Abstract
Many recent studies have addressed the mechanisms operating during the early stages of speciation, but surprisingly few studies have tested theoretical predictions on the evolution of strong reproductive isolation (RI). To help address this gap, we first undertook a quantitative review of the hybrid zone literature for flowering plants in relation to reproductive barriers. Then, using Populus as an exemplary model group, we analysed genome-wide variation for phylogenetic tree topologies in both early- and late-stage speciation taxa to determine how these patterns may be related to the genomic architecture of RI. Our plant literature survey revealed variation in barrier complexity and an association between barrier number and introgressive gene flow. Focusing on Populus, our genome-wide analysis of tree topologies in speciating poplar taxa points to unusually complex genomic architectures of RI, consistent with earlier genome-wide association studies. These architectures appear to facilitate the ‘escape’ of introgressed genome segments from polygenic barriers even with strong RI, thus affecting their relationships with recombination rates. Placed within the context of the broader literature, our data illustrate how phylogenomic approaches hold great promise for addressing the evolution and temporary breakdown of RI during late stages of speciation.
This article is part of the theme issue ‘Towards the completion of speciation: the evolution of reproductive isolation beyond the first barriers'.
Keywords: speciation, hybridization, reproductive isolation, gene flow, topology discordance, recombination rate
1. Introduction
Current research on speciation genomics strives to tackle two central questions in evolutionary biology: what is the origin and evolution of reproductive barriers in the genomes of diverging populations? And, how do divergent populations or species respond when challenged by hybridization upon secondary contact [1–4]? Theory predicts that speciation may occur in the face of ongoing or episodic gene flow [5]. A rapidly increasing number of speciation genomic studies have started to address divergence with gene flow (DWGF) in a range of different species [6,7], which has been greatly facilitated by advances in second- and third-generation sequencing technologies [8–14]. Hence, speciation genomics has developed into a vibrant research field [3,4,15–17], fuelling debates on topics of fundamental, philosophical and applied interest.
Rapid barrier evolution during DWGF has been predicted by population geneticists for decades and has become widely known as the ‘coupling’ of individual barrier loci, resulting in mutually strengthened total barriers to gene flow [3,18–21]. It is thought that coupling creates coincidence among the effects of single barrier loci (and thus the traits encoded by them), which may lead to a substantial, but often incomplete, barrier to gene flow [21]. This leads to a ‘grey zone’ of speciation [22] which may well be responsible for many of the great challenges experienced by taxonomists and systematic biologists in previous decades and centuries. Genetic contact (hybridization) among divergent lineages at these advanced stages of speciation can result in a range of hotly debated outcomes [13,21,23,24]. These may include both heterosis (hybrid vigour) and hybrid breakdown due to genomic incompatibilities including the breakdown of genomic co-adaptation [25–27].
Differentiation between populations and ultimately speciation yields complex patterns of divergence along the genome [28,29]. Theory predicts that individual barrier loci can result in peaks of divergence between species [12,30], but in reality, the interplay of linked selection, variation of recombination rates and density of functional sites results in a complex landscape of peaks and troughs, which may be independent of reproductive isolation (RI). For example, background selection in regions of low recombination and with a high density of functional sites can also lower diversity within species, resulting in divergence peaks between species [31,32]. Nonetheless, several studies have reported a positive correlation between introgression and recombination rate [29,33]. These patterns are consistent with highly polygenic barriers to gene flow and the more efficient removal of introgressed variation in regions of low recombination [34]. However, it remains unclear whether the influence of linked selection on introgressed variation diminishes with time since divergence or whether it holds for organisms with other life histories with high rates of effective recombination.
To this end, hybridizing species have become highly appreciated ‘natural labs’ for studying speciation [35–37]. This holds true for hybrids formed either during primary divergence or upon secondary contact, and whether the genetic transitions seen in these zones fit with clinal or ‘geographic mosaic’ evolutionary models [2]. Divergent yet hybridizing taxa can also serve as precious sources of recombinant crosses for studying the genomic architecture of RI and inter-population trait differences [36,38–40]. In addition, hybrid zones enable the impact of introgression on genomic patterns of divergence to be investigated at recent time scales by comparing parapatric populations flanking hybrid zones with allopatric populations [12]. At deeper time scales, studying hybridizing taxa also makes it possible to address important questions regarding the sorting of ancestral variation in young or emerging species, past episodes of gene flow and how this may relate to the evolution of RI [11,33,41]. This is greatly facilitated by recent conceptual developments in merging the analytical toolkits of population genomics and phylogenomics [14,41]. This approach may be particularly useful for organismal groups that maintain leaky reproductive barriers across species complexes for many generations—and thus for millions of years—such as perennial plants with relatively large effective population sizes (Ne), far-ranging pollen and seed dispersal, and the ability to maintain viable genotypes in populations by clonal reproduction [42,43].
Among different study systems for studying speciation in plants, Populus has become a perennial model group because of its ecological and economic importance and favourable genetic attributes such as small genome size (less than 500 Mb; 2C = 1.1 pg in the case of Populus trichocarpa), diploidy throughout the genus (2n = 38), ‘porous’ species barriers [44–46] and a well curated and annotated genome assembly [47]. Species of the genus are widespread across the Northern Hemisphere [48]. Several studies have attempted to resolve phylogenetic relationships of species in this genus [49,50], most notably a recent study using resequenced genomes [51]. Obligate outcrossing (dioecy), abundant wind-pollination, and mixed sexual and vegetative reproductive strategies in poplars have led to extensive introgression among species and relatively large effective population size (Ne) [52–55], which complicates phylogenetic inference.
Recent work on speciation genomics in Populus has revealed several patterns relevant to understanding the speciation continuum. Firstly, linked selection and recombination rate variation appear to have pervasive effects on genome-wide patterns of genetic diversity and divergence among poplar species, as exemplified by interspecific contrasts involving the two more closely related species Populus tremula, Populus tremuloides and the more distantly related P. trichocarpa. These effects are moderated by important demographic factors and events, such as temporal and interspecific changes in Ne experienced by these temperate tree species in response to climatic cycles [54,55]. At greater levels of divergence (1.73–1.90 Myr), a landmark study by Ma et al. [56] revealed the likely determinants of genome-wide patterns of diversity in the two Eurasian desert poplar species, Populus euphratica and Populus pruinosa, pointing to important roles for the divergent sorting of ancestral polymorphisms and divergent ecological selection. Finally, studies of the highly divergent Eurasian taxa Populus alba and P. tremula have shown that despite greater than 2.8 Myr of divergence [8], strong post-zygotic barriers due to genomic incompatibilities [57,58] and variable pre-zygotic barriers [58], these taxa still form viable and fertile hybrids within large mosaic hybrid zones in areas of both sym- and parapatry [57,59]. Although these species thus represent a useful showcase example for research on the late stages of speciation, studies that examine genome-wide phylogenomic patterns for taxa pairs representing the early and late stages of speciation are required to better understand how the genomic architecture of RI varies across the stages of speciation.
Beyond particular organismal model groups, categorizing the stage of speciation is dependent on both understanding the level of gene exchange among divergent taxa and identifying the presence of reproductive barriers [60,61]. For plants, the great diversity of mating systems, reproductive strategies and life-history traits may interact to influence the tempo and speed of speciation. Thus, we begin by undertaking a broad analysis including 133 hybridizing species pairs to examine the number of pre- and post-zygotic barriers and how these relate to gene flow in flowering plants. Here, we test the prediction of higher levels of gene flow in species pairs with fewer reproductive isolating barriers. We then ‘zoom in’ on the genomic footprints of RI and introgressive gene flow in species of the ‘model forest tree’ genus Populus (poplars/aspens/cottonwoods). These include widespread, ecologically divergent Eurasian taxa that provide key examples of the evolutionary mechanisms operating during the late stages of speciation. We analyse 36 re-sequenced genomes from seven species of this Eurasian species complex to examine how the genomic architecture of RI and introgressive gene flow varies across the stages of speciation. Then we analyse the data in a phylogenomic context and examine genome-wide relationships among well sorted versus introgressed tree topologies and recombination rates during both the early and late stages of speciation. Our purpose is to determine how genome-wide phylogenomic patterns (genome-wide tree topologies) are mediated by the genomic architecture of RI and the recombination landscape, and test whether these relations hold across the speciation continuum. Using tree typology weighting and phylogenetic tests for introgression, we compare the amount of gene flow and the relation between introgressed typologies and recombination rate, on five anciently diverged (late-stage speciation) and five recently diverged (early-stage speciation) species. Taken together, this broad to narrow approach provides novel insights into the processes and outcomes of DWGF from the early to late stages of speciation.
2. Material and methods
(a). Plant literature survey
To investigate the interaction between the presence of pre- and post-zygotic reproductive isolating barriers and gene flow, we collated data on hybridization in 133 species pairs, representing 72 genera and 41 plant families (for full description of methods, see Pickup et al. [62]). Following Abbott [63], we categorized gene flow into four categories: very low, low, high and variable (different among hybridizing populations) based on criteria and descriptions outlined by Pickup et al. [62], which were based on quantitative information on the frequency of hybrids and backcrosses (see also electronic supplementary material, ‘plant literature survey: categorization of gene flow’). For each taxon pair, we identified the presence (1) or absence (0) of each of a set of pre-zygotic and post-zygotic barriers (but we did not attempt to quantify their strength) based on Abbott [63] and descriptions or quantitative assessments from each individual study. Pre-zygotic barriers were: (i) geography (spatial isolation of parental species), (ii) habitat divergence (divergent habitat preference), (iii) divergent flowering phenology, (iv) divergent floral structure, (v) pollinator preference, (vi) mating system and (vii) pollen competition. Mating system (vi) was classified as a pre-zygotic barrier for taxon pairs with divergent mating systems. These include: (i) taxon pairs with a predominantly outcrossing self-compatible species and a highly selfing self-compatible species, (ii) pairs where both taxa are selfing and (iii) pairs including a self-incompatible and self-compatible species (see Pickup et al. [62] for details). Post-zygotic barriers were: (i) reduced hybrid viability, (ii) cyto-nuclear interactions, (iii) intrinsic genomic incompatibilities (the interaction between alleles results in lower fitness of individuals), and (iv) extrinsic (ecological context-dependent) incompatibilities, which require divergent ecological environments for the two populations and selection against maladapted hybrids in both environments. A χ2 contingency test was used to examine if the categories of gene flow (high versus low; combining low, very low and low variable) were associated with the total number of reproductive isolating barriers (combining pre- and post-zygotic barriers) for 123 species pairs where gene flow could be categorized (see Pickup et al. [62]). Statistical analysis was conducted in R and tested at α = 0.05.
(b). Poplar species and populations sequenced de novo for this study
According to the most commonly used classification of Populus, the genus comprises six sections and 29 species [48]. De novo sequence data collection for this study was focused on seven closely related species from section Populus (aspens and white poplars) that provide examples of large Ne and large geographical distribution versus small Ne and narrow distributions, sympatric versus parapatric versus allopatric distribution. Among these, P. alba (white poplar) and P. tremula (Eurasian aspen) are the two most widespread taxa, the former being widely distributed across large parts of southern Eurasia and North Africa, and the latter extending all the way from Scotland to eastern Russia and from northern Scandinavia to the Mediterranean [64]. The two species are at a late stage of speciation, as indicated by partial pre-zygotic and strong post-zygotic reproductive barriers [57,58] and an estimated divergence time of greater than 2.8 Myr [8]. Nevertheless, they still hybridize within large ‘geographical mosaic’ hybrid zones across a broad zone of overlap in Europe and Asia [8,59,65,66]. This species pair serves as a showcase example for the late stage of speciation in this study.
Among the other, more narrowly distributed species, Populus davidiana (the Chinese aspen) is distributed from the central to the northeastern part of China, while the Himalayan aspen Populus rotundifolia is narrowly endemic to the high-altitude regions of the Qinghai–Tibetan plateau. The two species are thought to have undergone recent parapatric, ecological speciation in the face of gene flow [67]. This species pair thus serves as a showcase example for the early stages of speciation in this study. Among the remaining species sampled and sequenced de novo, Populus adenopoda grows in warm and moist subtropical areas of south and east China [68], whereas Populus qiongdaoensis is a rare species only known from Hainan island. Publicly available data for the widespread North American trembling aspen P. tremuloides were included for comparative purposes.
Our sampling for de novo genome sequencing included 36 accessions from these seven ingroup species and two outgroup taxa from section Tacamahaca, P. trichocarpa (black cottonwood) and Populus balsamifera (balsam poplar) (electronic supplementary material, table S1). We collected three to five individuals for each species. For species collected in China, genomic DNA was extracted from silica-dried leaves by using the plant DNeasy mini kit (Qiagen, Germany). To increase the quality of total DNA, we used NucleoSpin gDNA clean-up kits for purification of DNA extracts. All libraries were 2× 150 bp paired-end sequenced on an Illumina HiSeq 3000 sequencer at the Institute of Genetics, University of Berne, Switzerland. Illumina HiSeq paired-end reads for P. tremuloides and the two outgroup species were downloaded from NCBI using the NCBI SRA toolkit under accession numbers PRJNA299390 and PRJNA276056. Further details about sampling locations and distributions are provided in electronic supplementary material, table S1. The reads of each individual were mapped to the P. trichocarpa reference genome using BWA [69]. Details about sequence data processing, variant calling and single nucleotide polymorphism (SNP) quality filtering are provided as electronic supplementary material.
(c). Phylo- and population genomic data analyses
To assess population structure in our whole-genome dataset, principal component analysis (PCA) was carried out based on biallelic SNPs using PLINK [70]. As an alternative means of depicting genetic relationships, a neighbour-joining (NJ) tree was constructed using PHYLIP v. 3.696 (http://evolution.genetics.washington.edu/phylip.html) and visualized using FigTree v. 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Owing to the limits of concatenation methods to infer a species tree, especially for species with large effective population size, we constructed a species tree using MP-EST v. 1.5 [71] based on the multi-species coalescent, established statistical support by bootstrapping and estimated divergence times using MCMCTree software in the PAML package [72] as described in the electronic supplementary material. To infer species’ demographic histories including Ne changes and the relative timing of species splits, we employed SMC++ v. 1.12.1, which combines a coalescent HMM approach with the computational efficiency of the site frequency spectrum for demographic inference [73]. This approach can use unphased data and has been shown to produce robust results in both the recent and ancient past.
To select poplar taxa at late and early stages of speciation, respectively, we explored the sharing of identity-by-descent (IBD) blocks between pairs of species using BEAGLE v. 4.1 [74] and the parameter settings: window=100 000; overlap = 10 000; ibdtrim = 50; ibdlod = 10, impute = false. To examine variation in genealogies along the genome and identify regions whose evolutionary history deviates from the species tree, we used topology weighting by iterative sampling of subtrees (TWISST) [75] to infer the weights (i.e. frequencies) of all different possible tree topologies for windows along the genome. Data were phased and imputed with BEAGLE v. 4.1 [76] and non-overlapping windows of 50 SNPs were used for inferring trees using PhyML [77]. In order to test the effect of different levels of divergence on tree topologies, we selected five anciently diverged (=late-stage speciation) and five recently diverged (=early-stage speciation) taxa for TWISST analysis. The five late-stage species included the well-studied hybridizing species pair P. alba and P. tremula introduced earlier, and the five early-stage species included the Chinese aspen P. davidiana and the Himalayan aspen P. rotundifolia. Based on Zheng et al. [67] and our own NJ analysis, we separated P. davidiana into two local taxa according to geography, central and northeastern China. All Python scripts used for this analysis can be downloaded at https://github.com/simonhmartin/twisst. Weightings for all topologies were plotted across chromosomes with loess span value set to 0.03. Chromosome-level averages of topology weights were compared with local recombination rates in P. tremula [54] in windows of 100 kb.
To gain deeper insights into the ancient and recent admixture events presumably responsible for the observed genome-wide patterns of topology weights for anciently and recently diverged species (above), we examined patterns of IBD tract sharing (above) and inferred ancient and recent admixture using D-statistics to test for gene flow [78]. To estimate the extent and direction of gene flow for late-stage speciation taxa, we conducted DFOIL five-taxon tests in 10 kb windows along the genome [78] using P. trichocarpa as an outgroup. For early-stage speciation taxa, we quantified gene flow using four-taxon D-statistics and P. alba as an outgroup; four-taxon tests were deemed sufficient here since our focus was on a single pair of species, P. davidiana and P. rotundifolia.
3. Results and discussion
(a). Relationships between reproductive barriers and introgressive gene flow in flowering plants
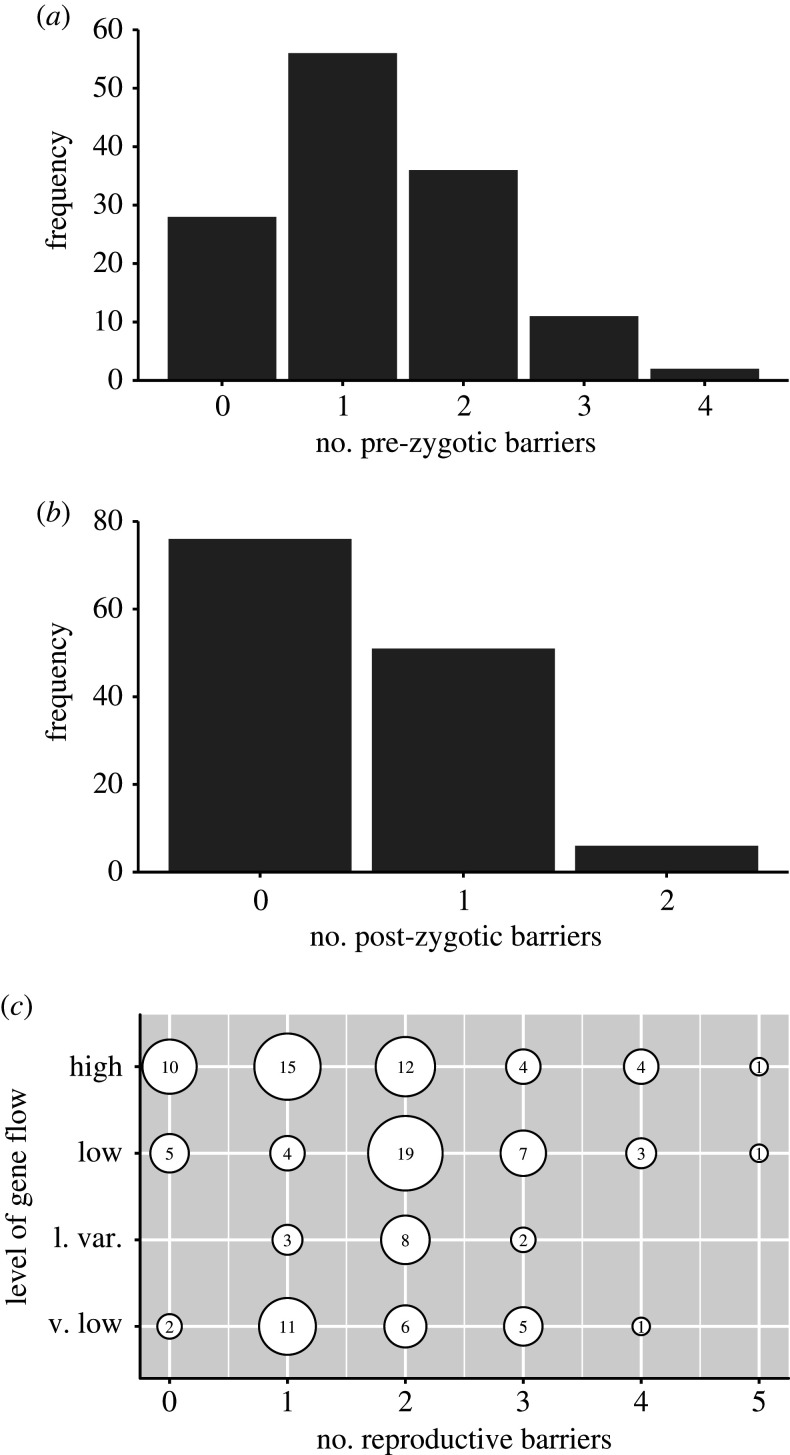
Of the 133 species pairs examined in our literature survey of flowering plants, 105 (78.9%) reported the presence of one or more pre-zygotic reproductive isolating barriers (figure 1a). The highest proportion had a single pre-zygotic barrier (n = 56, 42.1%) followed by the presence of two barriers (n = 36, 27.1%, figure 1a). Fewer taxon pairs had three pre-zygotic barriers (n = 11, 8.3%), and only two pairs (1.5%) recorded four pre-zygotic barriers. In contrast with the high prevalence of pre-zygotic barriers, fewer than half (42.9%) of the taxon pairs recorded post-zygotic reproductive barriers. Overall, there were also fewer post-zygotic barriers, with most taxon pairs recording only one barrier (n = 51, 38.3%; figure 1b). Although these analyses only examined the presence or absence of a barrier—rather than its strength—they provide an important overview of how the number of reproductive isolating barriers varies across plant taxa.
Figure 1.
The number of (a) pre-zygotic and (b) post-zygotic reproductive isolating barriers for 133 angiosperm species pairs. (c) The association between the number of reproductive isolating barriers (pre- and post-zygotic) and categories of gene flow for the 133 taxa pairs. l. var., low variable; v. low: very low.
To assess the prediction that reproductive isolating barriers are related to introgressive gene flow [1,2,6,23,63], we examined if there was an association between the total number of barriers (combining pre- and post-zygotic) and the categories of gene flow (high versus low) for the taxon pairs included in our survey. If reproductive isolating barriers are important for the degree of introgressive gene flow, then we would expect higher gene flow for hybridizing taxa with fewer barriers. Indeed, we found a significant negative association between the gene flow categories (high versus low) and the number of reproductive barriers (χ2 = 9.5793, d.f. = 1, N = 123, p = 0.048) (figure 1c). Although there are caveats to this approach (given that presence/absence does not quantify barrier strength [61] and there was not adequate replication to enable a phylogenetically controlled analysis), our results provide some insights into the potential variation in speciation stage across hybridizing plant taxa. Moreover, plants exhibit extensive variation in both life history and mating system [62,79,80]. These differences in life history may mediate the strength of this association between reproductive barriers and gene flow.
Case studies of closely related groups of species can provide further data on the processes underlying RI [6,7]. For example, within our literature survey, there were five hybridizing taxon pairs within the genus Populus that are all similar in life history (woody trees) and mating system (dioecious). Of these, P. alba and P. tremula provide an example of late-stage speciation, with these two taxa exhibiting a number of different reproductive isolating barriers, including habitat divergence, intrinsic incompatibilities and cyto-nuclear incompatibilities [43,57,58,65]. In comparison, P. davidiana and P. rotundifolia are two recently diverged species that inhabit distinct environments, and which provide an excellent example for the study of RI in the early stage of speciation [67].
(b). Early versus late stages of speciation in Populus: novel insights from whole-genome phylogenomics of a poplar species complex
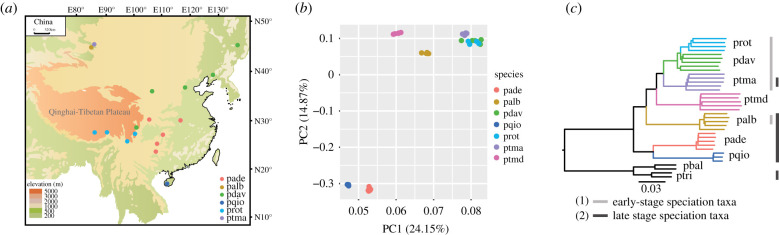
Whole-genome resequencing and reference-mapping of 36 individuals from seven ingroup and two outgroup taxa onto the P. trichocarpa genome assembly resulted in an average of 91.7% genomic regions covered, with an average coverage depth of 26.48×, yielding 7 026 036 high-quality SNPs (electronic supplementary material, table S2). Populus davidiana (the Chinese aspen) and P. rotundifolia (the Himalayan aspen), the two most recently derived species, were not monophyletic in NJ analysis. However, the three sequenced individuals of P. davidiana from central China were placed together with P. rotundifolia sampled in sym-/parapatry in the same geographical region (figure 2), rather than with conspecific individuals of P. davidiana from northeastern China, where its sister taxon P. rotundifolia is absent. This is suggestive of hybridization between these species in central China where they co-occur, thus also corroborating recent findings obtained with far more intensive biogeographic sampling but much sparser sampling of the genome [67]. Our two showcase species for late-stage speciation, P. alba and P. tremula, on the other hand, were clearly separated in PCA and NJ analysis (figure 2).
Figure 2.
Sample set of individuals from a Populus (poplar and aspen) species complex used for whole-genome phylogenomics. (a) Sampling locations of six Eurasian Populus species. (b) PCA of SNP data from resequenced genomes for seven Populus species, including the six Eurasian species (a) and one North American species, P. tremuloides. (c) Rooted NJ tree based on the genomic data, with P. trichocarpa and P. balsamifera as outgroups. (1) and (2) highlight the taxa selected for focused phylogenomic analysis in early stages of speciation (1) and late stages of speciation (2) as determined by IBD tract sharing (figure 3a). Species abbreviations: pade, P. adenopoda; palb, P. alba; pdav, P. davidiana; pqio, P. qiongdaoensis; prot, P. rotundifolia; ptma, P. tremula; ptmd, P. tremuloides; pbal, P. balsamifera; ptri, P. trichocarpa.
Our coalescent-based, dated species tree (electronic supplementary material, figure S1 and table S3) broadly reflected genetic relationships seen in the NJ tree and in a recent large-scale phylogenomic study of Populus [51], and demographic analysis using the site frequency spectrum and SMC++ complemented this coalescent-based analysis (electronic supplementary material, figure S2). SMC++ indicated an initial reduction in Ne in all species, coincident with the divergence of the major lineages in section Populus followed by population recovery to varying degrees (electronic supplementary material, figure S2). The results also reflected species splits seen in our coalescent-species tree, with Ne curves for P. alba and P. tremula splitting much further back in time than those for P. davidiana and P. rotundifolia. As expected, the Ne trajectories for P. alba and P. tremula separated more recently than those for P. alba and the North American aspen P. tremuloides, consistent with reports of hybridization and introgression between the partially sym-/parapatric Eurasian species P. alba and P. tremula [8,45,59,65].
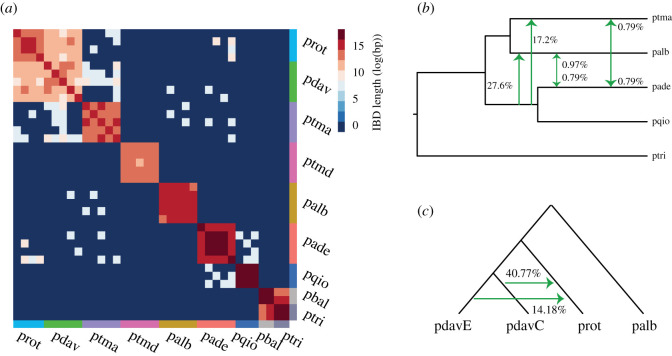
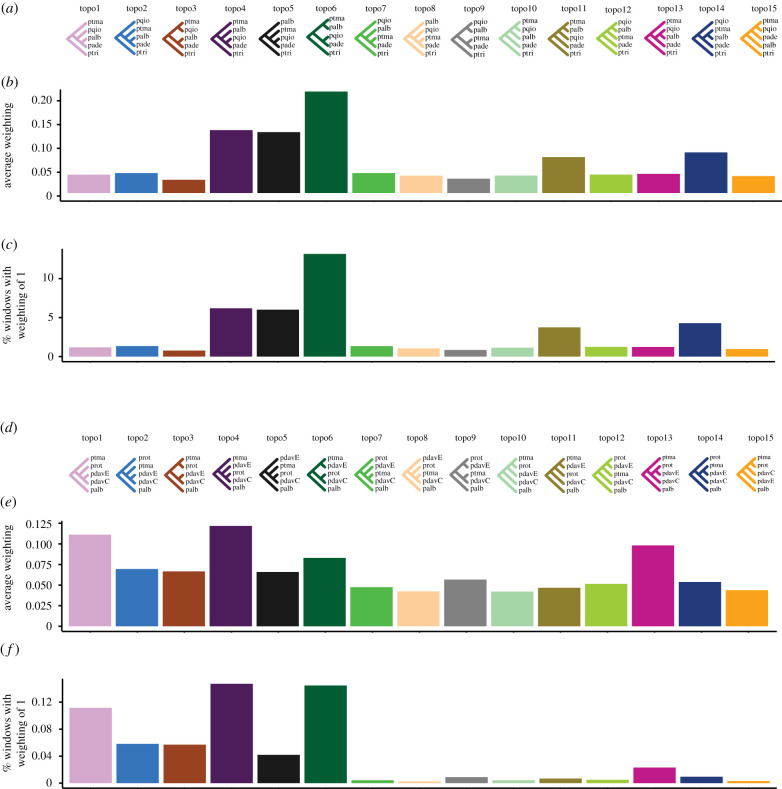
Genome-wide patterns of IBD tract sharing (figure 3a) allowed us to select groups of both early- and late-stage speciation taxa for subsequent phylogenomic analyses and contrasts. We selected five more recently diverged taxa with weak barriers [67], including P. davidiana and P. rotundifolia, and five more anciently diverged taxa with strong barriers [57,58], including P. alba and P. tremula, for genome-wide analyses of tree topologies using TWISST (figure 4). We found a high percentage of discordant tree topologies, especially in early-stage speciation taxa (figure 4), indicating extensive introgression or incomplete lineage sorting (ILS). Of the 15 possible topologies in late-stage speciation taxa, the three most common ones ordered by their frequency were topo6 (green), topo4 (purple) and topo5 (black), and topo6 was consistent with the species tree. As expected, more than 10% of genome windows reflecting the species tree (topo6) had completely sorted genealogies in these late-stage speciation taxa (indicated by ‘% windows with a weighting of 1’). The high weightings of genealogies topo4 and topo5 are indicative of either ILS or ancient introgressive gene flow involving P. tremula, P. alba, and the ancestor of P. adenopoda and P. qiongdaoensis. The ancient gene flow hypothesis was supported by DFOIL five-taxon tests (figure 3b,c), which have been validated to function even with high levels of ILS [78]. This is broadly consistent with widespread interspecific gene flow in Populus detected in a recent large-scale phylogenomic study [51].
Figure 3.
Patterns of haplotype sharing and introgressive gene flow. (a) The total length (log) of identical-by-descent (IBD) haplotypes shared between individuals. (b) DFOIL five-taxon analysis for five early-stage speciation taxa, and (c) four-taxon analysis for four early-stage speciation taxa. Green arrows indicate the direction of gene flow between species; numbers along arrows represent the proportion of genome windows with evidence for gene flow between taxa. The analyses were based on 10 kb windows, retaining values with p < 0.01. Taxon abbreviations follow figure 2.
Figure 4.
Topology weighting reveals widespread phylogenetic discordance in both early- and late speciation poplar taxa. (a) The 15 rooted topologies of late-stage speciation taxa: pade, pqio, ptma, palb and ptri as outgroup. (b,e) Average weighting of each topology. (c,f) The percentage of windows exhibiting complete lineage sorting for each topology. (d) The 15 rooted topologies of early-stage speciation taxa: pdavC, pdavE, prot, ptma and palb as outgroup. pdavC and pdavE are two populations representing two different phylogeographic lineages of P. davidiana from central and northeastern China, respectively. Taxon abbreviations follow figure 2.
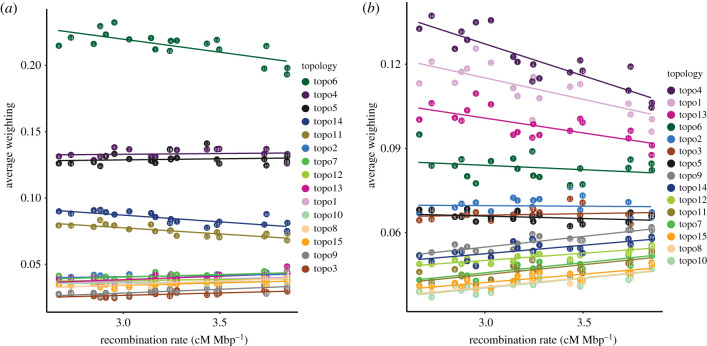
Under linked selection encompassing both directional selection and background selection against deleterious mutations, we would expect the weights of TWISST species tree topologies to be highest with low recombination rates, while the weights of introgression topologies (or admixture-related parameters more generally) should be released from this constraint or even increase with recombination [33]. This is analogous to the expectation that in the presence of hybrid incompatibilities, introgressed ancestry in populations is more likely to persist in regions of high recombination [81]. In line with this expectation, we observed the expected increase in species tree weights with reduced recombination rates for both early-stage and late-stage speciation taxa (figure 5; electronic supplementary material, table S4; topo6). The weights of putative introgression topologies in late-stage speciation taxa, however, did not show the expected increase for greater recombination rates (figure 5; electronic supplementary material, table S4; topo4 and topo5). Rather, these topologies received appreciable weights across all observed recombination rates. This is consistent with a breakdown in correlation between recombination rate and shared haplotype length in deeply divergent Populus spp. [51], and suggests that introgressed segments are able to escape barrier loci and linked selection over time, especially in species with high recombination rates.
Figure 5.
Average weightings per chromosome for all 15 topologies, plotted against average recombination rate (centimorgans per Mbp). Coloured circles represent poplar chromosomes 1–19. (a,b) Relationship between weightings of each topology and recombination rate in late-stage and early-stage speciation taxa, respectively, including linear fit. Recombination rate estimates for P. tremula were obtained with LDhat based on 100 kb windows [55]. The tree topologies for late- and early-stage speciation taxa are shown in figure 4a,d. Correlation and regression statistics are shown in electronic supplementary material, table S4.
Outcrossing, wind-pollinated trees such as poplar and aspen species exhibit fairly large Ne (greater than 100 000) and low levels of linkage disequilibrium (LD), consistent with high levels of effective recombination [52]. The decay of LD along chromosomes is even more rapid in species with continuous distributions such as P. tremula than in floodplain poplar species with more patchy distributions [53,55,82]. In such high recombination genomes, it should be easier to escape barrier loci [18,83] compared with other organisms with smaller Ne and slower LD decay [14,33]. Also, like other long-lived outcrossing perennial plant species, poplars harbour large amounts of standing genetic variation. This results in complex population genomic signatures of local adaptation, frequently involving subtle allele frequency shifts at many loci [10,66,84]. Importantly, these intraspecific patterns are mirrored by polygenic architectures of fitness-related trait differences between hybridizing species, including our two showcase species for late-stage speciation studied here, P. alba and P. tremula [39]. In fact, the observed relationships of tree topology weights with recombination rate in strongly divergent species [57,58] are consistent with the polygenic, complex architecture of fitness-related trait differences recently identified by ‘admixture mapping’ genome-wide association studies in hybrids [39]. Genomic regions supporting the species tree topology in late-stage speciation taxa apparently accumulated owing to linked selection across the genome. Nevertheless, this pattern is also expected to arise as a result of background selection or selective sweeps unrelated to reproductive barriers, effectively lowering Ne for chromosomal regions with low recombination rates [31,32]. Despite these confounding signals, recent simulation studies have shown that background selection alone may not be sufficient to explain recombination rate-dependent divergence landscapes in Ficedula flycatchers [85] and monkeyflowers [29], and such modelling approaches using more extensive population genomic data will be useful to further characterize the architecture of RI in deeply divergent poplars.
In our five selected early-stage speciation taxa, the introgression topology, topo4 (purple)—in which the locally parapatric populations P. rotundifolia and central P. davidiana were sister taxa—received even higher weightings than the species tree, topo1 (pink) (figure 4). The introgression topology also received consistently higher weightings in well delimited chromosome segments along the genome (electronic supplementary material, figure S3), which is reminiscent of haplotype signatures commonly observed with introgressive gene flow [40,86]. Accordingly, P. rotundifolia and P. davidiana exhibited extensive sharing of long IBD tracts (figure 3). This might also explain the conspicuous negative correlation between introgressed topology weightings (topo4) and recombination rate seen for these species (figure 5; electronic supplementary material, table S4), with increased weightings at low recombination rates. Increased introgression is not a priori expected in low recombination regions [33,81]. The high introgressed topology weightings at low recombination rates (figure 5) can alternatively result from insufficient time to break up long haplotypes stemming from recent introgressive gene flow. A strong positive correlation of the introgression tree and recombination rates as seen in other systems [87] may also be masked by extensive levels of ILS among windows supporting topo4, as suggested by the high frequency of the ‘mirrored’ topology, grouping together eastern P. davidiana and P. rotundifolia (topo13, magenta; figure 4). Topo13 also showed a weak negative correlation with recombination rate, highlighting how extensive standing variation in species with large Ne may slow down formation of strong reproductive barriers.
4. Conclusion
Our literature survey of hybridizing flowering plant species points to important roles for both pre- and post-zygotic barriers in plant speciation, and indicates that barrier complexity (i.e. the number of different barriers) is linked to an overall reduction in gene flow. Future efforts should explore how different aspects of life-history traits and mating systems (for which plants exhibit extraordinary variation; [62]) mediate the strength of this association, and how plants, animals and fungi differ in this regard. The model tree genus Populus offers suitable taxon pairs or groups for addressing the evolution of strong RI during plant speciation; this includes late-stage speciation taxa that are strongly isolated by multiple barriers, but which nevertheless form fertile hybrids. An important future task will be to assess the cumulative action of different pre- and post-zygotic barriers in this group, and how their effects become coupled towards the development of strong RI [4,21]. Each single barrier effect may have a simple or polygenic basis, and some traits may affect multiple barriers [88]. Thus, we anticipate that understanding the evolution of strong RI will benefit greatly from advances in high-throughput phenotyping and the quantitative evolutionary genomics of multivariate trait space.
Our phylogenomic data for a poplar species complex mirrored those from our literature survey, with stronger divergence and greatly reduced IBD tract sharing for late-stage speciation taxa separated by multiple barriers, in contrast with pronounced IBD sharing and topology discordance for early-stage taxa separated mainly by a weak eco-geographic barrier. Genome-wide variation in phylogenetic tree topologies based on 36 sequenced genomes highlights the potential role of both ancient and recent introgressive gene flow for the genomic composition of extant poplar species. This is in addition to ILS, which we must expect to be present at these evolutionary time scales [51]. While the weightings (frequencies) of species tree topologies—and their relationships with recombination rate variation along the genome—were broadly consistent with polygenic barriers and linked selection pinpointed by other studies on Populus spp. [54,55], the lack of a strong relationship of putatively introgressed topologies with recombination rates highlights the complexities of barrier formation in this group [39,89]. Complex architectures are expected to arise from a number of factors including (i) high levels of recombination and rapid LD decay along chromosomes in poplars [52,53,55], (ii) long generation times accentuated by the ability of viable genotypes to persist as clones [43], and (iii) large Ne, which enables these completely outcrossing, wind-pollinated tree species to hold extraordinary levels of standing genetic variation. For early-stage speciation taxa, the genome-wide topology/recombination rate relationship pointed to a protracted speciation process and the absence of strong barriers because of the apparent presence of both long introgressed haplotype tracts and high levels of ILS. A similarly protracted process may have been at work for late-stage speciation taxa, supported by an extended period of genetic exchange between the ancestor of P. adenopoda and P. qiongdoaensis and both P. tremula and P. alba. Indeed, phylogenomic approaches based on tree topology variation appear to lend themselves to studies of the evolution of strong RI during speciation and the extended time scales this may take. In species complexes of poplars, it appears that despite a polygenic basis of barriers, numbers of barrier loci are still too low (relative to recombination rates and individual selection coefficients) to facilitate strong coupling [86] and thus to prevent the escape of locally adaptive alleles. We hope this work will encourage more studies exploring discordance and concordance between patterns of RI seen through the lenses of different, complementary approaches available to speciation geneticists addressing different time scales.
Supplementary Material
Acknowledgements
We thank members of J.L.’s lab for collecting samples, Michael Barfuss and Elfi Grasserbauer for help in the laboratory, the Next Generation Sequencing Platform of the University of Berne for sequencing, the Vienna Scientific Cluster (VSC) for access to computational resources, and Claus Vogel and members of the PopGen Vienna graduate school for helpful discussions.
Data accessibility
Flowering plant literature analysis made use of the database introduced by Pickup et al. [62], and an updated data set and R scripts are available from Dryad at https://doi.org/10.5061/dryad.h9w0vt4fw [90]. Extensive biological sample and sequencing statistics are uploaded as electronic supplementary material, tables. All raw read data were uploaded to NCBI and can be found under Bioproject ID PRJNA612655.
Authors' contributions
Study conceived and designed by C.L. together with H.S., J.H., J.L. and D.L.F. Sample collection and laboratory work conducted by H.S. and J.L. Phylogenomic data analysis by H.S. and J.H. Interpretation of phylogenomic results was undertaken by H.S., J.H., C.L., D.L.F. and P.K.I. M.P. and D.L.F. compiled and analysed the plant literature database. C.L., H.S., J.H., D.L.F. and M.P. drafted the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by a fellowship from the China Scholarship Council (CSC) to H.S., Swiss National Science Foundation (SNF) grant no. 31003A_149306 to C.L., doctoral programme grant W1225-B20 to a faculty team including C.L., and the University of Vienna.
References
- 1.Abbott R, et al. 2013. Hybridization and speciation. J. Evol. Biol. 26, 229–246. ( 10.1111/j.1420-9101.2012.02599.x) [DOI] [PubMed] [Google Scholar]
- 2.Gompert Z, Mandeville EG, Buerkle CA. 2017. Analysis of population genomic data from hybrid zones. Annu. Rev. Ecol. Evol. Syst. 48, 207–229. ( 10.1146/annurev-ecolsys-110316-022652) [DOI] [Google Scholar]
- 3.Nosil P, Feder JL, Flaxman SM, Gompert Z. 2017. Tipping points in the dynamics of speciation. Nat. Ecol. Evol. 1, 1 ( 10.1038/s41559-016-0001) [DOI] [PubMed] [Google Scholar]
- 4.Ravinet M, Faria R, Butlin RK, Galindo J, Bierne N, Rafajlovic M, Noor MAF, Mehlig B, Westram AM. 2017. Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. J. Evol. Biol. 30, 1450–1477. ( 10.1111/jeb.13047) [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein J. 1981. Skepticism toward Santa Rosalia, or why are there so few kinds of animals? Evolution 35, 124–138. ( 10.1111/j.1558-5646.1981.tb04864.x) [DOI] [PubMed] [Google Scholar]
- 6.Coyne J, Orr H. 2004. Speciation. Oxford, UK: Oxford University Press. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 7.Nosil P. 2012. Ecological speciation. Oxford, UK: Oxford University Press. [Google Scholar]
- 8.Christe C, Stolting KN, Paris M, Fraisse C, Bierne N, Lexer C. 2017. Adaptive evolution and segregating load contribute to the genomic landscape of divergence in two tree species connected by episodic gene flow. Mol. Ecol. 26, 59–76. ( 10.1111/mec.13765) [DOI] [PubMed] [Google Scholar]
- 9.Ellegren H, et al. 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491, 756–760. ( 10.1038/nature11584) [DOI] [PubMed] [Google Scholar]
- 10.Evans LM, et al. 2014. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat. Genet. 46, 1089–1096. ( 10.1038/ng.3075) [DOI] [PubMed] [Google Scholar]
- 11.Novikova PY, et al. 2016. Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat. Genet. 48, 1077–1082. ( 10.1038/ng.3617) [DOI] [PubMed] [Google Scholar]
- 12.Tavares H, et al. 2018. Selection and gene flow shape genomic islands that control floral guides. Proc. Natl Acad. Sci. USA 115, 11 006–11 011. ( 10.1073/pnas.1801832115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Heliconius Genome Consortium. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98. ( 10.1038/nature11041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Belleghem SM, et al. 2017. Complex modular architecture around a simple toolkit of wing pattern genes. Nat. Ecol. Evol. 1, 52 ( 10.1038/s41559-016-0052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feder JL, Egan SP, Nosil P. 2012. The genomics of speciation-with-gene-flow. Trends Genet. 28, 342–350. ( 10.1016/j.tig.2012.03.009) [DOI] [PubMed] [Google Scholar]
- 16.Gompert Z, Egan SP, Barrett RD, Feder JL, Nosil P. 2017. Multilocus approaches for the measurement of selection on correlated genetic loci. Mol. Ecol. 26, 365–382. ( 10.1111/mec.13867) [DOI] [PubMed] [Google Scholar]
- 17.Seehausen O, et al. 2014. Genomics and the origin of species. Nat. Rev. Genet. 15, 176–192. ( 10.1038/nrg3644) [DOI] [PubMed] [Google Scholar]
- 18.Barton N, Bengtsson BO. 1986. The barrier to genetic exchange between hybridising populations. Heredity (Edinb.) 57, 357–376. ( 10.1038/hdy.1986.135) [DOI] [PubMed] [Google Scholar]
- 19.Barton NH, de Cara MA.. 2009. The evolution of strong reproductive isolation. Evolution 63, 1171–1190. ( 10.1111/j.1558-5646.2009.00622.x) [DOI] [PubMed] [Google Scholar]
- 20.Bierne N, Welch J, Loire E, Bonhomme F, David P. 2011. The coupling hypothesis: why genome scans may fail to map local adaptation genes. Mol. Ecol. 20, 2044–2072. ( 10.1111/j.1365-294X.2011.05080.x) [DOI] [PubMed] [Google Scholar]
- 21.Butlin RK, Smadja CM. 2018. Coupling, reinforcement, and speciation. Am. Nat. 191, 155–172. ( 10.1086/695136) [DOI] [PubMed] [Google Scholar]
- 22.Roux C, Fraisse C, Romiguier J, Anciaux Y, Galtier N, Bierne N. 2016. Shedding light on the grey zone of speciation along a continuum of genomic divergence. PLoS Biol. 14, e2000234 ( 10.1371/journal.pbio.2000234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barton NH, Gale KS. 1993. Genetic analysis of hybrid zones. In Hybrid zones and the evolutionary process (ed. Harrison RG.), pp. 13–45. New York, NY: Oxford University Press. [Google Scholar]
- 24.Martin SH, Jiggins CD. 2017. Interpreting the genomic landscape of introgression. Curr. Opin. Genet. Dev. 47, 69–74. ( 10.1016/j.gde.2017.08.007) [DOI] [PubMed] [Google Scholar]
- 25.Bar-Zvi D, Lupo O, Levy AA, Barkai N. 2017. Hybrid vigor: the best of both parents, or a genomic clash? Curr. Opin. Syst. Biol. 6, 22–27. ( 10.1016/j.coisb.2017.08.004) [DOI] [Google Scholar]
- 26.Gavrilets S. 2003. Perspective: models of speciation: what have we learned in 40 years? Evolution 57, 2197–2215. ( 10.1111/j.0014-3820.2003.tb00233.x) [DOI] [PubMed] [Google Scholar]
- 27.Lindtke D, Buerkle CA. 2015. The genetic architecture of hybrid incompatibilities and their effect on barriers to introgression in secondary contact. Evolution 69, 1987–2004. ( 10.1111/evo.12725) [DOI] [PubMed] [Google Scholar]
- 28.Han F, Lamichhaney S, Grant BR, Grant PR, Andersson L, Webster MT. 2017. Gene flow, ancient polymorphism, and ecological adaptation shape the genomic landscape of divergence among Darwin's finches. Genome Res. 27, 1004–1015. ( 10.1101/gr.212522.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stankowski S, Chase MA, Fuiten AM, Rodrigues MF, Ralph PL, Streisfeld MA. 2019. Widespread selection and gene flow shape the genomic landscape during a radiation of monkeyflowers. PLoS Biol. 17, e3000391 ( 10.1371/journal.pbio.3000391) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeaman S, Aeschbacher S, Burger R. 2016. The evolution of genomic islands by increased establishment probability of linked alleles. Mol. Ecol. 25, 2542–2558. ( 10.1111/mec.13611) [DOI] [PubMed] [Google Scholar]
- 31.Charlesworth B. 2012. The effects of deleterious mutations on evolution at linked sites. Genetics 190, 5–22. ( 10.1534/genetics.111.134288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cruickshank TE, Hahn MW. 2014. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol. Ecol. 23, 3133–3157. ( 10.1111/mec.12796) [DOI] [PubMed] [Google Scholar]
- 33.Martin SH, Davey JW, Salazar C, Jiggins CD. 2019. Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol. 17, e2006288 ( 10.1371/journal.pbio.2006288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edelman NB, et al. 2019. Genomic architecture and introgression shape a butterfly radiation. Science 366, 594–599. ( 10.1126/science.aaw2090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barton NH, Hewitt GM. 1985. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 16, 113–148. ( 10.1146/annurev.es.16.110185.000553) [DOI] [Google Scholar]
- 36.Buerkle CA, Lexer C. 2008. Admixture as the basis for genetic mapping. Trends Ecol. Evol. 23, 686–694. ( 10.1016/j.tree.2008.07.008) [DOI] [PubMed] [Google Scholar]
- 37.Harrison RG, Larson EL. 2016. Heterogeneous genome divergence, differential introgression, and the origin and structure of hybrid zones. Mol. Ecol. 25, 2454–2466. ( 10.1111/mec.13582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brelsford A, Toews DPL, Irwin DE. 2017. Admixture mapping in a hybrid zone reveals loci associated with avian feather coloration. Proc. R. Soc. B 284, 20171106 ( 10.1098/rspb.2017.1106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bresadola L, Caseys C, Castiglione S, Buerkle CA, Wegmann D, Lexer C. 2019. Admixture mapping in interspecific Populus hybrids identifies classes of genomic architectures for phytochemical, morphological and growth traits. New Phytol. 223, 2076–2089. ( 10.1111/nph.15930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pallares LF, Harr B, Turner LM, Tautz D. 2014. Use of a natural hybrid zone for genomewide association mapping of craniofacial traits in the house mouse. Mol. Ecol. 23, 5756–5770. ( 10.1111/mec.12968) [DOI] [PubMed] [Google Scholar]
- 41.Pease JB, Haak DC, Hahn MW, Moyle LC. 2016. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14, e1002379 ( 10.1371/journal.pbio.1002379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckenwalder JE. 1984. Natural intersectional hybridization between North American species of Populus (Salicaceae) in sections Aigeiros and Tacamahaca. II. Taxonomy. Can. J. Bot. 62, 325–335. ( 10.1139/b84-051) [DOI] [Google Scholar]
- 43.Macaya-Sanz D, Heuertz M, Lindtke D, Vendramin GG, Lexer C, Gonzalez-Martinez SC. 2016. Causes and consequences of large clonal assemblies in a poplar hybrid zone. Mol. Ecol. 25, 5330–5344. ( 10.1111/mec.13850) [DOI] [PubMed] [Google Scholar]
- 44.Martinsen GD, Whitham TG, Turek RJ, Keim P. 2001. Hybrid populations selectively filter gene introgression between species. Evolution 55, 1325–1335. [DOI] [PubMed] [Google Scholar]
- 45.Rajora OP, Dancik BP. 1992. Genetic characterization and relationships of Populus alba, P. tremula, and P. × canescens, and their clones. Theor. Appl. Genet. 84, 291–298. ( 10.1007/BF00229485) [DOI] [PubMed] [Google Scholar]
- 46.Suarez-Gonzalez A, Hefer CA, Christe C, Corea O, Lexer C, Cronk QC, Douglas CJ. 2016. Genomic and functional approaches reveal a case of adaptive introgression from Populus balsamifera (balsam poplar) in P. trichocarpa (black cottonwood). Mol. Ecol. 25, 2427–2442. ( 10.1111/mec.13539) [DOI] [PubMed] [Google Scholar]
- 47.Tuskan GA, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr, Gray). Science 313, 1596–1604. ( 10.1126/science.1128691) [DOI] [PubMed] [Google Scholar]
- 48.Stettler RF, Bradshaw HD Jr, Heilman PE, Hinckley TM. 1996. Biology of Populus and its implications for management and conservation. Ottawa, Canada: NRC Research Press. [Google Scholar]
- 49.Cervera MT, Storme V, Soto A, Ivens B, Van Montagu M, Rajora OP, Boerjan W.. 2005. Intraspecific and interspecific genetic and phylogenetic relationships in the genus Populus based on AFLP markers. Theor. Appl. Genet. 111, 1440–1456. ( 10.1007/s00122-005-0076-2) [DOI] [PubMed] [Google Scholar]
- 50.Hamzeh M, Périnet P, Dayanandan S. 2006. Genetic relationships among species of Populus (Salicaceae) based on nuclear genomic data. J. Torrey Bot. Soc. 133, 519–527. ( 10.3159/1095-5674(2006)133519:grasop]2.0.co;2) [DOI] [Google Scholar]
- 51.Wang M, et al. 2019. Phylogenomics of the genus Populus reveals extensive interspecific gene flow and balancing selection. New Phytol. 225, 1370–1382. ( 10.1111/nph.16215) [DOI] [PubMed] [Google Scholar]
- 52.Ingvarsson PK. 2008. Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula. Genetics 180, 329–340. ( 10.1534/genetics.108.090431) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slavov GT, et al. 2012. Genome resequencing reveals multiscale geographic structure and extensive linkage disequilibrium in the forest tree Populus trichocarpa. New Phytol. 196, 713–725. ( 10.1111/j.1469-8137.2012.04258.x) [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Street NR, Scofield DG, Ingvarsson PK. 2016. Variation in linked selection and recombination drive genomic divergence during allopatric speciation of European and American aspens. Mol. Biol. Evol. 33, 1754–1767. ( 10.1093/molbev/msw051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Street NR, Scofield DG, Ingvarsson PK. 2016. Natural selection and recombination rate variation shape nucleotide polymorphism across the genomes of three related Populus species. Genetics 202, 1185–1200. ( 10.1534/genetics.115.183152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma T, et al. 2018. Ancient polymorphisms and divergence hitchhiking contribute to genomic islands of divergence within a poplar species complex. Proc. Natl Acad. Sci. USA 115, E236–E243. ( 10.1073/pnas.1713288114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christe C, Stolting KN, Bresadola L, Fussi B, Heinze B, Wegmann D, Lexer C. 2016. Selection against recombinant hybrids maintains reproductive isolation in hybridizing Populus species despite F1 fertility and recurrent gene flow. Mol. Ecol. 25, 2482–2498. ( 10.1111/mec.13587) [DOI] [PubMed] [Google Scholar]
- 58.Lindtke D, Gompert Z, Lexer C, Buerkle CA. 2014. Unexpected ancestry of Populus seedlings from a hybrid zone implies a large role for postzygotic selection in the maintenance of species. Mol. Ecol. 23, 4316–4330. ( 10.1111/mec.12759) [DOI] [PubMed] [Google Scholar]
- 59.Zeng YF, Zhang JG, Duan AG, Abuduhamiti B. 2016. Genetic structure of Populus hybrid zone along the Irtysh River provides insight into plastid-nuclear incompatibility. Scient. Rep. 6, 28043 ( 10.1038/srep28043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kisel Y, Barraclough TG. 2010. Speciation has a spatial scale that depends on levels of gene flow. Am. Nat. 175, 316–334. ( 10.1086/650369) [DOI] [PubMed] [Google Scholar]
- 61.Lowry DB, Modliszewski JL, Wright KM, Wu CA, Willis JH. 2008. The strength and genetic basis of reproductive isolating barriers in flowering plants. Phil. Trans. R. Soc. B 363, 3009–3021. ( 10.1098/rstb.2008.0064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pickup M, Brandvain Y, Fraisse C, Yakimowski S, Barton NH, Dixit T, Lexer C, Cereghetti E, Field DL. 2019. Mating system variation in hybrid zones: facilitation, barriers and asymmetries to gene flow New Phytol. 224, 1035–1047. ( 10.1111/nph.16180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abbott RJ. 2017. Plant speciation across environmental gradients and the occurrence and nature of hybrid zones. J. Syst. Evol. 55, 238–258. ( 10.1111/jse.12267) [DOI] [Google Scholar]
- 64.Dickmann D, Kuzovkina Y. 2008. Poplars and willows in the world. In Poplars and willows in the world, meeting the needs of society and the environment. International Poplar Commission Working Paper no. IPC/9-2 (eds JG Isebrands, J Richardson), pp. 9–12. Rome, Italy: Food and Agricultural Organization. [Google Scholar]
- 65.Lexer C, Joseph JA, van Loo M, Barbara T, Heinze B, Bartha D, Castiglione S, Fay MF, Buerkle CA. 2010. Genomic admixture analysis in European Populus spp. reveals unexpected patterns of reproductive isolation and mating. Genetics 186, 699–712. ( 10.1534/genetics.110.118828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stölting KN, Nipper R, Lindtke D, Caseys C, Waeber S, Castiglione S, Lexer C. 2013. Genomic scan for single nucleotide polymorphisms reveals patterns of divergence and gene flow between ecologically divergent species. Mol. Ecol. 22, 842–855. ( 10.1111/mec.12011) [DOI] [PubMed] [Google Scholar]
- 67.Zheng H, Fan L, Milne RI, Zhang L, Wang Y, Mao K. 2017. Species delimitation and lineage separation history of a species complex of aspens in China. Front. Plant Sci. 8, 375 ( 10.3389/fpls.2017.00375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fan L, Zheng H, Milne RI, Zhang L, Mao K. 2018. Strong population bottleneck and repeated demographic expansions of Populus adenopoda (Salicaceae) in subtropical China. Ann. Bot. 121, 665–679. ( 10.1093/aob/mcx198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, 1303.3997 [q-bio.GN].
- 70.Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. ( 10.1086/519795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu L, Yu L, Edwards SV. 2010. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol. Biol. 10, 302 ( 10.1186/1471-2148-10-302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 73.Terhorst J, Kamm JA, Song YS. 2017. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 49, 303–309. ( 10.1038/ng.3748) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Browning BL, Browning SR. 2013. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194, 459–471. ( 10.1534/genetics.113.150029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin SH, Van Belleghem SM.. 2017. Exploring evolutionary relationships across the genome using topology weighting. Genetics 206, 429–438. ( 10.1534/genetics.116.194720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Browning SR, Browning BL. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097. ( 10.1086/521987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guindon S, Lethiec F, Duroux P, Gascuel O. 2005. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33, W557–W559. ( 10.1093/nar/gki352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pease JB, Hahn MW. 2015. Detection and polarization of introgression in a five-taxon phylogeny. Syst. Biol. 64, 651–662. ( 10.1093/sysbio/syv023) [DOI] [PubMed] [Google Scholar]
- 79.Charlesworth D. 2006. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2, e64 ( 10.1371/journal.pgen.0020064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goodwillie C, Kalisz S, Eckert CG. 2005. The evolutionary enigma of mixed mating systems in plants: occurrence, theoretical explanations, and empirical evidence. Annu. Rev. Ecol. Evol. Syst. 36, 47–79. ( 10.1146/annurev.ecolsys.36.091704.175539) [DOI] [Google Scholar]
- 81.Schumer M, et al. 2018. Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science 360, 656–660. ( 10.1126/science.aar3684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lexer C, Buerkle CA, Joseph JA, Heinze B, Fay MF. 2007. Admixture in European Populus hybrid zones makes feasible the mapping of loci that contribute to reproductive isolation and trait differences. Heredity 98, 74–84. ( 10.1038/sj.hdy.6800898) [DOI] [PubMed] [Google Scholar]
- 83.Uecker H, Setter D, Hermisson J. 2015. Adaptive gene introgression after secondary contact. J. Math. Biol. 70, 1523–1580. ( 10.1007/s00285-014-0802-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Carvalho D, et al. 2010. Admixture facilitates adaptation from standing variation in the European aspen (Populus tremula L.), a widespread forest tree. Mol. Ecol. 19, 1638–1650. ( 10.1111/j.1365-294X.2010.04595.x) [DOI] [PubMed] [Google Scholar]
- 85.Rettelbach A, Nater A, Ellegren H. 2019. How linked selection shapes the diversity landscape in Ficedula flycatchers. Genetics 212, 277–285. ( 10.1534/genetics.119.301991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kruuk LE, Baird SJ, Gale KS, Barton NH. 1999. A comparison of multilocus clines maintained by environmental adaptation or by selection against hybrids. Genetics 153, 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Edmands S, Timmerman CC. 2003. Modeling factors affecting the severity of outbreeding depression. Conserv. Biol. 17, 883–892. ( 10.1046/j.1523-1739.2003.02026.x) [DOI] [Google Scholar]
- 88.Smadja CM, Butlin RK. 2011. A framework for comparing processes of speciation in the presence of gene flow. Mol. Ecol. 20, 5123–5140. ( 10.1111/j.1365-294X.2011.05350.x) [DOI] [PubMed] [Google Scholar]
- 89.McKown AD, Guy RD, Klapste J, Geraldes A, Friedmann M, Cronk QC, El-Kassaby YA, Mansfield SD, Douglas CJ. 2014. Geographical and environmental gradients shape phenotypic trait variation and genetic structure in Populus trichocarpa. New Phytol. 201, 1263–1276. ( 10.1111/nph.12601) [DOI] [PubMed] [Google Scholar]
- 90.Shang H, Hess J, Pickup M, Field DL, Ingvarsson PK, Liu J, Lexer C. 2020. Data from: Evolution of strong reproductive isolation in plants: broad-scale patterns and lessons from a perennial model group Dryad Digital Repository. ( 10.5061/dryad.h9w0vt4fw) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Shang H, Hess J, Pickup M, Field DL, Ingvarsson PK, Liu J, Lexer C. 2020. Data from: Evolution of strong reproductive isolation in plants: broad-scale patterns and lessons from a perennial model group Dryad Digital Repository. ( 10.5061/dryad.h9w0vt4fw) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Flowering plant literature analysis made use of the database introduced by Pickup et al. [62], and an updated data set and R scripts are available from Dryad at https://doi.org/10.5061/dryad.h9w0vt4fw [90]. Extensive biological sample and sequencing statistics are uploaded as electronic supplementary material, tables. All raw read data were uploaded to NCBI and can be found under Bioproject ID PRJNA612655.