Abstract
Purpose
Leber hereditary optic neuropathy (LHON) and autosomal dominant optic atrophy (ADOA) are the two commonest forms of hereditary optic neuropathy. The aim of this study was to comprehensively investigate the incidence and spectrum of mutations in patients with suspected hereditary optic neuropathy by combining mitochondrial DNA (mtDNA) genome-wide and targeted exon sequencing.
Methods
A cohort of 1101 subjects were recruited to participate in the study, comprising 177 families (177 probands and their family members, a total of 537 subjects, including 254 patients) and 164 sporadic cases with suspected hereditary optic neuropathy, and 400 unrelated control subjects for genetic analysis: all subjects (including control subjects) underwent a comprehensive ophthalmologic examination and were subjected to sequencing analysis of mtDNA genome-wide and targeted exon. Overall, targeted exon sequencing was used to screen 792 genes associated with common hereditary eye diseases, and the mtDNA genome-wide were screened by next-generation sequencing.
Results
We found variants detected in 168 (40.2%, 168/418) of the 418 patients screened. Among these, 132 cases (78.6%, 132/168) were detected with known LHON disease-causing mtDNA variants; 40 cases (23.8%, 40/168) were detected with nuclear DNA (ntDNA) variants, which included 36 cases (21.4%, 36/168) with detected OPA1 mutations, 4 patients (2.4%, 4/168) with detected OPA3 mutations, and 2 patients (1.2%, 2/168) with detected TMEM126A homozygous mutation. Coexistence variation (mtDNA/mtDNA [n = 16], ntDNA/ntDNA [n = 4], mtDNA/ntDNA [n = 7]) was found in 27 patients (16.4%, 27/165), including mtDNA/ntDNA coexistence variation that was detected in seven patients. Among these ntDNA mutations, 38 distinct disease-causing variants, including autosomal recessive heterozygous mutations, were detected, which included 22 novel variants and two de novo variants. Total haplogroup distribution showed that 34.5% (29/84) and 28.6% (24/84) of the affected subjects with m.11778G>A belonged to haplogroup D and M, with a high frequency of subhaplogroups D4, D5, and M7.
Conclusions
The LHON-mtDNA mutations are the commonest genetic defects in this Chinese cohort, followed by the OPA1 mutations. To our knowledge, this is the first comprehensive study of LHON, ADOA, and autosomal recessive optic atrophy combined with mtDNA genome-wide and targeted exon sequencing, as well as haplogroup analysis, in a large cohort of Chinese patients with suspected hereditary optic neuropathy. Our findings provide a powerful basis for genetic counseling in patients with suspected hereditary optic neuropathy.
Translational Relevance
We applied mtDNA genome-wide sequencing combined with panel-based targeted exon sequencing to explore the pathogenic variation spectrum and genetic characteristics of patients with suspected hereditary optic neuropathy, providing a comprehensive research strategy for clinical assistant diagnosis, treatment, and genetic counseling.
Keywords: Leber hereditary optic neuropathy, autosomal dominant optic atrophy, autosomal recessive optic atrophy, mtDNA, ntDNA, panel-based targeted exon sequencing
Introduction
Hereditary optic neuropathy is a group of hereditary heterogeneous diseases involving function change in the optic nerve and retinal ganglion cells (RGC), which causes variable vision decline.1 There are several modes of inheritance for these disorders, including Leber hereditary optic neuropathy (LHON, OMIM535000) and autosomal dominant optic atrophy (ADOA, OMIM165500), which are the two commonest forms of these disorders caused by mutations in mitochondrial DNA (mtDNA) and nuclear DNA (ntDNA).2–4 Autosomal recessive optic atrophy (AROA) is a rare mode of inheritance, and several reports of this recessive form have been linked to TMEM126A (OMIM612988), RTN4IP1 (OMIM610502), ACO2 (OMIM100850), and YME1L1 (OMIM617302) genes.5–7,22
LHON is the commonest type of maternal hereditary disorder caused by structural and functional abnormalities of mtDNA, with an estimated prevalence of 1:50000 worldwide.8,9 It was first described by Theodor Leber as a maternally hereditary disorder of optic nerve degeneration that is usually occurring in patients between the age of 18 and 35, with acute or subacute visual loss of the central region and preferentially affects men.10,11 LHON is a heterogeneous disorder caused by mtDNA mutations. The composition of mtDNA includes 13 protein-coding genes, 2 ribosomal RNAs, and 22 tRNAs. Over 90% of LHON is caused by one of three primary disease-causing variants, m.3460G>A, m.11778G>A, and m.14484T>C, which involve the MT-ND1, MT-ND4, and MT-ND6 gene encoding three subunits of the respiratory chain complex I, respectively.8,11,12 Secondary mutations (such as MT-ND1 m.3394T>C and MT-ND4 m.11696G>A) often interact with primary mutations to affect the phenotypic expression of LHON, which is present in the LHON family, but at the same time appears in the control subjects at a lower frequency than LHON patients.13,14 These mtDNA mutations associated with LHON occurred in heterogeneous (mixed wild-type and mutant) or homogenous (mutant-only) forms.
ADOA is a disorder characterized by degenerative optic nerve fibers, mainly involving RGCs and axons of optic nerve formation. The estimated prevalence of the disease ranges from 1:12000 to 1:50000, characterized by early-onset loss of bilateral vision, defects in color vision, and typical temporal pale optic nerve.15 Although several genes have been reported to be associated with ADOA, most studies are still associated with genetic mutations in OPA1 (OMIM605290) and OPA3 (OMIM606580) gene encoding mitochondrial proteins, most of which are anchored to the mitochondrial inner membrane and are ubiquitously expressed.16–18 Previous studies of patients with ADOA have shown that most are caused by mutations in the OPA1 gene, accounting for approximately 60% to 80%. The OPA1 gene encodes mitochondrial proteins located in the mitochondrial inner membrane and regulates the stability of the mitochondrial network, mitochondrial bioenergetic output, and isolation of proapoptotic cytochrome c oxidase molecules in the mitochondrial cristae spaces, and more than 300 mutations have been detected in the previous reports.15,19–21
AROA is a rare disease that affects RGCs and the nervous system. To date, it has been reported that this form of AROA is related to 4 causative genes, including 3 variants detected in the transmembrane protein 126A gene (TMEM126A, OMIM612988) in 8 families, 14 variants detected in the reticulon 4-interacting protein 1 gene (RTN4IP1, OMIM610502) in 17 families, 7 variants detected in the aconitase 2 gene (ACO2, OMIM100850), and 1 variant detected in the YME1 Like 1 ATPase gene (YME1L1, OMIM617302) in 1 family.5–7,22–24 There are overlapping clinical and pathological features in almost all affected patients, in which RGCs are prone to mitochondrial dysfunction.25 Among these causative genes, the TMEM126A gene has been reported to be specifically associated with nonsyndromic AROA. The TMEM126A gene spans a genomic region of 8.5kb and consists of five exons, and it has been mapped to chromosome 11q14.1-q21 within a critical interval associated with optic atrophy identified by homozygosity mapping. In contrast to the OPA1 (OMIM605290) mutations, Hanein et al.5 found no fragmentation of the mitochondrial network and/or depletion of the mtDNA in patient fibroblasts caused by the TMEM26A mutation, which indicates that there may be no functional correlation between TMEM26A and OPA1. Thus far, three variants of the TMEM26A gene have been associated with AROA, including a recurrent R55X homozygous nonsense mutation that has been detected in multiple families. Among these families includes four families with autosomal recessive nonsyndromic optic atrophy and one family with optic atrophy combined auditory neuropathy, which suggestion that auditory neuropathy might be a key feature of TMEM126A associated optic neuropathy.5,26
Despite several previous studies that have described the genetic and clinical features of hereditary optic neuropathy, the overall status of LHON, ADOA, and AROA mutations remains to be determined in Chinese patients with suspected hereditary optic neuropathy. In this study, we report the results of a comprehensive molecular analysis of 418 patients with suspected hereditary optic neuropathy and detected molecular variability in nuclear and mitochondrial genomes based on next-generation sequencing.
Materials and Methods
Subjects and Ethics Statement
A cohort of 1101 subjects, including 177 families (177 probands and their relatives) and 164 sporadic subjects with suspected hereditary optic neuropathy (total patients: 418; total probands: 341) and 400 unrelated control subjects were recruited at the Eye and Ear, Nose, and Throat (ENT) Hospital of Fudan University from February 2016 to June 2019. The study was permitted by the ethics committee of the Eye and ENT Hospital of Fudan University and adhered to the tenets of the Declaration of Helsinki. Individuals participating in the study signed informed consent.
Clinical Assessment
The clinical criteria for the inclusion of patients in this study included a decrease in central vision unrelated to refraction, and a different degree of optic nerve atrophy observed through fundus examination. We eliminated all known causes, such as oppression, inflammation, glaucoma, and drug-induced optic nerve atrophy. All participants in the study underwent complete ophthalmologic examinations, comprising best-corrected visual acuity (BCVA) measurement, slit-lamp biomicroscopy examination, intraocular pressure detection (IOP, Goldmann tonometry; Haag-Streit, UK), full-field electroretinography, fundus photograph (Canon CR6-45NM fundus camera; Canon, Tokyo, Japan), and swept-domain optical coherence tomography (Heidelberg Engineering Inc., Heidelberg, Germany). In addition, information associated with the disease, including family history, age of onset, and decreased subjective visual acuity, were collected. The diagnosis of suspected hereditary optic neuropathy was assessed by professional neuroophthalmologists.
Targeted Exon Sequencing and Next-Generation Sequencing
Peripheral blood samples from all participants were collected at the Eye and ENT Hospital of Fudan University. The mtDNA and ntDNA were extracted from whole blood using the MGIEasy Mitochondrial Whole Genome Amplification Kit (MGI, Inc., Shenzhen, China) and FlexiGene DNA Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer's protocols, respectively. In this study, customized panel-based next-generation sequencing (NGS) and mtDNA genome-wide sequencing were performed for all participants. We collaborated with MGI-Shenzhen (Shenzhen, Guangdong, China) to design the Target_Eye_792_V2 chip, which has exon capture and untranslated regions of 792 genes associated with common hereditary eye diseases (Supplementary Table S1).27,28 On average, the mean coverage depth was more than 300X for ntDNA, the mean coverage depth was more than 30X for mtDNA, and the coverage of target region was close to 99.9% using the MGISEQ-2000 (DNBSEQ-G400) platform (MGI, Inc., Shenzhen, China).
Bioinformatics Analysis
The target exon sequencing reads are aligned to the reference human genome (USCS hg38) using the Burrows–Wheeler Aligner (BWA) version 0.7.10 (for more information, see github.com/lh3/bwa). All the mutations detected by sequencing were annotated using the 1000 Genomes Project (http://1000genomes.org/), dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/), ExAC (http://exac.broadinstitute.org), and ESP6500 (http://evs.gs.washington.edu/EVS/) databases, screening for mutations with minor allele frequency <0.1% to detect possible deleterious variations. In addition, four online tools, Sorting Intolerant from Tolerant (http://sift.jcvi.org/), MutationTaster software (http://www.mutationtaster.org/), FATHMM software (http://fathmm.biocompute.org.uk/), and LRT (http://www.genetics.wustl.edu/jflab/) were used to predict potential pathogenic variants. Then, the Clinvar (https://www.ncbi.nlm.nih.gov/clinvar/), HGMD (http://www.hgmd.cf.ac.uk/ac/index.php), and Online Mendelian Inheritance in Man (OMIM, http://www.omim.org/) were used to interpret and prioritize the variants, combined with potential deleterious impacts, genotype-phenotype relationships, and variants report. According to the American College of Medical Genetics (ACMG) and genomics guidelines, variants were classified into five categories, including pathogenic, likely pathogenic, novel variants of uncertain clinical significance, likely benign, and benign. Sanger sequencing was performed to confirm the candidate variants.29,30
The mtDNA genome-wide sequencing results were compared with the published mtDNA sequences (GenBank Accession No. NC_012920), and the mtDNA haplogroup status and potential pathogenic variation were both determined. The “Top 19” primary LHON mutations reported by MITOMAP (https://www.mitomap.org/foswiki/bin/view/MITOMAP/MutationsLHON) in the literature were screened in all subjects, as well as 19 other candidate LHON mutations and 21 secondary LHON mutations. Conservatism of mitochondrial genome variation was obtained by analysis using ClustalX (see http://www.clustal.org/clustal2/).
Results
Clinical Presentation and Genetic Findings
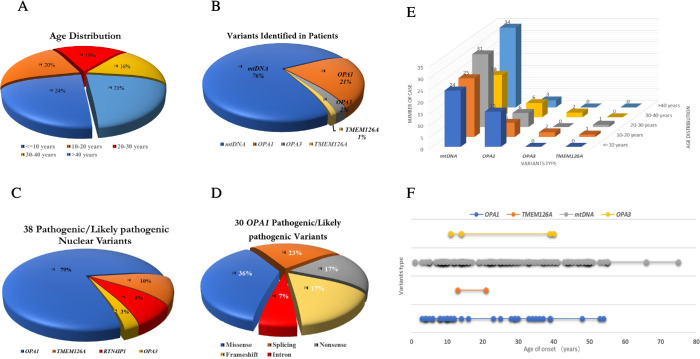
In this cohort, a total of 418 patients with hereditary optic neuropathy were recruited in the study, of whom most were unrelated probands (341/418). The average age of the patients was 25.7 ± 16.5 years (range, 0.83–66; median, 23), most of whom were under 30 years old, accounting for (63%; 263/418) of the total number of patients (Fig. 1A), and all participants were from all over the Chinese region. Among these patients, 263 were men and 155 were women, with an average age of 23.9 ± 15.0 (range, 0.83–62; median, 22) years and 28.6 ± 18.3 (range, 2–66; median, 27) years, respectively (Table 1). The average ages of the two groups of dominant optic atrophy and LHON patients were calculated, and the results showed that the age of onset of dominant optic atrophy in patients was younger than with LHON, with an average age of 19.8 ± 14.9 (range, 3–54; median, 12) years and 27.1 ± 15.7 (range, 1–66; median, 25) years, respectively. In addition, 400 unrelated control subjects were included from different regions of China, with an average age of 48.5 ± 15.0 (range, 22–92; median, 48) years, who showed no abnormalities through physical, neurologic, and ophthalmic examinations.
Figure 1.
Basic information of clinical presentation and genetic finding of the ntDNA in the patients. (A) The age distribution of the total patients, including age ≤10 years (n = 99), 10 to 20 years (n = 86), 20 to 30 years (n = 78), 30 to 40 years (n = 67), and >40 years (n = 88). (B) The patients received a confirmed genetics diagnosis in this cohort, including mtDNA (n = 132), OPA1 (n = 36), OPA3 (n = 4), and homozygous TMEM126A (n = 2) variants. (C) Thirty-eight different pathogenic/likely pathogenic nuclear variants were detected in this cohort, including OPA1 gene (n = 30), TMEM126A gene (n = 4), RTN4IP1 gene (n = 3), and OPA3 gene (n = 1). (D) Thirty different OPA1 pathogenic/likely pathogenic variants were identified, including missense (n = 11), splicing (n = 7), nonsense (n = 5), frameshift (n = 5), and intron (n = 2) variants. (E, F) Multidimensional comparison between detection of variability in patients and age distribution.
Table 1.
The Demography and the Genetic Screening Results of this Study
| Patients (%) | Total (Families/Sporadic) | Male (Families/Sporadic) | Female (Families/Sporadic) | Ratio (Male to Female) |
|---|---|---|---|---|
| Total screened (100%) | 418 (254/164) | 263 (154/109) | 155 (100/55) | 1.7:1 |
| LHON mutation (31.6%, 132/418) | 132 (84/48) | 90 (55/35) | 42 (29/13) | 2.1:1 |
| OPA1 mutation (8.6%, 36/418) | 36 (30/6) | 22 (18/4) | 14 (12/2) | 1.6:1 |
| OPA3 mutation (1.0%, 4/418) | 4 (3/1) | 3 (3/0) | 1 (0/1) | 3.0:1 |
| TMEM126A mutation (0.5%, 2/418) | 2 (2/0) | 1 (1/0) | 1 (1/0) | 1.0:1 |
| Total detected (40.2%, 168/418) | 168 (113/55) | 111 (72/39) | 57 (41/16) | 2.0:1 |
Overall, a total of 168 patients (40.2%; 168/418) received confirmed mtDNA or ntDNA variants, and the percentage of patients diagnosed with confirmed genetic variants in the families (44.5%; 113/254) was significantly higher than that of sporadic patients (33.5%; 55/164) (Table 1). As shown in Figure 1B, a total of 132 individuals (76%; 132/174) were detected with LHON-mtDNA mutations, 36 individuals were detected with OPA1 gene mutations, 4 individuals were detected with OPA3 gene mutations, and 2 patients were detected with homozygous TMEM126A gene mutations. We performed a multidimensional comparison between the type of variation and the age distribution of the patients examined. The results showed that the detection rate of mtDNA mutations was almost the same in patients of different ages, but OPA1 mutations were mainly concentrated in children under 10 years of age (Figs. 1E, 1F). Coexisting variants were detected in 27 patients (16.1%; 27/168), including 16 mtDNA/mtDNA variants (59.2%; 16/27), 4 ntDNA/ntDNA variants (14.8%, 4/27), and 7 mtDNA/ntDNA variants (25.9%, 7/27). Among these coexisting variations, the commonest form was ND4 m.11778G>A+ND6 m.14502T>C, followed by ND4 m.11778G>A+ND1 m.3394T>C, or ND6 m.14484T>C+ND1 m.3394T>C, all three forms have been reported.
mtDNA LHON Mutations
As shown in Table 1, LHON-mtDNA mutations were detected in 132 patients (31.6%, 132/418), of which 84 were families subjects (63.6%, 84/132) and 48 were sporadic subjects (36.3%, 48/132); the ratio of male to female was 2.1:1. Molecular and clinical features of LHON-positive individuals identified in the study are shown in Table 2. The top 3 primary LHON disease-causing mutation were detected in 96 patients, of which 84 patients detected m.11778G>A mutation, 7 patients detected m.3460G>A mutation, and 5 patients detected m.14484T>C mutation. In addition, 11 other candidate LHON disease-causing mutations were found in 52 patients, and 5 putative LHON mutations were detected in 40 patients. The primary top 3 LHON mutations accounted for 72.7% (96/132) of the patients with LHON mutations, and 16 mtDNA/mtDNA coexisting mutations accounted for 12.1% (16/132) (Table 3).
Table 2.
Molecular and Clinical Features of LHON-Positive Patients Identified in this Study
| Gene | Mutation | Conservation Index | N of 418 Patients | N of 400 Controls | Clinvar | Polymorphism Phenotyping | Clinical Significance | Age of Diagnosed Mean ± Standard Deviation (Range; Median) | The Most Common Haplogroup (Num; %) | Previous Report |
|---|---|---|---|---|---|---|---|---|---|---|
| ND4 | 11778G>A | 1 | 84 | 2 | Pathogenic | Probably damaging | Pathogenic | 28.7 ± 16.0 (0.83–66; 25.5) | D4 (20; 23.8%); M7 (12; 14.3%) | Yes |
| ND1 | 3460G>A | 0.9111 | 7 | 0 | Pathogenic | Probably damaging | Pathogenic | 21.6 ± 11.0 (8–36; 23) | F1 (3; 42.9%); M7 (2; 28.6%) | Yes |
| ND6 | 14484T>C | 0.83 | 5 | 0 | Pathogenic | Probably damaging | Pathogenic | 36.8 ± 17.5 (12–55; 40) | M9 (2; 40%) | Yes |
| Total of the “top 3” primary LHON-disease causing mutations | 28.6 ± 15.9 (1–66;15) | D4 (20; 20.8%); M7 (14; 14.6%) | ||||||||
| ND1 | 3394T>C | 0.9333 | 8 | 2 | Pathogenic | Benign | Risk factors; Pathogenic | – | – | Yes |
| ND1 | 3635G>A | 0.9333 | 2 | 0 | Pathogenic | Probably damaging | Pathogenic | – | – | Yes |
| ND1 | 4136A>G | 0.9778 | 2 | 0 | Pathogenic | Probably damaging | Pathogenic | – | – | Yes |
| ND3 | 10197G>A | 0.9556 | 1 | 0 | Pathogenic | Probably damaging | Pathogenic | – | – | Yes |
| ND4 | 10680G>A | 0.9333 | 1 | 0 | Pathogenic | Benign | Pathogenic | – | – | Yes |
| ND4 | 11253T>C | 0.4222 | 1 | 0 | Pathogenic | Benign | Risk factors; Pathogenic | – | – | Yes |
| ND4 | 11696G>A | 0.25 | 2 | 0 | Pathogenic | Benign | Risk factors; Pathogenic | – | – | Yes |
| ND5 | 12338T>C | 0.83 | 16 | 2 | Pathogenic | Unknown | Risk factors; Pathogenic | – | – | Yes |
| ND6 | 14502T>C | 0.7778 | 11 | 1 | Pathogenic | Benign | Risk factors; Pathogenic | – | – | Yes |
| COX3 | 9438G>A | 0.9333 | 1 | 0 | Pathogenic | – | Risk factors; Pathogenic | – | – | Yes |
| tRNA-Thr | 15951A>G | 0.78 | 7 | 0 | Pathogenic | Probably damaging | Risk factors; Pathogenic | – | – | Yes |
| Total of the other candidate LHON disease-causing mutations | 24.5 ± 14.8 (3–55; 22) | M10 (11; 21.2%); F2 (11; 21.2%) | ||||||||
| ND1 | 4216T>C | 0.2444 | 4 | 3 | Conflicting of pathogenicity | Benign | Risk factors | – | – | Yes |
| ND2 | 4917A>G | 0.9111 | 1 | 2 | Uncertain significance | Benign | Risk factors | – | – | Yes |
| ND5 | 12811T>C | 0.5556 | 16 | 17 | Uncertain significance | Benign | Risk factors | – | – | Yes |
| ND5 | 13708G>A | 0.3333 | 17 | 17 | Conflicting of pathogenicity | Benign | Risk factors | – | – | Yes |
| COX3 | 9804G>A | 0.9333 | 2 | 1 | Conflicting of pathogenicity | – | Risk factors | – | – | Yes |
| Total of the putative LHON mutation. | ||||||||||
Table 3.
Coexistence Variation Identified in the Patients
| Mutation | N of 418 Patients (%) | Age of Diagnosed Mean ± Standard Deviation (Range; Median) | Haplogroup (num; %) |
|---|---|---|---|
| ND4 m.11778G>A+ND4 m.11696G>A | 1 | 25 | D4j |
| ND4 m.11778G>A+ND6 m.14502T>C | 5 | 30.8 ± 17.6 (9–54; 30) | M10a1 |
| ND4 m.11778G>A+tRNA-Thr m.15951A>G | 3 | 19.3 ± 15.5 (8–37; 13) | D4h |
| ND4 m.11778G>A+ND1 m.3394T>C | 3 | 26 ± 14.8 (16–43; 19) | M9a1a2 |
| ND6 m.14484T>C+ND1 m.3394T>C | 3 | 29.7 ± 19.1 (12–50; 27) | M9a1a1 (2); M7b1a1 (1) |
| ND1 m.4136A>G+COX3 m.9438G>A | 1 | 55 | N9a7 |
| Coexistence variation of mtDNA/mtDNA | 16(3.8%) | 28.7 ± 16.1 (8–55; 26) | M7/9/10 (11; 68.8%); D4 (4; 25%); N9 (1; 6.2%) |
| OPA1 c.1682-1G>A+OPA3 c.123C>G | 2 | 13 ± 4.2 (10–16; 13) | D4a (1); B5a2a2 (1) |
| OPA1 c.1471dup(p.Ile491Asnfs*9) /c.1472T>A+TMEM126A c.236C>G,Het | 2 | 25 ± 19.8 (11–39; 25) | B4h1 |
| Coexistence variation of ntDNA/ntDNA | 4 (1%) | 19 ± 13.6 (10–39; 13.5) | B (3; 75%); D4 (1; 25%) |
| ND1 m.12338T>C+OPA1 c.2203G>T | 2 | 29 ± 26.9 (10–48; 29) | F2a1 (1); H5g (1) |
| ND1 m.12338T>C+RTN4IP1 c.308G>A, Het | 1 | 10 | F2 |
| ND1 m.4136A>G+OPA1 c.1835_1836delGA | 1 | 4 | G2a1 |
| ND4 m.10680G>A+OPA1 c.2661+1G>A | 1 | 5 | A17 |
| ND4 m.11778G>A+TMEM126A c.163C>T,Het | 1 | 16 | D5a2a |
| ND4 m.11253T>C+RTN4IP1 c.1015C>T,Het | 1 | 38 | G1 |
| Coexistence variation of mtDNA/ntDNA | 7(1.7%) | 18.7 ± 17.3 (4–48; 10) | F2 (2; 28.6); G (2; 28.6);D (1; 14.3%); A (1; 14.3%); H (1; 14.3%) |
| Total patients of coexistence variation | 27(6.5%) | 24.7 ± 16.3 (4–55; 19) | Top 3 haplogroup: M10a (5; 18.5%); M9a (5; 18.5%); D4 (5; 18.5%) |
The m.11778G>A mutation was detected in 114 individuals, including 84 patients and 30 unaffected family members. The affected individuals presented bilateral painless visual impairment along with scotomas (central or cecocentral) and pale optic disc. Family members who harbor the primary m.11778G>A mutation were considered as unaffected individuals when they did not show any symptoms of visual impairment at the time of sample collection. Among these patients who harbor the primary m.11778G>A mutation, 56 patients were male (66.7%, 56/84) and 28 were female (33.3, 28/84); the ratio of male to female was 2:1. When compared with the females, the penetrance of the disease was much higher in males, with 91.8% of males (56/61) who carried the primary m.11778G>A mutation developing blindness, whereas 52.8% of females (28/53) presented with loss of vision. The observation was consistence with previous European and Chinese studies, indicating that gender factors have a strong impact on the increased risk of visual impairment in males compared with females who harbor the m.11778G>A mutation in Chinese cohort.31,32
OPA1 and OPA3 Mutations
Thirty-eight distinct pathogenic/likely pathogenic nuclear variants were detected in this cohort (Supplementary Table S2), including OPA1 gene (n = 30, 79%), TMEM126A gene (n = 4, 10%), RTN4IP1 gene (n = 3, 8%), and OPA3 gene (n = 1, 3%) (Fig. 1C). We identified 30 distinct pathogenic/likely pathogenic variants of the OPA1 gene in 8.6% (36/418) of patients, including missense (n = 11, 36%), splicing (n = 7, 23%), nonsense (n = 5, 17%), frameshift (n = 5, 17%), and intron (n = 2, 7%) variants (Fig. 1D). Among the 30 OPA1 mutations, we classified 16 variants as novel variants because these had not been registered in the Human Gene Mutation Database or Clinvar database and have not been reported in the literature. In addition, when the variants were not detected in the patient's biological parents, we defined these as de novo variants. In this cohort, two de novo missense variants were detected in the OPA1 gene (1.43%, n = 2/140 families of trios), c.320C>A (p.S107X), and c.1499G>A (p.R500H) were detected in unrelated patients, respectively.
One recurrent OPA3 mutation was detected in four (1.0%) of the 418 patients screened. Of these four patients, two of them were from the same family (F74), and the other two were from independent families (F159, F305). The variant c.123C>G (p.I41M) was reported to be detected in a 12-year-old boy with a BCVA of 0.1. Ophthalmoscopy examination revealed bilateral optic disc pallor and diffuse defects in the retinal nerve fiber layer (RNFL). His twin brothers also harbors the same mutation, as well as visual acuity defects and optic atrophy.33 Moreover, the clinical significance of these variants was defined according to ACMG and genomics guidelines; the bioinformatics tools analysis results of the 17 missense variants are demonstrated in Supplementary Figure S1.
TMEM126A and RTN4IP1 Mutations
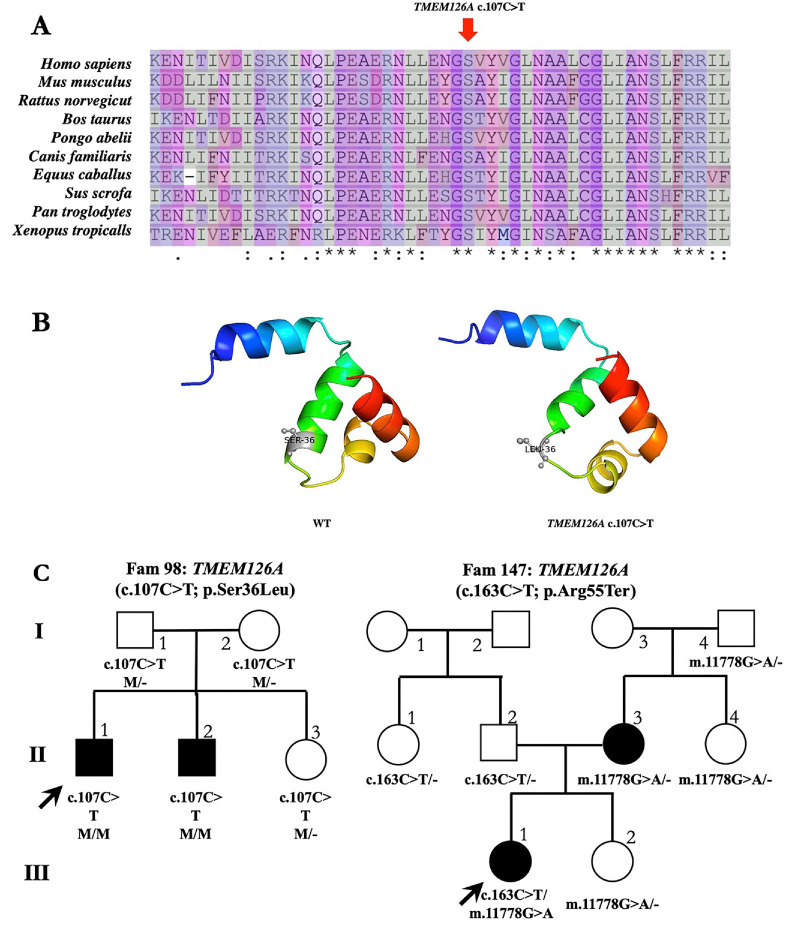
One missense mutation (c.107C>T, p.S36L) of the TMEM126A gene was identified in five members of family 98 (F98), the probands and his affected biological brother harbor homozygous state, and his parents and sister harbor heterozygous state (Fig. 2C). This missense variant affects a highly evolutionarily conserved region (Fig. 2A) and changes the amino acid from serine to leucine, and the variant segregates with the disease within the family. The SWISS-MODEL was used to predict the three-dimensional structure of this amino acid in the mutant and wild-type proteins (Fig. 2B), and we observed a difference between the mutated and wild-type structures of transmembrane protein 126A. There have not been any other LHON-mtDNA and ADOA and AROA genes detected in the two affected subjects of this family. Studies have reported that the two affected siblings from Iraqi descent detected the p.S36L variant and categorized the variant as unclear significance.34 However, according to the ACMG and genomics guidelines, the p.S36L variant is cosegregated with the disease in the family and affects evolutionary conserved amino acid residues. Therefore it should be categorized as pathogenic or likely pathogenic variants, so we regard it as a candidate causative factor for the patients with optic atrophy in this family.
Figure 2.
Detection and evaluation of mutations in the TMEM126A gene. (A) Multiple sequence alignment of candidate missense mutations from different species to explore the conservation of these mutations. The red arrow represents variants states. (B) SWISS-MODEL was used to predict the three-dimensional structure of both the mutant and wild-type proteins. (C) Pedigrees of the families with mutations. Squares indicate men and circles women; black and white symbols represent affected and unaffected individuals, respectively. The proband is marked with an arrow, and the asterisks indicate those members enrolled in this study.
Moreover, we detected three additional heterozygous states of the TMEM126A gene in affected and unaffected individuals (c.163C>T, p.Arg55Ter; c.62T>G, p.Ile21Ser; c.236C>G, p.Thr79Arg), including the detection of a recurrent nonsense mutation (p.Arg55Ter) in three individuals of a family (F147).5,26 Among the three mutation carriers, the proband was accompanied by an acute onset of optic neuropathy, and the other two carriers were asymptomatic fathers and aunts. Recurrent nonsense mutations (p.Arg55Ter) detected in the proband were inherited from his father. At the same time, LHON-mtDNA mutation m.11778G>A was also detected in the proband and his four family members, and the proband's mtDNA mutation was inherited from his mother (Fig. 2C). The proband showed more severe clinical manifestations than the infected mother, and the onset age was earlier (acute onset at the age of 16 years), and the vision dropped sharply in early childhood. However, the affected mother did not have a similar history of the disease; she was late onset and vision decrease was mild. The effect of the coexistence of mtDNA variants (m.11778G>A) and TMEM126A heterozygous variants (c.163C>T, p.Arg55Ter) on clinical manifestation is unknown, and its underlying mechanism remains to be explored.
In three families with early-onset recessive optic neuropathy, we identified three heterozygous missense states in the RTN4IP1 gene (c.1015C>T, p.R339C; c.308G>A, p.R103H; c.962G>C, p.G321A), which encodes a mitochondrial ubiquinol oxydo-reductase. Among these three mutations, two of them were novel heterozygous mutations for the c.1015C>T and c.962G>C substitution on a different haplotype F and C7 (F120 and F148), another one is a heterozygous variant of haplotype F (F8), which has been reported to detect homozygous and heterozygous variation of the mutation in four families.24
Haplogroup Distribution
Our comprehensive study of Chinese LHON patients confirmed that LHON individuals with m.11778G>A variants had different mtDNA haplotypes, which is different from previously reported observations that preferentially associate with Western European continent haplogroup J and increase clinical penetrance of the J2 subhaplogroup, indicating their possible influence on clinical expression.31,35 Comprehensive haplogroup analysis showed that 34.5% (29/84) and 28.6% (24/84) of the affected subjects belonged to haplogroup D and M, with a high frequency of subhaplogroups D4, D5, and M7 (Supplementary Table S3). These data suggest that such haplotypes are more likely to produce similar clinical manifestations in East Asian populations, and the same situation may exist in other mitochondrial diseases among Chinese populations. Moreover, we performed mitochondrial haplogroup comparison between the 400 control subjects and 418 patients, the result indicating that there is no difference between these two cohorts (Supplementary Table S4).
Discussion
To our knowledge, this is the first study screening for LHON, ADOA, and AROA in a large cohort of Chinese patients with suspected hereditary optic neuropathy. Comprehensive molecular screening for patients with suspected hereditary optic neuropathy might contribute to the high overall mutation detection rate (40.2%, 168/418). Our results indicate that LHON-mtDNA mutations are definitely the most common cause in Chinese patients, and mtDNA variants were found in 31.6% (132/418) of patients. The LHON-mtDNA mutation detection rate is higher than the incidence of OPA1 mutations, which were detected in less than 10% (8.6%, 36/418) of patients. However, the detection rate of LHON-mtDNA mutations is 31.6% (132/418), which is a discrepancy to 52.1% mutation rate (271/520) based on another Chinese cohort.33 In additional, these finding are in sharp contrast to the results of 980 unrelated patients in the large French population, which showed that the OPA1 mutation was the commonest mutation detected in 30% of probands, whereas the LHON-mtDNA mutation was only detected in 13% of patients.20 We believe that the difference in the detection rate with the French population is due to different ethnic backgrounds, and the difference in the detection rate between studies based on the Chinese population is due to different entry criteria. Our enrollment included a broader group of patients with suspected optic neuropathy, which may lead to a low detection rate, but the results show that mtDNA or OPA1 mutations are indeed detected in some patients with clinical symptoms that are not obvious. In clinical applications, a comprehensive examination helps us to diagnose patients with suspected optic neuropathy. The prevalence of LHON-mtDNA and OPA1, OPA3 mutations was similar to the results of one independent large Chinese cohort study from a domestic study group. In this large-scale cohort study, mutations in the mtDNA were detected in 346 probands (38.3%, 346/903) who were suspected with LHON from 903 Chinese families. G11778A, T14484C, and G3460A mutations were detected in 312 (90.2%), 30, and 4 probands, respectively.36,37 However, the top three LHON-mtDNA mutations were detected in 28.2% (96/341) of the screened probands in our cohort study, G11778A, T14484C, and G3460A mutations were detected in 84 (87.5%), 5, and 7 probands, which accounted for 72.7% (96/132) of all identified LHON-mtDNA mutations. Consistent with several previous reports, OPA1 is the commonest disease-causing gene in Chinese ADOA patients, OPA3 gene mutations were rare in Chinese cohort, and we only detected one missense variant c.123C>G (p.I41M) in four patients in our cohort study.36 In addition to detecting of the two most common forms of optic neuropathy in LHON-mtDNA and ADOA, four variants of the TMEM126A gene and three variants of the RTN4IP1 gene were also detected in this cohort study.
Except for the three commonest LHON-mtDNA mutations (11778G>A, 14484T>C, 3460G>A), 11 other candidate LHON disease-causing mutations were detected in 12.4% (52/418) of patients, and more than 6.5% (27/418) of patients were detected to harbor two variants (one of three coexisting variant forms). In several previous studies, the coexisting variants forms of mtDNA/mtDNA or mtDNA/ntDNA or ntDNA/ntDNA detected in one patient was very rare. In several recent studies, six unrelated patients harboring m.14484T>C and m.14502T>C mutations were detected in a large cohort study, which included 1218 Han Chinese patients; in another Chinese cohort study of 520 unrelated patients, nine coexisting variants were detected, including one patient harboring two common primary mutations (m.3460G>A and m.11778G>A); the remaining eight patients harbor one of the top three LHON mutations combined with one of the two rare primary LHON mutations (m.11696A>G and m.14502T>C).17,38 In the current study, a total of 27 patients were found to have one of three coexisting variants, including 16 mtDNA/mtDNA variants (3.8%, 16/418; 28.7 ± 16.1), 4 ntDNA/ntDNA variants (1%, 4/418; 19 ± 13.6), and 7 mtDNA/ntDNA variants (1.7%, 7/418; 18.7 ± 17.3) (Table 3). To our knowledge, this is the first report of the simultaneous detection of three forms of coexisting variants detected in Chinese cohort studies. In particular, the mtDNA/ntDNA variant forms are very unique, and novel forms of mutations might cause more severe impact on subjects harboring the coexist variants. Among the seven mtDNA/ntDNA variants, one patient was found harboring m.11778G>A and c.163C>T mutations, compared with the mother who only detected the m.11778G>A mutation, the probands who detected m.11778G>A and c.163C>T mutations showed an earlier acute onset.
Conclusions
LHON-mtDNA mutations are the commonest genetic defects in this Chinese cohort with suspected hereditary optic neuropathy, followed by the OPA1 and OPA3 mutations with ADOA, and TMEM126A mutations with the rare AROA. Comprehensive molecular screening for patients with suspected hereditary optic neuropathy is essential to assist in diagnosing the patient's disease categories and customize molecular therapies. Three forms of coexisting variation may have potential effects on disease manifestation and disease progression, a task that requires further exploration and in-depth research.
Supplementary Material
Acknowledgments
The authors thank all of the patients and families who agreed to participate in this study. In addition, the authors thank BGI Shenzhen for their technical support, and the staff at the Eye, Ear, Nose, and Throat Hospital of Fudan University for their assistance. Finally, the authors are grateful to Fang Chen, Ji-Hong Wu, and Jie Huang for their invaluable contributions to this work.
Supported by the National Natural Science Foundation of China (Grant NSFC 61373048), Shenzhen Municipal Government of China (No. JCYJ20170412152854656), the Non-Profit Central Research Institute Fund of Chinese Academy of Medical Sciences 2018PT32019, Shanghai Municipal Science and Technology Major Projects (Grant 2018SHZDZX05), the Natural Science Foundation of Guangdong Province (Grant 2018A0303100012), and a grant from the Research Grants Council of the Hong Kong Special Administrative Region, China (CityU 11256116 and CityU 11210119).
FC, J-HW, JH, J-KL, WL, and F-JG conceived and designed this study. FJG, FYH, SHZ, and JHW recruited patients, performed clinical examinations and interpretation. FJG, FYH, SHZ, JHW, S-FQ, JH, L-LL, and L-SW collected the clinical samples and clinical data. WL, J-KL, FC, YQ, and Z-WW analyzed the sequencing data. J-KL, WL, and F-JG wrote and revised the manuscript.
Data availability: The data that support the findings of this study have been deposited in the CNSA (https://db.cngb.org/cnsa/) of CNGBdb with accession number CNP0000453. The datasets are available from the corresponding author on reasonable request.
Disclosure: J.-K. Li, None; W. Li, None; F.-J. Gao, None; S.-F. Qu, None; F.-Y. Hu, None; S.-H. Zhang, None; L.-L. Li, None; Z.-W. Wang, None; Y. Qiu, None; L.-S. Wang, None; J. Huang, None; J.-H. Wu, None; F. Chen, None
References
- 1. Neuhann T, Rautenstrauss B.. Genetic and phenotypic variability of optic neuropathies. Expert Rev Neurother. 2013; 13: 357–367. [DOI] [PubMed] [Google Scholar]
- 2. Delettre C., Lenaers G, Griffoin JM, et al.. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000; 26: 207–210. [DOI] [PubMed] [Google Scholar]
- 3. Reynier P, Amati-Bonneau P, Verny C, et al.. OPA3 gene mutations responsible for autosomal dominant optic atrophy and cataract. J Med Genet. 2004; 41: e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carelli V, Ross-Cisneros FN, Sadun AA. Mitochondrial dysfunction as a cause of optic neuropathies. Prog Retin Eye Res. 2004; 23: 53–89. [DOI] [PubMed] [Google Scholar]
- 5. Hanein S, Perrault I, Roche O, et al.. TMEM126A, encoding a mitochondrial protein, is mutated in autosomal-recessive nonsyndromic optic atrophy. Am J Human Genet. 2009; 84: 493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Metodiev MD, Gerber S, Hubert L, et al.. Mutations in the tricarboxylic acid cycle enzyme, aconitase 2, cause either isolated or syndromic optic neuropathy with encephalopathy and cerebellar atrophy. J Med Genet. 2014; 51: 834–838. [DOI] [PubMed] [Google Scholar]
- 7. Zou X, Guo X, Su H, et al.. Whole exon sequencing identifies two novel mutations in the reticulon 4–interacting protein 1 gene in a Chinese family with autosomal recessive optic neuropathies. J Mol Neurosci. 2019; 68: 640–646. [DOI] [PubMed] [Google Scholar]
- 8. Yen MY, Wang AG, Wei YH. Leber's hereditary optic neuropathy: a multifactorial disease. Prog Retin Eye Res. 2006; 25: 381–396. [DOI] [PubMed] [Google Scholar]
- 9. Maresca A, Morgia CL, Caporali L, et al.. The optic nerve: a “mito-window” on mitochondrial neurodegeneration. Mol Cell Neurosci. 2013; 55: 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harding AE, Sweeney MG, Govan GG, et al.. Pedigree analysis in Leber hereditary optic neuropathy families with a pathogenic mtDNA mutation. Am J Human Genet. 1995; 57: 77. [PMC free article] [PubMed] [Google Scholar]
- 11. Wallace DC, Singh G, Lott MT, et al.. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988; 242: 1427–1430. [DOI] [PubMed] [Google Scholar]
- 12. Ruiz-Pesini E, Lott MT, Procaccio V, et al.. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007; 35: D823–D828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang M, Zhou X, Li C, et al.. Mitochondrial haplogroup M9a specific variant ND1 T3394C may have a modifying role in the phenotypic expression of the LHON-associated ND4 G11778A mutation. Mol Genet Metab. 2010; 101: 192–199. [DOI] [PubMed] [Google Scholar]
- 14. Xie S, Zhang J, Sun J, et al.. Mitochondrial haplogroup D4j specific variant m.11696G > a(MT-ND4) may increase the penetrance and expressivity of the LHON-associated m.11778G > a mutation in Chinese pedigrees. DNA Seq. 2017; 28: 434–441. [DOI] [PubMed] [Google Scholar]
- 15. Yu-Wai-Man P, Griffifiths PG, Burke A, et al.. The prevalence and natural history of dominant optic atrophy due to OPA1 mutations. Ophthalmology. 2010; 117: 1538–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lenaers G, Hamel C, Delettre C, et al.. Dominant optic atrophy. Orphanet J Rare Dis. 2012; 7: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Satoh M, Hamamoto T, Seo N, Kagawa Y, Endo H. Differential sublocalization of the dynamin-related protein OPA1 isoforms in mitochondria. Biochem Biophys Res Commun. 2003; 300: 482–493. [DOI] [PubMed] [Google Scholar]
- 18. Huizing M, Dorward H, Ly L, et al.. OPA3, mutated in 3-methylglutaconic aciduria type III, encodes two transcripts targeted primarily to mitochondria. Mol Genet Metab. 2010; 100: 149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thiselton DL, Alexander C, Taanman JW, et al.. A comprehensive survey of mutations in the opa1 gene in patients with autosomal dominant optic atrophy. Invest Ophthalmol Vis Sci. 2002; 43: 1715–1724. [PubMed] [Google Scholar]
- 20. Ferre M, Bonneau D, Milea D, et al.. Molecular screening of 980 cases of suspected hereditary optic neuropathy with a report on 77 novel OPA1 mutations. Hum Mutat. 2009; 30: E692–E705. [DOI] [PubMed] [Google Scholar]
- 21. Ferré M, Caignard A, Milea D, et al.. Improved locus-specific database for OPA1 mutations allows inclusion of advanced clinical data. Human Mutat. 2015; 36: 20–25. [DOI] [PubMed] [Google Scholar]
- 22. Hartmann B, Wai T, Hu H, et al.. Homozygous YME1L1mutation causes mitochondriopathy with optic atrophy and mitochondrial network fragmentation. Elife. 2016; 5: e16078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Charif M, Nasca A, Thompson K, et al.. Neurologic phenotypes associated with mutations in RTN4IP1 (OPA10) in children and young adults. JAMA Neurol. 2018; 75: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Claire A, Pierre-Olivier G, Yasmina TA, et al.. Recessive mutations in RTN4IP1 cause isolated and syndromic optic neuropathies. Am J Human Genet. 2015; 97: 754–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu-Wai-Man P, Votruba M, Burté F, et al.. A neurodegenerative perspective on mitochondrial optic neuropathies. Acta Neuropathol. 2016; 132: 789–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meyer E, Michaelides M, Tee LJ, et al.. Nonsense mutation in TMEM126A causing autosomal recessive optic atrophy and auditory neuropathy. Mol Vis. 2010; 16: 650–664. [PMC free article] [PubMed] [Google Scholar]
- 27. Gao FJ, Li JK, Chen H, et al.. Genetic and clinical findings in a large cohort of Chinese patients with suspected retinitis pigmentosa. Ophthalmology. 2019; 126: 1549–1556. [DOI] [PubMed] [Google Scholar]
- 28. Li W, Wang Z, Sun Y, et al.. A start codon mutation of the TSPAN12 gene in Chinese families causes clinical heterogeneous familial exudative vitreoretinopathy. Mol Genet Genomic Med. 2019; 7: e00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnston JJ, Biesecker LG.. Databases of genomic variation and phenotypes: existing resources and future needs. Human Mol Genet. 2013; 22: R27–R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stenson PD, Ball EV, Mort M, et al.. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics. 2012; Chapter 1: Unit1.13. [DOI] [PubMed] [Google Scholar]
- 31. Hudson G, Carelli V, Spruijt L, et al.. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA–haplogroup background. Am J Human Genet. 2007; 81: 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ji Y, Zhang AM, Jia Xi, et al.. Mitochondrial DNA haplogroups M7b1′2 and M8a affect clinical expression of Leber hereditary optic neuropathy in Chinese families with the m.11778G→A mutation. Am J Human Genet. 2008; 83: 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen J, Xu K, Zhang X, et al.. Mutation screening of mitochondrial DNA as well as OPA1 and OPA3 in a Chinese cohort with suspected hereditary optic atrophy. Invest Ophthal Vis Sci. 2014; 55: 6987–6995. [DOI] [PubMed] [Google Scholar]
- 34. Kloth K, Synofzik M, Kernstock C, et al.. Novel likely pathogenic variants in TMEM126A identified in non-syndromic autosomal recessive optic atrophy: two case reports. BMC Med Genet. 2019; 20: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Torroni A, Petrozzi M, D'Urbano L, et al.. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Human Genet. 1997; 60: 1107. [PMC free article] [PubMed] [Google Scholar]
- 36. Yu-Wai-Man P, Shankar S P, Biousse V, et al.. Genetic screening for OPA1 and OPA3 mutations in patients with suspected inherited optic neuropathies. Ophthalmology. 2011; 118: 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jia X, Li S, Xiao X, et al.. Molecular epidemiology of mtDNA mutations in 903 Chinese families suspected with Leber hereditary optic neuropathy. J Human Genet. 2006; 51: 851–856. [DOI] [PubMed] [Google Scholar]
- 38. Liang M, Jiang P, Li F, et al.. Frequency and spectrum of mitochondrial ND6 mutations in 1218 Han Chinese subjects with Leber's hereditary optic neuropathy. Invest Ophthalmol Vis Sci. 2014; 55: 1321–1331. [DOI] [PubMed] [Google Scholar]
- 39. Almind GJ, Ek J, Rosenberg T, et al.. Dominant optic atrophy in Denmark–report of 15 novel mutations in OPA1, using a strategy with a detection rate of 90%. BMC Med Genet. 2012; 13: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y, Xu M, Liu X, et al.. Targeted next-generation sequencing extends the mutational spectrums for OPA1 mutations in Chinese families with optic atrophy. Mol Vis. 2019; 25: 912–920. [PMC free article] [PubMed] [Google Scholar]
- 41. Chen J, Riazifar H, Guan MX, et al.. Modeling autosomal dominant optic atrophy using induced pluripotent stem cells and identifying potential therapeutic targets. Mitochondrion. 2015; 24: S30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cohn AC, Toomes C, Potter C, et al.. Autosomal dominant optic atrophy: penetrance and expressivity in patients with OPA1 mutations. Am J Ophthalmol. 2007; 143: 656–662. [DOI] [PubMed] [Google Scholar]
- 43. Nakamura M, Lin J, Ueno S, et al.. Novel mutations in the OPA1 gene and associated clinical features in Japanese patients with optic atrophy. Ophthalmology. 2006; 113: 483–488.e1. [DOI] [PubMed] [Google Scholar]
- 44. Alexander C, Votruba M, Pesch UEA, et al.. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000; 26: 211–215. [DOI] [PubMed] [Google Scholar]
- 45. Li H, Jones EM, Li H, et al.. Clinical and genetic features of eight Chinese autosomal-dominant optic atrophy pedigrees with six novel OPA1 pathogenic variants. Ophthalmic Genet. 2018; 39: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liskova P, Tesarova M, Dudakova L, et al.. OPA1 analysis in an international series of probands with bilateral optic atrophy. Acta Ophthalmol. 2017; 95: 363–369. [DOI] [PubMed] [Google Scholar]
- 47. Stewart JD, Hudson G, Yu-Wai-Man P, et al.. OPA1 in multiple mitochondrial DNA deletion disorders. Neurology. 2008; 71: 1829–1831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.