Abstract
Genetic and genomic architectures of traits under selection are key factors influencing evolutionary responses. Yet, knowledge of their impacts has been limited by a widespread assumption that most traits are controlled by unlinked polygenic architectures. Recent advances in genome sequencing and eco-evolutionary modeling are unlocking the potential for integrating genomic information into predictions of population responses to environmental change. Using eco-evolutionary simulations, we demonstrate that hypothetical single-locus control of a life history trait produces highly variable and unpredictable harvesting-induced evolution relative to the classically applied multilocus model. Single-locus control of complex traits is thought to be uncommon, yet blocks of linked genes, such as those associated with some types of structural genomic variation, have emerged as taxonomically widespread phenomena. Inheritance of linked architectures resembles that of single loci, thus enabling single-locus-like modeling of polygenic adaptation. Yet, the number of loci, their effect sizes, and the degree of linkage among them all occur along a continuum. We review how linked architectures are often associated, directly or indirectly, with traits expected to be under selection from anthropogenic stressors and are likely to play a large role in adaptation to environmental disturbance. We suggest using single-locus models to explore evolutionary extremes and uncertainties when the trait architecture is unknown, refining parameters as genomic information becomes available, and explicitly incorporating linkage among loci when possible. By overestimating the complexity (e.g., number of independent loci) of the genomic architecture of traits under selection, we risk underestimating the complexity (e.g., nonlinearity) of their evolutionary dynamics.
Keywords: climate change, evolutionary simulation, genetic architecture, linkage disequilibrium, recombination rate, structural genomic variation
Eco-evolutionary Responses Hinge on Genetic and Genomic Architecture
Predicting the responses of populations and species to anthropogenic disturbance is a major challenge, and one that requires urgent attention given the current climate and biodiversity crises (IPCC 2018; IPBES 2019). Advances in second- (high throughput) and third- (long read) generation sequencing have produced a wealth of sequence and structural genomic data on non-model organisms. These data have great potential to inform eco-evolutionary models of the responses of natural populations to a variety of selection pressures (Hoffmann et al. 2015; Coulson et al. 2017; Bay et al. 2017a). Such models can inform current management strategies and facilitate planning for future environmental conditions and associated ecosystem structures.
Key parameters influencing evolutionary responses are the genetic/genomic architectures underlying adaptive traits. These refer to how a trait is controlled by one or more genes and interactions among alleles (e.g., number and effect sizes of contributing loci, dominance, epistasis, pleiotropy), structural arrangement (e.g., inversions, fusions, translocations, duplications), position, and linkage among loci. These characteristics contribute to the inheritance models for genes underlying adaptive traits that are used in evolutionary predictions.
Here, we discuss some effects of genetic (the number of loci and their effect sizes) and genomic (the degree of linkage among loci) architectures on predictions of evolutionary responses to environmental disturbance. Single or unlinked loci have received comparatively more attention than linked genomic architectures in this regard (Bay et al. 2017a; Kardos and Luikart 2020). We demonstrate that hypothetical single locus control of a life-history trait under harvesting-induced selection generates a more variable evolutionary response compared to an unlinked polygenic scenario. We then suggest that linked polygenic architectures resemble those of single large-effect loci, yet exist along a continuum of linkage disequilibrium (LD), and are likely to play a large role in adaptation to rapid environmental change. We show that linked architectures underlie diverse traits in natural populations that are directly or indirectly under environmental selection. Finally, we discuss some barriers to modeling such architectures and challenges they present to conservation and management. More broadly, we aim to promote the integration of genomic data into eco-evolutionary modeling of responses to environmental change.
Large-Effect Loci Alter Evolutionary Predictions Compared With Traditional Polygenic Models
The degree to which a trait is controlled primarily by a single locus or multiple loci will influence its evolution in response to environmental stressors. Traditional evolutionary models have focused on the fixation dynamics of single-locus traits suddenly exposed to selection (Orr and Unckless 2014). As single-locus control of complex traits has been considered rare (Feder and Walser 2005), eco-evolutionary models of complex non-model organisms often incorporate a standard inheritance model of 10 or 20 unlinked loci (e.g., Kuparinen and Hutchings 2012). Multilocus (e.g., 100+ loci) models based on genomic single-nucleotide polymorphism (SNP) data have also been employed more recently to predict the capacity of a population to evolve in pace with global climate change (Bay et al. 2017a, 2018). Bay et al. (2017b) described a potential framework for genomic predictions of adaptive responses to environmental change, termed “evolutionary response architectures,” but focused on unlinked polygenic control of climate-associated traits.
The assumption that single genetic variants accounting for large amounts of phenotypic variation are rare is being challenged as more refined statistical genomics enable their discovery (Hoban et al. 2016). Large-effect loci have been documented in plants (Kivimäki et al. 2007; Baxter et al. 2010), insects (Reed et al. 2011), mammals (Johnston et al. 2013; Kardos et al. 2015; Jones et al. 2018; Barrett et al. 2019), birds (Toews et al. 2019; Merritt et al. 2020), and fishes (Colosimo et al. 2004; Lampert et al. 2010; Barson et al. 2015; Thompson et al. 2019). For example, the optix gene controls wing pattern in Heliconius butterflies (Reed et al. 2011), the RXFP2 gene controls horn architecture in Soay sheep (Johnston et al. 2013), and the Agouti locus controls coat color polymorphism in deer mice (Barrett et al. 2019). The implications of these variants for eco-evolutionary model predictions can be severe. After the discovery that the vgll3 gene is responsible for 40% of the variation in age at maturity in Atlantic salmon (Salmo salar) (Barson et al. 2015), Kuparinen and Hutchings (2017) demonstrated that hypothetical single-locus control of this key, sexually dimorphic life-history trait generates divergent and disruptive fisheries-induced evolution relative to that predicted by the classically applied, commonly assumed multi-locus model. As demonstrated in Box 1, these chaotic dynamics are largely driven by the single-locus control, not the sexual dimorphism, which indicates that they could occur for a broad range of traits. This finding is consistent with recent simulations demonstrating increased variability in population viability under rapid directional environmental change when the trait under selection is under single-locus rather than multi-locus control (Kardos and Luikart 2020). It further suggests that recovery following relaxation of selection is also highly variable.
Box 1. Drift dominates evolutionary trait dynamics of single-locus architectures.
We revisited the eco-evolutionary model developed by Kuparinen and Hutchings (2017) for Atlantic salmon. That model showed the evolution of age at maturity in response to fishing, assuming that it was controlled by a single locus with sexually dimorphic expression. Our objective was to compare single- and multi-locus architectures in a hypothetical species without sexually dimorphic trait expression, to illustrate how genetic architecture affects trait evolution in a more general scenario absent of sexual dimorphism. To this end, we took simple averages of the sex-specific probabilities for the male and female age at maturity in Atlantic salmon. By doing this, a male and a female carrying the same single-locus genotype have the same probabilities to mature at the ages of 1 sea winter (SW), 2 SW, and 3 SW (Table 2).
Table 2.
Probabilities for maturation in the single-locus scenario. The values are obtained as averages from the sex-specific probabilities reported by Barson et al. (2015)
| Timing of maturity | Homozygote (11) | Heterozygote (10 or 01) | Homozygote (00) |
|---|---|---|---|
| 2 SW → 3 SW | 0.5100 | 0.6130 | 0.9090 |
| 1 SW → 2 SW | 0.7950 | 0.2325 | 0.5660 |
Eco-evolutionary Model. The model simulates the annual demographic processes of mortality (both natural and fishing), maturation, and reproduction at the level of individuals. Parameters for these processes are based on Atlantic salmon except that the probability of maturing is not sex specific (Table 2). In practice, for each individual, its survival from the previous to the next time step is determined based on binomial trials (one for natural mortality and one for fishing mortality, when applicable). For the single-locus scenario, maturation is determined based on a binomial trial using the probabilities shown in Table 3. For the multilocus scenario, maturation is based on the sum across 10 loci with 2 alleles in each (allele sum is thus 0–20) coupled with phenotypic variability sampled from a normal distribution. Sums <6.66, 6.67–13.33, and >13.34 cause maturation at the age of 1 SW, 2 SW, and 3 SW, respectively. For each mature female, a mature male is sampled randomly and the number of eggs produced is based on empirical estimates (Table 3). The number of eggs surviving up to grilse is sampled from binomial distributions (probabilities given in Table 3). Genotypes of the newborn are sampled from the alleles of their parents and sex is assigned randomly with a 50:50 sex ratio. After these processes, the simulation proceeds to the next time step. Heritability was not held constant, but allowed to fluctuate with variation in allele frequencies. For further details about the simulation model, see Kuparinen and Hutchings (2017).
Table 3.
Demographic and fishing parameters for each age class, based on Atlantic salmon
| Parameter | Smolt (3 years) | 1 SW | 2 SW (immature/ mature) | 3 SW+ (immature/ mature) | References |
|---|---|---|---|---|---|
| Survival | 0.015 (egg to smolt) | 0.07 (smolt to grilse) | 0.9/0.2 | 0.9/0.2 | Hutchings and Jones (1998); Kuparinen and Hutchings (2017) |
| Fishing selectivity | — | 0.15 | 0.5 | 1 | Kuparinen and Hutchings (2017) |
| Eggs | — | 3040 | 7560 | 10 200 | Legault (2004) |
Simulation Design. To illustrate the conceptual differences between the single- and multilocus scenarios, we simulated 10 independent evolutionary trajectories for both scenarios and tracked the average age at maturity for each simulation time step. The simulations involved 3 phases: 1) pristine conditions in the absence of fishing (500 years, the first 400 of which were discarded as burn-in), 2) exposure to selective fishing mortality at the rate of 0.2 (selectivity given in Table 3), and 3) recovery in the absence of fishing.
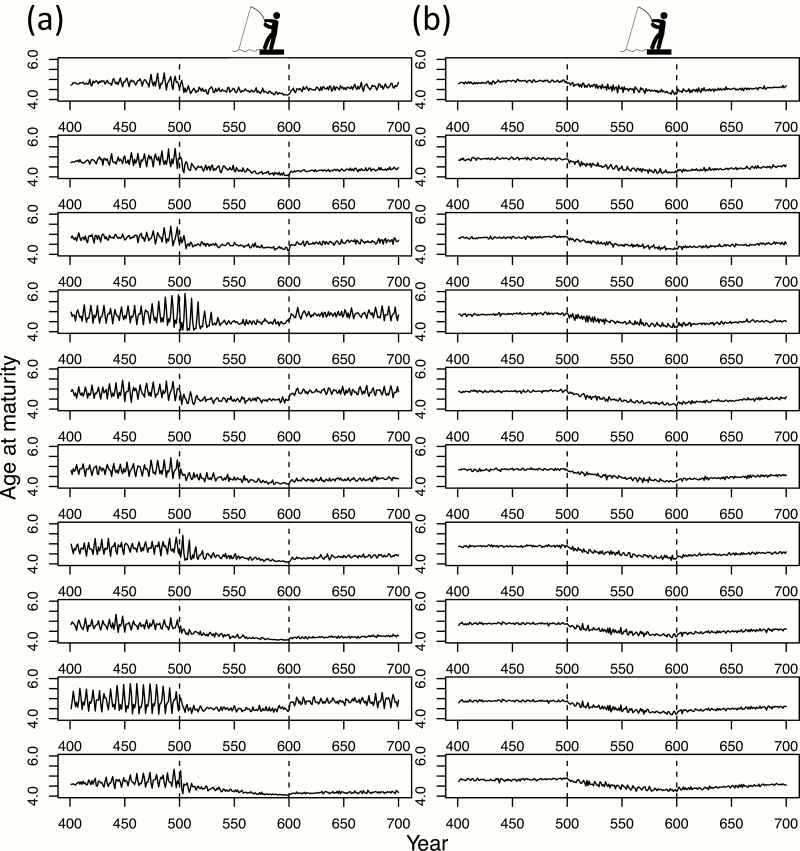
Results. The simulations demonstrate that single-locus control of age at maturity generates increased variability in this trait compared with the multi-locus scenario and that the response to, and recovery from, fishing are also highly varied (Figure 3). In the single-locus scenario, oscillations driven by heightened genetic drift leading to chaotic dynamics are evident under pristine conditions, reduced at varying rates under fishing pressure, and markedly return in only 3 of 10 simulations (Figure 3a). In contrast, the multilocus scenario does not generate chaotic dynamics and exhibits little variability both within and between simulations (Figure 3b). Consequently, single-locus control causes largely divergent and disruptive evolution of age at maturity with a wide variety of possible evolutionary trajectories and greater trait variability within trajectories, whereas polygenic control results in unidirectional evolution toward earlier maturation.
Figure 3.
The evolution of mean age at maturity in response to fishing for a hypothetical anadromous fish population under (a) single-locus and (b) multilocus scenarios for genetic architecture of the trait. Model parameters are based on Atlantic salmon except that the probability of maturing is not sex-specific for a given genotype. The beginning and end of the fishing period are indicated by dashed vertical lines. Each row represents one replicate simulation (N = 10).
Conclusion. Single-locus control of age at maturity results in highly unpredictable evolutionary and ecological responses to fishing-induced selection on this trait, relative to the commonly assumed multilocus control, due to greater phenotypic stochasticity (genetic drift). This appears to be the case with (Kuparinen and Hutchings 2017) and without (Figure 3) sexually dimorphic expression.
Inheritance of Linked Polygenic Architectures Resembles that of Single Loci
Particularly for complex traits, such as those contributing to growth, behavior, or environmental responses, polygenic control might indeed be the norm (Savolainen et al. 2013; Palumbi et al. 2014; Bay et al. 2017a). Yet, blocks of tightly linked putatively adaptive genes that undergo reduced or no recombination are taxonomically widespread (Nosil et al. 2009; Rogers et al. 2011; Yeaman 2013; Küpper et al. 2016; Wellenreuther and Bernatchez 2018; Pearse et al. 2019).
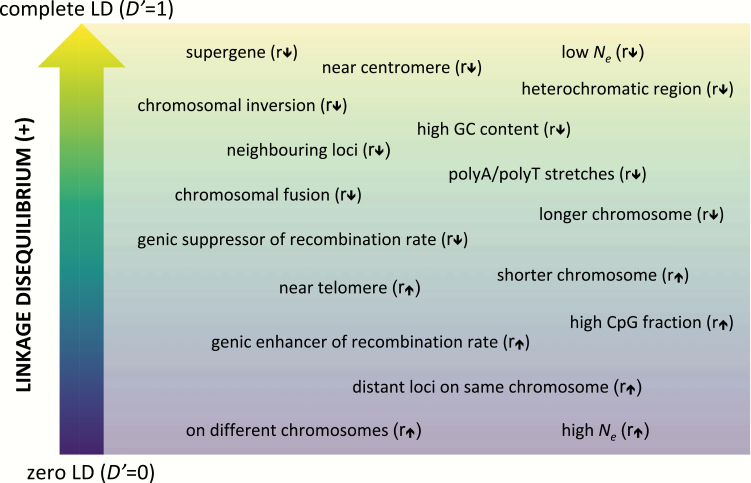
There are several mechanisms by which linked clusters might evolve in response to selection, but all are characterized by a reduction in recombination (Yeaman 2013). This is because when linkage captures an advantageous allelic combination, selection will favor a reduced recombination rate in that region to avoid splitting up complementary alleles (Charlesworth and Charlesworth 1979; Kirkpatrick and Barton 2006; Bürger and Akerman 2011; Yeaman and Whitlock 2011). Low recombination rates can be achieved by genic modifiers that decrease the frequency of crossovers during meiosis or genomic rearrangements that alter gene order, suppress recombination, and/or generate unbalanced gametes in heterozygotes (Figure 1; Butlin 2005; Ortiz-Barrientos et al. 2016). Recombination rate is also negatively correlated with epistasis, chromosome length, and proximity among loci (Figure 1; Kong et al. 2002; Butlin 2005). Sequence content can have both positive (CpG content) and negative (GC, polyA/polyT, and heterochomatin content) effects on recombination rate (Figure 1; Kong et al. 2002).
Figure 1.
Positive LD exists on a spectrum influenced by several factors potentially affecting recombination rate (r) among loci. D’ is a normalized metric of LD. For positive LD it is represented by the function D’ = (xaa − pa × qa)/min(xab, xba), whereby x is haplotype frequency and p and q are allele frequencies for two polymorphic loci with 2 alleles (a, b) each. Rather than a mechanistically accurate diagram, consider this figure as a roughly organized corkboard onto which the various factors and conditions have been pinned.
The lower the rate of recombination, the greater the degree of LD (nonrandom association of alleles at different loci). LD is also inversely proportional to effective population size through its relationship to genetic drift (Sved 1971; Waples et al. 2016), which could have notable consequences for small populations, as they would tend to harbor loci in higher LD. When LD is high, the inheritance pattern of a genomic region containing multiple loci resembles that of a single locus, such that complete linkage among genes would result in their coinheritance (Figure 2). Although, historically, precise estimates of LD have been challenging to obtain and are therefore not readily available for most species or populations, accessible high-throughput sequencing of non-model organisms is rapidly eliminating this barrier (Box 2). Other barriers to integrating linkage into eco-evolutionary models include a lack of understanding of the potential fitness effects of regions of high LD (e.g., partial sterility of heterozygotes, accumulation of deleterious mutations; Box 2).
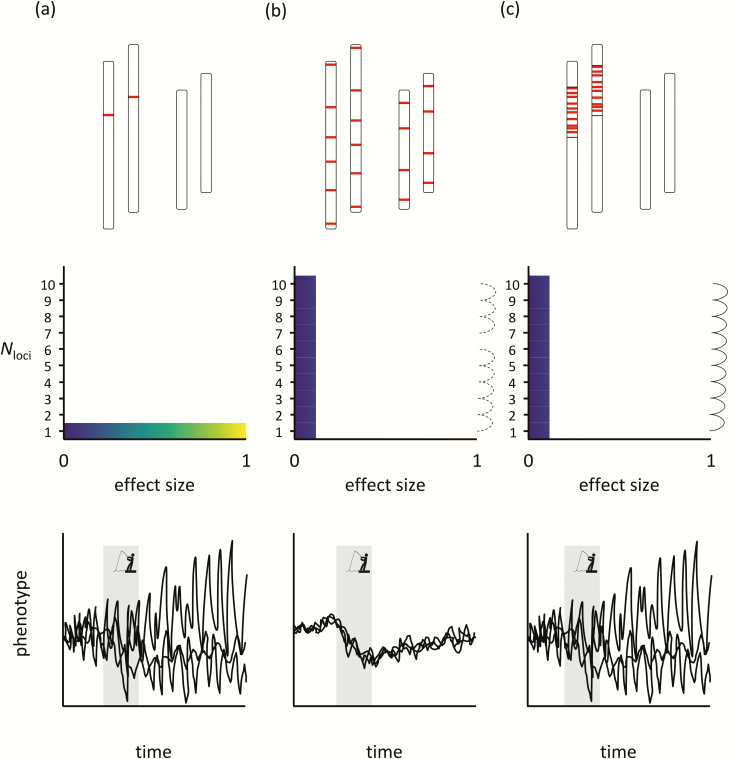
Figure 2.
Hypothetical genomic trait architectures (top row), corresponding model parameters (middle row), and evolutionary simulations (bottom row) for a population of diploid individuals with 2 chromosomes that is under a temporary period of directional selection (gray bars in the bottom row): (a) a single locus of large effect, (b) 10 loci of small effect with negligible LD , and (c) 10 loci of small effect with strong LD. In the top row, bars indicate the position of individual loci along a chromosome. Model parameters include the number of loci (Nloci), their effect sizes, and the degree of LD among them (when applicable), whereby continuous dashed lines indicate negligible levels of LD and continuous solid lines indicate strong LD. Individual black lines represent hypothetical replicate simulations (N = 3).
Box 2. Estimates of linkage disequilibrium and its effects on fitness are needed.
Estimates of Linkage Disequilibrium Are Becoming Widely Accessible
A major challenge to modeling the evolution of linked genomic architectures in diverse taxa has been the availability of relevant estimates for LD. LD depends on the distance and epistasis among loci, chromosome size, position of loci along a chromosome, local sequence content, other sources of recombination rate and gene conversion variation (e.g., genic modifiers and structural genomic polymorphisms), and demographic factors such as effective population size (Figure 1; Kong et al. 2002; Butlin 2005; Peñalba and Wolf 2020). LD can be estimated from individual-level population-scale genomic data (Gianola et al. 2013; Bilton et al. 2018; Ragsdale and Gravel 2020). For example, population-based inference of LD can be obtained by using multiple genome alignments to calculate the statistical association of alleles, ideally accounting for demographic history (Peñalba and Wolf 2020). One advantage of this approach is that the genomic data required are already available for many systems, which also explains the frequent use of such LD estimates as proxies for inferring population recombination rates (Peñalba and Wolf 2020).
More direct estimates of recombination and gene conversion rates can be obtained using pedigree-based and gamete-based approaches (e.g., Korunes and Noor 2019; Rowan et al. 2019; reviewed by Peñalba and Wolf 2020). Though they can be challenging to implement in some systems (e.g., wild populations lacking pedigree information and species without external fertilization for pedigree-based and gamete-based methods, respectively), recent technological advances have made them feasible for many non-model species (Peñalba and Wolf 2020). However, biological variation in recombination rates is abundant within and among chromosomes, individuals, sexes, populations, and species, the cataloging of which is still in its infancy (Peñalba and Wolf 2020). Importantly, even if accurate estimates of LD and/or recombination rate are obtained for a particular set of environmental conditions, recombination rates are often plastic (Stevison et al. 2017), complicating estimates of LD under environmental change. Nonetheless, the rapid advancement of this field will continue to eliminate the barriers associated with parameterizing LD in eco-evolutionary models.
Linked Architectures Have Fitness Effects Besides Those of the Target Phenotype
Besides effects of the trait that is the target of selection, other aspects of linked genomic architectures impact fitness and, consequently, the evolutionary trajectory of the architecture and trait. For example, linked architectures can prevent the purging of deleterious mutations, reducing the fitness of homozygotes (Jay et al. 2019). Conversely, heterozygotes for linked architectures can experience partial sterility due to the production of inviable gametes during meiosis, although strong evidence for this appears limited to plants (Hoffmann and Rieseberg 2008). Whether species outside of Diptera can displace crossovers away from the breakpoints of chromosomal rearrangements, thus altering their patterns of inheritance, is also poorly understood, as is taxonomic variation in other types of recombination modifiers. Furthermore, there is some probability of developing genetic incompatibilities at linked loci, which could be modeled explicitly. Finally, a better understanding of how eco-evolutionary feedbacks shape genomic architectures is needed.
Linked Polygenic Architectures Are Ubiquitous and Exist Along a Continuum of LD
Linked regions are often identified in population genomic studies as “genomic islands of divergence” (regions that exhibit greater differentiation than expected under neutrality; Wu 2001), although the degree of linkage varies depending on the mechanism of recombination suppression (Figure 1). For example, genic modifiers might only partially reduce recombination, leading to low or moderate levels of LD (Butlin 2005).
In contrast, extreme cases of tightly linked coadapted gene complexes associated with discrete complex phenotypes, known as supergenes, underlie key life-history traits in a variety of species (Schwander et al. 2014). Supergenes are often associated with structural genomic variation, which underlies complex phenotypes and adaptive processes in a wide variety of non-model taxa (Wellenreuther et al. 2019; Mérot et al. 2020a). Nonrecombining sex chromosomes are extreme examples of supergenes extending entire chromosomes, many of which evolved through structural genomic mutations (e.g., the mammalian Y chromosome evolved through a series of inversions following the formation of the Sex-Determining Region Y gene) (Lahn & Page 1999; Bachtrog 2013). There is emerging evidence that structural genomic variants might comprise the most important source of genomic variation in natural populations (Mérot et al. 2020a), as they have been found to account for several times more variation, in terms of the number of affected nucleotides, than SNPs (e.g., 3× in the Australasian snapper [Chrysophrys auratus; Catanach et al. 2019], 12× in Homo sapiens [Pang et al. 2010]).
One of the most well studied types of structural variant is the chromosomal inversion (Sturtevant 1921; Wellenreuther et al. 2019). Inversions prevent recombination within the inverted region by displacing crossovers away from breakpoints during meiosis (at least in Diptera flies) or by producing lethal meiotic products (if the inversion does not include the centromere) or inviable gametes (if the inversion spans the centromere) in heterokaryotypes, resulting in the selective recovery of nonrecombinant chromosomes (Rieseberg 2001; Hoffmann and Rieseberg 2008; Wellenreuther and Bernatchez 2018).
Other types of structural variation, such as chromosomal fusions and fissions, translocations, and copy number variants (CNVs), can also generate unbalanced gametes (Rieseberg 2001), which is expected to have similar implications for recombination rate reduction. Chromosomal fusions reduce recombination to a lesser extent than inversions, but to varying degrees in both heterozygotes and fused homozygotes (Bidau et al. 2001; Guerrero and Kirkpatrick 2014). There is also at least one example of a complex CNV maintaining linkage among candidate genes associated with multiple traits, effectively acting as a supergene (Tigano et al. 2018). Although the precise mechanisms for reducing recombination are not yet clear for many types of structural variation, the field is poised for major advances (Mérot et al. 2020a).
Linked Architectures Play a Large Role in Adaptation to Rapid Change
Linked architectures are hypothesized to facilitate rapid adaptation by enabling inheritance of coadapted gene complexes. Instead of accumulating beneficial alleles over multiple generations, they come as a package that has the potential to spread rapidly through a population, similar to a single large-effect gene (Kirkpatrick and Barrett 2015). Therefore, although the extent of recombination in linkage blocks can vary, a linked genomic architecture would enable single-locus-like modeling of polygenic adaptation. Their prevalence, especially the rising ubiquity of structural variation, necessitates a reassessment of common assumptions regarding the degree to which genes contributing to a polygenic trait are likely to be physically linked and/or experience reduced recombination.
“Mixed-Effect” Architectures Only Partially Alleviate Uncertainty
There are numerous examples of single loci (e.g., Carter 1977; Daetwyler et al. 2014; Carlson et al. 2016; Barrett et al. 2019) and blocks of tightly linked loci (e.g., supergenes; Schwander et al. 2014) controlling alternative phenotypes in a seemingly discrete, “monogenic” manner. Yet, mixed architectures consisting of a large-effect locus, supergene, or haploblock in addition to numerous small-effect, potentially unlinked, loci are likely common as well (herein, referred to as “mixed-effect architectures”). Small-effect variants accompanying those of large effect are challenging to detect using common approaches (e.g., genome-wide and gene–environment associations) given the difficulties of distinguishing weak signatures of selection from demographic processes or selection on other traits (Hoban et al. 2016; Stephan 2016). Thus, whether large-effect variants are truly monolithic is especially difficult to confirm outside of domesticated and model species owing to diverse genomic backgrounds and environmental influences.
Sinclair-Waters et al. (2020) recently characterized the mixed-effect architecture of age-at-maturity in Atlantic salmon, using an extensive genome-wide association study of 11,166 males from a single aquaculture strain, combined with high-density SNP arrays and pedigree information. Including the previously known large-effect vgll3 and six6 loci, they identified 120 genes contributing to age-at-maturity with various effect sizes. One would expect such mixed-effect architectures to exhibit evolutionary dynamics intermediate to those of single locus and highly polygenic scenarios: the addition of many small-effect loci along with a large-effect locus should reduce the stochasticity and trait variance within and between populations. Indeed, Kardos and Luikart (2020) simulated a range of mixed-effect architectures underlying a phenotypic response to a sudden environmental shift and obtained intermediate levels of average phenotype, population viability, and extinction rate relative to the single locus and highly polygenic models. Therefore, although mixed-effect architectures seem to generate somewhat more predictable evolutionary dynamics compared with single-locus architectures, they are unlikely to completely alleviate the concerning degree of stochasticity that seems to be characteristic of architectures with major effect loci. Therefore, the evolution of age-at-maturity in Atlantic salmon is still likely to exhibit increased variability relative to the classical multi-locus model of genomic architecture, but unlikely to be as extremely varied as the hypothetical single-locus scenario presented previously (Kuparinen and Hutchings 2017).
Unfortunately, such large-scale, high-throughput approaches as used for Atlantic salmon (Sinclair-Waters et al. 2020) are not feasible for most non-model species, which typically lack pedigree information and sufficient sample sizes and genomic resources, especially when they are of conservation concern. When the precise architecture is unknown, the genomic background (e.g., population or ecotype) in which a large-effect variant occurs can be considered as to whether it potentially influences expression of the focal variant for the trait of interest, but only if the variant is polymorphic within different genomic backgrounds. If the genomic background alters trait expression within particular genotypes of the focal variant (and independently of environmental factors), then there must be additional loci affecting the trait. How best to model mixed-effect architectures when they can be characterized requires further investigation and we encourage research in this direction. Nonetheless, in the next section, we seek to highlight the broad potential of modeling tightly linked architectures as single loci for the purpose of predicting responses of natural populations to environmental disturbance. Acknowledging the prevalence and ubiquity of large-effect linked architectures, in addition to single loci of large effect and mixed-effect architectures, is a critical next step towards modeling the full spectrum of genomic architectural complexity.
Linked Genomic Architectures Underlie Diverse Traits in Natural Populations That Are Directly or Indirectly Under Environmental Selection
In recent years, linked genomic architectures have been associated with a variety of adaptive traits in natural populations (Table 1). Although inversions appear to be the most commonly studied (Wellenreuther and Bernatchez 2018; Wellenreuther et al. 2019), chromosomal fusions (Wellband et al. 2019) and complex architectures involving multiple rearrangements (Tigano et al. 2018; Pearse et al. 2019) are also associated, directly or indirectly, with traits relevant for adaptation. In some cases, SNPs located in proximity to one another are found to be in LD and the structural architecture is yet to be determined (e.g., Micheletti et al. 2018). Considering that the cataloguing of structural genomic variation is still in its infancy and that our understanding of recombination rate variation is lacking (Mérot et al. 2020a), we adopt an inclusive approach regarding the examples discussed.
Table 1.
Genomic regions in linkage disequilibrium that are associated with environmental or life-history traits under selection. Variant names and/or locations are specified differently across taxa and depending on the type of genomic information available. Specific names/locations are not provided if there are more than 5 variants per row
| Species name | Common name | Variant architecture | Variant names and/or locations | Associated phenotype | References |
|---|---|---|---|---|---|
| Plants | |||||
| Boechera stricta | Drummond’s rockcress | Inversion | LG1 | Environmental adaptation (water regime) | Lee et al. (2017) |
| Helianthus spp. | Sunflower | Haploblocks and inversions | genome-wide (N = 37) | Environmental adaptation (climate) and reproduction (morphology) | Todesco et al. (2020) |
| Mimulus guttatus | Monkeyflower | Inversion | DIV1 on chromosome 8 | Perenniality | Lowry and Willis (2010); Twyford and Friedman (2015); Coughlan and Willis (2019) |
| Zea mays ssp. mays | Highland maize | Inversion | Inv4m | Environmental adaptation (highland) | Crow et al. (2019) |
| Invertebrates | |||||
| Anopheles spp. | Mosquito | Inversions | genome-wide (N=17) | Environmental adaptation (latitudinal clines, altitude-associated habitat) | Ayala et al. (2014, 2017) |
| Apis mellifera | East African honeybee | Inversions | r7, r9 | Environmental adaptation (altitude- associated habitat) | Wallberg et al. (2017); Christmas et al. (2019) |
| Coelopa frigida | Seaweed fly | Inversion | Chromosome 1 | Environmental adaptation (latitudinal clines) | Mérot et al. (2018); Mérot et al. (2020b) |
| Drosophila melanogaster | Fruit fly | Inversions | In(3R)Payne (3RP), In(3L) P, In(3R)C, In(2L)t | Environmental adaptation (latitudinal and altitudinal clines) | Krimbas and Powell (1992); Anderson et al. (2005); Rane et al. (2015); Kapun and Fabian (2016); Kapun and Flatt (2019) |
| Drosophila mojavensis | Fruit fly | Inversions | Chromosome 2 (N = 7) | Environmental adaptation (desert and/ or host plant) | Guillén and Ruiz (2012) |
| Drosophila pseudoobscura | Fruit fly | Inversions | AR, PP, CH, ST, TL | Environmental adaptation (latitudinal clines) | Schaeffer (2008); Fuller et al. (2016, 2017) |
| Littorina saxatilis | Rough periwinkle | Inversions, putative inversions | Genome-wide (N = 17) | Environmental adaptation (wave vs. crab ecotype, low-shore vs. high shore) | Faria et al. (2019); Morales et al. (2019) |
| Fishes | |||||
| Gadus morhua | Atlantic cod | Inversions | LG02, LG07, LG12 | Environmental adaptation (latitudinal and salinity clines) | Bradbury et al. (2010); Berg et al. (2015); Sodeland et al. (2016); Barth et al. (2017, 2019) |
| Double inversion | LG01 | Migratory behavior | Berg et al. (2016); Kirubakaran et al. (2016); Berg et al. (2017); Kess et al. (2019) | ||
| Menidia menidia | Atlantic silverside | Block of differentiation | Chromosome 24 | Environmental adaptation (latitudinal clines), growth | Therkildsen et al. (2019) |
| Salmo salar | Atlantic salmon | Fusion | Ssa08/Ssa29 | Environmental adaptation (summer precipitation) | Wellband et al. (2019) |
| Linked genomic region | Ssa09 | Sea age at maturity, run timing | Johnston et al. (2014); Barson et al. (2015); Cauwelier et al. (2018) | ||
| Oncorhynchus mykiss | Rainbow trout | Double inversion | Omy05 | Migratory behavior (anadromous vs. resident and fluvial vs. adfluvial), environmental adaptation (latitudinal and temperature clines) | Pearse et al. (2014); Arostegui et al. (2019); Pearse et al. (2019) |
| Oncorhynchus nerka | Sockeye salmon | Linked genomic region | Ssa09 homolog | Environmental adaptation (stream- vs. lake-spawning ecotypes) | Veale and Russello (2017) |
| Clupea harengus | Atlantic herring | Inversion | Chromosome 12 | Environmental adaptation (latitudinal clines), spawning time | Barrio et al. (2016); Lamichhaney et al. (2017); Fuentes-Pardo et al. (2019); Pettersson et al. (2019) |
| Oncorhynchus tshawytscha | Chinook salmon | Linked genomic region | Ots28 | Timing of arrival to spawning grounds | Narum et al. (2018) |
| Ammodytes tobianus | Lesser sandeel | Haploblock | Unknown (13 linked SNPs) | Putative environmental adaptation (sea bottom temperature) | Jiménez-Mena et al. (2020) |
| Oncorhynchus mykiss | Steelhead trout | Linked genomic region | Omy28 | Migration timing | Micheletti et al. (2018) |
| Mallotus villosus | Capelin | Fusion | Chromosomes 2 and 9 | Environmental adaptation (spawning site, temperature) | Cayuela et al. (2020) |
| Gasterosteus aculeatus | Three-spined stickleback | Inversions | Chromosomes 1, 11, and 21 | Environmental adaptation (marine vs. freshwater) | Jones et al. (2012) |
| Pleuronectes platessa | European plaice | Putative inversions | SV19, SV21 | Environmental adaptation (latitudinal and salinity clines) | Le Moan et al. (2019a, 2019b) |
| Aves | |||||
| Uria aalge | Common murre | Complex copy number variant | Scaffold 72 | Plumage coloration, thermal adaptation | Tigano et al. (2018) |
Linked Architectures Are Favored by Selection Under Gene Flow
Theoretical work has implicated the genetic and genomic architecture of adaptive traits as the key element in determining whether they will be lost under gene flow (Bürger & Akerman 2011; Yeaman and Whitlock 2011; Yeaman 2013; Aeschbacher and Bürger 2014; Akerman and Bürger 2014). Alleles with large effect sizes are less likely to be overwhelmed by gene flow (Yeaman and Otto 2011), as are tightly linked polygenic architectures because they effectively act as a single large-effect locus (Griswold 2006; Yeaman and Otto 2011). Gene flow increases the risk of breaking up coadapted alleles, thereby selecting for reduced recombination and increased linkage (Nosil et al. 2009; Tigano and Friesen 2016).
The evolution of linked architectures under gene flow might explain why they appear to be common in species with high dispersal capabilities: flying insects, birds, and fishes (Table 1; Wellenreuther and Bernatchez 2018). Many examples of extremely tight linkage derive from systems in which closely related species or ecotypes are living in sympatry (Nosil et al. 2009; Hooper 2016). Yet, high gene flow is pervasive in the natural world, including many plants with high seed dispersal and marine organisms with pelagic early life stages. Therefore, linked architectures are likely common.
Linked Architectures Are Often Directly Associated With Environmental Adaptation
Linked architectures associated with adaptation to local environmental variables seem particularly common among flies and fishes (Table 1; Wellenreuther and Bernatchez 2018). Flies in the genus Drosophila provide numerous examples of inversions associated with environmental adaptation, including those exhibiting latitudinal (Krimbas and Powell 1992; Anderson et al. 2005; Rane et al. 2015; Fuller et al. 2016; Kapun and Fabian 2016; Fuller et al. 2017; Kapun and Flatt 2019) and altitudinal (Kapun and Flatt 2019) clines, as well as adaptation to a desert environment (possibly through the host plant; Guillén and Ruiz 2012). Of particular interest from a human health perspective, environmentally structured inversion polymorphisms are common among Anopheles spp. mosquitos, potentially enhancing the adaptability and vector potency of these primary malaria vectors (Ayala et al. 2014, 2017). Inversions in Anopheles spp. vary in karyotype frequencies across latitudinal clines and between mountain forests and lowland savannahs (Ayala et al. 2017), and are associated with aridity tolerance.
Inversions and blocks of differentiation (which may or may not be associated with inversions) appear to underlie adaptation to salinity (Jones et al. 2012; Berg et al. 2015) and vary in frequency across latitudinal clines in marine fishes, suggesting an association with temperature or growing season length (Pettersson et al. 2019; Therkildsen et al. 2019; Kess et al. 2020). Chromosomal translocations and fusions are common among salmonids (Phillips 2005). Their adaptive significance, if any, is not generally known, although a fusion in Atlantic salmon (Ssa08/Ssa29) is associated with summer precipitation in a Canadian river system (Wellband et al. 2019).
Although we have focused on adaptation to abiotic environmental variables, traits controlled by linked architectures can also be associated with the biotic environment. For example, cryptic coloration in timena stick insects (Lindtke et al. 2017; Lucek et al. 2019) and mimicry patterns in butterflies (Joron et al. 2013; Nishikawa et al. 2015), associated with a large haploblock and inversions, respectively, are under selection via local host plants and predators associated with particular environments. Changes in these environments can alter camouflage substrates and predator distributions. Adaptation will therefore depend partly on the evolutionary dynamics of the linked regions.
Linked Architectures Are Also Indirectly Associated With Environmental Adaptation
Architectures that link genes controlling several types of co-adapted traits can result in indirect selection on traits associated with environmental adaptation. The inversions in Anopheles spp. associated with differences in aridity tolerance (Cheng et al. 2018) are also associated with morphology and behavior (reviewed by Ayala et al. 2014). Inversions underlie alternative reproductive phenotypes in some birds (Thomas et al. 2008; Horton et al. 2014; Küpper et al. 2016; Zinzow-Kramer et al. 2015), simultaneously controlling morphological (e.g., plumage coloration), behavioral (e.g., mating tactic), and life history (e.g., maturation, growth rate) traits. Common murres (Uria aalge) have a complex CNV maintaining linkage among genes associated with plumage coloration and thermal tolerance despite random mating (Tigano et al. 2018).
Alternate behavioral or life history strategies often impose different environmental selection pressures. Fish populations that migrate between freshwater and saltwater for reproduction and feeding require different temperature and salinity adaptations compared to resident populations that do not migrate. Consequently, linked architectures in marine and freshwater fishes are associated with coexisting migratory ecotypes experiencing different environments (Table 1; Pearse et al. 2014; Berg et al. 2016; Kirubakaran et al. 2016; Arostegui et al. 2019; Kess et al. 2019; Pearse et al. 2019). For example, a double inversion in steelhead/rainbow trout (Oncorhynchus mykiss) varies in frequency between anadromous (maturing at sea) and resident populations, as well as between fluvial (maturing in rivers) and adfluvial (maturing in lakes) populations, and exhibits latitudinal- and temperature-associated frequency clines (Pearse et al. 2014; Arostegui et al. 2019; Pearse et al. 2019).
Variation in reproductive timing can require adaptations to different environmental conditions, such as different temperatures experienced during early life (Oomen and Hutchings 2015, 2016), which could favor architectures that link genes associated with environmental and life history traits. An inversion in Atlantic herring (Clupea harengus) is associated with both temperature and timing of reproduction (Fuentes-Pardo et al. 2019; Pettersson et al. 2019), whereas linked regions in Chinook salmon (O. tshawytscha; Narum et al. 2018) and steelhead (Micheletti et al. 2018) are associated with the timing of arrival to spawning grounds.
Many social traits in insects, birds, fishes, and plants are controlled by linked architectures due to bidirectional influences between social behavior and genome architecture (reviewed by Rubenstein et al. 2019). For example, a supergene containing multiple chromosomal rearrangements underlies several social traits in the fire ant (Solenopsis invicta; Huang et al. 2018). The connection between social traits and linked architectures has broad relevance for predicting responses to environmental change, as sociality itself is often under environmental selection. For example, thermal stress and resource scarcity select for more or less sociality in different species and contexts (Doering et al. 2018; Kao et al. 2020). Further, social traits are often correlated with phenotypes that might be under selection from anthropogenic stressors such as climate change and harvesting (e.g. alternative mating tactics and growth rate in swordtail fish [Xiphophorus spp.; Lampert et al. 2010]).
Theory predicts that supergenes are also likely to arise when there is coevolution between social traits and dispersal, because dispersal will be selected against in benevolent individuals so that they tend to interact with relatives and selected for in selfish individuals so that they tend to interact with nonrelatives (Mullon et al. 2018; Rubenstein et al. 2019). Therefore, linkage between genes for dispersal traits (e.g., locomotion, physiology) and social behavior is expected to evolve under a variety of circumstances (Rubenstein et al. 2019). As dispersal is one of the primary mechanisms of organismal responses to environmental change, control by linked architectures will likely alter predictions of responses to disturbance in a wide array of taxa.
Therefore, selection on diverse traits could indirectly impose selection on traits associated with environmental adaptation and incorporating linkage when modeling environmental responses has broad taxonomic utility.
Flexible Ecogenetic Models Can Reflect a Diversity of Genomic Architectures
The best modeling strategy will depend on what is known regarding the genomic trait architecture. For most traits, the precise architecture is not known and is often estimated to consist of between 10 and 100 unlinked loci of equal effects (e.g., Kuparinen and Hutchings 2012). Given the rising prevalence of major effect loci and tightly linked architectures, exploring both extremes—single locus, representing both major effect and tightly linked loci, and highly polygenic, unlinked loci—is warranted. In reality, mixed architectures are likely common and are expected to produce intermediate levels of stochasticity (Kardos and Luikart 2020). Yet, the output of these extreme scenarios will be informative about the range and distribution of possible outcomes and the sensitivity of the model to the genomic architecture in a particular case (e.g., life history or selection regime). As genomic information becomes available, more precise estimates of the number of loci, their effect sizes, and the degree of linkage among them can be incorporated into eco-evolutionary models.
Conclusion
The challenges of obtaining estimates of LD are being rapidly overcome by high-throughput sequencing and advances in statistical genomics (Peñalba and Wolf 2020; Box 2). Our understanding of the fitness effects of linked genomic architectures is also poised for great improvements in the near future due to increased quality and accessibility of genomic resources for non-model species and, consequently, heightened interest in the topic (Wellenreuther et al. 2019; Mérot et al. 2020a; Box 2). Nonetheless, simple approximations can be obtained in the meantime by treating tightly linked genomic architectures as single loci of large effect. Although this approach is likely too simplistic for some purposes, it has been shown to be more powerful for detecting genotype–environment associations with linked haploblocks compared to characterizing genotypes based on SNPs within the blocks (Todesco et al. 2020). Ultimately, we view single-locus approximations of tightly linked architectures as a useful counterpoint to the conventional way of thinking about and modeling polygenic architectures (i.e., the quantitative genetics paradigm, which should be continually revisited in light of genomic data; Nelson et al. 2013). Therefore, when the genomic architecture is not known, a precautionary approach considers the greater variability and higher uncertainty that appears to be characteristic of a single-locus scenario (also see Kardos and Luikart 2020). Rapid developments in the fields of recombination rate variation and the population genomics of structural variants will continue to improve predictions borne from genomic data.
Implementing genomic architecture into spatially or temporally explicit conservation and management plans presents additional considerations. For example, variable rates of gene flow within species could result in different genomic architectures underlying adaptation at different spatial and temporal scales (Nosil et al. 2009; Oomen 2019). The same trait could be under selection at both scales, as contrasting genomic architectures can produce similar phenotypic outcomes (Therkildsen et al. 2019). The diversity of genomic architectures underlying the same or different traits also complicates the process of delineating evolutionarily significant units for conservation, particularly when the relative fitness consequences of trait variation is unclear (Waples and Lindley 2018; Waples et al. 2020). Nonetheless, it is clear that we must consider linked genomic architectures underlying adaptive traits when predicting the consequences of environmental disturbance to natural populations. Otherwise, by overestimating the complexity (e.g., number of independent loci) of the genomic architecture of traits under selection, we risk underestimating the complexity (e.g., nonlinearity) of their evolutionary dynamics.
Funding
This work was supported by a James S. McDonnell Foundation 21st Century Postdoctoral Fellowship Award to R.A.O.; the Academy of Finland to A.K.; the European Research Council (grant number COMPLEX-FISH 770884) to A.K.; the Natural Sciences and Engineering Research Council of Canada Discovery Grant to J.A.H.; the Killam Trusts to J.A.H.; and Loblaw Companies Limited to J.A.H. The present study reflects only the authors’ view and the European Research Council is not responsible for any use that may be made of the information it contains.
Acknowledgments
We thank Robin Waples for the invitation to submit this manuscript and feedback on it, as well as Marty Kardos and an anonymous reviewer for helpful comments. We also thank Marine Brieuc and Mitchell Newberry for useful discussions on aspects of the manuscript.
References
- Aeschbacher S, Bürger R. 2014. The effect of linkage on establishment and survival of locally beneficial mutations. Genetics. 197:317–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman A, Bürger R. 2014. The consequences of gene flow for local adaptation and differentiation: a two-locus two-deme model. J Math Biol. 68:1135–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AR, Hoffmann AA, McKechnie SW, Umina PA, Weeks AR. 2005. The latitudinal cline in the In(3R)Payne inversion polymorphism has shifted in the last 20 years in Australian Drosophila melanogaster populations. Mol Ecol. 14:851–858. [DOI] [PubMed] [Google Scholar]
- Arostegui MC, Quinn TP, Seeb LW, Seeb JE, McKinney GJ. 2019. Retention of a chromosomal inversion from an anadromous ancestor provides the genetic basis for alternative freshwater ecotypes in rainbow trout. Mol Ecol. 28:1412–1427. [DOI] [PubMed] [Google Scholar]
- Ayala D, Acevedo P, Pombi M, Dia I, Boccolini D, Costantini C, Simard F, Fontenille D. 2017. Chromosome inversions and ecological plasticity in the main African malaria mosquitoes. Evolution. 71:686–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D, Ullastres A, González J. 2014. Adaptation through chromosomal inversions in Anopheles. Front Genet. 5:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y chromosome evolution: emerging insights into processes of Y chromosome degeneration. Nat Rev Genet. 14:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RDH, Laurent S, Mallarino R, Pfeifer SP, Xu CCY, Foll M, Wakamatsu K, Duke-Cohan JS, Jensen JD, Hoekstra HE. 2019. Linking a mutation to survival in wild mice. Science. 363:499–504. [DOI] [PubMed] [Google Scholar]
- Barrio AM, Lamichhaney S, Fan G, Rafati N, Pettersson M, Zhang H, Dainat J, Ekman D, Höppner M, Jern P, et al. 2016. The genetic basis for ecological adaptation of the Atlantic herring revealed by genome sequencing. eLife. 5:e12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barson NJ, Aykanat T, Hindar K, Baranski M, Bolstad GH, Fiske P, Jacq C, Jensen AJ, Johnston SE, Karlsson S, et al. 2015. Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature. 528:405–408. [DOI] [PubMed] [Google Scholar]
- Barth JMI, Berg PR, Jonsson PR, Bonanomi S, Corell H, Hemmer-Hansen J, Jakobsen KS, Johannesson K, Jorde PE, Knutsen H, et al. 2017. Genome architecture enables local adaptation of Atlantic cod despite high connectivity. Mol Ecol. 26:4452–4466. [DOI] [PubMed] [Google Scholar]
- Barth JMI, Villegas-Ríos D, Freitas C, Moland E, Star B, André C, Knutsen H, Bradbury I, Dierking J, Petereit C, et al. 2019. Disentangling structural genomic and behavioural barriers in a sea of connectivity. Mol Ecol. 28:1394–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Brazelton JN, Yu D, Huang YS, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz JO, Nordborg M, et al. 2010. A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet. 6:e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bay RA, Harrigan RJ, Underwood VL, Gibbs HL, Smith TB, Ruegg K. 2018. Genomic signals of selection predict climate-driven population declines in a migratory bird. Science. 359:83–86. [DOI] [PubMed] [Google Scholar]
- Bay RA, Rose N, Barrett R, Bernatchez L, Ghalambor CK, Lasky JR, Brem RB, Palumbi SR, Ralph P. 2017a. Predicting responses to contemporary environmental change using evolutionary response architectures. Am Nat. 189:463–473. [DOI] [PubMed] [Google Scholar]
- Bay RA, Rose NH, Logan CA, Palumbi SR. 2017b. Genomic models predict successful coral adaptation if future ocean warming rates are reduced. Sci Adv. 3:e1701413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg PR, Jentoft S, Star B, Ring KH, Knutsen H, Lien S, Jakobsen KS, André C. 2015. Adaptation to low salinity promotes genomic divergence. Genome Biol Evol. 7:1644–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg PR, Star B, Pampoulie C, Bradbury IR, Bentzen P, Hutchings JA, Jentoft S, Jakobsen KS. 2017. Trans-oceanic genomic divergence of Atlantic cod ecotypes is associated with large inversions. Heredity (Edinb). 119:418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg PR, Star B, Pampoulie C, Sodeland M, Barth JM, Knutsen H, Jakobsen KS, Jentoft S. 2016. Three chromosomal rearrangements promote genomic divergence between migratory and stationary ecotypes of Atlantic cod. Sci Rep. 6:23246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidau CJ, Giménez MD, Palmer CL, Searle JB. 2001. The effects of Robertsonian fusions on chiasma frequency and distribution in the house mouse (Mus musculus domesticus) from a hybrid zone in northern Scotland. Heredity (Edinb). 87:305–313. [DOI] [PubMed] [Google Scholar]
- Bilton TP, McEwan JC, Clarke SM, Brauning R, van Stijn TC, Rowe SJ, Dodds KG. 2018. Linkage disequilibrium estimation in low coverage high-throughput sequencing data. Genetics. 209:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury IR, Hubert S, Higgins B, Borza T, Bowman S, Paterson IG, Snelgrove PV, Morris CJ, Gregory RS, Hardie DC, et al. 2010. Parallel adaptive evolution of Atlantic cod on both sides of the Atlantic Ocean in response to temperature. Proc Biol Sci. 277:3725–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger R, Akerman A. 2011. The effects of linkage and gene flow on local adaptation: a two-locus continent-island model. Theor Popul Biol. 80:272–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin RK. 2005. Recombination and speciation. Mol Ecol. 14:2621–2635. [DOI] [PubMed] [Google Scholar]
- Carlson DF, Lancto CA, Zang B, Kim ES, Walton M, Oldeschulte D, Seabury C, Sonstegard TS, Fahrenkrug SC. 2016. Production of hornless dairy cattle from genome-edited cell lines. Nat Biotechnol. 34:479–481. [DOI] [PubMed] [Google Scholar]
- Carter CO. 1977. Monogenic disorders. J Med Genet. 14:316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanach A, Crowhurst R, Deng C, David C, Bernatchez L, Wellenreuther M. 2019. The genomic pool of standing structural variation outnumbers single nucleotide polymorphism by threefold in the marine teleost Chrysophrys auratus. Mol Ecol. 28:1210–1223. [DOI] [PubMed] [Google Scholar]
- Cauwelier E, Gilbey J, Sampayo J, Stradmeyer L, Middlemas SJ. 2018. Identification of a single genomic region associated with seasonal river return timing in adult Scottish Atlantic salmon (Salmo salar), using a genome-wide association study. Can J Fish Aquat Sci. 75:1427–1435. [Google Scholar]
- Cayuela H, Rougemont Q, Laporte M, Mérot C, Normandeau E, Dorant Y, Tørresen OK, Hoff SNK, Jentoft S, Sirois P, et al. 2020. Shared ancestral polymorphisms and chromosomal rearrangements as potential drivers of local adaptation in a marine fish. Mol Ecol. 1–20. doi: 10.1111/mec.15499 [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. 1979. Selection on recombination in a multi-locus system. Genetics. 91:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Tan JC, Hahn MW, Besansky NJ. 2018. Systems genetic analysis of inversion polymorphisms in the malaria mosquito Anopheles gambiae. Proc Natl Acad Sci USA. 115:E7005–E7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmas MJ, Wallberg A, Bunikis I, Olsson A, Wallerman O, Webster MT. 2019. Chromosomal inversions associated with environmental adaptation in honeybees. Mol Ecol. 28:1358–1374. [DOI] [PubMed] [Google Scholar]
- Colosimo PF, Peichel CL, Nereng K, Blackman BK, Shapiro MD, Schluter D, Kingsley DM. 2004. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2:E109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan JM, Willis JH. 2019. Dissecting the role of a large chromosomal inversion in life history divergence throughout the Mimulus guttatus species complex. Mol Ecol. 28:1343–1357. [DOI] [PubMed] [Google Scholar]
- Coulson T, Kendall BE, Barthold J, Plard F, Schindler S, Ozgul A, Gaillard JM. 2017. Modeling adaptive and nonadaptive responses of populations to environmental change. Am Nat. 190:313–336. [DOI] [PubMed] [Google Scholar]
- Crow T, Ta J, Nojoomi S, Aguilar-Rangel MR, Torres Rodríguez JV, Gates D, Rellan-Alvarez R, Sawers R, Runcie D, et al. 2019. Gene regulatory effects of a large chromosomal inversion in highland maize. bioRxiv. 861583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler HD, Capitan A, Pausch H, Stothard P, van Binsbergen R, Brøndum RF, Liao X, Djari A, Rodriguez SC, Grohs C, et al. 2014. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat Genet. 46:858–865. [DOI] [PubMed] [Google Scholar]
- Doering GN, Scharf I, Moeller HV, Pruitt JN. 2018. Social tipping points in animal societies in response to heat stress. Nat Ecol Evol. 2:1298–1305. [DOI] [PubMed] [Google Scholar]
- Faria R, Chaube P, Morales HE, Larsson T, Lemmon AR, Lemmon EM, Rafajlović M, Panova M, Ravinet M, Johannesson K, et al. 2019. Multiple chromosomal rearrangements in a hybrid zone between Littorina saxatilis ecotypes. Mol Ecol. 28:1375–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder ME, Walser JC. 2005. The biological limitations of transcriptomics in elucidating stress and stress responses. J Evol Biol. 18:901–910. [DOI] [PubMed] [Google Scholar]
- Fuentes-Pardo AP, Bourne C, Singh R, Emond K, Pinkham L, McDermid JL, Andersson L, Ruzzante DE. 2019. Adaptation to seasonal reproduction and thermal minima-related factors drives fine-scale divergence despite gene flow in Atlantic herring populations. bioRxiv. 578484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller ZL, Haynes GD, Richards S, Schaeffer SW. 2016. Genomics of natural populations: how differentially expressed genes shape the evolution of chromosomal inversions in Drosophila pseudoobscura. Genetics. 204:287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller ZL, Haynes GD, Richards S, Schaeffer SW. 2017. Genomics of natural populations: evolutionary forces that establish and maintain gene arrangements in Drosophila pseudoobscura. Mol Ecol. 26:6539–6562. [DOI] [PubMed] [Google Scholar]
- Gianola D, Qanbari S, Simianer H. 2013. An evaluation of a novel estimator of linkage disequilibrium. Heredity (Edinb). 111:275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold CK. 2006. Gene flow’s effect on the genetic architecture of a local adaptation and its consequences for QTL analyses. Heredity (Edinb). 96:445–453. [DOI] [PubMed] [Google Scholar]
- Guerrero RF, Kirkpatrick M. 2014. Local adaptation and the evolution of chromosome fusions. Evolution. 68:2747–2756. [DOI] [PubMed] [Google Scholar]
- Guillén Y, Ruiz A. 2012. Gene alterations at Drosophila inversion breakpoints provide prima facie evidence for natural selection as an explanation for rapid chromosomal evolution. BMC Genomics. 13:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban S, Kelley JL, Lotterhos KE, Antolin MF, Bradburd G, Lowry DB, Poss ML, Reed LK, Storfer A, Whitlock MC. 2016. Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am Nat. 188:379–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Griffin P, Dillon S, Catullo R, Rane R, Byrne M, Jordan R, Oakeshott J, Weeks A, Joseph L, et al. 2015. A framework for incorporating evolutionary genomics into biodiversity conservation and management. Clim Change Resp. 2:1. [Google Scholar]
- Hoffmann AA, Rieseberg LH. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 39:21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper DM. 2016. Range overlap drives chromosome inversion fixation in Passerine birds. bioRxiv. 053371. [Google Scholar]
- Horton BM, Moore IT, Maney DL. 2014. New insights into the hormonal and behavioural correlates of polymorphism in white-throated sparrows, Zonotrichia albicollis. Anim Behav. 93:207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-C, Dang VD, Chang N-C, Wang J. 2018. Multiple large inversions and breakpoint rewiring of gene expression in the evolution of the fire ant social supergene. Proc R Soc B Biol Sci. 285:20180221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings JA, Jones MEB. 1998. Life history variation and growth rate thresholds for maturity in Atlantic salmon, Salmo salar. Can J Fish Aquat Sci. 55:22–47. [Google Scholar]
- IPBES . 2019. Summary for policymakers of the global assessment report on biodiversity and ecosystem services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services. In: Díaz S, Settele J, Brondízio ES, et al., editors. IPBES secretariat, Bonn, Germany. p. 56. [Google Scholar]
- IPCC. 2018. Summary for policymakers. In: Masson-Delmotte V, Zhai P, Pörtner H-O, et al., editors. Global warming of 1.5°C. An IPCC Special Report on the impacts of global warming of 1.5°C above pre-industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change. In press. [Google Scholar]
- Jay P, Chouteau M, Whibley A, Bastide H, Llaurens V, Parrinello H, Joron M. 2019. Mutation accumulation in chromosomal inversions maintains wing pattern polymorphism in a butterfly. bioRxiv. 736504. [Google Scholar]
- Jiménez-Mena B, Le Moan A, Christensen A, van Deurs M, Mosegaard H, Hemmer-Hansen J, Bekkevold D. 2020. Weak genetic structure despite strong genomic signal in lesser sandeel in the North Sea. Evol Appl. 13:376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, Gratten J, Berenos C, Pilkington JG, Clutton-Brock TH, Pemberton JM, Slate J. 2013. Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature. 502:93–95. [DOI] [PubMed] [Google Scholar]
- Johnston SE, Orell P, Pritchard VL, Kent MP, Lien S, Niemelä E, Erkinaro J, Primmer CR. 2014. Genome-wide SNP analysis reveals a genetic basis for sea-age variation in a wild population of Atlantic salmon (Salmo salar). Mol Ecol. 23:3452–3468. [DOI] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. ; Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team . 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 484:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Mills LS, Alves PC, Callahan CM, Alves JM, Lafferty DJR, Jiggins FM, Jensen JD, Melo-Ferreira J, Good JM. 2018. Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science. 360:1355–1358. [DOI] [PubMed] [Google Scholar]
- Joron M, Frezal L, Jones RT, Chamberlain NL, Lee SF, Haag CR, Whibley A, Becuwe M, Baxter SW, Ferguson L, et al. 2013. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature. 477:203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao AB, Hund AK, Santos FP, Young J-G, Bhat D, Garland J, Oomen RA, McCreery HF. 2020. Changes in group size during resource shifts reveal drivers of sociality across the tree of life. bioRxiv. 2020.03.17.994343. [Google Scholar]
- Kapun M, Fabian DK, Goudet J, Flatt T. 2016. Genomic evidence for adaptive inversion clines in Drosophila melanogaster. Mol Biol Evol. 33:1317–1336. [DOI] [PubMed] [Google Scholar]
- Kapun M, Flatt T. 2019. The adaptive significance of chromosomal inversion polymorphisms in Drosophila melanogaster. Mol Ecol. 28:1263–1282. [DOI] [PubMed] [Google Scholar]
- Kardos M, Luikart G. 2020. The genetic architecture of fitness drives population viability during rapid environmental change. bioRxiv. 660803. [DOI] [PubMed] [Google Scholar]
- Kardos M, Luikart G, Bunch R, Dewey S, Edwards W, McWilliam S, Stephenson J, Allendorf FW, Hogg JT, Kijas J. 2015. Whole-genome resequencing uncovers molecular signatures of natural and sexual selection in wild bighorn sheep. Mol Ecol. 24:5616–5632. [DOI] [PubMed] [Google Scholar]
- Kess T, Bentzen P, Lehnert SJ, Sylvester EVA, Lien S, Kent MP, Sinclair-Waters M, Morris CJ, Regular P, Fairweather R, et al. 2019. A migration-associated supergene reveals loss of biocomplexity in Atlantic cod. Sci Adv. 5:eaav2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kess T, Bentzen P, Lehnert SJ, Sylvester EVA, Lien S, Kent MP, Sinclair-Waters M, Morris C, Wringe B, Fairweather R, et al. 2020. Modular chromosome rearrangements reveal parallel and nonparallel adaptation in a marine fish. Ecol Evol. 10:638–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barrett B. 2015. Chromosome inversions, adaptive cassettes and the evolution of species’ ranges. Mol Ecol. 24:2046–2055. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. 2006. Chromosome inversions, local adaptation and speciation. Genetics. 173:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirubakaran TG, Grove H, Kent MP, Sandve SR, Baranski M, Nome T, De Rosa MC, Righino B, Johansen T, Otterå H, et al. 2016. Two adjacent inversions maintain genomic differentiation between migratory and stationary ecotypes of Atlantic cod. Mol Ecol. 25:2130–2143. [DOI] [PubMed] [Google Scholar]
- Kivimäki M, Kärkkäinen K, Gaudeul M, Løe G, Agren J. 2007. Gene, phenotype and function: GLABROUS1 and resistance to herbivory in natural populations of Arabidopsis lyrata. Mol Ecol. 16:453–462. [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. 2002. A high-resolution recombination map of the human genome. Nat Genet. 31:241–247. [DOI] [PubMed] [Google Scholar]
- Korunes KL, Noor MAF. 2019. Pervasive gene conversion in chromosomal inversion heterozygotes. Mol Ecol. 28:1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas CB, Powell JR. 1992. Drosophila inversion polymorphism. Boca Raton (FL): CRC Press. [Google Scholar]
- Kuparinen A, Hutchings JA. 2012. Consequences of fisheries-induced evolution for population productivity and recovery potential. Proc Biol Sci. 279:2571–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuparinen A, Hutchings JA. 2017. Genetic architecture of age at maturity can generate divergent and disruptive harvest-induced evolution. Philos Trans R Soc Lond B Biol Sci. 372:20160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper C, Stocks M, Risse JE, Dos Remedios N, Farrell LL, McRae SB, Morgan TC, Karlionova N, Pinchuk P, Verkuil YI, et al. 2016. A supergene determines highly divergent male reproductive morphs in the ruff. Nat Genet. 48:79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. 1999. Four evolutionary strata on the human X chromosome. Science. 286:964–967. [DOI] [PubMed] [Google Scholar]
- Lamichhaney S, Fuentes-Pardo AP, Rafati N, Ryman N, McCracken GR, Bourne C, Singh R, Ruzzante DE, Andersson L. 2017. Parallel adaptive evolution of geographically distant herring populations on both sides of the North Atlantic Ocean. Proc Natl Acad Sci USA. 114:E3452–E3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert KP, Schmidt C, Fischer P, Volff JN, Hoffmann C, Muck J, Lohse MJ, Ryan MJ, Schartl M. 2010. Determination of onset of sexual maturation and mating behavior by melanocortin receptor 4 polymorphisms. Curr Biol. 20:1729–1734. [DOI] [PubMed] [Google Scholar]
- Le Moan A, Bekkevold D, Hemmer-Hansen J. 2019a. Evolution at two time-frames: ancient and singular origin of two structural variants involved in local adaptation of the European plaice (Pleuronectes platessa). bioRxiv. 662577. [Google Scholar]
- Le Moan A, Gaggiotti O, Henriques R, Martinez P, Bekkevold D, Hemmer-Hansen J. 2019b. Beyond parallel evolution: when several species colonize the same environmental gradient. bioRxiv. 662569. [Google Scholar]
- Lee CR, Wang B, Mojica JP, Mandáková T, Prasad KVSK, Goicoechea JL, Perera N, Hellsten U, Hundley HN, Johnson J, et al. 2017. Young inversion with multiple linked QTLs under selection in a hybrid zone. Nat Ecol Evol. 1:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legault CM. 2004. Salmon PVA: a population viability analysis model for Atlantic salmon in the Maine distinct population segment. Northeast Fisheries Science Center Reference Document 04-02. Woods Hole, MA: US. Department of Commerce. [Google Scholar]
- Lindtke D, Lucek K, Soria-Carrasco V, Villoutreix R, Farkas TE, Riesch R, Dennis SR, Gompert Z, Nosil P. 2017. Long-term balancing selection on chromosomal variants associated with crypsis in a stick insect. Mol Ecol. 26:6189–6205. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8:e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucek K, Gompert Z, Nosil P. 2019. The role of structural genomic variants in population differentiation and ecotype formation in Timema cristinae walking sticks. Mol Ecol. 28:12274–1237. [DOI] [PubMed] [Google Scholar]
- Mérot C, Berdan EL, Babin C, Normandeau E, Wellenreuther M, Bernatchez L. 2018. Intercontinental karyotype – environment parallelism supports a role for a chromosomal inversion in local adaptation in a seaweed fly. Proc R Soc B Biol Sci. 285:20180519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérot C, Oomen RA, Tigano A, Wellenreuther M. 2020a. A Roadmap for understanding the evolutionary significance of structural genomic variation. Trends Ecol Evol. 35:561–572. [DOI] [PubMed] [Google Scholar]
- Mérot C, Llaurens V, Normandeau E, Bernatchez L, Wellenreuther M. 2020b. Balancing selection via life-history trade-offs maintains an inversion polymorphism in a seaweed fly. Nat Comm. 11: 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt JR, Grogan KE, Zinzow-Kramer WM, Sun D, Ortlund EA, Yi SV, Maney DL. 2020. A behavioral polymorphism caused by a single gene inside a supergene. bioRxiv. 2020.01.13.897637. [Google Scholar]
- Micheletti SJ, Hess JE, Zendt JS, Narum SR. 2018. Selection at a genomic region of major effect is responsible for evolution of complex life histories in anadromous steelhead. BMC Evol Biol. 18:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales HE, Faria R, Johannesson K, Larsson T, Panova M, Westram AM, Butlin RK. 2019. Genomic architecture of parallel ecological divergence: beyond a single environmental contrast. Sci Adv. 5:eaav9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullon C, Keller L, Lehmann L. 2018. Social polymorphism is favoured by the co-evolution of dispersal with social behaviour. Nat Ecol Evol. 2:132–140. [DOI] [PubMed] [Google Scholar]
- Narum SR, Di Genova A, Micheletti SJ, Maass A. 2018. Genomic variation underlying complex life-history traits revealed by genome sequencing in Chinook salmon. Proc R Soc B Biol Sci. 285:20180935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RM, Pettersson ME, Carlborg Ö. 2013. A century after Fisher: time for a new paradigm in quantitative genetics. Trends Genet. 29:669–676. [DOI] [PubMed] [Google Scholar]
- Nishikawa H, Iijima T, Kajitani R, Yamaguchi J, Ando T, Suzuki Y, Sugano S, Fujiyama A, Kosugi S, Hirakawa H, et al. 2015. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat Genet. 47:405–409. [DOI] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. 2009. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 18:375–402. [DOI] [PubMed] [Google Scholar]
- Oomen RA. 2019. The genomic basis and spatial scale of variation in thermal responses of Atlantic cod (Gadus morhua) [doctoral thesis]. [ Halifax, (NS)]: Dalhousie University. [Google Scholar]
- Oomen RA, Hutchings JA. 2015. Variation in spawning time promotes genetic variability in population responses to environmental change in a marine fish. Conserv Physiol. 3:cov027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomen RA, Hutchings JA. 2016. Genetic variation in plasticity of life-history traits between Atlantic cod (Gadus morhua) populations exposed to contrasting thermal regimes. Can J Zool. 94:257–264. [Google Scholar]
- Orr HA, Unckless RL. 2014. The population genetics of evolutionary rescue. PLoS Genet. 10:e1004551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Engelstädter J, Rieseberg LH. 2016. Recombination rate evolution and the origin of species. Trends Ecol Evol. 31:226–236. [DOI] [PubMed] [Google Scholar]
- Palumbi SR, Barshis DJ, Traylor-Knowles N, Bay RA. 2014. Mechanisms of reef coral resistance to future climate change. Science. 344:895–898. [DOI] [PubMed] [Google Scholar]
- Pang AW, MacDonald JR, Pinto D, Wei J, Rafiq MA, Conrad DF, Park H, Hurles ME, Lee C, Venter JC, et al. 2010. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 11:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse DE, Barson NJ, Nome T, Gao G, Campbell MA, Abadía-Cardoso A, Anderson EC, Rundio DE, Williams TH, Naish KA, et al. 2019. Sex-dependent dominance maintains migration supergene in rainbow trout. Nat Ecol Evol. 3:1731–1742. [DOI] [PubMed] [Google Scholar]
- Pearse DE, Miller MR, Abadıa-Cardoso A, Garza JC. 2014. Rapid parallel evolution of standing variation in a single, complex,genomic region is associated with life history in steelhead/rainbow trout. Proc R Soc B Biol Sci. 281:20140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalba JV, Wolf JBW. 2020. From molecules to populations: appreciating and estimating recombination rate variation. Nat Rev Genet. 21: 476– 492. [DOI] [PubMed] [Google Scholar]
- Pettersson ME, Rochus CM, Han F, Chen J, Hill J, Wallerman O, Fan G, Hong X, Xu Q, Zhang H, et al. 2019. A chromosome-level assembly of the Atlantic herring – detection of a supergene and other signals of selection. bioRxiv. 668384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RB. 2005. Chromosome morphology. In: Cadrin SX, Friedland KD, Waldman JR, editors. Stock identification methods. Amsterdam (Netherlands): Elsevier. p. 273–294. [Google Scholar]
- Ragsdale AP, Gravel S. 2020. Unbiased estimation of linkage disequilibrium from unphased data. Mol Biol Evol. 37: 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane RV, Rako L, Kapun M, Lee SF, Hoffmann AA. 2015. Genomic evidence for role of inversion 3RP of Drosophila melanogaster in facilitating climate change adaptation. Mol Ecol. 24:2423–2432. [DOI] [PubMed] [Google Scholar]
- Reed RD, Papa R, Martin A, Hines HM, Counterman BA, Pardo-Diaz C, Jiggins CD, Chamberlain NL, Kronforst MR, Chen R, et al. 2011. Optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science. 333:1137–1141. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol Evol. 16:351–358. [DOI] [PubMed] [Google Scholar]
- Rogers SM, Mee JA, Bowles E. 2011. The consequences of genomic architecture on ecological speciation in postglacial fishes. Curr Zool. 59:53–71. [Google Scholar]
- Rowan BA, Heavens D, Feuerborn TR, Tock AJ, Henderson IR, Weigel D. 2019. An ultra high-density Arabidopsis thaliana crossover map that refines the influences of structural variation and epigenetic features. Genetics. 213:771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein DR, Ågren JA, Carbone L, Elde NC, Hoekstra HE, Kapheim KM, Keller L, Moreau CS, Toth AL, Yeaman S, et al. 2019. Coevolution of genome architecture and social behavior. Trends Ecol Evol. 34:844–855. [DOI] [PubMed] [Google Scholar]
- Savolainen O, Lascoux M, Merilä J. 2013. Ecological genomics of local adaptation. Nat Rev Genet. 14:807–820. [DOI] [PubMed] [Google Scholar]
- Schaeffer SW. 2008. Selection in heterogeneous environments maintains the gene arrangement polymorphism of Drosophila pseudoobscura. Evolution. 62:3082–3099. [DOI] [PubMed] [Google Scholar]
- Schwander T, Libbrecht R, Keller L. 2014. Supergenes and complex phenotypes. Curr Biol. 24:R288–R294. [DOI] [PubMed] [Google Scholar]
- Sinclair-Waters M, Ødegård J, Korsvoll SA, Moen T, Lien S, Primmer CR, Barson NJ. 2020. Beyond large-effect loci: large-scale GWAS reveals a mixed large-effect and polygenic architecture for age at maturity of Atlantic salmon. Genet Sel Evol. 52:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeland M, Jorde PE, Lien S, Jentoft S, Berg PR, Grove H, Kent MP, Arnyasi M, Olsen EM, Knutsen H. 2016. “Islands of Divergence” in the atlantic cod genome represent polymorphic chromosomal rearrangements. Genome Biol Evol. 8:1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W. 2016. Signatures of positive selection: from selective sweeps at individual loci to subtle allele frequency changes in polygenic adaptation. Mol Ecol. 25:79–88. [DOI] [PubMed] [Google Scholar]
- Stevison LS, Sefick S, Rushton C, Graze RM. 2017. Recombination rate plasticity: revealing mechanisms by design. Philos Trans R Soc B Biol Sci. 372:20160459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. 1921. A case of rearrangement of genes in drosophila. Proc Natl Acad Sci USA. 7:235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved JA. 1971. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor Popul Biol. 2:125–141. [DOI] [PubMed] [Google Scholar]
- Therkildsen NO, Wilder AP, Conover DO, Munch SB, Baumann H, Palumbi SR. 2019. Contrasting genomic shifts underlie parallel phenotypic evolution in response to fishing. Science. 365:487–490. [DOI] [PubMed] [Google Scholar]
- Thomas JW, Cáceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, Maney DL, Martin CL. 2008. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics. 179:1455–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson TQ, Bellinger MR, O’Rourke SM, Prince DJ, Stevenson AE, Rodrigues AT, Sloat MR, Speller CF, Yang DY, Butler VL, et al. 2019. Anthropogenic habitat alteration leads to rapid loss of adaptive variation and restoration potential in wild salmon populations. Proc Natl Acad Sci USA. 116:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigano A, Friesen VL. 2016. Genomics of local adaptation with gene flow. Mol Ecol. 25:2144–2164. [DOI] [PubMed] [Google Scholar]
- Tigano A, Reiertsen TK, Walters JR, Friesen VL. 2018. A complex copy number variant underlies differences in both colour plumage and cold adaptation in a dimorphic seabird. bioRxiv. 507384. [Google Scholar]
- Todesco M, Owens GL, Bercovich N, Légaré J-S, Soudi S, Burge DO, Huang K, Ostevik KL, Drummond EBM, Imerovski I, et al. 2020. Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature. doi: 10.1038/s41586-020-2467-6 [DOI] [PubMed] [Google Scholar]
- Toews DPL, Taylor SA, Streby HM, Kramer GR, Lovette IJ. 2019. Selection on VPS13A linked to migration in a songbird. Proc Natl Acad Sci USA. 116:18272–18274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyford AD, Friedman J. 2015. Adaptive divergence in the monkey flower Mimulus guttatus is maintained by a chromosomal inversion. Evolution. 69:1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veale AJ, Russello MA. 2017. An ancient selective sweep linked to reproductive life history evolution in sockeye salmon. Sci Rep. 7:1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A, Schöning C, Webster MT, Hasselmann M. 2017. Two extended haplotype blocks are associated with adaptation to high altitude habitats in East African honey bees. PLoS Genet. 13:e1006792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RK, Larson WA, Waples RS. 2016. Estimating contemporary effective population size in non-model species using linkage disequilibrium across thousands of loci. Heredity (Edinb). 117:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS, Lindley ST. 2018. Genomics and conservation units: the genetic basis of adult migration timing in Pacific salmonids. Evol Appl. 11:1518–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS, Naish KA, Primmer CR. 2020. Conservation and management of salmon in the age of genomics. Annu Rev Anim Biosci. 8:117–143. [DOI] [PubMed] [Google Scholar]
- Wellband K, Mérot C, Linnansaari T, Elliott JAK, Curry RA, Bernatchez L. 2019. Chromosomal fusion and life history-associated genomic variation contribute to within-river local adaptation of Atlantic salmon. Mol Ecol. 28:1439–1459. [DOI] [PubMed] [Google Scholar]
- Wellenreuther M, Bernatchez L. 2018. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol Evol. 33:427–440. [DOI] [PubMed] [Google Scholar]
- Wellenreuther M, Mérot C, Berdan E, Bernatchez L. 2019. Going beyond SNPs: the role of structural genomic variants in adaptive evolution and species diversification. Mol Ecol. 28:1203–1209. [DOI] [PubMed] [Google Scholar]
- Wu C-I. 2001. The genic view of the process of speciation. J Evol Biol. 14:851–865. [Google Scholar]
- Yeaman S, Otto SP. 2011. Establishment and maintenance of adaptive genetic divergence under migration, selection, and drift. Evolution. 65:2123–2129. [DOI] [PubMed] [Google Scholar]
- Yeaman S. 2013. Genomic rearrangements and the evolution of clusters of locally adaptive loci. Proc Natl Acad Sci USA. 110:E1743–E1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman S, Whitlock MC. 2011. The genetic architecture of adaptation under migration-selection balance. Evolution. 65:1897–1911. [DOI] [PubMed] [Google Scholar]
- Zinzow-Kramer WM, Horton BM, McKee CD, Michaud JM, Tharp GK, Thomas JW, Tuttle EM, Yi S, Maney DL. 2015. Genes located in a chromosomal inversion are correlated with territorial song in white-throated sparrows. Genes Brain Behav. 14:641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]