Abstract
Vaccines are the most effective means available for preventing infectious diseases. However, vaccine-induced immune responses are highly variable between individuals and between populations in different regions of the world. Understanding the basis of this variation is, thus, of fundamental importance to human health. Although the factors that are associated with intra- and inter-population variation in vaccine responses are manifold, emerging evidence points to a key role for the gut microbiome in controlling immune responses to vaccination. Much of this evidence comes from studies in mice, and causal evidence for the impact of the microbiome on human immunity is sparse. However, recent studies on vaccination in subjects treated with broad-spectrum antibiotics have provided causal evidence and mechanistic insights into how the microbiota controls immune responses in humans.
Keywords: vaccines, microbiome, human immunology, systems vaccinology
In this review, de Jong et al. examine recent studies on vaccination in humans treated with broad-spectrum antibiotics that provided evidence indicating the microbiota controls vaccine immunity. Since antibiotics and vaccines represent widely used medical interventions, these findings have important implications for clinical practice and public health.
Main Text
Introduction
Humans are inhabited by trillions of diverse microorganisms, known collectively as the microbiome. The terms microbiome and microbiota are often used interchangeably, with the former referring to the aggregate of genomes from all the microorganisms in the body and the latter referring to the specific microorganisms contained in the body. In addition to bacteria, the microbiota also consists of viruses, fungi, protozoa, and archaea (Pfeiffer and Virgin, 2016; Robinson and Pfeiffer, 2014). The microbiota performs a wide range of essential and beneficial functions, including controlling mucosal immunity, breaking down nutrients, and preventing pathogen colonization (Kundu et al., 2017). The colonization of microbes starts at birth and continues through the first years of life, establishing a symbiotic relationship with the host that lasts a lifetime. The impact of the microbiota on the immune system is well established and has been reviewed elsewhere (Belkaid and Hand, 2014; Belkaid and Harrison, 2017; Levy et al., 2017).
Research during the past decade using animal models has revealed a major impact of the microbiota on diverse physiological processes such as metabolism (Nicholson et al., 2012; San-Cristobal et al., 2020; Sonnenburg and Bäckhed, 2016), cardiovascular function (Zhao and Wang, 2020), central nervous system function (Carabotti et al., 2015) as well as on susceptibilities to inflammatory disorders, such as allergic (Mitre et al., 2018; Patrick et al., 2020) and autoimmune diseases (Kostic et al., 2014; Scher et al., 2013). The microbiota has also been linked to the efficacy of anti-programed cell death protein 1 (anti-PD-1)-based cancer immunotherapy (Gopalakrishnan et al., 2018; Routy et al., 2018).
Despite growing evidence of a connection between the microbiota and the immune system, its impact on immunity to vaccination remains poorly understood. Recent studies have demonstrated a profound impairment in vaccine-induced antibody responses during microbiota perturbation. Yet, much of this evidence comes from studies in animal models, such as germ-free mice or mice treated with antibiotics. Even though vaccines and antibiotics represent the most widely used and important public health interventions, surprisingly little is known about the interaction between them. In this review, we discuss the impact of the microbiota on vaccine immunogenicity and efficacy. This review will primarily focus on gut bacteria, as this is by far the most studied aspect of the microbiota.
First, we summarize the known knowns and known unknowns about how the microbiota, which is widely distributed in diverse tissues, can act locally or at distal sites in the body. Then, we discuss how vaccine efficacies vary in populations throughout the world and between individuals and consider the ways in which the microbiota could contribute toward this variation. Much of the experimental evidence for this comes from studies in mice and correlative studies in humans but establishing causality in humans has been challenging. However, emerging studies using systems vaccinology approaches to study vaccine responses in healthy humans that were given antibiotics are providing causal evidence for the role of the microbiota on human immunity (Hagan et al., 2019). We discuss these studies and the mechanistic insights into how the microbiota controls the immune system. We conclude by considering the major challenges that need to be addressed to understand the complex and dynamic interplay between the human microbiota and the host response to vaccines, especially in the very young and the very old, and how this knowledge can be exploited to advance novel vaccines.
Games Microbiota Play
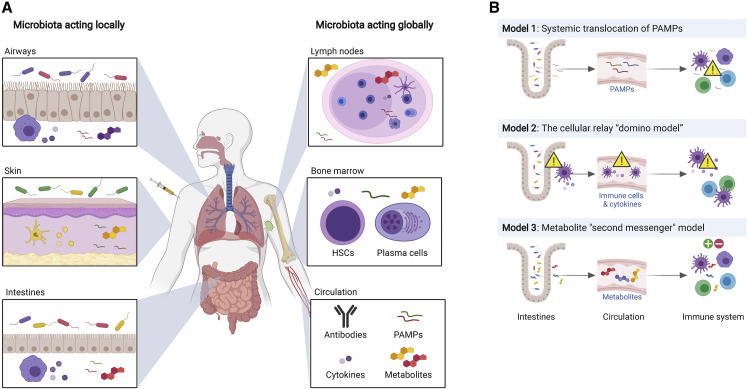
Microbial communities are widely distributed in the gut, lung, skin, and other epithelial surfaces. The microbiota can influence host responses locally at the site, or act at a distance, and exercise profound systemic influence (Figure 1 A). This, in turn, could potentially impact immune responses to vaccination. The mechanisms by which the microbiota exerts local or systemic effects are considered below.
Figure 1.
The Microbiota Exerts Local and Global Immune Influence Through a Variety of Mechanisms
(A) The microbiota can influence host responses locally at the site, such as the airways, skin, and intestines, or act at a distance and exert profound influences systemically in, for example, lymph nodes, bone marrow, or the circulation.
(B) The microbiota can influence immune reactions in distal locations in several ways. Model 1 depicts systemic translocation of bacterial products such as LPS from mucosal sites. Model 2 depicts a “domino effect” mechanism, where signals from the microbiota are delivered to cells in the vicinity, which then circulate throughout the body and relay this information. Model 3 describes the effects of microbiota on distant locations via secretion of microbiota-derived metabolites. HSCs, hematopoietic stem cells. PAMPs, pathogen-associated molecular patterns.
Concept 1: Acting Locally
Gut Microbiota
The alimentary canal presents a very diverse set of niches for colonization by the microbiota. Local interaction with the immune system primarily occurs in the small and large intestines, which harbor large numbers of B and T cells as well as antigen-presenting cells. These cells can detect many of the biomolecules produced by the microbiota, including short-chain fatty acids (SCFAs) (Macia et al., 2015), tryptophan metabolites (Li et al., 2011), bacterial DNA (Hall et al., 2008), vitamin A (Al Nabhani et al., 2019), sphingolipids (An et al., 2014), polysaccharide A (Mazmanian et al., 2005), and muramyl dipeptide (Jiang et al., 2013). Some of these compounds are passively or actively transported across the epithelial lining of the gut and can be detected and acted upon by immune cells in the lamina propria. Antigen-specific immunity is generated by dendritic cells that continuously sample the intestinal lumen (Macpherson and Uhr, 2004), and even simple bacterial cell adhesion to the gut epithelium has the potential to modulate the immune system (Atarashi et al., 2015). Naturally, the responses to such stimuli are context dependent and have the potential to elicit complex changes in the gut immune system.
Imprinting of the gut immune system by the microbiota might also affect its capabilities to respond to pathogens. The most commonly administered oral vaccines are live-attenuated versions of microorganisms that replicate in the gastrointestinal tract, such as polio, cholera, typhoid fever, and rotavirus. Understanding the local interplay between gut microbiota and the immune system is vital to improve the efficacy of oral vaccines. Oral and systemic antibiotics can also have a profound effect on the gut microbiome and therefore potentially on responsiveness to (oral) vaccines. Consistent with this idea, low microbiota diversity early in life has been associated with differences in immune phenotype (Olin et al., 2018) and differences in gut microbiota composition have been correlated to vaccine response (discussed below).
Skin Microbiota
Compared with the gut, the community of skin microbes is less diverse and are fewer in number. However, there is still communication with the skin immune system. Bacterial cues from the skin can trigger interleukin-1 (IL-1) signaling (Naik et al., 2012) and steer the recruitment of innate lymphoid cells (Kobayashi et al., 2019). Identifying specific signals has been difficult, but the secretomes of Staphylococcus epidermidis and Staphylococcus aureus provoked opposing effects on immune activation, indicating phylum-specific effects (Laborel-Preneron et al., 2015). Lipopeptides binding the Toll-like receptor-2/6 (TLR-2/6) heterodimer have also shown to be immunosuppressive in this context (Skabytska et al., 2014). Microbiota-host interaction at the skin has the potential to modify immune function, as illustrated by the connection between the microbiota and various immune-related skin disorders (Stacy and Belkaid, 2019) and could potentially impact immunity to vaccination.
Airway Microbiota
While the lungs were long believed to be sterile, sequencing-based methods and new techniques of bacterial cell culture have revealed that the luminal surface harbors a microbiota, albeit a less diverse one than that of the gut (Dickson et al., 2016). So far, the live-attenuated influenza vaccine is the only vaccine administered through the intranasal route. However, several vaccines for respiratory pathogens, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are being developed (World Health Organization, 2020), and will require the appropriate quality and quantity of mucosal antibody response, and T cell response in the lung, to be effective. These mucosal responses could conceivably be influenced by the lung microbiota. For example, plasma cells and tissue-resident memory T cells (TRMs) in the lung may derive signals from lung bacterial products that enhance their survival and/or function. In this context, our recent study demonstrates that TRMs in the vaginal tissues provide signals to neighboring cells, including myeloid cells, to enhance antiviral responses in such cells (Arunachalam et al., 2020). The extent to which the local microbiota could impinge on such TRM-innate interactions and whether such interactions are pervasive in other tissues remain to be seen.
Concept 2: Acting Globally
In addition to affecting their local milieu, microbes can also influence immune reactions in anatomical locations distal from the site of colonization. This can conceivably happen through several mechanisms (Figure 1B): (1) translocation of bacterial products, such as lipopolysaccharides (LPSs) from mucosal sites to the systemic circulation (Sandler and Douek, 2012), (2) a “domino effect” mechanism, where signals from the microbiota are delivered to cells in the vicinity, which then circulate throughout the body and relay this information (perhaps through cytokines, metabolites, or other molecules), and (3) via dissemination of microbiota-derived metabolites (metabolite second messenger model). Consistent with this idea, microbiota-derived metabolites can be identified in various tissues and, thus, have the potential to be detected by the immune system at those sites (Uchimura et al., 2018). Distal immune stimulation has been reported in various tissues such as the bone marrow (Clarke et al., 2010; Shi et al., 2011), the liver (Li et al., 2017a, ), the peritoneum (Abt et al., 2012), and the spleen (Kim et al., 2016b). Bacterial antigens disseminated to the spleen and mesenteric lymph nodes can trigger the production of IgG, which provides systemic protection against bacterial infection (Zeng et al., 2016).
Another fascinating example of how the microbiota could act globally comes from recent studies that suggest that the response to HIV, and perhaps other viruses, could be imprinted by prior exposure to antigenically cross-reactive microbiota-derived antigens (Williams et al., 2018). Haynes and colleagues showed that HIV vaccine-induced CD4+ T and B cell responses could originate from a pool of intestinal cross-reactive immune cells. When they examined anti-HIV responses in ileum B cells and probed their relationship to commensal bacteria, remarkably, a majority (82%) of the ileum HIV anti-gp41 antibodies cross-reacted with commensal bacteria, and of those, 43% showed non-HIV-1 antigen polyreactivity (Trama et al., 2014).
Variations in Vaccine Efficacy
Vaccine efficacies can vary widely between individuals in a given region (Praharaj et al., 2015). For example, the magnitude of hemagglutinin inhibition titers induced by vaccination with the inactivated seasonal influenza vaccine can vary by more than 100-fold between individuals in a single cohort (Nakaya et al., 2015). Furthermore, the magnitude of neutralizing antibody titers and CD8+ effector T cell responses induced by vaccination of humans with the live-attenuated yellow fever vaccine 17D, one of the most successful vaccines ever developed (Pulendran, 2009), can range more than 10-fold among individuals (Querec et al., 2009).
Vaccine responses can also vary widely between people in different parts of the world. For example, the protection against tuberculosis by the Bacillus Calmette–Guérin (BCG) vaccine varies from 0% to 80%, with a higher response rate in Europe than in Africa (Fine, 1995; Hur et al., 2014). Also, vaccines against poliomyelitis, rotavirus, malaria, and yellow fever provide less protection in Africa and Asia as compared with Europe or the USA (Hanlon et al., 1987; Muyanja et al., 2014; Sissoko et al., 2017; Tate et al., 2012). Indeed, since their introduction, a recurring observation has been that immune responses to oral vaccines may be lower and less consistent in low- to middle-income countries (LMICs) compared with high-income countries (Praharaj et al., 2015). The oral rotavirus vaccines and oral polio vaccines (OPVs) have historically performed poorly in LMICs (Greenberg and Estes, 2009). More recent studies with the newly licensed vaccines Rotarix and Rotateq have shown that they have lower efficacy in LMICs in Asia, Africa, and Latin America (Armah et al., 2010; Madhi et al., 2010; Patel et al., 2009; Zaman et al., 2010).
Vaccine immunogenicity can be influenced by diverse factors such as host genetics, nutritional status, immunological imprinting as a result of prior exposure to the pathogen, and maternal antibodies (Praharaj et al., 2015). In addition, the prevalence of chronic infections such as tuberculosis, HIV, or parasites can also have an impact (Clarke and Desselberger, 2015; Qadri et al., 2013). Importantly, direct immunomodulatory effects of chronic infections by pathogens and parasites, such as helminths, Plasmodium, and hepatitis C virus, whose prevalence varies considerably worldwide, has also shown to influence immune responses (Stelekati and Wherry, 2012). There is now increasing evidence that the microbiome is also a major determinant of immunity to vaccination (Hagan et al., 2019). This is especially relevant since there is an important demographic shift in the developing countries, in which entire populations change their lifestyle from a traditional one to an urban lifestyle, which could lead to major changes in the microbiome (Pulendran, 2014). Below, we summarize the evidence that the gut microbiome can influence vaccine responses and discuss potential mechanisms through which this can occur. We will focus our discussion on bacteria, since other constituents of the microbiota, such as the viruses and fungi, are much less studied in the context of vaccination.
Experimental Evidence in Mice
Numerous studies in mice have illustrated potent and complex roles for the microbiota in modulating immune responses to mucosal or systemic infections. In a model of oral rotavirus infection, Gewirtz and colleagues showed that germ-free and antibiotics-treated mice had reduced viral infection and enhanced serum and mucosal rotavirus-specific antibody responses, suggesting that the microbiota suppressed antibody responses (Uchiyama et al., 2014). In contrast, segmented filamentous bacteria were sufficient to prevent and cure mice of rotavirus infection and the associated diarrhea (Shi et al., 2019). Another study showed that the virus-specific CD4+ and CD8+ T cells and antibody responses following respiratory influenza virus infection were critically dependent on the microbiota (Ichinohe et al., 2011). In the case of systemic infections or lung infections, Artis and colleagues demonstrated that antibiotics-treated mice displayed impaired innate and adaptive antiviral immune responses and delayed viral clearance after exposure to systemic lymphocytic choriomeningitis virus (LCMV) or mucosal influenza virus (Abt et al., 2012). Transcriptome profiling of macrophages isolated from antibiotics-treated mice revealed decreased expression of genes associated with antiviral immunity and defective responses to type I and type II interferons. They also exhibited an impaired capacity to limit viral replication. In a recent study in mice, Kim et al. demonstrated that SCFAs produced by the gut microbiota as fermentation products of dietary fiber enhanced plasma cell differentiation and antibody responses (Kim et al., 2016b). Mice with low SCFA production due to reduced dietary fiber consumption or microbial insufficiency were defective in homeostatic and pathogen-specific antibody responses, resulting in greater susceptibility to infection. Collectively, these data suggest that the gut microbiota modulates innate and adaptive immune responses to oral and systemic infections.
In the case of immune responses to vaccination, in a model of oral immunization using heat-labile enterotoxin of enterotoxigenic Escherichia coli as an adjuvant (LT R192G/ L211A), Belkaid and colleagues showed that depletion of the gut microbiota was associated with reduced Th1 and Th17 responses to the antigen (Hall et al., 2008; Norton et al., 2011). In another study by Nunez and colleagues, the microbiota was discovered to be critical for inducing antigen-specific immunoglobulin G (IgG) production after intranasal immunization with the model antigen human serum albumin plus cholera toxin as an adjuvant (Kim et al., 2016a) (Table 1 ).
Table 1.
A Selection of Recent Evidence for the Potential Importance of Microbiota in Immunity to Vaccination
| Model | Vaccine | Host | Study Outcomes | Reference |
|---|---|---|---|---|
| Animal Studies |
TIV, OPV | Mice | TLR5-mediated sensing of flagellin from gut microbiota had an adjuvant effect on TIV and OPV. No effect with adjuvanted vaccines or live-attenuated yellow fever vaccine. | (Oh et al., 2014) |
| CT | Mice | The mucosal adjuvant activity of CT was mediated through the recognition of symbiotic bacteria by Nod2 in CD11c-expressing phagocytes. | (Kim et al., 2016a) | |
| BCG, MenB, MenC, PCV13, Hexa | Mice | Antibiotics-induced dysbiosis in infant (but not adult) mice leads to impaired antibody responses and elevated ex vivo cytokine recall responses. | (Lynn et al., 2018) | |
| TIV | Rhesus macaques | Subclinical CMV infection resulted in increase in butyrate-producing bacteria and lower antibody responses to influenza vaccination. | (Santos Rocha et al., 2018) | |
| Correlative human studies | RV | Ghanaian and Dutch infants | Microbiome composition was different between RV responders and non-responders. Ghanaian responders were more similar to Dutch infants than to non-responders. | (Harris et al., 2017) |
| RV | Pakistani and Dutch infants | RV response correlated with a higher relative abundance of bacteria belonging to Clostridium cluster XI and Proteobacteria. | (Harris et al., 2018a) | |
| RV, OPV | Indian infants | No differences in microbiome composition between RV responders and non-responders. Co-administered OPV reduced the response to RV. | (Parker et al., 2018) | |
| OPV | Indian infants | Enteric viruses have a greater impact on OPV response than the bacterial microbiota, especially for recent enterovirus infections. | (Praharaj et al., 2019) | |
| BCG, TT, HBV, OPV | Bangladeshi infants | High abundance of stool Actinobacteria, including Bifidobacterium, was associated with higher responses to oral and parenteral vaccines and with higher CD4+ T cell and antibody responses 2 years after vaccination. | (Huda et al., 2014, 2019) | |
| HIV | Swiss adults | The immunogenicity of HIV vaccine was correlated with microbiota clusters. | (Cram et al., 2019) | |
| Causation studies in humans | OPV | Indian infants | Antibiotics did not improve the immunogenicity of OPV, despite the reduction of biomarkers of enteropathy and pathogenic intestinal bacteria. | (Grassly et al., 2016) |
| RV, Pneumo23, TT | Dutch adults | Narrow-spectrum antibiotics resulted in higher day-7 anti-RV IgA boosting and increased RV-antigen shedding but no different absolute titers. The antibiotics did not affect pneumococcal or TT vaccination. | (Harris et al., 2018b) | |
| TIV | American adults | Antibiotics-induced microbiome loss impaired antibody response in subjects with low pre-existing immunity. | (Hagan et al., 2019) |
TIV, trivalent inactivated influenza vaccine; OPV, oral polio vaccine; CT, cholera toxoid; BCG, Bacillus Calmette–Guérin; MenB, Bexsero meningococcal serogroup B vaccine; MenC, NeisVac-C meningococcal serogroup C vaccine; PCV13, the Prevenar 13-valent pneumococcal conjugate vaccine; Hexa, the INFANRIX Hexa combination vaccine, which contains antigens from hepatitis B, diphtheria, tetanus, acellular pertussis, Haemophilus influenzae type b, and inactivated poliomyelitis virus; RV, rotavirus vaccine; TT, tetanus toxoid; HBV, hepatitis B vaccine; Pneumo23, polysaccharide pneumococcal vaccine.
We demonstrated that germ-free or antibiotic-treated mice mount substantially reduced IgG and IgM antibody responses to the seasonal influenza vaccine (Oh et al., 2014). This vaccine does not contain any exogenous adjuvants. The genesis of this study came from our previous work in humans, in which we had observed that vaccination with the seasonal influenza vaccine induced expression of TLR5 (a pattern recognition receptor for flagellin) in peripheral blood mononuclear cells (PBMCs) 3–7 days after vaccination. The level of TLR5 expression correlated with the hemagglutination inhibition titers (a measure of antibody response to the influenza vaccine) 4 weeks later (Nakaya et al., 2015). This led to the hypothesis that TLR5 was involved in the regulation of the immune response to the seasonal influenza vaccine. We tested this hypothesis and showed that TLR5 knockout mice were impaired in their ability to mount antibody responses to vaccination with the seasonal influenza vaccine (Oh et al., 2014).
Since TLR5 is a sensor for bacterial flagellin, this result raised the possibility that flagellin produced by the microbiota might augment antibody responses to influenza vaccination. We thus immunized germ-free mice, or mice that had been treated with broad-spectrum antibiotics for several weeks, with the seasonal influenza vaccine and observed a marked reduction in the influenza-specific IgG and IgM antibody responses at days 7 and 14 after vaccination. In addition, there were substantially reduced antigen-specific plasma cell responses, demonstrating that the microbiota did indeed enhance the antibody response to vaccination. Finally, antibody responses were restored in antibiotic-treated or germ-free mice by oral reconstitution of the gut microbiota with a flagellated, but not with a non-flagellated, strain of Escherichia coli.
This finding indicated that flagellin derived from the microbiota could act as a natural adjuvant for the influenza vaccine, by signaling via TLR5. To determine whether this dependence on the microbiota was a peculiarity of the response to the influenza vaccine, or whether responses to other vaccines were also affected, we immunized germ-free or antibiotics-treated mice with the live-attenuated yellow fever vaccine YF-17D (which contains its own endogenous adjuvants that trigger TLR and RIG-I-like receptor signaling), the Tdap vaccine (tetanus-diphtheria-pertussis adjuvanted with alum), the recombinant hepatitis B vaccine (with alum), the recombinant HIV Env vaccine (adjuvanted with alum), and the inactivated polio vaccine (IPOL, which does not contain any adjuvant). Interestingly, only the response stimulated by the non-adjuvanted IPOL vaccine was dependent on the microbiota. These results suggest that the microbiota was essential for augmenting antibody responses to non-adjuvanted vaccines and that the microbiota was in effect acting as an endogenous adjuvant.
Evidence for a role for the microbiota in modulating vaccine responses in early life was recently provided by a study by Lynn et al., who demonstrated that antibiotic-driven intestinal dysbiosis in mice, specifically in early life, leads to significantly impaired antibody responses to five different adjuvanted and live vaccines (Lynn et al., 2018). Restoration of the microbiota rescues these impaired responses. Interestingly, in contrast to the attenuated antibody responses, immunized mice exposed to early-life antibiotics display significantly enhanced T cell cytokine recall responses upon ex vivo restimulation with the vaccine antigen. These results indicate that, in mice, antibiotic-driven dysregulation of the gut microbiota in early life can modulate immune responses to vaccines that are routinely administered to infants.
Correlative Evidence in Humans
In support of the notion that the microbiota influences vaccine responses in humans, several studies have demonstrated correlations between particular species of gut bacteria and vaccine immunity. Recently, the link between the gut microbiota and impaired vaccine responses has been studied in Ghana, Pakistan, and Bangladesh. In both Ghanaian and Pakistani infants, the microbiota composition was different between responders and non-responders after oral rotavirus vaccination (Harris et al., 2017, 2018a). Interestingly, the microbiota of responders from both Ghana and Pakistan was more comparable to that of Dutch infants than non-responders (Harris et al., 2017, 2018a). These comparisons with Dutch infants emphasize how certain bacterial phyla could be connected to vaccine efficacy in diverse populations. A study performed among Bangladeshi infants showed that Bifidobacterium abundance in early infancy was correlated with the immune response to the oral polio vaccine, as well as to the parenteral vaccines BCG, tetanus toxoid, and hepatitis B virus vaccine (Huda et al., 2019, 2014). Interestingly Bifidobacterium abundance at vaccination also correlated positively with CD4+ T cell and antibody responses after 2 years, indicating that the microbiota can be linked to the durability of the immune response (Huda et al., 2019). A recent HIV vaccination study in Switzerland also showed a correlation between microbiota composition and parenteral vaccine immunogenicity in a high-income country (Cram et al., 2019). However, it should be noted that the interpretation of correlations between individual taxa and immune responses to vaccination is difficult and the results probably indirect.
Human Intervention Studies
Probiotics Studies
Probiotics Are Preparation of Live Microbes that Are Consumed with the Intention of Promoting Good Health. Several studies have assessed the impact of probiotic administration on immunity to vaccination in humans (Praharaj et al., 2015). The effect of probiotics on oral rotavirus vaccine response has been studied in a pig model, where the probiotic resulted in enhanced antibody responses and protection by the vaccine as compared with control piglets who remained bacteriologically sterile (Kandasamy et al., 2014). Administration of probiotics to infants have yielded variable results, depending on the antigen, strain of probiotic, and geographical region (Isolauri et al., 1995; Matsuda et al., 2011; Soh et al., 2010). Although there are many efforts evaluating the effect of probiotics on the immune response to vaccines in adults, they share with the studies in children the problems of variability in antigens, probiotics strains, and populations, as well as the limitations of power imposed by the small numbers of participants in most studies. Additionally, the degree to which probiotic organisms actually colonize the host is often not investigated, rendering the interpretation of such experiments difficult.
Antibiotics Studies
A few antibiotic intervention studies in humans have recently been performed. In an attempt to improve the immunogenicity of the oral polio vaccine, Indian infants were treated with azithromycin before vaccination (Grassly et al., 2016). Treatment with this broad-spectrum antibiotic did not improve the immunogenicity of the vaccine, even though biomarkers of environmental enteropathy and the prevalence of pathogenic intestinal bacteria were reduced. However, the polio vaccine response did correlate with lower concurrent enterovirus infections. While showing the effect of the virome on polio vaccine responses, the contributions of bacteria cannot yet be excluded as the type and duration of the antibiotic treatment might be important.
In a second human antibiotic intervention study, different antibiotic treatment regimens were compared (Harris et al., 2018b). By giving the narrow-spectrum antibiotic vancomycin, the study aimed to amplify Gammaproteobacteria and reduce Bacteroidetes, and this change in microbiota composition was hypothesized to boost rotavirus vaccine responses based on earlier work. The narrow-spectrum antibiotic treatment was compared with broad-spectrum antibiotics treatment (vancomycin, ciprofloxacin, and metronidazole) and untreated controls. Even though the rotavirus vaccine is usually given to infants, this intervention study was performed with adult volunteers for ethical reasons. Both the broad-spectrum and narrow-spectrum antibiotic treatment did not affect absolute vaccine IgA titers. However, the narrow-spectrum antibiotic enhanced IgA boosting at day 7 and both antibiotic treatments increased rotavirus shedding, indicating better replication and take of the vaccine. Despite the enhancing effect of the antibiotics on the oral rotavirus vaccine, no effect was seen on the parenteral unadjuvanted pneumococcal polysaccharide vaccine and parenteral adjuvanted tetanus toxoid vaccine.
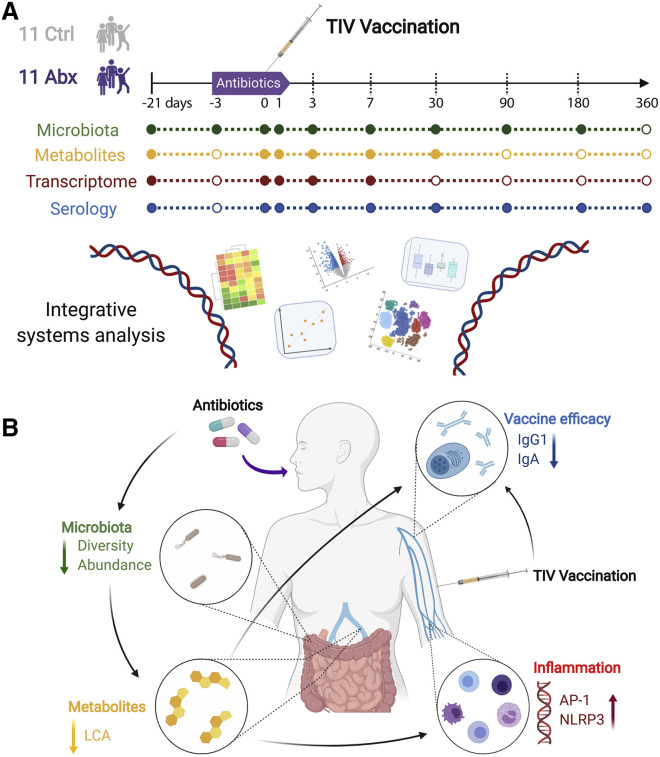
Recently, we used a systems vaccinology approach (Hagan et al., 2019) to investigate the impact of antibiotics-driven microbiota perturbation on immunity to vaccination in humans (Figure 2 A). In this study, healthy adults were administered broad-spectrum antibiotics (vancomycin, neomycin, and metronidazole) for 5 days and then immunized with the seasonal influenza vaccine 1 day prior to the cessation of antibiotics. Within 3 to 5 days from the start of antibiotics administration, there was a profound 10,000-fold reduction in the number of bacteria per gram of stool, but this rapidly returned to the baseline level within a few days. Also, there was a long-lasting diminution in bacterial diversity. Despite such drastic effects on the gut microbiota, there was no significant reduction in neutralizing antibody titers after vaccination.
Figure 2.
Antibiotics Impair the Vaccine Response in Healthy Adults
(A) Outline of the study by Hagan et al. 11 subjects were treated with antibiotics for 5 days and vaccinated with the trivalent influenza vaccine on the fourth day. These were then compared with 11 vaccinated controls untreated with antibiotics. Fecal and blood samples were collected at regular intervals.
(B) Results from the study by Hagan et al. Administration of antibiotics led to reduced microbial diversity and abundance and a consequential reduction in secondary bile acids. This in turn led to increased inflammation and a diminished vaccine response. TIV, trivalent influenza vaccine; LCA, litocholic acid.
Most humans have pre-existing immunity to seasonal influenza due to previous infection or vaccination. This pre-existing immunity is mediated by immunological memory, which results in a more vigorous and rapid response to recall immunization and might thus not be dependent on the natural adjuvant effect of the microbiota. In contrast, primary immune responses to antigens that had not been previously seen by the immune system may be more dependent on adjuvant signals from the microbiota. To test this hypothesis, we conducted a second clinical trial in individuals who had had no exposure to influenza vaccination or infection during the preceding 3 years. These subjects also had markedly lower pre-vaccination anti-influenza antibody titers than the first cohort had. In this second study, antibiotics-treated individuals demonstrated significantly lower levels of H1N1-specific IgG1 and IgA antibody titers and an impaired ability to neutralize the H1N1 influenza strain. Interestingly, the impairment of IgG1 and IgA titers was only observed against the H1N1 strain and not against the H3N2 or B strains. The reasons for this are unclear, but it is possible that adults have higher immune memory to these subtypes and were able to withstand the effects of the antibiotics. Thus, these results suggest that in the absence of pre-existing immunity, antibody-driven microbiota dysbiosis could lead to a significant impairment in the antibody response to seasonal influenza (Figure 2B).
To assess the impact of antibiotics on the innate immune response, we performed a transcriptional analysis of PBMCs isolated prior to antibiotics administration or at days 0, 1, 3, 7, and 14 after vaccination. This analysis revealed that antibiotics administration resulted in transcriptional signatures of enhanced innate immune responses, including dendritic cell activation and enhanced inflammation. In particular, there was an enhanced activity of transcriptional signatures associated with the transcription factors activating protein 1 (AP-1, comprising FOS and JUN) and nuclear receptor 4A1 (NR4A), which orchestrate inflammatory responses. Furthermore, we identified putative target genes of AP-1 and NR4A and observed strong correlations between these transcription factors and several of their respective target genes. Consistent with these transcriptional signatures, flow cytometric analysis revealed enhanced frequencies of activated myeloid dendritic cells. Curiously, signatures of AP-1 and NR4A activation were also induced in healthy elderly subjects, but not healthy young subjects, in a previous study we had performed (Nakaya et al., 2015). Thus, antibiotics-induced microbiota dysbiosis in healthy young adults altered their transcriptional response signature to seasonal influenza vaccination, to resemble that of elderly subjects, in terms of enhanced expression of the AP-1 and NR4A signature, which are indicative of a pro-inflammatory state (Figure 2B).
Antibiotics administration also induced a profound perturbation of the plasma metabolome. Strikingly, there was a reduction in the concentration of secondary bile acids, such as lithocholic acid (LCA), which was reduced by 1,000-fold, consistent with the established role of the microbiota in converting primary bile to secondary bile (Ridlon et al., 2014). Given that secondary bile acids have been shown to inhibit inflammatory responses (Bakke and Sun, 2018; Staudinger et al., 2001), in particular NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome activation (Guo et al., 2016), we investigated whether there was a connection between the reduction in secondary bile acids and enhanced inflammatory signatures, including signatures of NLRP3 and AP-1 activation, and found a strong correlation. Furthermore, a recent study in mice showed how two derivatives of LCA, 3-oxoLCA and IsoalloLCA, modulated the balance between pro- and anti-inflammatory responses, by reducing Th17 cell differentiation and increasing regulatory T cell (Treg) differentiation in the lamina propria (Hang et al., 2019). These data together suggest a potential mechanism by which the microbiota can regulate secondary bile acid production and consequently inflammatory responses in humans (Figure 2B).
Given the enhanced inflammatory signature in the elderly (Nakaya et al., 2015), to what extent the dysbiosis-secondary bile acid-inflammation axis contributes to enhanced inflammation in the elderly deserves further exploration. Although the impact of age-related changes in the microbiota on bile acid metabolism is not well established, there is evidence that the gut microbiota of elderly subjects is less capable of producing other immunomodulatory metabolites, such as SCFAs (Rampelli et al., 2013). These upregulated pathways (AP-1 and NR4A signaling) are also known to be triggered by bacterial LPS, and gut permeability has been shown to increase with age in mice (Thevaranjan et al., 2017). Further studies should, thus, explore by which mechanisms gut dysbiosis modulates the immune system in the elderly to determine how age-associated changes in the microbiota can contribute to chronic inflammation and immunosenescence.
Interestingly, the AP-1 and NR4A inflammatory signatures, or the changes in the secondary bile acid metabolites, did not correlate with the antibody response to influenza vaccination. Therefore, in order to determine the critical drivers of the antibody response, we constructed a multiscale multi-response network (Li et al., 2017b) in which we performed an integrative analysis of orthogonal datasets. This analysis suggested that the influenza-specific IgG1 response was significantly associated with two metabolic clusters highly enriched in fatty acid metabolism, which is emerging as an important orchestrator of immune responses (Ganeshan and Chawla, 2014). These results highlight the capability of the microbiota to exert diverse effects on immune function, not only through direct interaction with immune cells in the gut but also through indirect mechanisms such as regulating the systemic availability of critical metabolites.
Future Perspectives
The importance of including the microbiota in exploring the conceptual framework of the immune system cannot be overstated. While elegant mechanistic studies in mice and other animal models have highlighted the profound impact of the microbiota on diverse physiological systems and pathologies, the relevance of such mechanisms in the human model is poorly understood. Causal evidence for the role of the microbiota on modulating human physiology and susceptibility to disease is sparse.
Vaccines are powerful tools for probing the human immune system (Pulendran, 2014), as they allow the immune system to be perturbed in a very synchronized way. Moreover, the tools of systems biology allow the delineation of the cellular and molecular changes that occur in response to vaccination, with an exquisite degree of precision. Therefore, future studies should be aimed at using a systems vaccinology approach in exploring the impact of the microbiota on immunity to vaccination. The results of the aforementioned studies in humans have provided the first mechanistic insights into the effects of the microbiota on vaccine immunity in humans. Such investigation must be extended to infants, and the elderly population in which vaccine efficacy is a general lower, and the effects of non-genetic factors, such as antibiotics, diet, and comorbidities, deserve examination.
Impact of the Microbiota on Vaccine Immunity at the Extremes of Age
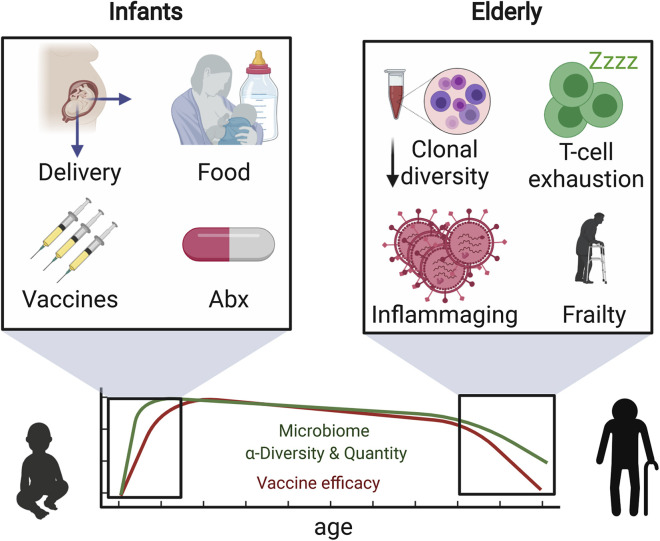
Early human life is a period of rapid change, borne out of the necessity to adapt to life outside the womb. Following the initial stress of the delivery process, the immune system must cope with a range of foreign antigens from the bacteria settling on all epithelial surfaces, foreign particles introduced into the lungs with the initiation of breathing, and food in the form of breast milk (Figure 3 ). During this period, there is also a high risk of contracting infections, which are sometimes treated with antibiotics that can wreak havoc in the nascent microbiota. After birth, the newborn baby is confronted with a plethora of diverse immunological stimuli, including vaccines against as many as twelve different pathogens. In the United States, the Center for Disease Control and Prevention recommended immunization schedule for children in the first year of life includes vaccines against hepatitis B, rotavirus, diphtheria, tetanus, and pertussis, Haemophilus influenzae type B, pneumococcus, polio, influenza, meningococcus, measles, mumps, and rubella. These constitute a wide array of immunological stimuli of viral and bacterial origins as well as aluminum-based adjuvants. Despite the widespread use of these vaccines, the molecular and cellular nature of the innate and adaptive responses induced by these immunizations in children is poorly understood.
Figure 3.
Unique Environmental Factors and Biological Changes in the Very Young and the Very Old that Can Be Detrimental to Vaccine Efficacy
Newborn children and elderly people undergo physiological changes and are exposed to environmental stimuli that can be detrimental to their immune system. Simultaneously, they often experience decreased microbial diversity. These factors interplay to make them less responsive to vaccination.
To match the onslaught of immunological stimuli, the newborn immune system moves through a series of phenotypical changes, following a stereotypic progression (Olin et al., 2018). These changes occur at different rates in different children and are affected by factors such as mode of delivery, preterm birth, and diet (Kollmann et al., 2017). Being able to predict vaccine efficacy based on immune status, or being able to design vaccines specifically tailored to the newborn immune system, would be major accomplishments.These challenges require a deeper understanding of the dynamics of the molecular networks that drive immunity to vaccination in children. Future studies should be aimed at using systems biology approaches to study the impact of the microbiota on immunity to vaccination in the neonatal and infant populations in diverse parts of the world.
At the other end of life, improvements in public health, and the advent of antibiotics and vaccination, have led to a staggering increase in life expectancy. These shifting demographics in the age distribution and the associated increase in the incidence of obesity and metabolic disorders pose new challenges for vaccination, as emerging evidence suggests that age (Duraisingham et al., 2013) and the metabolic state of individuals can exert major influences on the immune system (Hotamisligil, 2006). As stated above, the gut microbiota of elderly subjects appears impaired in its capacity to produce beneficial metabolites such as SCFAs (Rampelli et al., 2013), and this may affect the immunological state in the elderly. Also, gut permeability has been shown to increase with age in mice (Thevaranjan et al., 2017), and this could result in bacterial products, such as lipopolysaccharides or flagellin gaining systemic access and imprinting a low, tonic inflammatory state. Further studies in elderly humans, using systems-based approaches should, thus, explore the mechanisms by which gut dysbiosis modulates the age-associated decline in immune function known as immunosenescence.
Although the scientific field of vaccine development is over 200 years old, we, unfortunately, are still not able to develop a successful vaccine against any given pathogen that gives full protection in all individuals. The irregularities in vaccine responses between individuals and populations give us clues toward a better understanding of what factors determine efficacy. This could prove critical for the development of vaccines against the infectious diseases that so far have resisted our attempts. The current SARS-CoV-2 pandemic provides a salient example of a case where a deeper understanding of the molecular mechanisms underlying vaccine immunity could help focus the vaccine development efforts. The toolkit of systems vaccinology holds part of the promise of gaining such knowledge. There is certainly much left to discover in this burgeoning field, and the new discoveries are certain to provide creative opportunties to harness the microbiome in promoting robust and durable protective immunity induced by vaccination.
Acknowledgments
This work was supported by NIH grants HIPC U19AI090023 (to B.P.), Emory-UGA CEIRS contract HHSN272201400004C (to B.P. and principal investigator Walt Orenstein), U19AI057266 (to B.P. and principal investigator Rafi Ahmed), the Sean Parker Cancer Institute, the Soffer endowment (B.P.), Open Philanthropy endowment (B.P.), and the Violetta Horton endowment (B.P.), the Wenner-Gren Foundations (A.O.), and the Rubicon grant of the Netherlands Organisation for Scientific Research/ZonMW (S.J).
References
- Abt M.C., Osborne L.C., Monticelli L.A., Doering T.A., Alenghat T., Sonnenberg G.F., Paley M.A., Antenus M., Williams K.L., Erikson J., et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Nabhani Z., Dulauroy S., Marques R., Cousu C., Al Bounny S., Déjardin F., Sparwasser T., Bérard M., Cerf-Bensussan N., Eberl G. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity. 2019;50:1276–1288.e5. doi: 10.1016/j.immuni.2019.02.014. [DOI] [PubMed] [Google Scholar]
- An D., Oh S.F., Olszak T., Neves J.F., Avci F.Y., Erturk-Hasdemir D., Lu X., Zeissig S., Blumberg R.S., Kasper D.L. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armah G.E., Sow S.O., Breiman R.F., Dallas M.J., Tapia M.D., Feikin D.R., Binka F.N., Steele A.D., Laserson K.F., Ansah N.A., et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;376:606–614. doi: 10.1016/S0140-6736(10)60889-6. [DOI] [PubMed] [Google Scholar]
- Arunachalam P.S., Charles T.P., Joag V., Bollimpelli V.S., Scott M.K.D., Wimmers F., Burton S.L., Labranche C.C., Petitdemange C., Gangadhara S., et al. T cell-inducing vaccine durably prevents mucosal SHIV infection even with lower neutralizing antibody titers. Nat. Med. 2020;26:932–940. doi: 10.1038/s41591-020-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Ando M., Kamada N., Nagano Y., Narushima S., Suda W., Imaoka A., Setoyama H., Nagamori T., et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakke D., Sun J. Ancient nuclear receptor VDR With new functions: microbiome and inflammation. Inflamm. Bowel Dis. 2018;24:1149–1154. doi: 10.1093/ibd/izy092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y., Hand T.W. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y., Harrison O.J. Homeostatic immunity and the microbiota. Immunity. 2017;46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabotti M., Scirocco A., Maselli M.A., Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015;28:203–209. [PMC free article] [PubMed] [Google Scholar]
- Clarke E., Desselberger U. Correlates of protection against human rotavirus disease and the factors influencing protection in low-income settings. Mucosal Immunol. 2015;8:1–17. doi: 10.1038/mi.2014.114. [DOI] [PubMed] [Google Scholar]
- Clarke T.B., Davis K.M., Lysenko E.S., Zhou A.Y., Yu Y., Weiser J.N. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram J.A., Fiore-Gartland A.J., Srinivasan S., Karuna S., Pantaleo G., Tomaras G.D., Fredricks D.N., Kublin J.G. Human gut microbiota is associated with HIV-reactive immunoglobulin at baseline and following HIV vaccination. PLoS One. 2019;14:e0225622. doi: 10.1371/journal.pone.0225622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R.P., Erb-Downward J.R., Martinez F.J., Huffnagle G.B. The microbiome and the respiratory tract. Annu. Rev. Physiol. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingham S.S., Rouphael N., Cavanagh M.M., Nakaya H.I., Goronzy J.J., Pulendran B. Systems biology of vaccination in the elderly. Curr. Top. Microbiol. Immunol. 2013;363:117–142. doi: 10.1007/82_2012_250. [DOI] [PubMed] [Google Scholar]
- Fine P.E. Variation in protection by BCG: implications of and for heterologous immunity. Lancet. 1995;346:1339–1345. doi: 10.1016/s0140-6736(95)92348-9. [DOI] [PubMed] [Google Scholar]
- Ganeshan K., Chawla A. Metabolic regulation of immune responses. Annu. Rev. Immunol. 2014;32:609–634. doi: 10.1146/annurev-immunol-032713-120236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassly N.C., Praharaj I., Babji S., Kaliappan S.P., Giri S., Venugopal S., Parker E.P., Abraham A., Muliyil J., Doss S., et al. The effect of azithromycin on the immunogenicity of oral poliovirus vaccine: a double-blind randomised placebo-controlled trial in seronegative Indian infants. Lancet Infect. Dis. 2016;16:905–914. doi: 10.1016/S1473-3099(16)30023-8. [DOI] [PubMed] [Google Scholar]
- Greenberg H.B., Estes M.K. Rotaviruses: from pathogenesis to vaccination. Gastroenterology. 2009;136:1939–1951. doi: 10.1053/j.gastro.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Xie S., Chi Z., Zhang J., Liu Y., Zhang L., Zheng M., Zhang X., Xia D., Ke Y., et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 2016;45:802–816. doi: 10.1016/j.immuni.2016.09.008. [DOI] [PubMed] [Google Scholar]
- Hagan T., Cortese M., Rouphael N., Boudreau C., Linde C., Maddur M.S., Das J., Wang H., Guthmiller J., Zheng N.Y., et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178:1313–1328.e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J.A., Bouladoux N., Sun C.M., Wohlfert E.A., Blank R.B., Zhu Q., Grigg M.E., Berzofsky J.A., Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang S., Paik D., Yao L., Kim E., Trinath J., Lu J., Ha S., Nelson B.N., Kelly S.P., Wu L., et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. 2019;576:143–148. doi: 10.1038/s41586-019-1785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon P., Hanlon L., Marsh V., Byass P., Shenton F., Hassan-King M., Jobe O., Sillah H., Hayes R., M'Boge B.H. Trial of an attenuated bovine rotavirus vaccine (RIT 4237) in Gambian infants. Lancet. 1987;1:1342–1345. doi: 10.1016/s0140-6736(87)90649-0. [DOI] [PubMed] [Google Scholar]
- Harris V.C., Armah G., Fuentes S., Korpela K.E., Parashar U., Victor J.C., Tate J., de Weerth C., Giaquinto C., Wiersinga W.J., et al. Significant correlation between the infant gut microbiome and rotavirus vaccine response in Rural Ghana. J. Infect. Dis. 2017;215:34–41. doi: 10.1093/infdis/jiw518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris V., Ali A., Fuentes S., Korpela K., Kazi M., Tate J., Parashar U., Wiersinga W.J., Giaquinto C., de Weerth C., de Vos W.M. Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes. 2018;9:93–101. doi: 10.1080/19490976.2017.1376162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris V.C., Haak B.W., Handley S.A., Jiang B., Velasquez D.E., Hykes B.L., Jr., Droit L., Berbers G.A.M., Kemper E.M., van Leeuwen E.M.M., et al. Effect of antibiotic-mediated microbiome modulation on rotavirus vaccine immunogenicity: A human, randomized-control proof-of-Concept Trial. Cell Host Microbe. 2018;24:197–207.e4. doi: 10.1016/j.chom.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Huda M.N., Ahmad S.M., Alam M.J., Khanam A., Kalanetra K.M., Taft D.H., Raqib R., Underwood M.A., Mills D.A., Stephensen C.B. Bifidobacterium abundance in early infancy and vaccine response at 2 years of age. Pediatrics. 2019;143:e20181489. doi: 10.1542/peds.2018-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huda M.N., Lewis Z., Kalanetra K.M., Rashid M., Ahmad S.M., Raqib R., Qadri F., Underwood M.A., Mills D.A., Stephensen C.B. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134:e362–e372. doi: 10.1542/peds.2013-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur Y.G., Gorak-Stolinska P., Lalor M.K., Mvula H., Floyd S., Raynes J., Ben-Smith A., Fitchett J.R., Flanagan K.L., Burl S., et al. Factors affecting immunogenicity of BCG in infants, a study in Malawi, The Gambia and the UK. BMC Infect. Dis. 2014;14:184. doi: 10.1186/1471-2334-14-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T., Pang I.K., Kumamoto Y., Peaper D.R., Ho J.H., Murray T.S., Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isolauri E., Joensuu J., Suomalainen H., Luomala M., Vesikari T. Improved immunogenicity of oral D x RRV reassortant rotavirus vaccine by Lactobacillus casei GG. Vaccine. 1995;13:310–312. doi: 10.1016/0264-410x(95)93319-5. [DOI] [PubMed] [Google Scholar]
- Jiang W., Wang X., Zeng B., Liu L., Tardivel A., Wei H., Han J., MacDonald H.R., Tschopp J., Tian Z., Zhou R. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J. Exp. Med. 2013;210:2465–2476. doi: 10.1084/jem.20122490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy S., Chattha K.S., Vlasova A.N., Rajashekara G., Saif L.J. Lactobacilli and bifidobacteria enhance mucosal B cell responses and differentially modulate systemic antibody responses to an oral human rotavirus vaccine in a neonatal gnotobiotic pig disease model. Gut Microbes. 2014;5:639–651. doi: 10.4161/19490976.2014.969972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Kim Y.G., Seo S.U., Kim D.J., Kamada N., Prescott D., Chamaillard M., Philpott D.J., Rosenstiel P., Inohara N., Núñez G. Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin. Nat. Med. 2016;22:524–530. doi: 10.1038/nm.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Qie Y., Park J., Kim C.H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016;20:202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Voisin B., Kim D.Y., Kennedy E.A., Jo J.H., Shih H.Y., Truong A., Doebel T., Sakamoto K., Cui C.Y., et al. Homeostatic control of sebaceous glands by innate lymphoid cells regulates commensal bacteria equilibrium. Cell. 2019;176:982–997.e16. doi: 10.1016/j.cell.2018.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmann T.R., Kampmann B., Mazmanian S.K., Marchant A., Levy O. Protecting the newborn and young infant from infectious diseases: lessons from immune ontogeny. Immunity. 2017;46:350–363. doi: 10.1016/j.immuni.2017.03.009. [DOI] [PubMed] [Google Scholar]
- Kostic A.D., Xavier R.J., Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P., Blacher E., Elinav E., Pettersson S. Our gut microbiome: the evolving inner self. Cell. 2017;171:1481–1493. doi: 10.1016/j.cell.2017.11.024. [DOI] [PubMed] [Google Scholar]
- Laborel-Preneron E., Bianchi P., Boralevi F., Lehours P., Fraysse F., Morice-Picard F., Sugai M., Sato'o Y., Badiou C., Lina G., et al. Effects of the Staphylococcus aureus and Staphylococcus epidermidis secretomes isolated from the skin microbiota of atopic children on CD4+ T cell activation. PLoS One. 2015;10:e0141067. doi: 10.1371/journal.pone.0141067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Kolodziejczyk A.A., Thaiss C.A., Elinav E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017;17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- Li F., Hao X., Chen Y., Bai L., Gao X., Lian Z., Wei H., Sun R., Tian Z. The microbiota maintain homeostasis of liver-resident γδT-17 cells in a lipid antigen/CD1d-dependent manner. Nat. Commun. 2017;7:13839. doi: 10.1038/ncomms13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Sullivan N.L., Rouphael N., Yu T., Banton S., Maddur M.S., McCausland M., Chiu C., Canniff J., Dubey S., et al. Metabolic phenotypes of response to vaccination in humans. Cell. 2017;169:862–877.e17. doi: 10.1016/j.cell.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Innocentin S., Withers D.R., Roberts N.A., Gallagher A.R., Grigorieva E.F., Wilhelm C., Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Lynn M.A., Tumes D.J., Choo J.M., Sribnaia A., Blake S.J., Leong L.E.X., Young G.P., Marshall H.S., Wesselingh S.L., Rogers G.B., Lynn D.J. Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe. 2018;23:653–660.e5. doi: 10.1016/j.chom.2018.04.009. [DOI] [PubMed] [Google Scholar]
- Macia L., Tan J., Vieira A.T., Leach K., Stanley D., Luong S., Maruya M., Ian McKenzie C., Hijikata A., Wong C., et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- Macpherson A.J., Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- Madhi S.A., Cunliffe N.A., Steele D., Witte D., Kirsten M., Louw C., Ngwira B., Victor J.C., Gillard P.H., Cheuvart B.B., et al. Effect of human rotavirus vaccine on severe diarrhea in African infants. N. Engl. J. Med. 2010;362:289–298. doi: 10.1056/NEJMoa0904797. [DOI] [PubMed] [Google Scholar]
- Matsuda F., Chowdhury M.I., Saha A., Asahara T., Nomoto K., Tarique A.A., Ahmed T., Nishibuchi M., Cravioto A., Qadri F. Evaluation of a probiotics, Bifidobacterium breve BBG-01, for enhancement of immunogenicity of an oral inactivated cholera vaccine and safety: a randomized, double-blind, placebo-controlled trial in Bangladeshi children under 5 years of age. Vaccine. 2011;29:1855–1858. doi: 10.1016/j.vaccine.2010.12.133. [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Liu C.H., Tzianabos A.O., Kasper D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mitre E., Susi A., Kropp L.E., Schwartz D.J., Gorman G.H., Nylund C.M. Association Between use of acid-suppressive medications and antibiotics during infancy and allergic diseases in early childhood. JAMA Pediatr. 2018;172:e180315. doi: 10.1001/jamapediatrics.2018.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyanja E., Ssemaganda A., Ngauv P., Cubas R., Perrin H., Srinivasan D., Canderan G., Lawson B., Kopycinski J., Graham A.S., et al. Immune activation alters cellular and humoral responses to yellow fever 17D vaccine. J. Clin. Invest. 2014;124:3147–3158. doi: 10.1172/JCI75429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S., Bouladoux N., Wilhelm C., Molloy M.J., Salcedo R., Kastenmuller W., Deming C., Quinones M., Koo L., Conlan S., et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya H.I., Hagan T., Duraisingham S.S., Lee E.K., Kwissa M., Rouphael N., Frasca D., Gersten M., Mehta A.K., Gaujoux R., et al. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity. 2015;43:1186–1198. doi: 10.1016/j.immuni.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Norton E.B., Lawson L.B., Freytag L.C., Clements J.D. Characterization of a mutant Escherichia coli heat-labile toxin, LT(R192G/L211A), as a safe and effective oral adjuvant. Clin. Vaccine Immunol. 2011;18:546–551. doi: 10.1128/CVI.00538-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J.Z., Ravindran R., Chassaing B., Carvalho F.A., Maddur M.S., Bower M., Hakimpour P., Gill K.P., Nakaya H.I., Yarovinsky F., et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olin A., Henckel E., Chen Y., Lakshmikanth T., Pou C., Mikes J., Gustafsson A., Bernhardsson A.K., Zhang C., Bohlin K., Brodin P. Stereotypic immune system development in newborn children. Cell. 2018;174:1277–1292.e14. doi: 10.1016/j.cell.2018.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker E.P.K., Praharaj I., Zekavati A., Lazarus R.P., Giri S., Operario D.J., Liu J., Houpt E., Iturriza-Gómara M., Kampmann B., et al. Influence of the intestinal microbiota on the immunogenicity of oral rotavirus vaccine given to infants in south India. Vaccine. 2018;36:264–272. doi: 10.1016/j.vaccine.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M., Pedreira C., De Oliveira L.H., Tate J., Orozco M., Mercado J., Gonzalez A., Malespin O., Amador J.J., Umaña J., et al. Association between pentavalent rotavirus vaccine and severe rotavirus diarrhea among children in Nicaragua. JAMA. 2009;301:2243–2251. doi: 10.1001/jama.2009.756. [DOI] [PubMed] [Google Scholar]
- Patrick D.M., Sbihi H., Dai D.L.Y., Al Mamun A., Rasali D., Rose C., Marra F., Boutin R.C.T., Petersen C., Stiemsma L.T., et al. Decreasing antibiotic use, the gut microbiota, and asthma incidence in children: evidence from population-based and prospective cohort studies. Lancet Respir. Med. 2020:30052–30057. doi: 10.1016/S2213-2600. [DOI] [PubMed] [Google Scholar]
- Pfeiffer J.K., Virgin H.W. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science. 2016;351:aad5872. doi: 10.1126/science.aad5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praharaj I., John S.M., Bandyopadhyay R., Kang G. Probiotics, antibiotics and the immune responses to vaccines. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20140144. doi: 10.1098/rstb.2014.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praharaj I., Parker E.P.K., Giri S., Allen D.J., Silas S., Revathi R., Kaliappan S.P., John J., Prasad J.H., Kampmann B., et al. Influence of nonpolio enteroviruses and the bacterial gut microbiota on oral poliovirus vaccine response: a study from South India. J. Infect. Dis. 2019;219:1178–1186. doi: 10.1093/infdis/jiy568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat. Rev. Immunol. 2009;9:741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- Pulendran B. Systems vaccinology: probing humanity's diverse immune systems with vaccines. Proc. Natl. Acad. Sci. USA. 2014;111:12300–12306. doi: 10.1073/pnas.1400476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadri F., Bhuiyan T.R., Sack D.A., Svennerholm A.M. Immune responses and protection in children in developing countries induced by oral vaccines. Vaccine. 2013;31:452–460. doi: 10.1016/j.vaccine.2012.11.012. [DOI] [PubMed] [Google Scholar]
- Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D., Pirani A., Gernert K., Deng J., Marzolf B., et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelli S., Candela M., Turroni S., Biagi E., Collino S., Franceschi C., O'Toole P.W., Brigidi P. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging (Albany, NY) 2013;5:902–912. doi: 10.18632/aging.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J.M., Kang D.J., Hylemon P.B., Bajaj J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C.M., Pfeiffer J.K. Viruses and the microbiota. Annu. Rev. Virol. 2014;1:55–69. doi: 10.1146/annurev-virology-031413-085550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillère R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- San-Cristobal R., Navas-Carretero S., Martínez-González M.Á., Ordovas J.M., Martínez J.A. Contribution of macronutrients to obesity: implications for precision nutrition. Nat. Rev. Endocrinol. 2020;16:305–320. doi: 10.1038/s41574-020-0346-8. [DOI] [PubMed] [Google Scholar]
- Sandler N.G., Douek D.C. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat. Rev. Microbiol. 2012;10:655–666. doi: 10.1038/nrmicro2848. [DOI] [PubMed] [Google Scholar]
- Santos Rocha C., Hirao L.A., Weber M.G., Méndez-Lagares G., Chang W.L.W., Jiang G., Deere J.D., Sparger E.E., Roberts J., Barry P.A., et al. Subclinical Cytomegalovirus infection is associated with altered host immunity, gut microbiota, and vaccine responses. J. Virol. 2018;92 doi: 10.1128/JVI.00167-18. e00167-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher J.U., Sczesnak A., Longman R.S., Segata N., Ubeda C., Bielski C., Rostron T., Cerundolo V., Pamer E.G., Abramson S.B., et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Jia T., Mendez-Ferrer S., Hohl T.M., Serbina N.V., Lipuma L., Leiner I., Li M.O., Frenette P.S., Pamer E.G. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity. 2011;34:590–601. doi: 10.1016/j.immuni.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Zou J., Zhang Z., Zhao X., Noriega J., Zhang B., Zhao C., Ingle H., Bittinger K., Mattei L.M., et al. Segmented filamentous bacteria prevent and cure rotavirus infection. Cell. 2019;179:644–658.e13. doi: 10.1016/j.cell.2019.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissoko M.S., Healy S.A., Katile A., Omaswa F., Zaidi I., Gabriel E.E., Kamate B., Samake Y., Guindo M.A., Dolo A., et al. Safety and efficacy of PfSPZ Vaccine against Plasmodium falciparum via direct venous inoculation in healthy malaria-exposed adults in Mali: a randomised, double-blind phase 1 trial. Lancet Infect. Dis. 2017;17:498–509. doi: 10.1016/S1473-3099(17)30104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabytska Y., Wölbing F., Günther C., Köberle M., Kaesler S., Chen K.M., Guenova E., Demircioglu D., Kempf W.E., Volz T., et al. Cutaneous innate immune sensing of Toll-like receptor 2–6 ligands suppresses T cell immunity by inducing myeloid-derived suppressor cells. Immunity. 2014;41:762–775. doi: 10.1016/j.immuni.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Soh S.E., Ong D.Q., Gerez I., Zhang X., Chollate P., Shek L.P., Lee B.W., Aw M. Effect of probiotic supplementation in the first 6 months of life on specific antibody responses to infant hepatitis B vaccination. Vaccine. 2010;28:2577–2579. doi: 10.1016/j.vaccine.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Sonnenburg J.L., Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy A., Belkaid Y. Microbial guardians of skin health. Science. 2019;363:227–228. doi: 10.1126/science.aat4326. [DOI] [PubMed] [Google Scholar]
- Staudinger J.L., Goodwin B., Jones S.A., Hawkins-Brown D., MacKenzie K.I., LaTour A., Liu Y., Klaassen C.D., Brown K.K., Reinhard J., et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelekati E., Wherry E.J. Chronic bystander infections and immunity to unrelated antigens. Cell Host Microbe. 2012;12:458–469. doi: 10.1016/j.chom.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J.E., Burton A.H., Boschi-Pinto C., Steele A.D., Duque J., Parashar U.D., WHO-coordinated Global Rotavirus Surveillance Network 2008 Estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect. Dis. 2012;12:136–141. doi: 10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- Thevaranjan N., Puchta A., Schulz C., Naidoo A., Szamosi J.C., Verschoor C.P., Loukov D., Schenck L.P., Jury J., Foley K.P., et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21:455–466.e4. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trama A.M., Moody M.A., Alam S.M., Jaeger F.H., Lockwood B., Parks R., Lloyd K.E., Stolarchuk C., Scearce R., Foulger A., et al. HIV-1 envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe. 2014;16:215–226. doi: 10.1016/j.chom.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura Y., Fuhrer T., Li H., Lawson M.A., Zimmermann M., Yilmaz B., Zindel J., Ronchi F., Sorribas M., Hapfelmeier S., et al. Antibodies set boundaries limiting microbial metabolite penetration and the resultant mammalian host response. Immunity. 2018;49:545–559.e5. doi: 10.1016/j.immuni.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama R., Chassaing B., Zhang B., Gewirtz A.T. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J. Infect. Dis. 2014;210:171–182. doi: 10.1093/infdis/jiu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams W.B., Han Q., Haynes B.F. Cross-reactivity of HIV vaccine responses and the microbiome. Curr. Opin. HIV AIDS. 2018;13:9–14. doi: 10.1097/COH.0000000000000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Draft landscape of COVID-19 candidate vaccines. 2020. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines
- Zaman K., Dang D.A., Victor J.C., Shin S., Yunus M., Dallas M.J., Podder G., Vu D.T., Le T.P., Luby S.P., et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;376:615–623. doi: 10.1016/S0140-6736(10)60755-6. [DOI] [PubMed] [Google Scholar]
- Zeng M.Y., Cisalpino D., Varadarajan S., Hellman J., Warren H.S., Cascalho M., Inohara N., Núñez G. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang Z. Gut microbiome and cardiovascular disease. Curr. Opin. Cardiol. 2020;35:207–218. doi: 10.1097/HCO.0000000000000720. [DOI] [PMC free article] [PubMed] [Google Scholar]