ABSTRACT
Bacterial endophytes are found in the internal tissues of plants and have intimate associations with their host. However, little is known about the diversity of medicinal plant endophytes (ME) or their capability to produce specialised metabolites that may contribute to therapeutic properties. We isolated 75 bacterial ME from 24 plant species of the Western Ghats, India. Molecular identification by 16S rRNA gene sequencing grouped MEs into 13 bacterial genera, with members of Gammaproteobacteria and Firmicutes being the most abundant. To improve taxonomic identification, 26 selected MEs were genome sequenced and average nucleotide identity (ANI) used to identify them to the species-level. This identified multiple species in the most common genus as Bacillus. Similarly, identity of the Enterobacterales was also distinguished within Enterobacter and Serratia by ANI and core-gene analysis. AntiSMASH identified non-ribosomal peptide synthase, lantipeptide and bacteriocin biosynthetic gene clusters (BGC) as the most common BGCs found in the ME genomes. A total of five of the ME isolates belonging to Bacillus, Serratia and Enterobacter showed antimicrobial activity against the plant pathogen Pectobacterium carotovorum. Using molecular and genomic approaches we have characterised a unique collection of endophytic bacteria from medicinal plants. Their genomes encode multiple specialised metabolite gene clusters and the collection can now be screened for novel bioactive and medicinal metabolites.
Keywords: endophytic bacteria, antimicrobials, Bacillus, medicinal plants, bacterial genomes, biosynthetic gene clusters
A collection of bacterial endophytes isolated from a number of medicinal plants of the Western Ghats, India were investigated for their capability to produce specialised metabolites that may contribute to therapeutic properties.
INTRODUCTION
Multiple countries use indigenous plants as traditional remedies for treatment of injury or disease. In the Indian traditional medicinal system of Ayurveda and other similar practices, leaves, roots, seeds and fruits are commonly used as alternative medicines. Garcinia indica (Baliga et al. 2011), Salacia chinensis (Deokate and Khadabadi 2012) and Alstonia scholaris (Ganjewala and Gupta 2013) are examples of Indian medicinal plant species described to have multiple therapeutic properties. Garcinia indica, commonly known as the kokum tree, produces fruits which are used in Ayurvedic medicine for its antimicrobial, antiulcer, anticancer and antiobesity properties, as well as being able to ease inflammatory and pain-related issues (Baliga et al. 2011). The roots of the Salacia chinensis herb tree have also been exploited for beneficial properties in treating tooth decay, ulcers, obesity and skin conditions (Deokate and Khadabadi 2012). Multiple parts of the Indian devil tree, Alstonia scholaris, such as leaves, follicles and latex show extensive antimicrobial and antioxidant properties (Ganjewala and Gupta 2013). Recently, the medicinal plant-associated microbiome, and especially the interaction between the complex community of endophytic microorganisms (endomicrobiome; Köberl et al. 2013) have been attributed to these antimicrobial (Martinez-Klimova, Rodríguez-Peña and Sánchez 2017) and bioactive properties through the metabolites they produce (Gouda et al. 2016; Ek-Ramos et al. 2019). Endophytic bacteria isolated from traditional Chinese medicinal plants used as anticancer therapy were screened for bioactivity and all isolates exhibited either cytotoxic, antibacterial or antifungal activities in at least one assay (Miller et al. 2012b).
Endophytic microorganisms (endophytes) are bacteria or fungi that colonise the intercellular and/or intracellular spaces of plants, often living in a symbiotic relationship (Hardoim et al. 2015). Endophytes are known to promote plant growth and nutrient gain, improve yield and aid the plant to survive in harsh conditions when under stress or attack from pathogens (Ryan et al. 2008; Hardoim et al. 2015; Santoyo et al. 2016). It is thought for this reason many endophytes produce a range of unique specialised metabolites, such as peptides, polyketides and alkaloids, to aid the plants immune response and prevent colonisation by pathogens and other microbes. Natural products from endophytes frequently possess bioactivities such as antimicrobial, antifungal, anticarcinogen, immunosuppressant and antioxidant (Zhang, Song and Tan 2006; Akinsanya et al. 2015; Sharma et al. 2020) and their investigation offers huge potential in identifying new pharmaceutical compounds.
However, whilst nearly all plants are thought to contain endophytes, very little is known about the diversity of endophytes in traditional Indian medicinal plant species. India is considered to be one of the 16 mega diversity countries in the world with around 17 500 higher plants species, of which 4050 plants are found in the Western Ghats (Pascal, Ramesh and De Franceschi 2004), many of which are used in the treatment of infection, disease, wounds and injuries (Ayyanar and Ignacimuthu2011). In this study we aimed to determine the culturable diversity of bacterial endophytes present within a large collection of plant species taken from the Western Ghats region. Plants were chosen based on their ethnobotanical usage, being endemic to the region or were found growing within biodiversity rich areas (Strobel and Daisy 2003). Endophytes were then isolated from the leaves (the main plant part of medicinal value) from 24 plant species, initially identified by 16S rRNA gene analysis, and then followed up with whole genome sequencing for finer resolution of their taxonomy. Selected endophytes were also investigated further for their specialised metabolite potential via a genome mining and antimicrobial bioactivity analysis.
MATERIALS AND METHODS
Sample site and plant material collection
The Western Ghats (or Sahyadri) is a mountain region that covers an area of around 140 000 km2 and 1600 km in length running parallel to the western coast of the Indian peninsula from the river Tapti in the North to Kanyakumari in the South (Reddy, Jha and Dadhwal 2016). It traverses parts of six states, Kerala, Tamil Nadu, Karnataka, Goa, Maharashtra and Gujarat and is a UNESCO World Heritage Site and one of the eight "hottest hotspots" of biological diversity in the world (Myers et al. 2000). It has non-equatorial tropical evergreen forests which hosts at least 325 globally threatened species of flora and fauna (UNESCO). The database on ethnomedicinal plants of Western Ghats lists 500 plants from 115 families that have been used to prepare around 600 different medicinal formulations as listed by the Indian Council of Medical Research (Project by SD Kholkute, 2005–2008, submitted to ICMR). However, it is estimated that the true number of medicinal plants in the Western Ghats is >700 species with many being endemic and listed as endangered in the International Union for Conservation of Nature (IUCN) Red List of threatened species (https://www.iucnredlist.org/).
Leaf samples (and one fruit sample) from 24 plant species (covering 19 plant genera; Figure S1a and Table S1, Supporting Information) were collected from two sites of the Western Ghats and one site of Mysore in Karnataka, India between 5th July and 28th August 2017. All samples were then transported to the laboratory in sterile polypropylene bags and processed within 24 h of collection. Each plant was identified by referring to literature, herbarium specimens, consulting with taxonomists and searching databases including The Western Ghats (India Biodiversity Portal), Sahyadri (Western Ghats Biodiversity Information System) and Digital Flora of Karnataka. Samples of the plant species were preserved in the Herbarium of Department of Studies in Microbiology, University of Mysore, India.
Isolation of endophytic bacteria
Samples were washed with distilled water and surface sterilised using the following procedure: 0.1% (w/v) HgCl2 solution for 1 min, sterile water for 1 min, 90% (v/v) ethanol for 2 min and finally washed again with sterile water. Leaf samples were then cut into segments of approximately 0.5 cm2 using a sterile scalpel and placed onto Luria-Bertani (LB) agar (HiMedia Laboratories, Mumbai, India) plates and incubated at ambient temperature in the dark. To ensure bacterial growth was only obtained from plant endophytes, one additional LB agar plate for each plant was also incubated with uncut surface sterilised leaves as a control (Martinez-Klimova, Rodríguez-Peña and Sánchez 2017). No growth from plant epiphytic bacteria was observed. During incubation, inoculated plates were frequently observed for bacterial growth at the cut-ends of the leaf tissue and emerging bacteria were transferred onto fresh LB agar. Bacterial endophytes were streaked, and individual colonies were selected and sub-cultured three times to obtain pure bacterial cultures on LB agar. Each bacterial isolate was transferred separately to LB agar slopes and stored at 4°C for further study. The cultures were maintained at the University of Mysore for characterisation and elucidation of bioactive compounds, while phylogenetic analysis and genome sequencing of endophytes was performed at Cardiff University. All bacterial cultures isolated in this study are available from the laboratory collection held at the Department of Studies in Microbiology, University of Mysore, India by request from the corresponding author.
For molecular characterisation analysis, bacterial isolates were revived on TSA (Tryptone Soy agar; Oxoid, Basingstoke, UK) plates at 30°C, sub-cultured three times and checked for purity, except isolates ME7 and ME8 which grew better on Reasoner's 2A agar (R2A agar, Oxoid). Pure cultures were stored at −80°C in 8% (v/v) dimethyl sulfoxide (DMSO) and tryptone soya broth (TSB) or R2A.
16S rRNA gene diversity and phylogenetic analysis of bacterial endophytes
DNA was extracted from 10 µL of an overnight culture (grown in TSB or R2A at 30°C) with 100 µL of 5% (w/v) Chelex 100 resin (Walsh, Metzger and Higuchi 1991) by undergoing two cycles of boiling and freezing (5 mins each) as described (Parkes et al. 2010). The crude DNA extract was then used as template in a 16S rRNA gene PCR with bacterial primers 27F and 907R (Webster et al. 2006). All 16S rRNA gene PCR amplicons were analysed by 1.2% (w/v) agarose gel electrophoresis, purified and sequenced at Eurofins Genomics (https://www.eurofinsgenomics.eu/en/home/) by Sanger sequencing with primer 27F. Sequence chromatograms were analysed using Chromas version 2.6.6 (http://technelysium.com.au) and mixed sequences (suggestive that some isolates were not pure) were removed from further analysis resulting in 75 pure endophytic bacterial isolates (see Table 1).
Table 1.
List of bacterial endophytes isolated from leaves of medicinal plant species sampled at different locations of the Western Ghats, Karnataka, India.
| Endophytic bacteriuma | Plant species isolated from | Sampling location | Identification by 16S rRNA gene similarity | Identification by average nucleotide identity (ANI) | Identification by Type strain Genome Server (TYGS) |
|---|---|---|---|---|---|
| ME1 | Memecylon malabaricum | Bisle Ghat region | Serratia sp. | ||
| ME3 | Memecylon malabaricum | Bisle Ghat region | Serratia sp. | ||
| ME4 | Aphanamixis polystachya | Bisle Ghat region | Bacillus sp. | ||
| ME5 | Aphanamixis polystachya | Bisle Ghat region | Bacillus sp. | Bacillus thuringiensis | Bacillus paranthracis |
| ME6 | Terminalia bellirica | Bisle Ghat region | Bacillus sp. | ||
| ME7 | Terminalia bellirica | Bisle Ghat region | Aureimonas sp. | Aureimonas sp. | Aureimonas sp. |
| ME8 | Terminalia bellirica | Bisle Ghat region | Aureimonas sp. | ||
| ME9 | Terminalia bellirica | Bisle Ghat region | Bacillus sp. | ||
| ME10 | Ventilago sp. | Bisle Ghat region | Enterobacter sp. | ||
| ME11 | Ventilago sp. | Bisle Ghat region | Bacillus sp. | ||
| ME12 | Terminalia paniculata | Bisle Ghat region | Curtobacterium sp. | Curtobacterium sp. | Curtobacterium sp. |
| ME13 | Terminaliapaniculata | Bisle Ghat region | Enterobacter sp. | Enterobacterbugandensis | Enterobacterbugandensis |
| ME14 | Aristolochia tagala | Bisle Ghat region | Enterobacter sp. | ||
| ME15 | Aristolochia tagala | Bisle Ghat region | Klebsiella sp. | ||
| ME16 | Aristolochia tagala | Bisle Ghat region | Enterobacter sp. | ||
| ME17A | Aristolochia tagala | Bisle Ghat region | Klebsiella sp. | ||
| ME18 | Aristolochia tagala | Bisle Ghat region | Enterobacter sp. | ||
| ME19 | Aristolochia tagala | Bisle Ghat region | Enterobacter sp. | ||
| ME20 | Aristolochia tagala | Bisle Ghat region | Bacillus sp. | ||
| ME21 | Aristolochia tagala | Bisle Ghat region | Klebsiella sp. | ||
| ME23 | Garcinia xanthochymus | Bisle Ghat region | Bacillus sp. | ||
| ME25 | Aphanamixis polystachya | Bisle Ghat region | Bacillus sp. | Bacillus taxi | Bacillus taxi |
| ME26 | Ventilago sp. | Bisle Ghat region | Curtobacterium sp. | Curtobacterium sp. | Curtobacterium sp. |
| ME27 | Aphanamixis polystachya | Bisle Ghat region | Acinetobacter sp. | Acinetobacter lactucae | Acinetobacter lactucae |
| ME28 | Salacia macrosperma | Bisle Ghat region | Bacillus sp. | ||
| ME29 | Salacia macrosperma | Bisle Ghat region | Bacillus sp. | ||
| ME30 | Garcinia xanthochymus | Bisle Ghat region | Klebsiella sp. | Klebsiella pneumoniae | Klebsiella pneumoniae |
| ME31 | Garcinia xanthochymus | Bisle Ghat region | Enterobacter sp. | ||
| ME32 | Ventilago sp. | Bisle Ghat region | Bacillus sp. | ||
| ME33 | Ventilago sp. | Bisle Ghat region | Brevibacillus sp. | ||
| ME34 | Ventilago sp. | Bisle Ghat region | Enterobacter sp. | Enterobacter bugandensis | Enterobacter bugandensis |
| ME35 | Terminalia bellirica | Bisle Ghat region | Bacillus sp. | Bacillus licheniformis | Bacillus licheniformis |
| ME36 | Terminalia paniculata | Bisle Ghat region | Bacillus sp. | ||
| ME38 | Ventilago sp. | Bisle Ghat region | Bacillus sp. | ||
| ME39 | Pterocarpus santalinus | Mysore | Bacillus sp. | Bacillus aryabhattai | Bacillus aryabhattai |
| ME40 | Garcinia indica | Mysore | Bacillus sp. | Bacillus megaterium | Bacillus sp. |
| ME42 | Pterocarpus santalinus | Mysore | Bacillus sp. | Bacillus aryabhattai | Bacillus aryabhattai |
| ME43 | Coscinium fenestratum | Mangaluru | Serratia sp. | Serratia marcescens | Serratia sp. |
| ME44 | Coscinium fenestratum | Mangaluru | Enterobacter sp. | Enterobacter asburiae | Enterobacter asburiae |
| ME45 | Coscinium fenestratum | Mangaluru | Enterobacter sp. | ||
| ME46 | Coscinium fenestratum | Mangaluru | Enterobacter sp. | ||
| ME47 | Coscinium fenestratum | Mangaluru | Serratia sp. | Serratia marcescens | Serratia sp. |
| ME51 | Coscinium fenestratum | Mangaluru | Stenotrophomonas sp. | ||
| ME53 | Coix lacryma-jobi | Mangaluru | Stenotrophomonas sp. | ||
| ME55 | Coix lacryma-jobi | Mangaluru | Stenotrophomonas sp. | Stenotrophomonas pavanii | Stenotrophomonas pavanii |
| ME56 | Coix lacryma-jobi | Mangaluru | Paenibacillus sp. | ||
| ME57 | Coix lacryma-jobi | Mangaluru | Klebsiella sp. | ||
| ME60 | Coix lacryma-jobi | Mangaluru | Bacillus sp. | ||
| ME62 | Coix lacryma-jobi | Mangaluru | Enterobacter sp. | ||
| ME63 | Coix lacryma-jobi | Mangaluru | Pseudomonas sp. | Pseudomonas sp. | Pseudomonas sp. |
| ME64 | Coix lacryma-jobi | Mangaluru | Enterobacter sp. | ||
| ME66 | Coix lacryma-jobi | Mangaluru | Klebsiella sp. | ||
| ME67 | Coix lacryma-jobi | Mangaluru | Stenotrophomonas sp. | ||
| ME68 | Salacia chinensis | Mangaluru | Klebsiella sp. | ||
| ME70 | Salacia chinensis | Mangaluru | Klebsiella sp. | ||
| ME71 | Salacia chinensis | Mangaluru | Stenotrophomonas sp. | ||
| ME72 | Salacia chinensis | Mangaluru | Klebsiella sp. | ||
| ME73 | Salacia chinensis | Mangaluru | Klebsiella sp. | Klebsiella variicola | Klebsiella variicola |
| ME74 | Salacia chinensis | Mangaluru | Enterobacter sp. | ||
| ME75 | Calophyllum inophyllum | Mangaluru | Bacillus sp. | Bacillus aryabhattai | Bacillus sp. |
| ME76 | Calophyllum inophyllum | Mangaluru | Bacillus sp. | Bacillus aryabhattai | Bacillus sp. |
| ME78 | Madhuca insignis | Mangaluru | Bacillus sp. | Bacillus thuringiensis | Bacillus cereus |
| ME79 | Madhuca insignis | Mangaluru | Klebsiella sp. | Klebsiella variicola | Klebsiella variicola |
| ME81 | Garcinia morella | Mangaluru | Erwinia sp. | Pantoea sp. | Pantoea sp. |
| ME83 | Apama siliquosa | Mangaluru | Klebsiella sp. | ||
| ME84 | Apama siliquosa | Mangaluru | Stenotrophomonas sp. | ||
| ME86 | Desmodium pulchellum | Mangaluru | Klebsiella sp. | Klebsiella variicola | Klebsiella variicola |
| ME87 | Barringtonia acutangula | Mangaluru | Enterobacter sp. | ||
| ME89 | Barringtonia acutangula fruit | Mangaluru | Enterobacter sp. | ||
| ME90 | Alstonia scholaris | Mangaluru | Enterobacter sp. | ||
| ME91 | Alstonia scholaris | Mangaluru | Enterobacter sp. | ||
| ME92 | Alstonia scholaris | Mangaluru | Enterobacter sp. | ||
| ME93 | Alstonia scholaris | Mangaluru | Pseudomonas sp. | ||
| ME94 | Alstonia scholaris | Mangaluru | Methylobacterium sp. | Methylobacterium radiotolerans | Methylobacterium radiotolerans |
| ME95 | Alstonia scholaris | Mangaluru | Pseudomonas sp. |
Genome sequenced medicinal plant endophytes (ME) are highlighted in bold font
Bacterial 16S rRNA gene sequences were analysed using Nucleotide BLAST implemented on the NCBI server (https://blast.ncbi.nlm.nih.gov) against the nucleotide collection (nr/nt) and the 16S ribosomal RNA sequences databases to identify closest relatives. Sequences were assigned to various operational taxonomic units (OTUs) by using BLASTClust (http://www.ncbi.nlm.nih.gov/) at 95% similarity, representing a genus level grouping (Schloss and Handelsman 2004). Diversity measurements including rarefaction curves, coverage, Shannon's and Simpson's indices of diversity and species richness (SChao1) were calculated using the Past software package v3.14 (Hammer, Harper and Ryan 2001).
All 16S rRNA gene sequences were aligned using MAFFT v7 online (Katoh, Rozewicki and Yamada 2019) with sequences retrieved from the database. Alignments were edited manually using BioEdit (Hall 1999) and phylogenetic trees were constructed using MEGA7 (Kumar, Stecher and Tamura 2016) by using the Maximum Likelihood method with the General Time Reversible model and Gamma distribution. Congruent trees were also obtained using other methods, including minimum evolution and LogDet distance, neighbour-joining with Jukes-Cantor algorithm.
Bacterial genome sequencing and assembly
Bacterial genomic DNA was extracted from isolates of interest (n = 26), identified by 16S rRNA gene sequencing, from a 3 mL overnight culture grown in TSB or R2A at 30°C. Cells were collected by centrifugation at 4000 rpm using ALC PK120 Centrifuge for 10 min, resuspended in 4M guanidinium Isothiocyanate and DNA extracted using an automated Maxwell® 16 Instrument with Tissue DNA Purification Kits (Promega UK Ltd, Southampton, UK) according to the manufacturer's instructions. DNA was quantified using a Qubit 3.0 Fluorometer, and libraries prepared for 250 bp nucleotide paired-end sequencing using the NEBNext® Ultra II DNA Library Prep Kit for Illumina. Genome libraries were then sequenced by an Illumina MiSeq platform.
Sequence reads were trimmed from Illumina adaptors using the TrimGalore v0.4.2 script (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and paired reads were merged with FLASH v1.2.11 (Magoc and Salzberg 2011). Genomes were assembled with SPAdes v3.13.0, and mis-assemblies corrected using Pilon v1.22 (Bankevich et al. 2012; Walker et al. 2014).
Bacterial genome and 16S rRNA gene sequences reported in this study have been submitted to the European Nucleotide Archive (ENA) under the project/study accession number PRJEB37902.
Species Identification of bacterial endophyte genomes
To allow species identification of the 26 bacterial endophyte genomes, the genus of each bacterial isolate was initially assigned by 16S rRNA gene comparison to the NCBI BLAST database coupled with genome identification using the taxonomic sequence classification system Kraken2 v2.0.6-beta and RefSeq complete bacterial genomes. Using this preliminary identification as a guide, full species assignment was then achieved by combining average nucleotide identity (ANI) and core-gene phylogenomics. The MinHash-based ANI tool FastANI (Jain et al. 2018) was used to identify RefSeq genomes of the same genus with high sequence similarity to each endophyte isolate. RefSeq genomes of each genus were downloaded using a NCBI genome download script available at GitHub (https://github.com/kblin/ncbi-genome-download). Up to 30 genomes with >90% sequence identity to each isolate, in addition to other endophyte isolates of the same genus, were passed to an alignment-based ANI tool, PyANI (Pritchard et al. 2016) for enhanced ANI accuracy. A core-gene phylogeny was constructed for each genus comprising genomes (Refseq and endophyte isolates) with >95% sequence identity to a given isolate, in addition to type strains and additional species representatives. Core-gene alignments were generated with Roary v3.13.0 (Page et al. 2015) implementing MAFFT v7.407 (Katoh and Standley 2013) and using genome annotations produced with Prokka v1.12. Maximum-likelihood phylogenetic trees were constructed using RaxML v8.2.12 with a general time reversible substitution model and gamma model of rate heterogeneity; and visualised with FigTree (http://tree.bio.ed.ac.uk/software/). In addition, for comparison genome sequences were also uploaded to the Type (strain) Genome Server (TYGS) bioinformatics platform available (https://tygs.dsmz.de) for whole genome-based taxonomic analysis (Meier-Kolthoff and Göker 2019). This platform provides both species assignment and digital DNA–DNA hybridisation (dDDH) values to the closest type strain genomes available.
Assessing biosynthetic gene cluster potential of whole-genome sequenced endophytes
To ascertain the biosynthetic gene cluster (BGC) potential of bacterial endophytes the genomes were analysed with the specialised metabolite predicting software antiSMASH v4.0 (Blin et al. 2017). Following BGC prediction, the sequences were extracted for de-replication to understand the overall biosynthetic diversity of the endophyte genome collection. BGC sequences were grouped according to genus and de-replicated using a pairwise k-mer-based comparison with Mash v2.2 (Ondov et al. 2016) and applying a maximum distance threshold of 0.24. De-replication was performed with the assumption that BGCs would not be shared between the different genera. The resulting distance network was visualised using Cytoscape v3.4.0 (Shannon 2003). Manual curation of the network was required to identify instances of BGCs split across multiple contigs and erroneously predicted hybrid BGCs due to close genomic locus proximity.
In vitro antagonism assays
Genome-sequenced medicinal plant endophytes (ME) were tested for antimicrobial activity against a small panel of human and plant pathogens (Pectobacterium carotovorum LMG 2464; Staphylococcus aureus NCTC 12981; Candida albicans SC5314) using an agar overlay inhibition assay as described (Mullins et al. 2019). In brief, ME isolates were grown overnight at 30°C on agar-solidified basal salts medium supplemented with glycerol (BSMG). After 24 h growth, a 10 µL-sized loopful of bacteria was resuspended in 1 mL phosphate buffered saline (PBS) buffer, spotted (3 µL volume) onto BSMG plates and incubated at 30°C for 48 h. ME isolates were killed by chloroform exposure for 2 mins, overlaid with pathogen-seeded half-strength iso-sensitest agar (Oxoid) supplemented with 0.2% (w/v) triphenyl tetrazolium chloride and incubated at 30°C or 37°C for 24 h.
RESULTS
Medicinal plants from the Western Ghats contain high diversity of bacterial endophytes
A total of 26 different medicinal plant samples (Table S1 and Figure S1a, Supporting Information) were taken from two sites in the Western Ghats and one site in Mysore, India. This represented one of the largest surveys of bacterial endophytes in Indian plants used for multiple medicinal purposes (Table S1, Supporting Information). The incubation of inoculated leaf tissue samples on LB agar readily enabled the growth of culturable endophytes from medicinal plants (Figure S1b, Supporting Information). During incubation, visible colonies were easily distinguishable on the edges of the leaf sections. After further subculture and incubation, 95 plant endophyte cultures were collected. These cultures were then further purified on TSA/R2A and checked for purity using 16S rRNA gene sequencing which resulted in 75 pure cultures of medicinal plant endophytes (designated as ME isolates). The assembled pure bacterial collection included 50 ME Gram-negative and 25 ME Gram-positive bacterial isolates (Table 1).
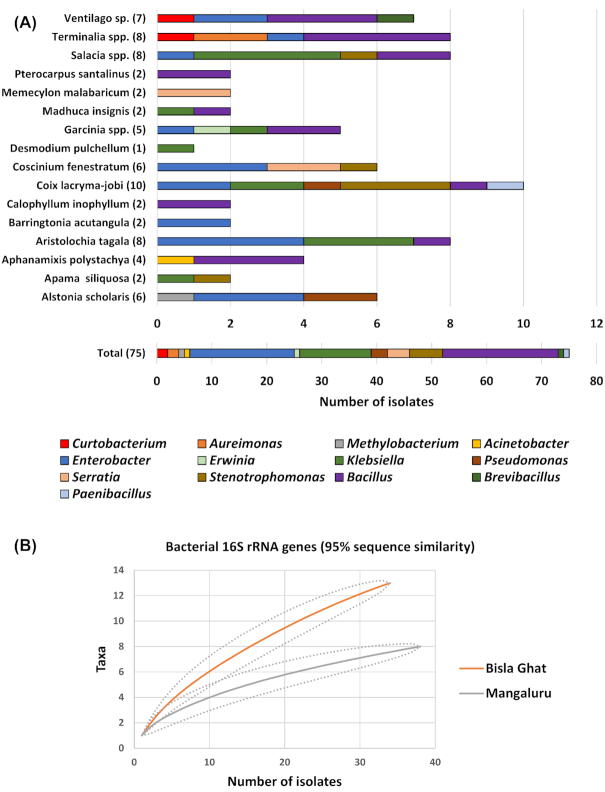
Overall, from the three locations sampled (Bisle Ghat, Mysore and Mangaluru) pure culturable endophytes were isolated from 20 plant species covering 16 plant genera (Fig. 1A; Table 1). Only three plant genera (four plant species: Nothapodytes nimmoniana, Garcinia gummi-gutta, Kingiodendron pinnatum and Dysoxylum binectariferum) were unsuccessful in ME pure culture isolation. Interestingly, diversity indices and rarefaction analysis calculated at the bacterial genus level (Fig. 1B; Table 2; Figure S2, Supporting Information) suggested that the endophyte population collected from Bisle Ghat (34 isolates) was more diverse and species rich than the populations collected at Mangaluru (38 isolates) or Mysore (three isolates). In addition, rarefaction curves (Figure S2, Supporting Information) and Good's coverage statistics (Table 2) suggest that the total culturable bacterial diversity has not yet been isolated from the medicinal plants investigated in this study and further analysis is necessary to identify the full range of bacterial endophytes present.
Figure 1.
Community composition and rarefaction curves for the Western Ghats medicinal plant endophyte (ME) collection isolated during this study. (A) Composition of the bacterial endophyte collection assigned at the taxonomic genus level based on 16S rRNA genes. Each bar represents the relative distribution of each bacterial genus isolated from different medicinal plant genera. Numbers in parentheses represent the number of ME obtained from each plant genera. (B) Rarefaction curves for bacterial endophyte 16S rRNA gene diversity. ME were isolated from leaves of plants from the Bisle Ghat and Mangaluru regions of the Western Ghats, India. Curves were plotted for 95% similarity for 16S rRNA genes. Note, since there were only three isolates from Mysore leaf samples, this endophyte collection is not included.
Table 2.
Diversity indices for bacterial endophyte 16S rRNA gene sequences using genus-level groupings (95% similarity).
| Diversity indices | All isolates | Bisle Ghat region | Mangaluru | Mysore |
|---|---|---|---|---|
| Number of isolates | 75 | 34 | 38 | 3 |
| Unique OTUs | 16 | 13 | 8 | 2 |
| Good's coverage (%) | 79 | 62 | 79 | 33 |
| Simpson's diversity index (1-D) | 0.72 | 0.82 | 0.60 | 0.44 |
| Shannon's diversity index (H') | 1.88 | 2.10 | 1.33 | 0.64 |
| SChao1 | 23 | 17 | 11 | 2 |
OTU, operational taxonomic unit.
S Chao1 represent the expected number of OTUs present in an environment if sampling were complete.
Shannon's and Simpson's indices are measures of species diversity and both increase with increasing genetic diversity.
Using 16S rRNA gene sequence similarity all medicinal plant endophytes were representatives of three bacterial phyla (Proteobacteria, 66%; Firmicutes 31%; Actinobacteria, 3%; Fig. 2; Figure S3, Supporting Information) belonging to the following 13 genera (Fig. 1A) in order of dominance: Bacillus, 28%; Enterobacter, 25.4%; Klebsiella, 17.3%; Stenotrophomonas, 8%; Serratia, 5.4%; Pseudomonas, 4%; Acinetobacter, 1.3%; Aureimonas, 1.3%; Curtobacterium, 1.3%; Brevibacillus, 1.3%; Erwinia/Pantoea, 1.3%; Methylobacterium, 1.3%; Paenibacillus, 1.3%.
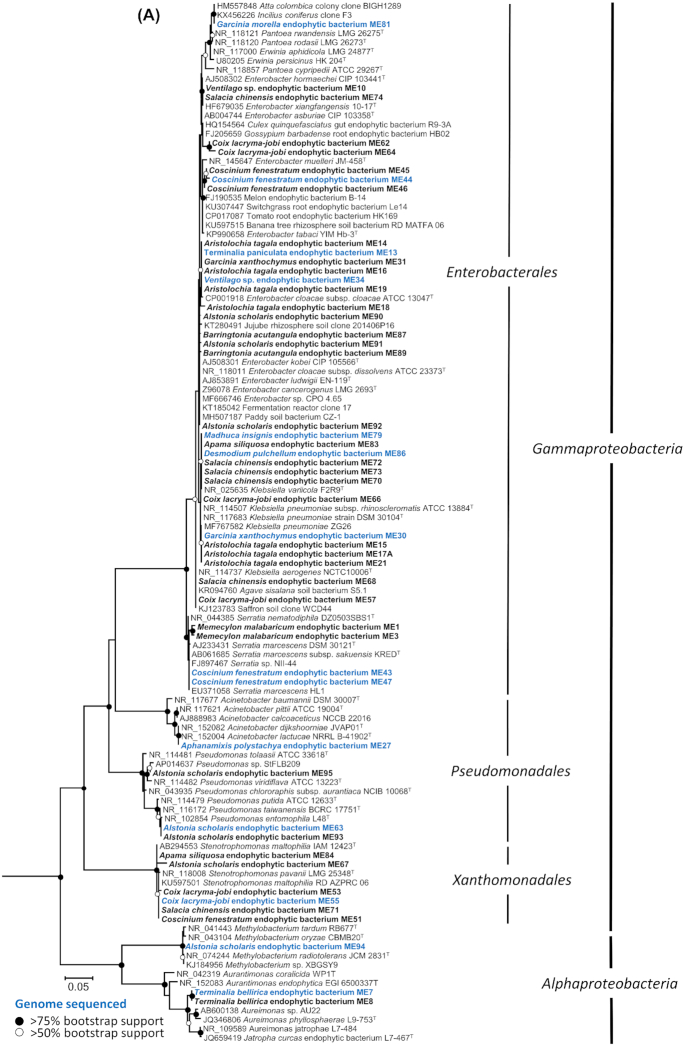
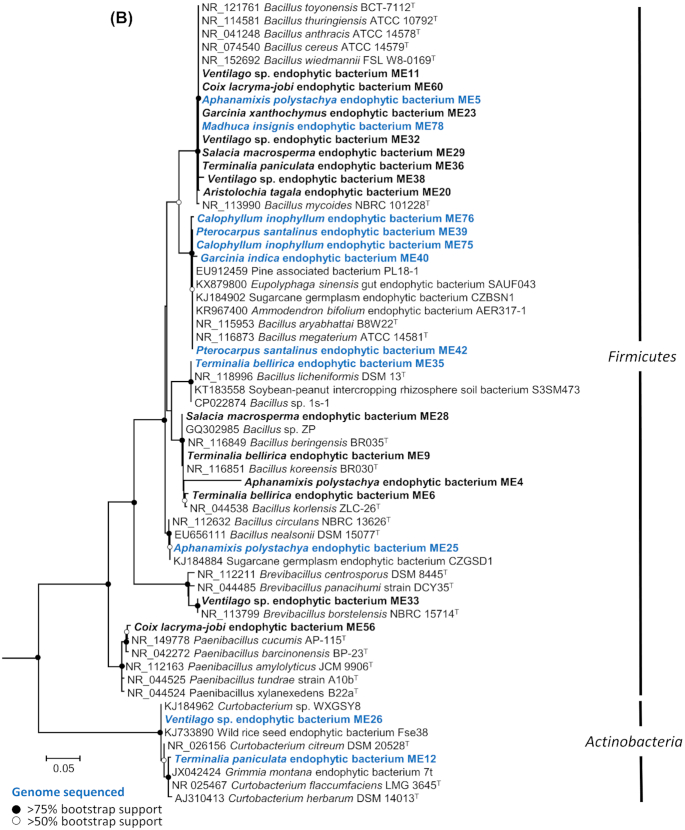
Figure 2.
Phylogenetic trees showing the relationship of medicinal plant endophyte (ME) 16S rRNA gene sequences to sequences from representative type species and other plant endophytes. (A)Proteobacteria (Gram-negative) and (B)Firmicutes and Actinobacteria (Gram-positive). Trees were constructed using Maximum Likelihood method based on the GTR model. A discrete Gamma distribution was used to model evolutionary rate differences among sites. All positions containing gaps and missing data were eliminated and there was a total of 627 and 695 positions in the final datasets, respectively. Deltaproteobacteria 16S rRNA gene sequences were used as outgroups in (A) Desulfobacter curvatus DSM 3379 (AF418175), Desulfuromonas acetoxidans DSM 684 (AAEW02000008), Desulfovibrio aerotolerans Dv06 (AY746987); and Proteobacteria 16S rRNA gene sequences were used as outgroups in (B) Methylobacterium radiotolerans JSCM 2831 ( NR_074244), Desulfovibrio aerotolerans Dv06 (AY746987), Enterobacter ludwigii EN-119T (AJ853891). Evolutionary analyses were conducted in MEGA7. The percentage of trees in which the associated taxa clustered together is shown next to the branches, based on 100 bootstraps. Nodes with black circles represent >75% bootstrap support; nodes with white circles represent >50% bootstrap support. The scale bar represents 5% sequence divergence. Sequences in bold represent ME isolates and sequences in bold blue represent ME isolates that had their genomes sequenced.
Figure 2.
continued
The grass species Coix lacryma-jobi was observed to contain the highest culturable diversity (ten isolates) of MEs (Fig. 1A; Table 1) with six different bacterial genera present (Enterobacter, Klebsiella, Pseudomonas, Stenotrophomonas, Bacillus and Paenibacillus). Contrastingly, the shrub Desmodium pulchellum had the lowest culturable diversity with only one bacterial isolate belonging to Klebsiella (ME86). Both plants were sampled from the Mangaluru location. For comparison, Aristolochia tagala, a climbing species found in forests of Asia had the highest culturable diversity (eight isolates) of MEs (Enterobacter, Klebsiella and Bacillus) identified from leaf samples taken from the Bisle Ghat (Fig. 1A; Table 1).
Multiple ME isolates were taxonomically related (based on 16S rRNA gene similarity) to previously known endophytes or bacteria isolated from soil and rhizosphere environments (Fig. 2). For example, the large collection of ME Enterobacter (19 isolates) are closely related (Fig. 2A) to endophytes from tomato, switchgrass, banana, jujube, cotton and rice paddy soils, and were isolated from a range of medicinal plants which include Ventilago sp., Salacia chinensis, Coix lacryma-jobi, Coscinium fenestratum, Aristolochia tagala, Terminalia paniculata, Garcinia xanthochymus, Alstonia scholaris and Barringtonia acutangular sampled from both the Bisle Ghat and Mangalaru locations. Similarly, the 21 ME isolates belonging to the genus Bacillus (12 different plant species and 3 locations) are related to endophytes from pine trees, sugar cane, legumes as well as insect guts (Fig. 2B). Many of these Bacillus isolates (ten isolates) were closely related to soil-borne bacteria within the Bacillus cereus group (Rasko et al. 2005; Carroll, Wiedmann and Kovac 2020).
Average nucleotide identity and dDDH reveal the predominant bacteria in the sequenced panel as Bacillus spp. and Enterobacteriaceae
Few endophytes have been genome sequenced as part of their collection and initial characterisation. A total of 26 selected medical plant endophytes were genome sequenced to increase the level to which they could be identified and characterised. The genus-level diversity of the genome sequenced endophytes as determined by genomic ANI and dDDH analysis (Table 1) were as follows: Bacillus (n = 9), Klebsiella (n = 4), Enterobacter (n = 3), Curtobacterium (n = 2), Serratia (n = 2), Aureimonas (n = 1), Stenotrophomonas (n = 1), Acinetobacter (n = 1), Pantoea (n = 1), Methylobacterium (n = 1) and Pseudomonas (n = 1). A summary of the genome assembly metrics is given in Table S2 (Supporting Information). Overall identification at the genus-level by genome analysis was in agreement with classification by 16S rRNA gene identification (Figs 2–5, Table 1, Figures S4–S9, Supporting Information) with the exception of isolate ME81 which was identified initially by 16S rRNA gene analysis as Erwinia (Fig. 2A) but subsequently classified by both genome analysis methods as Pantoea (Table 1, Figure S7, Supporting Information).
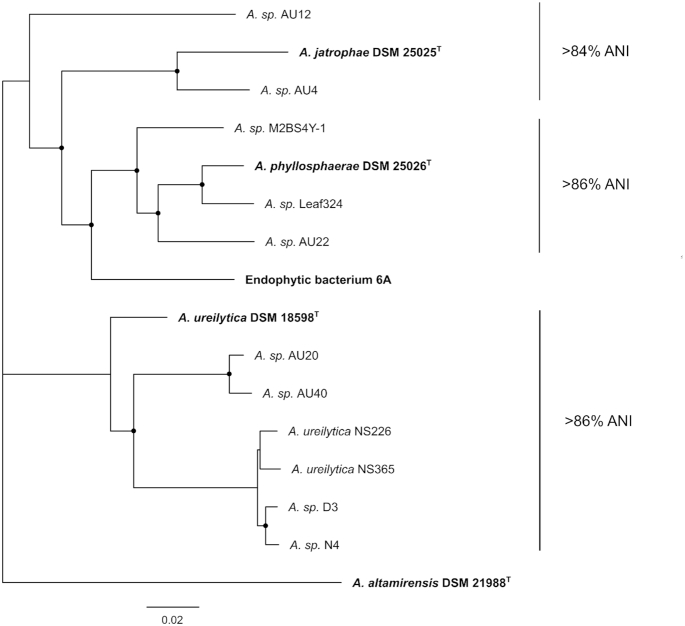
Figure 5.
Core-gene phylogeny of medicinal plant endophytes (ME) belonging to Aureimonas species. A 25 core-gene alignment generated by Roary was used to construct a maximum likelihood tree highlighting the placement of Aureimonas endophytes. Isolate ME7 was placed as a novel species close to the A. phyllosphaerae clade. The phylogenetic tree was constructed with GTR model with gamma substitution and supported by 100 bootstraps. Nodes with black circles represent >90% bootstrap support. Scale bar = substitutions per site.
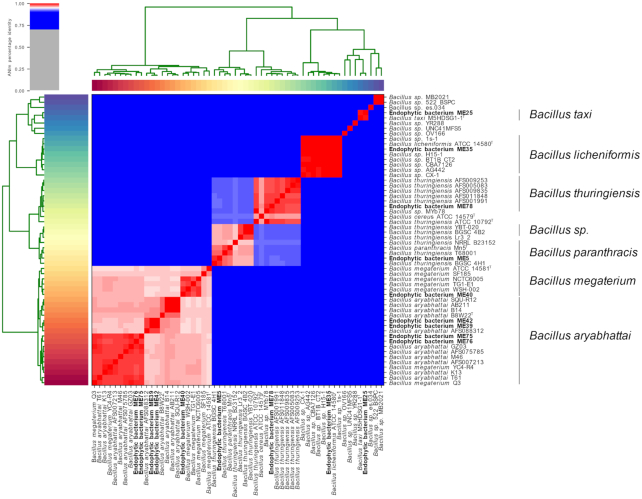
The species-level identity provided was consistent between ANI and dDDH for all isolates, excluding five Bacilli. A total of two Bacillus isolates, ME5 and ME78, were identified as Bacillus thuringiensis (98.8%) by ANI but were identified as Bacillus paranthracis and Bacillus cereus respectively by dDDH using TYGS. The remaining isolates, ME40, ME75 and ME76 could not be assigned a species-level identity by TYGS, but were identified as Bacillus megaterium (ME40, 95.8% identity), and Bacillus aryabhattai (ME75 and ME76, 96.3% identity) respectively by ANI. A heatmap providing a visual representation of the full diversity of Bacillus endophytes by ANI is shown in Fig. 3.
Figure 3.
Genome sequence taxonomic placement of Bacillus medicinal plant endophytes (ME) inferred by average nucleotide identity (ANI). Heatmap generated by the PyANI script, indicating the degree of nucleotide-level similarity between Bacillus species ME and their closest reference strains. ME are highlighted in bold font, whilst species type strains are denoted by T. Colour indicates the degree of nucleotide similarity, with red areas indicating >95% ANI, and darker shades of red indicating greater similarity. Blue indicates <95% ANI.
Core-gene analysis indicates a high-degree of intra-genus similarity for Enterobacter and Serratia endophytes, whilst highlighting the novelty of the Aureimonas sp. ME7
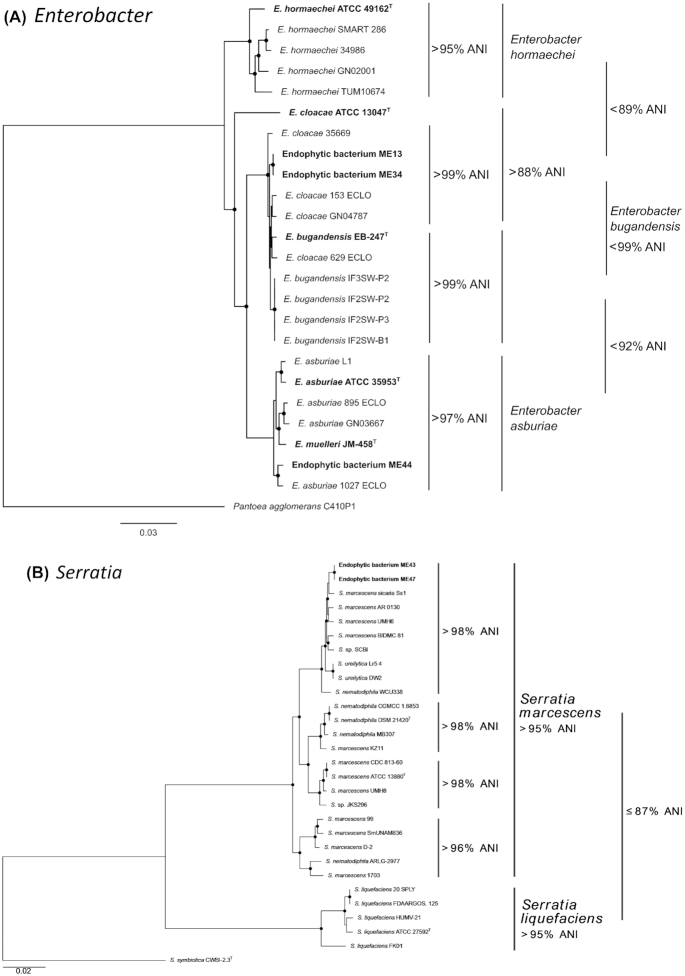
To increase the resolution of genomic taxonomy applied to the endophytic bacterial collection phylogenomic approaches were also applied on selected genera as follows. Core-gene phylogenetic analysis (Fig. 4) revealed three genera of interest, due to either the high degree of similarity between one or more sequenced endophytes (Enterobacter and Serratia) or unique phylogenetic placement supported by ANI, indicating a novel species group (Aureimonas). The analysis of Enterobacter genomes revealed that endophyte ME13, isolated from Terminalia paniculata in the Bisle Ghat region possessed the same core-genes as endophyte ME34, isolated from Ventilago sp. in the same region (Fig. 4A). The nearest neighbour of these isolates was the genome of Enterobacter cloacae 153_ECLO. This isolate, and both ME13 and ME34, were however distinct from the E. cloacae type strain, ATCC 13047T, both phylogenetically and in terms of ANI. Core-gene analysis of Serratia genomes revealed that endophytes ME43 and ME47, both of which were isolated from Coscinium fenestratum in the Mangalaru region were identical in terms of core-gene content (Fig. 4B). Additionally, both ME43 and ME47 possessed ≥95% identity in comparison to the Serratia marcescens type strain, ATCC 13880T (Fig. 4B).
Figure 4.
Average nucleotide identity (ANI) and core genome analysis of medicinal plant endophytes (ME) belonging to the order Enterobacterales: (A)Enterobacter and (B)Serratia. (A) Core-gene phylogeny of ME belonging to Enterobacter species. A 1912 core-gene alignment generated by Roary was used to construct a maximum likelihood tree highlighting the placement of Enterobacter endophytes. Isolates ME13 and ME34 placed within the E. cloacae species clade, whilst ME44 placed within the E. asburiae clade. (B) Core-gene phylogeny of ME belonging to Serratia species. A 255 core-gene alignment generated by Roary was used to construct a maximum likelihood phylogeny highlighting the placement of isolated Serratia endophytic bacteria. ME43 and ME47 were placed within the Serratia marcescens species clade with >95% ANI to other members of the species. Phylogenetic trees were constructed with GTR model with gamma substitution and supported by 100 bootstraps. Nodes with black circles represent >90% bootstrap support. Scale bar = substitutions per site.
Notably, core-gene analysis revealed Aureimonas sp. isolate ME7 as a novel endophyte, as shown by its unique phylogenetic placement (Fig. 5). All sequenced genomes obtained for the genus Aureimonas were extremely diverse, displaying deep phylogenetic branching and ANI values far below the established 95% threshold for species delineation (≥85% identity). The nearest neighbours for this isolate were all known endophytes and included Aureimonas sp. AU22 and Leaf324 from soybean and Arabidopsis thaliana, respectively.
Core-gene phylogenetic analyses for endophytes that did not belonging to genera of interest can be found in Figures S4–S9 (Supporting Information). In addition, since only a limited number of genomes are available for the Actinobacteria genus, Curtobacterium full-length 16S rRNA gene phylogenies were constructed instead (Figure S8, Supporting Information). Phylogenetic analysis demonstrated that endophyte ME12 (from Terminalia paniculate) was closely related (99% sequence similarity) to the plant pathogen Curtobacterium flaccumfaciens strains and that isolate ME26 from Ventilago sp. was related (99% sequence similarity) to novel endophytic Curtobacterium sp. WXGSY8 from sugarcane and Curtobacterium sp. ER1/6 from Citreus sinensis (sweet orange), a potential biocontrol strain (Garrido et al.2016). The frequent isolation of Curtobacterium as endophytes from asymptomatic citrus plants infested with the pathogen Xylella fastidiosa indicated that endophytic Curtobacterium species may help to resist infection (Rosenblueth and Martínez-Romero 2006).
Biosynthetic gene cluster prediction revealed both known and uncharacterised specialised metabolites
Following the prediction and curation of BGCs of the 26 sequenced endophyte genomes, a total of 102 distinct BGCs were identified across the 11 bacterial genera. These BGCs represented 15 known metabolite classes including siderophores, lassopeptides and non-ribosomal peptides (Table 3). Approximately 15% of BGCs could not be assigned a class and were collated under the antiSMASH category ‘Other’. The most prevalent classes were non-ribosomal peptides synthetases (NRPS), terpenes and bacteriocins representing approximately 45% of curated BGCs. The genus Bacillus with nine ME isolates contributed the majority of predicted BGCs to the endophyte biosynthetic potential, representing one-third of the 102 gene clusters (Table 3).
Table 3.
Summary of biosynthetic gene cluster (BGC) potential of medicinal plant endophytic bacteria. AntiSMASH was used to predict BGCs in the 26 whole-genome sequenced endophytic isolates. The predicted BGCS were curated and de-replicated to determine the number of distinct BGCs in the genome collection.
| Specialised metabolite class | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genus | Number of genomes | AntiSMASH predicted BGCs | Distinct BGCs | NRPS | Bacteriocin | T3PKS | NRPS-T1PKS | T1PKS | Thiopeptide | Lassopeptide | Lantipeptide | Terpene | Phosphonate | Arylpolyene | Butyrolactone | Homoserine lactone | Microcin | Siderophore | Other |
| Bacillus | 9 | 67 | 34 | 7 | 5 | 1 | 1 | 0 | 1 | 2 | 5 | 5 | 1 | 0 | 0 | 0 | 0 | 3 | 3 |
| Klebsiella | 4 | 17 | 6 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| Enterobacter | 3 | 15 | 6 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| Serratia | 2 | 33 | 10 | 4 | 1 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Curtobacterium | 2 | 12 | 8 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 4 |
| Acinetobacter | 1 | 6 | 7 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 0 |
| Aureimonas | 1 | 4 | 4 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Methylobacterium | 1 | 8 | 8 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 2 |
| Pantoea | 1 | 11 | 9 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 |
| Pseudomonas | 1 | 18 | 6 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 |
| Stenotrophomonas | 1 | 4 | 4 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Total | 26 | 195 | 102 | 20 | 12 | 4 | 4 | 2 | 6 | 2 | 5 | 13 | 1 | 6 | 3 | 7 | 1 | 6 | 10 |
BGC, biosynthetic gene cluster; NRPS, nonribosomal peptide synthase; T1PKS, Type 1 polyketide synthase; T3PKS, Type 3 polyketide synthase; NRPS-T1PKS, nonribosomal peptide synthase-Type 1 polyketide synthase hybrid
Only four hybrid non-ribosomal peptide synthetase-polyketide synthases (NRPS-PKS) were predicted, one of the hybrid BGCs from Klebsiella sp. ME86 possessed similarity to the yersiniabactin BGC, while the remaining three of these represented uncharacterised BGCs. Additional known BGCs identified in the endophyte genomes included the lassopeptide genes responsible for lichenicidin synthesis in Bacillus sp. ME35, and the NRPS required for acinetobactin synthesis in Acinetobacter sp. ME27. The majority of endophyte derived BGCs lacked homology to known specialised metabolite BGCs using the MiBIG database (Medema et al. 2015) via antiSMASH (Blin et al. 2017) that was applied.
Antimicrobial activity of medicinal plant endophytes
A total of five of the 26 genome-sequenced bacterial endophytes showed antimicrobial activity against the plant pathogen, Pectobacterium carotovorum (Figure S10, Supporting Information for examples). However, no zones of clearing were observed for the pathogens Staphylococcus aureus or Candida albicans by any ME tested. Isolates with clear antibacterial activity against the Gram-negative bacterium, P. carotovorum were identified as Bacillus aryabhattai (ME39), Bacillus sp. (ME40), Enterobacter asburiae (ME44) and Serratia sp. (ME43 and ME47). A total of four additional isolates showed weak antimicrobial activity against P. carotovorum: Bacillus aryabhattai (ME42), Bacillus sp. (ME75), Klebsiella pneumoniae (ME30) and Klebsiella variicola (ME73) (Figure S10, Supporting Information). Interestingly, several of the ME isolates with antibacterial activity and with predicted BGCs were obtained from medicinal plants used in traditional medicine for the treatment of wounds and/or known to have described antimicrobial activity. For example, Coscinium fenestratum (isolates ME43, ME44 and ME47) and Garcinia species (isolates ME30 and ME40) plant extracts have shown activity against Escherichia coli and other pathogenic bacteria (Nair et al. 2005; Baliga et al. 2011; Joseph, Dandin and Murthy Hosakatte 2016).
DISCUSSION
Medicinal plant endophyte diversity
Using a cultivation-based approach we have successfully isolated and identified 75 fast-growing cultivable bacteria that were associated with leaves of different plant species. Previously, endophytes have been reported from various other traditional medicinal plants; for example, Gynura procumbens (Bhore, Nithya and Loh 2010), Artemisia annua (Li et al. 2011), Tridax procumbens (Preveena and Bhore 2013), ginseng (Khan Chowdhury et al. 2017) and other traditional Chinese herbs (Miller et al. 2012a). However, to our knowledge, this study is unique in exploring a diverse range of bacterial isolates from a large collection (covering 24 plant species) of medicinal plants from the Western Ghats region of India. The identified bacterial endophytes belonged to four major taxa, Alphaproteobacteria, Gammaproteobacteria, Actinobacteria and Firmicutes, with isolates from the following genera: Bacillus, Enterobacter, Klebsiella, Stenotrophomonas, Serratia, Pseudomonas, Acinetobacter, Aureimonas, Curtobacterium, Brevibacillus, Pantoea, Methylobacterium and Paenibacillus. Previously, culture-based bacterial endophyte diversity analysis has shown that most culturable endophytes are Proteobacteria, followed by Actinobacteria, Bacteroidetes and Firmicutes (Rosenblueth and Martínez-Romero 2006; Khan Chowdhury et al. 2017). The same limited group of bacterial phyla were also found to predominate in the phyllosphere of different plants identified by a range of culture-independent approaches including metagenomic shotgun sequencing of total genomic DNA (Vorholt 2012). However, concerted efforts have been made to study Actinobacteria since they are a major source of natural antibiotics and metabolites (Passari et al. 2017; Ek-Ramos et al. 2019), while other bacterial phyla are a natural resource that are still relatively untapped.
A previous study observed that higher culturable endophytic bacterial diversity was associated with a higher likelihood of the host plant exhibiting antimicrobial properties (Egamberdieva et al. 2017). However, in contrast to bacterial diversity, this study also reported that the total bacterial cell numbers of colonizing microbes maybe higher in plants that have poor antimicrobial activity (Egamberdieva et al. 2017). Presumably, this is due to less stringent conditions encountered in these plants which allows for high numbers of colonizing bacteria to proliferate due to the lack of competition and lower concentrations of antimicrobials. In our study, only a tentative relationship was observed, linking high culturable microbial diversity to previously known medicinal properties for the treatment of bacteria-associated disease or known antibacterial activity. The medicinal plants with the highest culturable bacterial diversity, Terminalia spp., Ventilago sp. and Salacia spp. are used to treat bacterial diseases and leaf extracts of Coix lacryma-jobi and Coscinium fenestratum (Nair et al. 2005; Das et al. 2017) have been reported to have antimicrobial properties. However, we also observed plants with similar medicinal uses to have a low culturable diversity of ME isolates, namely Calophyllum inophyllum and Memecylon malabaricum (Table S1, Supporting Information). It should be noted that total bacterial numbers found within the leaves were not counted in our study. Further studies may be necessary to address the issue of achieving full culturable diversity, through focused efforts to isolate and count other endophytic community members including slow growing bacteria and fungi through the use of less complex and/or specific media (Eevers et al. 2015; Martinez-Klimova, Rodríguez-Peña and Sánchez 2017).
The nearest neighbours to endophytes of interest can be isolated from a variety of environments
Core-gene analysis demonstrates that the nearest neighbours of all endophytes in this study are not limited to association with plants, but are instead ubiquitous, and able to endure a plethora of environments. This is evidenced in the analysis of Enterobacter, where the nearest neighbours to endophytes ME13 and ME34 (Fig. 4A), namely E. cloacae 35 669 (Doijad et al. 2016), 153_ECLO, 629_ECLO and GN04787 (Matteoli et al. 2020), were all isolated from clinical infections (Fig. 4A). In contrast, the nearest neighbours Enterobacter bugandensis IF2SW-B1, IF2SW-P2, IF2SW-P3 and IF3SW-P2 were all recently isolated from the International Space Station (Singh et al. 2018). The similarity of the space station isolates to clinical isolate 153 ECLO has been commented upon previously (Singh et al. 2018) and is thus concordant with the analysis in this study. Members of the Enterobacter genus also comprise species that have been reported as plant beneficial organisms and these include, plant-growth promoting endophytes of Enterobacter asburiae on date palm (Yaish 2016), Enterobacter cloacae with citrus and banana plants (Araujo et al. 2002; Macedo-Raygoza et al. 2019) and Enterobacter sp. J49, a biofertilizer for peanut and maize (Ludueña et al. 2019).
A number of nearest phylogenomic neighbours to Serratia endophytes ME43 and ME47, including S. marcescens AR_0130, BIDMC 81 and UMH6 (Anderson et al. 2017), originate from the nosocomial environment, whilst S. marcescens sicaria Ss1 was isolated from the haemolymph of worker bees suffering from sepsis and implicated as a new pathogen of honey bees (Burritt et al. 2016). However, isolates of S. marcescens are known to fix nitrogen and act as plant growth promoting endophytic colonisers of rice roots and stems (Gyaneshwar et al. 2001). Nonclinical isolates of S. marcescens have also been used as biocontrol agents (Hallmann et al. 1997) and induce systemic resistance to fungal and viral pathogens (Press et al. 1997), as well as the production of the biologically active compound prodigiosin (Khanam and Chandra 2018).
Interestingly, the Aureimonas endophyte ME7, was related to bacteria originally isolated from surfaces and internal tissues of plants and identified as a unique species by ANI and core-gene analyses (see Fig. 5). Members of the genus, Aureimonas are increasingly being isolated from leaves of plants (Madhaiyan et al. 2013; Li et al. 2017; Tuo and Yan 2019) and thought to be involved in the cycling of carbon and nitrogen (Ikeda et al. 2010). The nearest phylogenomic neighbours for this isolate were Aureimonas sp. AU22 and Aureimonas sp. Leaf324 isolated from the stems of soybean, and the leaves of Arabidopsis thaliana respectively, whilst the nearest neighbouring type-strains, Aureimonas phyllosphaerae DSM 25024T and Aureimonas jatrophae DSM 25025T were both isolated from the leaves of Jatropha curcas (Madhaiyan et al. 2013), a small tree whose seed oil is widely used as biofuel, soap and medicine (Pandey et al. 2012).
Biosynthetic capacity of medicinal plant endophytes
Previous studies have investigated the NRPS and PKS diversity of medicinal plant bacterial and fungal endophytes through culture-independent PCR-based methods (Miller et al. 2012a). The benefits of this culture-independent approach included a lack of culture-bias and the ability to detect both fungal and bacterial NRPS and PKS potential. However, as noted, the limitations of a PCR screen were the inability to detect low level target DNA, and divergent sequence domains. Additional studies by Miller et al. (2012b) on Chinese medicinal plants obtained pure bacterial and fungal isolates that permitted cytotoxicity and antimicrobial phenotypic testing (Miller et al. 2012b). Although our culture-dependent isolation of endophytes was biased towards bacteria capable of growth on LB agar, the output of this study included draft whole-genome sequences and pure cultures of the isolated bacterial endophytes. This enabled both phenotypic testing of antimicrobial activity and genome mining for a multitude of biosynthetic gene clusters. A high proportion of the sequenced ME isolates possessed BGCs with NRPS and PKS-predictions (73% and 54%, respectively). This represents a significantly larger proportion than previously described endophyte collections (Miller et al. 2012b). However, this is partly biased by only examining the genome-sequenced portion of the collection, and the ability to predict type 3 polyketide synthase (T3PKS) and hybrid NRPS-PKS BGCs. The draft genomes described in our study enabled accurate taxonomic identification and resulted in the prediction of BGCs representing multiple metabolite classes, a contrast to existing work on medicinal plant endophytes.
The isolation of multiple endophytic bacterial isolates has previously been coupled to phenotypic assays of antimicrobial activity, tandem mass spectrometry analyses and PCR-based detection of conserved PKS and NRPS domains (Passari et al. 2017). This combinatory approach has led to promising leads of novel antimicrobial metabolites (Passari et al. 2017). Future work into the identification and isolation of metabolites of the endophyte collection in this study can be guided by the genomic insight into the biosynthetic origins of potential metabolites of these bacteria. Despite the identification of BGCs with high sequence similarity to previously characterised BGCs, most of the 102 biosynthetic gene clusters possessed no homology to published BGCs, and thus represent a novel source of pharmaceutically relevant products.
Potential use of medicinal plant endophytes as antimicrobial and biocontrol agents
ME isolates which showed antibacterial activity towards the plant pathogen, P. carotovorum belonged to the genera Bacillus (n = 4), Klebsiella (n = 2), Serratia (n = 2) and Enterobacter (n = 1). Previously, Bacillus endophytes have demonstrated activity against bacterial phytopathogens (Ryan et al. 2008; Santoyo et al. 2016; Chen et al. 2019), including P. carotovorum (Wang et al. 2019). For example, the strain Bacillus sp. NA-HTong-7, isolated from the stems of the medicinal plant, Dendrobium possessed activity against both fungal (Athelia rolfsii and Myrothecium roridum) and bacterial (P. carotovorum subsp. actinidiae) pathogens of Dendrobium species and has potential as a biocontrol agent (Wang et al. 2019). Whereas, other studies have reported that Bacillus species isolated from medicinal plants exhibited general antibacterial activity including that against Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa (Akinsanya et al. 2015; Beiranvand et al. 2017; Egamberdieva et al. 2017) and suggest that they are responsible for the plants therapeutic properties. For a comprehensive review of the use of endophytes as therapeutic agents in Asian medicinal plants see the recent paper by Sharma and colleagues (Sharma et al. 2020).
Bacillus species have been found to be one of the most abundant metabolite-producing Gram-positive bacterial endophytes (Frank, Saldierna-Guzmán and Shay 2017), and in this study Bacillus were responsible for a third of all distinct BGCs identified (Table 3). Bacillus species produce a wide variety of antimicrobial metabolites, including ribosomally synthesised antimicrobial peptides (e.g. bacteriocins, lantipeptides and lassopeptides), as well as non-ribosomally synthesised peptides and polyketides (Zhao and Kuipers 2016). The Bacillus ME isolates (ME39 and ME40), with good bioactivity against P. carotovorum found in this study were shown by genome mining to contain several complete BGCs that may contribute to antimicrobial activity. Bacillus aryabhattai ME39 was shown to carry both lantipeptide and bacteriocin BGCs, while Bacillus sp. ME40 carried a lassopeptide BGC with sequence similarity and gene synteny to the paeninodin BGC. While the paeninodin lassopeptide lacked antimicrobial activity against representatives of Actinobacteria, Firmicutes and Proteobacteria (Zhu et al. 2016), antagonism against P. carotovorum was not investigated. However, several endophytic bacterial peptides with antimicrobial activity have been reported (Zhao and Kuipers 2016).
Other BGCs of interest in the remaining isolates with clear antagonism included hybrid NRPS-PKS BGCs in Serratia sp. ME47 with no homology to characterised BGCs; and thiopeptide BGCs predicted in Enterobacter asburiae ME44, Serratias isolates ME43 and ME47. Most characterised thiopeptides display nanomolar potency toward Gram-positive bacteria by blocking protein translation, and the majority of them have been identified from Actinobacteria and Bacilli (Schwalen et al. 2018). However, thiopeptides were also identified by genome mining in Proteobacteria (Schwalen et al. 2018). Our study shows the potential of Bacillus and other bacterial endophytes as biological control agents of plant pathogenic bacteria, and that ME isolates could be used to produce peptide-based antimicrobial and/or other compounds for therapeutic use. In addition, this study adds support to the claims and reports that some species of medicinal plants of the Western Ghats possess antimicrobial properties and may explain their ethnomedicinal use.
CONCLUSIONS
This study identified multiple bacterial endophytes across a diverse array of medicinal plants from one of the World's ‘Hottest Hotspots’ of biodiversity, the Western Ghats (Myers et al. 2000). The draft genome assemblies obtained from these endophytes have permitted an insight into the biosynthetic diversity of these bacteria, whilst the isolation of pure cultures enables the future exploitation of the identified biosynthetic potential. To our knowledge, this represents one of the largest collections of isolates with draft genomes available from endophytic bacteria in a single study of medicinal plants. Identifying and understanding the medicinal plant endophytic microbial diversity therefore has potential for the discovery of new natural products.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for the assistance of the School of Biosciences Genomics Research Hub, Cardiff University for help with genome library preparation and Illumina sequencing. We thank Andrew Manson (University of Aberdeen) for additional technical support during his undergraduate training year. All analyses were done using the Medical Research Council (MRC) Cloud Infrastructure for Microbial Bioinformatics (CLIMB).
Notes
Department of Studies in Microbiology, University of Mysore, Mysore, Karnataka, 570006, India. Tel: +91 821 2419441; E-mail: raivittal@gmail.com
Contributor Information
Gordon Webster, Microbiomes, Microbes and Informatics Group, Organisms and Environment Division, School of Biosciences, Cardiff University, Cardiff, CF10 3AX, Wales, UK.
Alex J Mullins, Microbiomes, Microbes and Informatics Group, Organisms and Environment Division, School of Biosciences, Cardiff University, Cardiff, CF10 3AX, Wales, UK.
Edward Cunningham-Oakes, Microbiomes, Microbes and Informatics Group, Organisms and Environment Division, School of Biosciences, Cardiff University, Cardiff, CF10 3AX, Wales, UK.
Arun Renganathan, Department of Studies in Microbiology, University of Mysore, Karnataka, 570006, Mysore, India.
Jamuna Bai Aswathanarayan, Department of Studies in Microbiology, University of Mysore, Karnataka, 570006, Mysore, India.
Eshwar Mahenthiralingam, Microbiomes, Microbes and Informatics Group, Organisms and Environment Division, School of Biosciences, Cardiff University, Cardiff, CF10 3AX, Wales, UK.
Ravishankar Rai Vittal, Department of Studies in Microbiology, University of Mysore, Karnataka, 570006, Mysore, India.
AUTHOR CONTRIBUTIONS
We describe author contributions to the paper using the CRedit taxonomy. Conceptualisation: GW, AJM, ECO, EM and RRV; Data Curation: GW, AJM, ECO, AR, JBA, EM and RRV; Formal analysis: GW, AJM, ECO, AR, JBA, EM and RRV; Funding Acquisition: EM and RRV; Investigation: GW, AJM, ECO, AR, JBA and RRV; Methodology: AR, JBA, GW, AJM and ECO; Project Administration: EM and RRV; Resources: EM and RRV; Software: AJM, ECO and GW; Supervision: EM and RRV; Validation: GW, AJM, ECO, EM and RRV; Visualisation: GW, AJM, ECO and AR; Writing-Original Draft: GW, AJM, ECO, JBA, EM and RRV; Writing-Review & Editing: GW, AJM, ECO, AR, JBA, EM and RRV.
FUNDING
This work was supported by funding from a Cardiff University Incoming Visiting Fellowship award to EM and RRV. Additional support from the Biotechnology and Biological Sciences Research Council (BBSRC) is also acknowledged as follows: a genome mining award to EM (BB/L021692/1), and PhD studentships for ECO and AM (South West Doctoral Training Partnership, BB/M009122/1). ECO also acknowledges funding from Unilever R&D industrial CASE studentship awards during his doctoral studies. GW acknowledges funding from a Wellcome Trust Institutional Strategic Support Fund award. AJM, GW and EM also acknowledge current funding from BBSRC grant BB/S007652/1.
Conflicts of interest
None declared.
REFERENCES
- Akinsanya MA, Goh JK, Lim SP et al. Diversity, antimicrobial and antioxidant activities of culturable bacterial endophyte communities in Aloe vera. FEMS Microbiol Lett. 2015;362:fnv184. [DOI] [PubMed] [Google Scholar]
- Anderson MT, Mitchell LA, Zhao L et al. Capsule production and glucose metabolism dictate fitness during Serratia marcescens bacteremia. mBio. 2017;8:e00740–00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo WL, Marcon J, Maccheroni W et al. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol. 2002;68:4906–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyanar M, Ignacimuthu S.. Ethnobotanical survey of medicinal plants commonly used by Kani tribals in Tirunelveli hills of Western Ghats, India. J Ethnopharmacol. 2011;134:851–64. [DOI] [PubMed] [Google Scholar]
- Baliga MS, Bhat HP, Pai RJ et al. The chemistry and medicinal uses of the underutilized Indian fruit tree Garcinia indica Choisy (kokum): a review. Food Res Int. 2011;44:1790–9. [Google Scholar]
- Bankevich A, Nurk S, Antipov D et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiranvand M, Amin M, Hashemi-Shahraki A et al. Antimicrobial activity of endophytic bacterial populations isolated from medical plants of Iran. Iran J Microbiol. 2017;9:11–18. [PMC free article] [PubMed] [Google Scholar]
- Bhore SJ, Nithya R, Loh CY. Screening of endophytic bacteria isolated from leaves of Sambung Nyawa [Gynura procumbens (Lour.) Merr.] for cytokinin-like compounds. Bioinformation. 2010;5:191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Wolf T, Chevrette MG et al. antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burritt NL, Foss NJ, Neeno-Eckwall EC et al. Sepsis and hemocyte loss in honey bees (Apis mellifera) infected with Serratia marcescens strain Sicaria. PLoS One. 2016;11:e0167752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll LM, Wiedmann M, Kovac J. Proposal of a taxonomic nomenclature for the Bacillus cereus group which reconciles genomic definitions of bacterial species with clinical and industrial phenotypes. mBio. 2020;11:e00034–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Shi H, Heng J et al. Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiol Res. 2019;218:41–48. [DOI] [PubMed] [Google Scholar]
- Das S, Akhter R, Khandaker S et al. Phytochemical screening, antibacterial and anthelmintic activities of leaf and seed extracts of Coix lacryma-obi L. J Coast Life Med. 2017;5:360–4. [Google Scholar]
- Deokate UA, Khadabadi SS. Phytopharmacological aspects of Salacia chinensis. J Pharmacognosy Phytother. 2012;4:1–5. [Google Scholar]
- Doijad S, Imirzalioglu C, Yao Y et al. Enterobacter bugandensis sp. nov., isolated from neonatal blood. Int J Syst Evol Microbiol. 2016;66:968–74. [DOI] [PubMed] [Google Scholar]
- Eevers N, Gielen M, Sánchez-López A et al. Optimization of isolation and cultivation of bacterial endophytes through addition of plant extract to nutrient media. Micro Biotech. 2015;8:707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egamberdieva D, Wirth S, Behrendt U et al. Antimicrobial activity of medicinal plants correlates with the proportion of antagonistic endophytes. Front Microbiol. 2017;8:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek-Ramos MJ, Gomez-Flores R, Orozco-Flores AA et al. Bioactive products from plant-endophytic Gram-positive bacteria. Front Microbiol. 2019;10:463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank A, Saldierna-Guzmán J, Shay J. Transmission of bacterial endophytes. Microorganisms. 2017;5:E70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganjewala D, Gupta AK.. Study on phytochemical composition, antibacterial and antioxidant properties of different parts of Alstonia scholaris Linn. Adv Pharm Bull. 2013;3:379–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido LM, Alves JMP, Oliveira LS et al. Draft genome sequence of Curtobacteriumsp. strain ER1/6, an endophytic strain isolated from Citrus sinensis with potential to be used as a biocontrol agent. Genome Announc. 2016;4:e01264–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouda S, Das G, Sen SK et al. Endophytes: a treasure house of bioactive compounds of medicinal importance. Front Microbiol. 2016;7:1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyaneshwar P, James EK, Mathan N et al. Endophytic colonization of rice by a diazotrophic strain of Serratia marcescens. J Bact. 2001;183:2634–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann J, Quadt-Hallmann A, Mahaffee WF et al. Bacterial endophytes in agricultural crops. Can J Microbiol. 1997;43:895–914. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41: 95–98. [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9. [Google Scholar]
- Hardoim PR, Van Overbeek LS, Berg G et al. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79:293–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, Okubo T, Anda M et al. Community- and genome-based views of plant-associated bacteria: plant–bacterial interactions in soybean and rice. Plant Cell Physiol. 2010;51:1398–410. [DOI] [PubMed] [Google Scholar]
- Jain C, Rodriguez-R LM, Phillippy AM et al. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph KS, Dandin VS, Murthy Hosakatte N. Chemistry and biological activity of Garcinia xanthochymus: a review. J Biol Act Prod Nat. 2016;6:173–94. [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanam B, Chandra R.. Comparative analysis of prodigiosin isolated from endophyte Serratia marcescens. Lett Appl Microbiol. 2018;66:194–201. [DOI] [PubMed] [Google Scholar]
- Khan Chowdhury ME, Jeon J, Ok Rim S et al. Composition, diversity and bioactivity of culturable bacterial endophytes in mountain-cultivated ginseng in Korea. Sci Rep. 2017;7:10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köberl M, Schmidt R, Ramadan EM et al. The microbiome of medicinal plants: diversity and importance for plant growth, quality and health. Front Microbiol. 2013;4:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FN, Tuo L, Pan Z et al. Aureimonas endophytica sp. nov., a novel endophytic bacterium isolated from Aegiceras corniculatum. Int J Syst Evol Microbiol. 2017;67:2934–40. [DOI] [PubMed] [Google Scholar]
- Li J, Zhao GZ, Huang HY et al. Isolation and characterization of culturable endophytic actinobacteria associated with Artemisia annua L. Ant van Leeuwenhoek. 2011;101:515–27. [DOI] [PubMed] [Google Scholar]
- Ludueña LM, Anzuay MS, Angelini JG et al. Genome sequence of the endophytic strain Enterobacter sp. J49, a potential biofertilizer for peanut and maize. Genomics. 2019;111:913–20. [DOI] [PubMed] [Google Scholar]
- Macedo-Raygoza GM, Valdez-Salas B, Prado FM et al. Enterobacter cloacae, an endophyte that establishes a nutrient-transfer symbiosis with banana plants and protects against the black Sigatoka pathogen. Front Microbiol. 2019;10:804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhaiyan M, Hu CJ, Jegan Roy J et al. Aureimonas jatrophae sp. nov. and Aureimonas phyllosphaerae sp. nov., leaf-associated bacteria isolated from Jatropha curcas L. Int J Syst Evol Microbiol. 2013;63:1702–8. [DOI] [PubMed] [Google Scholar]
- Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Klimova E, Rodríguez-Peña K, Sánchez S. Endophytes as sources of antibiotics. Biochem Pharmacol. 2017;134:1–17. [DOI] [PubMed] [Google Scholar]
- Matteoli FP, Passarelli-Araujo H, Pedrosa-Silva F et al. Population structure and pangenome analysis of Enterobacter bugandensis uncover the presence of blaCTX-M-55, blaNDM-5 and blaIMI-1, along with sophisticated iron acquisition strategies. Genomics. 2020;112:1182–91. [DOI] [PubMed] [Google Scholar]
- Medema MH, Kottmann R, Yilmaz P et al. Minimum information about a biosynthetic gene cluster. Nat Chem Biol. 2015;11:625–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Göker M.. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KI, Qing C, Sze DM-Y et al. Culturable endophytes of medicinal plants and the genetic basis for their bioactivity. Microb Ecol. 2012b;64:431–49. [DOI] [PubMed] [Google Scholar]
- Miller KI, Qing C, Sze DMY et al. Investigation of the biosynthetic potential of endophytes in traditional Chinese anticancer herbs. PLoS One. 2012a;7:e35953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins AJ, Murray JAH, Bull MJ et al. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria. Nat Microbiol. 2019;4:996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG et al. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–8. [DOI] [PubMed] [Google Scholar]
- Nair GM, Narasimhan S, Shiburaj S et al. Antibacterial effects of Coscinium fenestratum. Fitoterapia. 2005;76:585–7. [DOI] [PubMed] [Google Scholar]
- Ondov BD, Treangen TJ, Melsted P et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Cummins CA, Hunt M et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey VC, Singh K, Singh JS et al. Jatropha curcas: a potential biofuel plant for sustainable environmental development. Renew Sust Energ Rev. 2012;16:2870–83. [Google Scholar]
- Parkes RJ, Sass H, Webster G et al. Methods for studying methanogens and methanogenesis in marine sediments. In: Timmis KN. (ed). Handbook of Hydrocarbon and Lipid Microbiology, pp. 3799–826.. Springer Berlin Heidelberg, Berlin, Heidelberg: 2010. [Google Scholar]
- Pascal JP, Ramesh BR, De Franceschi D. Wet evergreen forest types of the southern Western Ghats, India. Trop Ecol. 2004;45:281–92. [Google Scholar]
- Passari AK, Mishra VK, Singh G et al. Insights into the functionality of endophytic actinobacteria with a focus on their biosynthetic potential and secondary metabolites production. Sci Rep. 2017;7:11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press CM, Wilson M, Tuzun S et al. Salicylic acid produced by Serratia marcescens 90–166 is not the primary determinant of induced systemic resistance in cucumber or tobacco. Mol Plant Microbe Interact. 1997;10:761–8. [Google Scholar]
- Preveena J, Bhore SJ.. Identification of bacterial endophytes associated with traditional medicinal plant Tridax procumbens Linn. Anc Sci Life. 2013;32:173–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L, Glover RH, Humphris S et al. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8:12–24. [Google Scholar]
- Rasko DA, Altherr MR, Han CS et al. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol Rev. 2005;29:303–29. [DOI] [PubMed] [Google Scholar]
- Reddy CS, Jha CS, Dadhwal VK. Assessment and monitoring of long-term forest cover changes (1920–2013) in Western Ghats biodiversity hotspot. J Earth Syst Sci Reddy, Jha and Dadhwal. 2016;125:103–14. [Google Scholar]
- Rosenblueth M, Martínez-Romero E. Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact. 2006;19:827–37. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Germaine K, Franks A et al. Bacterial endophytes: recent developments and applications. FEMS Microbiol Lett. 2008;278:1–9. [DOI] [PubMed] [Google Scholar]
- Santoyo G, Moreno-Hagelsieb G, Del Carmen Orozco-Mosqueda M et al. Plant growth-promoting bacterial endophytes. Microbiol Res. 2016;183:92–99. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Status of the microbial census. Microbiol Mol Biol R. 2004;68:686–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalen CJ, Hudson GA, Kille B et al. Bioinformatic expansion and discovery of thiopeptide antibiotics. J Am Chem Soc. 2018;140:9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Malhotra B, Kharkwal H et al. Therapeutic agents from endophytes harbored in Asian medicinal plants. Phytochem Rev. 2020, DOI: 10.1007/s11101-020-09683-8. [Google Scholar]
- Singh NK, Bezdan D, Checinska Sielaff A et al. Multi-drug resistant Enterobacter bugandensis species isolated from the International Space Station and comparative genomic analyses with human pathogenic strains. BMC Microbiol. 2018;18:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol R. 2003;67:491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuo L, Yan X-R. Aureimonas flava sp. nov., a novel endophytic bacterium isolated from leaf of Acrostichum aureum. Int J Syst Evol Microbiol. 2019;69:846–51. [DOI] [PubMed] [Google Scholar]
- Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10:828–40. [DOI] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 1991;10:506–13. [PubMed] [Google Scholar]
- Wang S-S, Liu J-M, Sun J et al. Diversity of culture-independent bacteria and antimicrobial activity of culturable endophytic bacteria isolated from different Dendrobium stems. Sci Rep. 2019;9:10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster G, Parkes RJ, Cragg BA et al. Prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol Ecol. 2006;58:65–85. [DOI] [PubMed] [Google Scholar]
- Yaish MW. Draft genome sequence of endophytic bacterium Enterobacter asburiae PDA134, isolated from date palm (Phoenix dactylifera L.) roots. Genome Announc. 2016;4:e00848–00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HW, Song YC, Tan RX. Biology and chemistry of endophytes. Nat Prod Rep. 2006;23:753–71. [DOI] [PubMed] [Google Scholar]
- Zhao X, Kuipers OP.. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genomics. 2016;17:882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Hegemann JD, Fage CD et al. Insights into the unique phosphorylation of the lasso peptide paeninodin. J Biol Chem. 2016;291:13662–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.