.tmaid.2020.101647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grein J., et al. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parhizgar A.R. Introducing new antimalarial analogues of chloroquine and amodiaquine: a narrative review. Iran. J. Med. Sci. 2017;42:115–128. [PMC free article] [PubMed] [Google Scholar]
- 39.Haładyj E., et al. Antimalarials - are they effective and safe in rheumatic diseases? Rheumatologia. 2018;56:164–173. doi: 10.5114/reum.2018.76904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devaux C.A., et al. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;55:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delvecchio R., et al. Chloroquine, an endocytosis blocking agent, inhibits zika virus infection in different cell models. Viruses. 2016;8:322. doi: 10.3390/v8120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long J., et al. Antiviral therapies against Ebola and other emerging viral diseases using existing medicines that block virus entry. F1000Research. 2015;4:30. doi: 10.12688/f1000research.6085.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Bari M.A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017;5:e00293. doi: 10.1002/prp2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia H.P., et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W., et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keyaerts E., et al. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vincent M.J., et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou D., et al. COVID-19, a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J. Antimicrob. Chemother. 2020;75:1667–1670. doi: 10.1093/jac/dkaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao J., et al. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 51.Chen Z., et al. Efficacy of hydroxychloroquine in patients with COVID-19, results of a randomized clinical trial. medRxiv. 2020;7 2020.03.22.20040758. [Google Scholar]
- 52.Srinivasa A., et al. Increased incidence of gastrointestinal side effects in patients taking hydroxychloroquine: a brand-related issue? J. Rheumatol. 2017;44:368. doi: 10.3899/jrheum.161063. [DOI] [PubMed] [Google Scholar]
- 53.Lv Z., et al. HIV protease inhibitors: a review of molecular selectivity and toxicity. HIV/AIDS Res. Palliat. Care. 2015;7:95–104. doi: 10.2147/HIV.S79956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang D.A.W., Zhang J.Z.H. Full quantum mechanical study of binding of HIV-1 protease drugs. Int. J. Quantum Chem. 2005;103:246–257. [Google Scholar]
- 55.Chandwani A., Shuter J. Lopinavir/ritonavir in the treatment of HIV-1 infection: a review. Ther. Clin. Risk Manag. 2008;4:1023–1033. doi: 10.2147/tcrm.s3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sevrioukova I.F., Poulos T.L. Interaction of human cytochrome P4503A4 with ritonavir analogs. Arch. Biochem. Biophys. 2012;520:108–116. doi: 10.1016/j.abb.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sevrioukova I.F., Poulos T.L. Structure and mechanism of the complex between cytochrome P4503A4 and ritonavir. Proc. Natl. Acad. Sci. U. S. A. 2010;107 doi: 10.1073/pnas.1010693107. 18422–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hurst M., Faulds D. Lopinavir. Drugs. 2000;60:1371–1379. doi: 10.2165/00003495-200060060-00009. [DOI] [PubMed] [Google Scholar]
- 59.Kaplan S.S., Hicks C.B. Lopinavir/ritonavir in the treatment of human immunodeficiency virus infection. Expert Opin. Pharmacother. 2005;6:1573–1585. doi: 10.1517/14656566.6.9.1573. [DOI] [PubMed] [Google Scholar]
- 60.De Wilde A.H., et al. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu X., Wang X.-J. Potential inhibitors against (2019-nCoV coronavirus M protease from clinically approved medicines. J. Genet. Genomics. 2020;47:119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nukoolkarn V., et al. Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CLpro inhibitors. J. Theor. Biol. 2008;254:861–867. doi: 10.1016/j.jtbi.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020;251:117627. doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choy K.-T., et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao B., et al. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng C. Lopinavir/ritonavir did not shorten the duration of SARS CoV-2 shedding in patients with mild pneumonia in Taiwan. J. Microbiol. Immunol. Infect. 2020;53:488–492. doi: 10.1016/j.jmii.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Furuta Y., et al. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 2002;46:977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Furuta Y., et al. T-705 (favipiravir) and related compounds: novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 2009;82:95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Furuta Y., et al. Mechanism of action of T-705 against influenza virus. Antimicrob. Agents Chemother. 2005;49:981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sangawa H., et al. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob. Agents Chemother. 2013;57:5202–5208. doi: 10.1128/AAC.00649-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oestereich L., et al. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res. 2014;105:17–21. doi: 10.1016/j.antiviral.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 72.Chen C., et al. Favipiravir versus arbidol for COVID-19: a randomized clinical trial. medRxiv. 2020 doi: 10.1101/2020.03.17.20037432. Published online April 14, 2020. [DOI] [Google Scholar]
- 73.Cai Q., et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020 doi: 10.1016/j.eng.2020.03.007. Published online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blaising J., et al. Arbidol as a broad-spectrum antiviral: An update. Antiviral Res. 2014;107:84–94. doi: 10.1016/j.antiviral.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Proskurnina E.V., et al. Antioxidant potential of antiviral drug umifenovir. Molecules. 2020;25:1577. doi: 10.3390/molecules25071577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li M., et al. Antiviral activity of arbidol hydrochloride against herpes simplex virus I in vitro and in vivo. Int. J. Antimicrob. Agents. 2018;51:98–106. doi: 10.1016/j.ijantimicag.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 77.Xu K., et al. Clinical efficacy of arbidol in patients with 2019 novel coronavirus-infected pneumonia: a retrospective cohort study. SSRN Electron. J. 2020 ahead of print. [Google Scholar]
- 78.Boriskin Y., et al. Arbidol: a broad-spectrum antiviral compound that blocks viral fusion. Curr. Med. Chem. 2008;15:997–1005. doi: 10.2174/092986708784049658. [DOI] [PubMed] [Google Scholar]
- 79.Brooks M.J., et al. Antiviral activity of arbidol, a broad-spectrum drug for use against respiratory viruses, varies according to test conditions. J. Med. Virol. 2012;84:170–181. doi: 10.1002/jmv.22234. [DOI] [PubMed] [Google Scholar]
- 80.Kadam R.U., Wilson I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. U. S. A. 2017;114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanders J.M., et al. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020 doi: 10.1001/jama.2020.6019. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 82.Wang Z., et al. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020;71:769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tu Y.-F., et al. A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21:2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomas E., et al. Review the application and mechanism of action of ribavirin in therapy of hepatitis C. Antivir. Chem. Chemother. 2012;23:1–12. doi: 10.3851/IMP2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Graci J.D., Cameron C.E. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2006;16:37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paeshuyse J., et al. Ribavirin for the treatment of chronic hepatitis C virus infection: a review of the proposed mechanisms of action. Curr. Opin. Virol. 2011;1:590–598. doi: 10.1016/j.coviro.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 87.Khalili J.S., et al. Novel coronavirus treatment with ribavirin: groundwork for evaluation concerning COVID-19. J. Med. Virol. 2020;92:740–746. doi: 10.1002/jmv.25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gross A.E., Bryson M.L. Oral ribavirin for the treatment of noninfluenza respiratory viral infections: a systematic review. Ann. Pharmacother. 2015;49:1125–1135. doi: 10.1177/1060028015597449. [DOI] [PubMed] [Google Scholar]
- 89.Peiris J.S.M., et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Omrani A.S., et al. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect. Dis. 2014;14:1090–1095. doi: 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Booth C.M., et al. Clinical features and short-term outcomes of 144 patients with SARS in the Greater Toronto Area. J. Am. Med. Assoc. 2003;289:2801–2809. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 92.Kim G.W., et al. IL-6 inhibitors for treatment of rheumatoid arthritis: past, present, and future. Arch. Pharm. Res. 2015;38:575–584. doi: 10.1007/s12272-015-0569-8. [DOI] [PubMed] [Google Scholar]
- 93.Shimabukuro-Vornhagen A., et al. Cytokine release syndrome. J. Oncol. Sci. 2018;6:56. doi: 10.1186/s40425-018-0343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scheller J., et al. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta Mol. Cell Res. 2011;1813:878–888. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 95.Zhang C., et al. The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist Tocilizumab may be the key to reduce the mortality. Int. J. Antimicrob. Agents. 2020;55:105954. doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scheller J., et al. Interleukin-6: from basic biology to selective blockade of pro-inflammatory activities. Semin. Immunol. 2014;26:2–12. doi: 10.1016/j.smim.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 97.Hunter C.A., Jones S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 98.Liu B., et al. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimmun. 2020;6:102452. doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gao Y., et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J. Med. Virol. 2020;92:791–796. doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Navarro G., et al. Tocilizumab in rheumatoid arthritis: a meta-analysis of efficacy and selected clinical conundrums. Semin. Arthritis. Rheum. 2014;43:458–469. doi: 10.1016/j.semarthrit.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 101.Fu B., et al. Why tocilizumab could be an effective treatment for severe COVID-19? J. Transl. Med. 2020;18:164. doi: 10.1186/s12967-020-02339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Luo P., et al. Tocilizumab treatment in COVID-19, a single center experience. J. Med. Virol. 2020;92:814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bittmann S. COVID 19: camostat and the role of serine protease entry inhibitor TMPRSS2. J. Regen. Biol. Med. 2020;2:1–2. [Google Scholar]
- 104.Uno Y. Camostat mesilate therapy for COVID-19. Intern. Emerg. Med. 2020 doi: 10.1007/s11739-020-02345-9. ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kailas S., et al. Homology modeling and docking studies of TMPRSS2 with experimentally known inhibitors camostat mesylate, nafamostat and bromhexine hydrochloride to control SARS-coronavirus-2. chemRxiv. 2020 doi: 10.1016/j.imu.2021.100597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McKee D.L., et al. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gautret P., et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19, results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;56:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 109.Fohner A.E., et al. PharmGKB summary: macrolide antibiotic pathway, pharmacokinetics/pharmacodynamics. Pharmacogenet. Genomics. 2017;27:164–167. doi: 10.1097/FPC.0000000000000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Molina J.M., et al. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Mal. Infect. 2020;50:384. doi: 10.1016/j.medmal.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yuan J., et al. The correlation between viral clearance and biochemical outcomes of 94 COVID-19 infected discharged patients. Inflamm. Res. 2020;69:599–606. doi: 10.1007/s00011-020-01342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang Q., et al. Clinical trial analysis of (2019-nCoV therapy registered in China. J. Med. Virol. 2020;92:540–545. doi: 10.1002/jmv.25733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu C., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gordon C.J., et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020 doi: 10.1004/jbc.RA120.013679. Published online April 13. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fantini J., et al. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;55:105960. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mothay D., Ramesh K.V. Binding site analysis of potential protease inhibitors of COVID-19 using AutoDock. Virusdisease. 2020;31:194–199. doi: 10.1007/s13337-020-00585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dawood A.A. Mutated COVID-19 may foretell a great risk for mankind in the future. New Microbes New Infect. 2020;35 doi: 10.1016/j.nmni.2020.100673. Published online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tang X., Wu C., Li X., et al. On the origin and continuing evolution of SARS-CoV -2. Natl. Sci. Rev. 2020 doi: 10.1093/nsr/nwaa036. ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cava C., et al. In silico discovery of candidate drugs against covid-19. Viruses. 2020;12:404. doi: 10.3390/v12040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ou X., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Walls A.C., et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]