Abstract
Context
Detection of high-risk human papillomavirus (HR-HPV) in squamous cell carcinoma is important for classification and prognostication. In situ hybridization (ISH) is a commonly used HR-HPV–specific test that targets viral RNA or DNA. The College of American Pathologists (CAP) provides proficiency testing for laboratories performing HR-HPV ISH.
Objective
To compare the analytical performance of RNA- and DNA-based ISH methods on CAP HR-HPV proficiency tests.
Design
Data from the 2016–2018 CAP HPV ISH proficiency testing surveys were reviewed. These surveys consist of well-characterized samples with known status for HR-HPV, including 1 to 2 copies, 50 to 100 copies, 300 to 500 copies, and no copies of HR-HPV per cell.
Results
Ninety-five participants submitted 1268 survey results from 20 cores. Overall, RNA ISH had a significantly higher percentage of correct responses than DNA ISH: 97.4% (450 of 462) versus 80.6% (650 of 806) (P < .001). This disparity appears to be the consequence of a superior sensitivity of RNA ISH compared to DNA ISH for samples with 1 to 2 and with 50 to 100 copies of HR-HPV per cell: 95.2% (120 of 126) versus 53.8% (129 of 240), P < .001, respectively, and 100% (89 of 89) versus 76.3% (119 of 156), P < .001, respectively.
Conclusions
An assessment of CAP HR-HPV proficiency test performance indicates that RNA ISH shows significantly higher accuracy than DNA ISH owing to higher analytical sensitivity of RNA ISH in tumors with low (1–2 copies per cell) to intermediate (50–100 copies per cell) HR-HPV viral copy numbers. These data support the use of RNA over DNA ISH in clinical laboratories that perform HR-HPV testing as part of their testing algorithms.
High-risk human papillomavirus (HR-HPV) causes HPV-positive squamous cell carcinomas (SCCs) at multiple anatomic sites including the oropharynx, anus, and urogenital tracts. The detection of HR-HPV in tissue samples containing SCC can be important for their classification and prognostication. For example, the HR-HPV status of oropharyngeal squamous cell carcinoma (OPSCC) allows this tumor to be identified as distinct from HPV-negative OPSCC with a favorable prognosis and with potential impact on treatment.1–4
A range of tests can be applied to detect HR-HPV in tissue samples. Immunohistochemistry for p16 is a surrogate marker for HR-HPV and is commonly used because of its high sensitivity, low cost, widespread availability, and proven prognostic utility. However, it is often used in conjunction with HPV-specific nucleic acid tests owing to its lower specificity.5–8 For example, the recent College of American Pathologists (CAP) guideline for HPV testing of head and neck carcinomas recommends performing p16 immunohistochemistry for OPSCCs, but also recommends performing HPV-specific testing for a subset of metastatic p16-positive SCCs owing to the possibility of p16 overexpression unrelated to HR-HPV.9
In situ hybridization (ISH) is a frequently used HR-HPV–specific test. Technical ISH options available in clinical practice include targeting either viral DNA or RNA. HR-HPV DNA ISH suffers from poor analytical sensitivity.7,10–12 Although the analytical validity of RNA ISH has been less extensively studied than DNA ISH, several studies have suggested superior analytical sensitivity and specificity.5–8 Polymerase chain reaction has also been used by some laboratories for HR-HPV detection in tissue samples, but it is not widely used, in part, because of expense and because it requires more technical expertise than the other tests.7,13–15
The CAP is a well-known provider of proficiency testing to ensure that laboratories fulfill the Clinical Laboratory Improvement Amendments requirement of assessing analytical validity during initial development and ongoing clinical use of various tissue-based tests. As such, the CAP is uniquely positioned to compare the performance of different technical approaches to clinical testing in participating laboratories from across the world. The CAP Molecular Oncology Committee provides proficiency testing for laboratories performing HR-HPV ISH. The objective of the current study was to compare the analytical performance of DNA- and RNA-based ISH methods on CAP HR-HPV proficiency tests from 2016–2018.
MATERIALS AND METHODS
Data from the 2016–2018 CAP HPV ISH proficiency testing surveys (Survey name: ISH) were reviewed. These surveys, offered biannually, are composed of 1 tissue microarray with 4 cores of formalin-fixed, paraffin-embedded tissue. The HR-HPV–positive tissue cores are derived from xenografts of 1 of 3 cell lines with known status for HR-HPV, including SiHa (1–2 copies of HPV-16 per cell), HeLa (50–100 copies of HPV-18 per cell), and Caski (300–500 copies of HPV-16 per cell).16–18 The HR-HPV–negative core is derived from a human SCC specimen that is negative for HR-HPV by p16 immunohistochemistry, DNA ISH, and RNA ISH.
Participants were asked to perform ISH and interpret each core as positive or negative for HR-HPV. Each participant was also asked to describe the probe manufacturer, the probe target (DNA versus RNA), the HPV type(s) targeted, and the controls performed by his or her laboratory, including positive and negative tissue controls, DNA/RNA controls, and the use of a negative control probe.
Multivariate logistic models were used to statistically analyze the survey results and concordance rates. A concordant response was defined as a core participant result match to the sample’s intended HR-HPV status. The model to analyze performance differences between RNA and DNA ISH was fit with 3 performance factors: (1) ISH type (DNA or RNA), (2) copies/cell classification, and (3) the interaction term to compare the performance of DNA versus RNA ISH by the viral copies per cell. Two models to evaluate within ISH method performance were also used. These models were fit with 2 factors: (1) copies/cell classification and (2) probe manufacturer. The Bonferroni correction was applied to account for the performance factors’ multiple comparisons. Sensitivity and specificity results between ISH type and performance by institution location were analyzed by using χ2 tests. A paired t test was used to compare pairwise performance differences between laboratories that changed from DNA to RNA ISH during the course of the study. A significance level of .05 was used for all statistical analyses and all analyses were performed with the SAS 9.4 software (SAS Institute, Cary, North Carolina).
Several data adjustments were applied before the statistical analysis. Forty-two survey results were excluded owing to a missing ISH method or core result. An additional 10 survey results were also excluded for HPV-18–positive cores tested with RNA or DNA ISH probes reported to target only HPV-16. Owing to low frequency usage, Biocare Medical was classified in the “Other (DNA)” group along with participants who reported “Other” probe manufacturer and used DNA ISH. Similarly, RNA ISH participants who reported “Other” probe manufacturer were classified in the “Other (RNA)” group.
RESULTS
The analysis includes results from 95 participants who submitted 1268 survey results for 20 tissue cores in the following 5 ISH survey mailings: 2016-B, 2017-A, 2017-B, 2018-A, and 2018-B. Across all 5 surveys, there were 6 samples with 1 to 2 copies of HR-HPV per cell, 4 samples with 50 to 100 copies of HR-HPV per cell, 5 samples with 300 to 500 copies of HR-HPV per cell, and 5 samples that were negative for HR-HPV. Eighty-six percent (82 of 95) of participants were from the United States and the remaining participants were from Brazil (4), Saudi Arabia (3), Belgium (1), China (1), Colombia (1), Hong Kong (1), Thailand (1), and the United Arab Emirates (1). There was no difference in the percentage of participants who submitted the correct response for US versus non-US participants, 86.9% (966 of 1111) versus 85.4% (134 of 157), respectively (χ2 test; P = .58). Of the 88 participants with an institution classification in the CAP demographics database, 48.9% (43) were from academic (university or teaching) institutions, 33.0% (29) from independent/commercial reference laboratories, 12.5% (11) from nonhospitals, and 5.7% (5) from hospital/medical center laboratories.
The overall percentage of participants who submitted the correct response for the 1268 proficiency testing samples was 86.8% (1100). For the multivariate logistic model to test factors associated with performance differences between RNA and DNA ISH, there were statistically significant performance differences for the ISH target (DNA versus RNA) and HR-HPV copies/cell.
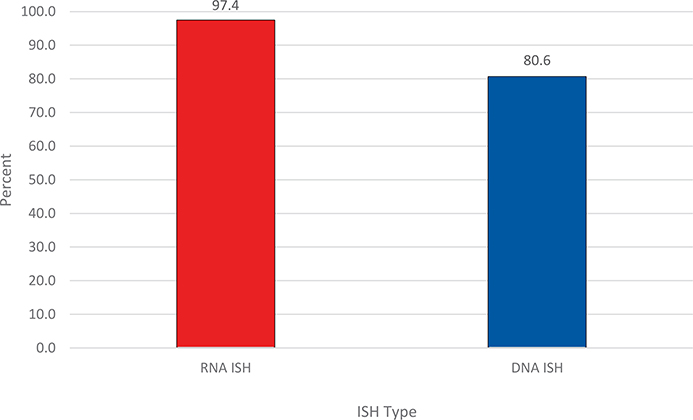
For ISH type, RNA ISH testing had a significantly higher percentage of participants who submitted the correct response than did DNA ISH, 97.4% (450 of 462) versus 80.6% (650 of 806) (P < .001), respectively (Figure 1; Table 1). The most specific performance differences were related to the copies of HR-HPV/cell.
Figure 1.
Overall percentage of proficiency testing participants who submitted the correct response for RNA and DNA ISH for high-risk human papillomavirus. P < .001. Abbreviation: ISH, in situ hybridization.
Table 1.
Summary of Performance Between High-Risk Human Papillomavirus (HR-HPV) RNA and DNA In Situ Hybridization (ISH)
| Performance Factor | No. of Responses | Correct No. (%) | P Valuea |
|---|---|---|---|
| ISH type | <.001 | ||
| RNA | 462 | 450 (97.4) | |
| DNA | 806 | 650 (80.6) | |
| HR-HPV copies/cell | |||
| Negative for HR-HPV | .99 | ||
| RNA | 117 | 116 (99.1) | |
| DNA | 203 | 199 (98.0) | |
| 1–2 copies/cell | <.001 | ||
| RNA | 126 | 120 (95.2) | |
| DNA | 240 | 129 (53.8) | |
| 50–100 copies/cell | <.001 | ||
| RNA | 89 | 89 (100.0) | |
| DNA | 156 | 119 (76.3) | |
| 300–500 copies/cell | .99 | ||
| RNA | 130 | 125 (96.2) | |
| DNA | 207 | 203 (98.1) | |
Boldface indicates statistical significance.
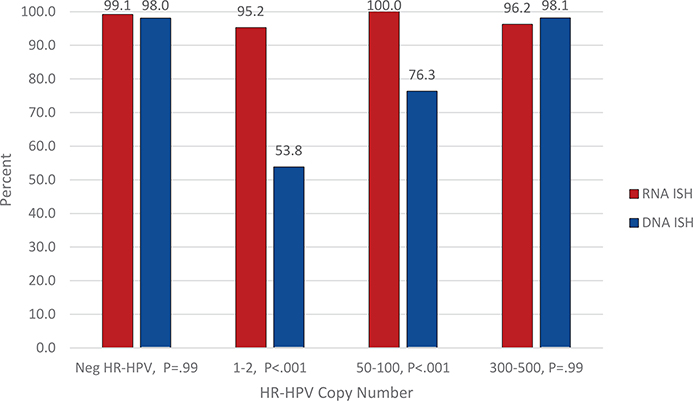
The ISH method comparisons are provided in Table 1 and graphically in Figure 2. When comparing RNA and DNA ISH, there were significant differences in HR-HPV detection for samples with 1 to 2 copies/cell (95.2% [120 of 126] versus 53.8% [129 of 240], P < .001, respectively) and 50 to 100 copies/cell (100% [89 of 89] versus 76.3% [119 of 156], P <.001, respectively). There was no significant difference between RNA and DNA ISH for samples with 300 to 500 copies of HR-HPV per cell, 96.2% (125 of 130) versus 98.1% (203 of 207) (P = .99), respectively. Likewise, there was no significant difference between RNA and DNA ISH for samples that were negative for HR-HPV, with negativity rates of 99.1% (116 of 117) versus 98.0% (199 of 203) (P = .99), respectively.
Figure 2.
Percentage of proficiency testing participants who submitted the correct response for RNA ISH versus DNA ISH by the HR-HPV copy number per cell. Abbreviations: HR-HPV, high-risk human papillomavirus; ISH, in situ hybridization.
There were also significant performance differences within ISH method. DNA ISH performed significantly better for samples with 300 to 500 copies/cell (98.1% [203 of 207]) and samples that were negative for HR-HPV (98.0% [199 of 203]) than for samples with 1 to 2 copies/cell (53.8% [129 of 240]), P < .001, and 50 to 100 copies/cell (76.3%, [119 of 156]), P < .001. DNA ISH also performed significantly better for samples with 50 to 100 copies/cell (76.3% [119 of 156]) versus samples with 1 to 2 copies/cell (53.8% [129 of 240]), P < .001. For RNA ISH, there were no significant performance differences based on the samples’ HR-HPV copies/cell (P = .23). The within ISH method results are provided in Table 2.
Table 2.
Summary of Performance Within High-Risk Human Papillomavirus (HR-HPV) RNA and DNA In Situ Hybridization (ISH) Methods for Viral Copies per Cell and Probe Manufacturer
| Performance Factor | No. of Responses | Correct No. (%) | P Valuea |
|---|---|---|---|
| RNA ISH | |||
| HR-HPV copies/cell | .23 | ||
| Negative for HR-HPV | 117 | 116 (99.1) | |
| 1–2 copies/cell | 126 | 120 (95.2) | |
| 50–100 copies/cell | 89 | 89 (100.0) | |
| 300–500 copies/cell | 130 | 125 (96.2) | |
| Probe manufacturerb | .42 | ||
| ACD | 272 | 270 (99.3) | |
| Ventana (RNA) | 158 | 149 (94.3) | |
| Other (RNA)b | 32 | 31 (96.9) | |
| DNA ISH | |||
| HR-HPV copies/cellc | <.001 | ||
| Negative for HR-HPV | 203 | 199 (98.0) | |
| 1–2 copies/cell | 240 | 129 (53.8) | |
| 50–100 copies/cell | 156 | 119 (76.3) | |
| 300–500 copies/cell | 207 | 203 (98.1) | |
| Probe manufacturerb | .47 | ||
| Enzo Biochem | 452 | 364 (80.5) | |
| Leica | 202 | 159 (78.7) | |
| Ventana (DNA) | 109 | 92 (84.4) | |
| Other (DNA)b | 39 | 32 (82.1) | |
Abbreviation: ACD, Advanced Cell Diagnostics.
Boldface indicates statistical significance.
“Other (RNA)” includes participants using RNA ISH who reported “Other” probe manufacturer. “Other (DNA)” includes Biocare Medical and participants using DNA ISH who reported “Other” probe manufacturer.
Negative for HR-HPV versus 1 to 2 copies/cell, P <.001. Negative for HR-HPV versus 50 to 100 copies/cell, P < .001; 1 to 2 copies/cell versus 50 to 100 copies/cell, P <.001; 1 to 2 copies/cell versus 300 to 500 copies/cell, P < .001; 50 to 100 copies/cell versus 300 to 500 copies/cell, P <.001.
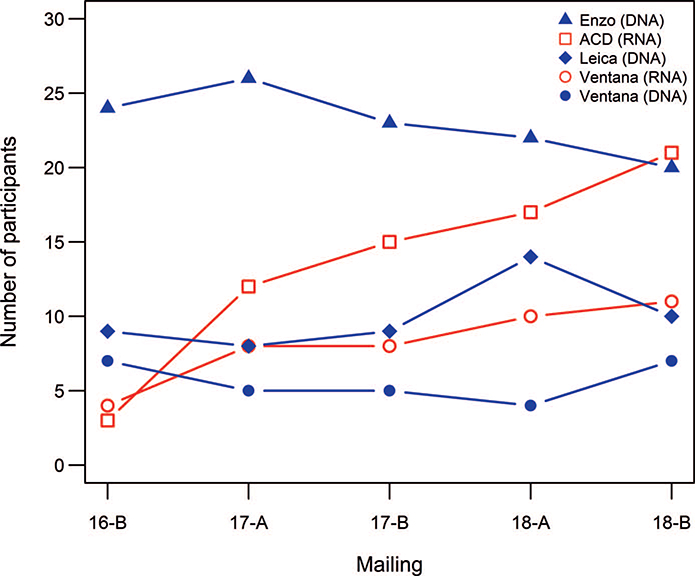
The number of participants using each probe manufacturer during the course of the study and by proficiency test mailing is listed in Table 3 and Figure 3, respectively. Performance categorized by the probe manufacturer is listed in Table 2. There were no significant performance differences between DNA-specific probe manufacturers (P = .47). Likewise, there were no significant performance differences between RNA-specific probe manufacturers (P = .42).
Table 3.
The Number of Participants Using Each Probe Manufacturera
| Probe Manufacturer | No. (%) n = 115 |
|---|---|
| Enzo (DNA) | 34 (29.6) |
| ACD (RNA) | 25 (21.7) |
| Leica (DNA) | 22 (19.1) |
| Ventana (RNA) | 16 (13.9) |
| Ventana (DNA) | 10 (8.7) |
| Other (DNA) | 4 (3.5) |
| Other (RNA) | 4 (3.5) |
Abbreviation: ACD, Advanced Cell Diagnostics.
Each participant may have used more than 1 probe manufacturer during the course of the study.
Figure 3.
Trend plot showing the number of participants using each specific probe manufacturer by proficiency test mailing. Abbreviation: ACD, Advanced Cell Diagnostics.
Overall, RNA ISH exhibited significantly higher analytical sensitivity than DNA ISH on proficiency testing samples, 96.8% versus 74.8% (P < .001), respectively. There was no statistically significant difference in analytical specificity between DNA and RNA ISH testing. These results are provided in Table 4.
Table 4.
Analytical Sensitivity and Specificity of RNA and DNA In Situ Hybridization (ISH) on Proficiency Testing Samples
|
Performance Results |
||||
| ISH Type | TP | TP + FN | Sensitivity, % (95% CI) | P Valuea |
| RNA | 334 | 345 | 96.8 (94.4–98.4) | <.001 |
| DNA | 451 | 603 | 74.8 (71.1–78.2) | |
| TN | TN + FP | Specificity, % (95% CI) | ||
| RNA | 116 | 117 | 99.1 (95.3–99.9) | .44 |
| DNA | 199 | 203 | 98.0 (95.0–99.5) | |
Abbreviations: FN, false negative; FP, false positive; TN, true negative; TP, true positive.
χ2 test. Boldface indicates statistical significance.
During the time course of the proficiency testing used for this study, 51 of the 95 laboratories (53.7%) used DNA probes only, 27 (28.4%) used RNA probes only, and 17 laboratories (17.9%) changed their test methodology from DNA to RNA ISH. In the subgroup of participants who changed from DNA to RNA ISH, there was a statistically significant improvement in performance with the mean percentage of correct results increasing 11.9% (P = .007; 95% CI, 3.8–20.1; SD = 15.8). There were no participants who switched from RNA ISH to DNA ISH.
Of the 95 laboratories that participated in the ISH proficiency test, 98.9% (94) reported performing at least 1 type of control sample and/or probe in conjunction with the assessment of clinical samples. Approximately 94% (88 of 94) of laboratories reported using positive or negative tissue controls, and just more than half of the laboratories (51.1%; 48) reported using a negative control probe. Of the laboratories performing RNA ISH and DNA ISH, 54.5% (24 of 44) and 31.8% (21 of 66) reported using an RNA or DNA control probe, respectively. There were no performance differences based on the type of control(s) used when adjusting for the ISH type, copies/cell, and manufacturer probe.
DISCUSSION
HPV-specific nucleic acid tests like ISH play a key role in the detection of HR-HPV in SCC, particularly in the resolution of a potential false-positive p16 immunohistochemical result. Importantly, the present study demonstrated that RNA ISH has significantly better overall sensitivity than DNA ISH for the determination of HR-HPV status in well-characterized proficiency testing samples. Our study found that the main difference is due to its superior analytical sensitivity in samples with low (1–2) to intermediate (50–100) HR-HPV copy numbers per cell. There was no significant difference between the analytical sensitivity of RNA and DNA ISH for samples with high (300–500) copy numbers. Likewise, there were no significant differences between the specificity of RNA and DNA ISH for HR-HPV–negative samples.
The most likely reason for the superior performance of RNA ISH compared to DNA ISH is the difference in the abundance of the target nucleic acids for each assay. For DNA ISH, samples with low viral copy numbers yield very small punctate signals that may be easily overlooked. Because RNA ISH targets the E6 and E7 RNA transcripts, these nucleic acids are typically more abundant and result in stronger, more robust signals that are more readily identified. In addition, the technical aspects of signal amplification used in the RNA ISH tests may play a role.
For the limited number of proficiency testing participants who transitioned from DNA to RNA ISH during the course of the study, the performance of each laboratory uniformly improved in a manner consistent with the performance differences between DNA and RNA ISH. This improvement suggests that the inferior performance by some laboratories is likely a consequence of the methodology used rather than the technical expertise of the laboratory itself.
Overall, 94 of 95 laboratories (98.9%) performed controls in conjunction with clinical testing. The performance of such controls is an accreditation requirement (CAP Checklist Items: ANP.22964, MIC.65800, MOL.39146) and demonstrates conformance to laboratory practices associated with high quality.19–21 The type of tissue controls and/or control probes used, however, was variable, but this was not associated with significant differences in performance.
Of note, there are several limitations of this study. First, the HR-HPV–positive samples used for this study are xenografts of cell lines and thus may not be entirely reflective of the range of samples seen in clinical practice. Specifically, the percentage of cases with 1 to 2 copies of HR-HPV per cell was 30% (6 of 20) and may be dissimilar to percentage distributions encountered in clinical practice. While the exact percentage of SCCs with low viral copy number is not precisely known, a study by Cerasuolo et al22 identified a wide range of low and high HR-HPV copy numbers in both cervical SCCs and OPSCCs, highlighting the need for assays that can accurately identify HR-HPV across that range. Furthermore, SCCs with low viral copy number may be just as transcriptionally active, if not more so, than cases with high viral copy numbers, further supporting the importance of detecting HR-HPV in these cases.22
Another potential drawback herein is that the analytical sensitivity and specificity calculations are based on the proficiency testing samples and may not necessarily reflect the sensitivity and specificity seen in clinical samples. Moreover, the samples used for proficiency testing are of high quality, with known specimen handling and are less likely to be susceptible to preanalytical problems that may affect RNA integrity, such as prolonged time to fixation and inadequate or prolonged fixation time. Lastly, the data for probe manufacturers are restricted in some cases by the number of participants and may not be an adequate representation of performance for those manufacturers.
CONCLUSIONS
HPV ISH is an important HPV-specific test methodology that complements p16 immunohistochemistry in the determination of HR-HPV status in patients with SCC. On CAP proficiency tests, RNA ISH for HR-HPV exhibits significantly higher accuracy than DNA ISH, a difference attributable to the higher analytical sensitivity of RNA ISH in tumors with low or intermediate HR-HPV viral copy numbers. These data support the use of RNA ISH over DNA ISH in clinical laboratories that perform HPV-specific testing of SCC as part of their testing algorithms.
Submit Your Research to the CAP20 Abstract Program.
Abstract and case study submissions to the College of American Pathologists (CAP) 2020 Abstract Program are now being accepted. Pathologists, laboratory professionals, and researchers in related fields are encouraged to submit original studies for possible poster presentation at the CAP20 meeting.
Submissions will be accepted until 5 p.m. Central time Tuesday, March 10, 2020. Accepted abstracts and case studies will appear on the Archives of Pathology & Laboratory Medicine Web site as a Web-only supplement to the September 2020 issue.
Visit the CAP20 Web site (www.thepathologistsmeeting.org) or the Archives Web site (www.archivesofpathology.org) for additional abstract program information including a link to the submission site.
Acknowledgments
The identification of specific products or scientific instrumentation is considered an integral part of the scientific endeavor and does not constitute endorsement or implied endorsement on the part of the authors, DoD, or any component agency. The views expressed in this article are those of the authors and do not reflect the official policy of the Department of Army/Navy/Air Force, Department of Defense, or US Government.
Footnotes
All others declare no potential conflicts of interest with the contents of this manuscript.
Contributor Information
Elaine S. Keung, Department of Pathology and Laboratory Medicine, Walter Reed National Military Medical Center, Bethesda, Maryland.
Rhona J. Souers, Biostatistics Department, College of American Pathologists, Northfield, Illinois.
Julia A. Bridge, Division of Molecular Pathology, The Translational Genomics Research Institute (TGen)/Ashion Laboratory, Phoenix, Arizona, and the Department of Pathology & Microbiology, University of Nebraska Medical Center, Omaha.
William C. Faquin, Department of Pathology, Massachusetts General Hospital, Harvard Medical School, Boston.
Rondell P. Graham, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota.
Meera R. Hameed, Department of Pathology, Memorial Sloan Kettering Cancer Center, New York, New York.
James S. Lewis, Jr, Department of Pathology, Microbiology, and Immunology, Vanderbilt University Medical Center, Nashville, Tennessee.
Jason D. Merker, Departments of Pathology and Laboratory Medicine & Genetics, Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill.
Patricia Vasalos, Proficiency Testing, College of American Pathologists, Northfield, Illinois.
Joel T. Moncur, Office of the Director, The Joint Pathology Center, Silver Spring, Maryland.
References
- 1.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011; 29(32):4294–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus–positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261–269. [DOI] [PubMed] [Google Scholar]
- 4.Ragin CCR, Taioli E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta-analysis. Int J Cancer. 2007;121(8):1813–1820. [DOI] [PubMed] [Google Scholar]
- 5.Bishop JA, Ma XJ, Wang H, et al. Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol. 2012; 36(12):1874–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Augustin J, Outh-Gauer S, Mandavit M, et al. Evaluation of the efficacy of the 4 tests (p16 immunochemistry, polymerase chain reaction, DNA, and RNA in situ hybridization) to evaluate a human papillomavirus infection in head and neck cancers: a cohort of 348 French squamous cell carcinomas. Hum Pathol. 2018;78:63–71. [DOI] [PubMed] [Google Scholar]
- 7.Mills AM, Dirks DC, Poulter MD, Mills SE, Stoler MH. HR-HPV E6/E7 mRNA in situ hybridization. Am J Surg Pathol. 2017;41(5):607–615. [DOI] [PubMed] [Google Scholar]
- 8.Ukpo OC, Flanagan JJ, Ma X, Luo Y, Thorstad WL, Lewis JS. High-risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol. 2011;35(9):1343–1350. [DOI] [PubMed] [Google Scholar]
- 9.Lewis JS, Beadle B, Bishop JA, et al. Human papillomavirus testing in head and neck carcinomas: guideline from the College of American Pathologists. Arch Pathol Lab Med. 2018;142(5):559–597. [DOI] [PubMed] [Google Scholar]
- 10.Mirghani H, Casiraghi O, Amen F, et al. Diagnosis of HPV-driven head and neck cancer with a single test in routine clinical practice. Mod Pathol. 2015; 28(12):1518–1527. [DOI] [PubMed] [Google Scholar]
- 11.Rooper LM, Gandhi M, Bishop JA, Westra WH. RNA in-situ hybridization is a practical and effective method for determining HPV status of oropharyngeal squamous cell carcinoma including discordant cases that are p16 positive by immunohistochemistry but HPV negative by DNA in-situ hybridization. Oral Oncol. 2016;55:11–16. [DOI] [PubMed] [Google Scholar]
- 12.Schlecht NF, Brandwein-Gensler M, Nuovo GJ, et al. A comparison of clinically utilized human papillomavirus detection methods in head and neck cancer. Mod Pathol. 2011;24(10):1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii T, Masumoto N, Saito M, et al. Comparison between in situ hybridization and real-time PCR technique as a means of detecting the integrated form of human papillomavirus 16 in cervical neoplasia. Diagn Mol Pathol. 2005; 14(2):103–108. [DOI] [PubMed] [Google Scholar]
- 14.Guo M, Baruch AC, Silva EG, et al. Efficacy of p16 and ProExC immunostaining in the detection of high-grade cervical intraepithelial neoplasia and cervical carcinoma. Am J Clin Pathol. 2011;135(2):212–220. [DOI] [PubMed] [Google Scholar]
- 15.Levi AW, Bernstein JI, Hui P, et al. A comparison of the Roche cobas HPV test with the Hybrid Capture 2 tests for the detection of high-risk human papillomavirus genotypes. Arch Pathol Lab Med. 2016;140(2):153–157. [DOI] [PubMed] [Google Scholar]
- 16.Meissner JD. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol. 1999;80(7):1725–1733. [DOI] [PubMed] [Google Scholar]
- 17.Mincheva A, Gissmann L, zur Hausen H. Chromosomal integration sites of human papillomavirus DNA in three cervical cancer cell lines mapped by in situ hybridization. Med Microbiol Immunol. 1987;176(5):245–256. [DOI] [PubMed] [Google Scholar]
- 18.Shibata DK, Arnheim N, Martin WJ. Detection of human papilloma virus in paraffin-embedded tissue using the polymerase chain reaction. J Exp Med. 1988; 167(1):225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CAP Accreditation Program. Anatomic Pathology Checklist. Northfield, IL: College of American Pathologists; 2018:38. [Google Scholar]
- 20.CAP Accreditation Program. Microbiology Checklist. Northfield, IL: College of American Pathologists; 2018:93. [Google Scholar]
- 21.CAP Accreditation Program. Molecular Pathology Checklist. Northfield, IL: College of American Pathologists; 2018:61. [Google Scholar]
- 22.Cerasuolo A, Annunziata C, Tortora M, et al. Comparative analysis of HPV16 gene expression profiles in cervical and in oropharyngeal squamous cell carcinoma. Oncotarget. 2017;8(21):34070–34081. [DOI] [PMC free article] [PubMed] [Google Scholar]