Abstract
Simpson's fossil-record inspired model of ‘adaptive zones’ proposes that evolution is dominated by small fluctuations within adaptive zones, occasionally punctuated by larger shifts between zones. This model can help explain why the process of population divergence often results in weak or moderate reproductive isolation (RI), rather than strong RI and distinct species. Applied to the speciation process, the adaptive zones hypothesis makes two inter-related predictions: (i) large shifts between zones are relatively rare, (ii) when large shifts do occur they generate stronger RI than shifts within zones. Here, we use ecological, phylogenetic and behavioural data to test these predictions in Timema stick insects. We show that host use in Timema is dominated by moderate shifts within the systematic divisions of flowering plants and conifers, with only a few extreme shifts between these divisions. However, when extreme shifts occur, they generate greater RI than do more moderate shifts. Our results support the adaptive zones model, and suggest that the net contribution of ecological shifts to diversification is dependent on both their magnitude and frequency. We discuss the generality of our findings in the light of emerging evidence from diverse taxa that the evolution of RI is not always the only factor determining the origin of species diversity.
This article is part of the theme issue ‘Towards the completion of speciation: the evolution of reproductive isolation beyond the first barriers’.
Keywords: host preference, phylogenetics, speciation, reproductive isolation, Timema stick insects
1. Introduction
A number of evolutionary models, such as Simpson's adaptive zones model of quantum evolution [1,2], propose that small evolutionary changes within adaptive zones (i.e. changes within a broad resource or habitat category, sensu [3,4]) are common, but that more extreme evolutionary change occurs rarely [5,6]. Most evidence for such models stems from deep macro-evolutionary timescales and high taxonomic levels, such as orders or families [1,2,5]. Thus, the processes and mechanisms generating these patterns are not well understood. For example, the roles of random drift, fluctuating selection and macro-mutation (e.g. ‘hopeful monsters’ [7]) in generating patterns consistent with these models remain unclear, but must be resolved to understand whether and which micro-evolutionary processes best explain broad-scale macro-evolutionary patterns.
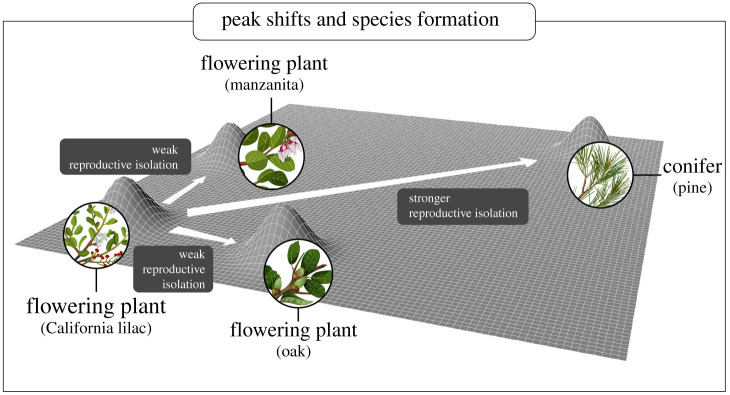
Here, we specifically apply the adaptive zones model to the speciation process, which often occurs by populations diverging into partially reproductively isolated ecotypes or subspecies, and eventually into strongly reproductively isolated species [8–15]. Such a differentiation process or ‘speciation continuum’ has been observed in fish [16–19], amphibians [20], birds [21], plants [22] and insects [13,14,23–25]. However, it is often unclear why populations differ in levels of reproductive isolation (RI) [9–11,26]. How such differences affect the diversification of a clade is then further complicated by the relative frequencies with which different levels of RI are reached. The adaptive zones hypothesis can be applied to explain this variation, making two inter-related predictions: (i) shifts between zones are relatively rare and (ii) when large shifts between zones do occur, they generate stronger RI than shifts within zones (figure 1).
Figure 1.
Ecological shifts and the process of speciation. A schematic depiction of how large peak shifts between flowering plant and conifer hosts, although relatively rare, generate greater RI than more moderate host shifts among flowering plant families.
Note that even without invoking the adaptive zones model per se, these predictions should hold; large ecological shifts that generate strong RI may be rare. Moreover, although we here test the adaptive zones model using discrete categories of ecological divergence, similar logic could be applied to continuous scenarios. Just as one may ask whether shifts between more extreme categories generate more RI, one could test whether more extreme quantitative shifts in ecology (e.g. temperature, elevation, aridity) generate more RI. For example, it has been shown that more extreme differentiation in quantitative ecological variables is associated with stronger RI across disparate plant and animal taxa, although this work did not consider phylogenetic shifts per se [27,28].
Testing these predictions is challenging because it requires integration of macro-evolutionary patterns, for example, at phylogenetic timescales, with data on micro-evolutionary processes and the evolution of RI. Most generally, such studies might help connect broad diversification patterns (i.e. defined as the net result of the speciation and extinction processes over time) to micro-evolutionary processes. We provide such a study here by integrating phylogenetically based inferences on rates of host shifts for greater than 100 host-associated populations of 11 Timema stick insect species with experimental estimates of host-plant preference. Because Timema feed, mate and spend most of their lives on their hosts [29,30], host preferences are likely to translate to premating isolation in nature. Thus, we here use results from host preference experiments in the laboratory as a proxy for RI, with the understanding that future work testing RI in nature is warranted. Notably, Timema feed on a very wide range of hosts [29,30], but the frequency of host shifts of different magnitude over the approximately 30 Myr old history of this group has yet to be quantified [25].
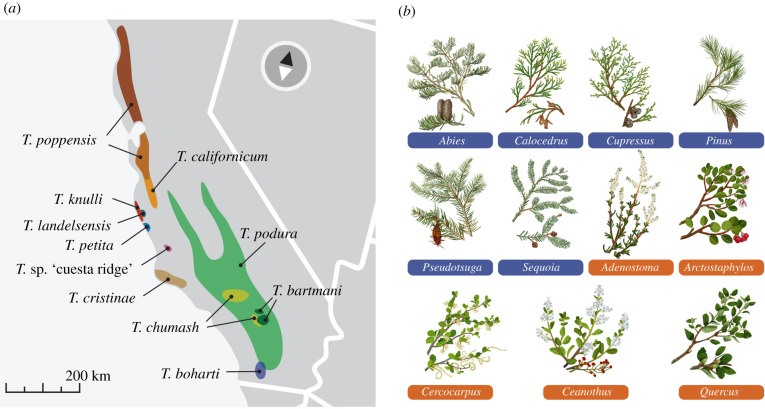
We thus here study Timema taxa that use a wide range of conifer (e.g. pine, cedar, redwood and fir) and flowering plant (e.g. oak, roses and manzanita) hosts (figure 2; electronic supplementary material, table S1; a host-plant population is defined as conspecific individuals collected from a common host genus at a geographical locality). In this context, we consider conifer and flowering plant hosts to represent different adaptive zones and thus shifts between them to be large relative to those within plant divisions, based on: (i) the fact that few insect species (or even sets of closely related species) use both these plant divisions as hosts [31] and (ii) the deep phylogenetic divergence between these two divisions and their great differences in chemistry, physical structure and evolutionary dynamics [32–36].
Figure 2.
Timema species and populations, and host-plant use. (a) Timema species ranges (from [25]). (b) Host-plant species used by Timema populations used in this study (conifers have blue labels, flowering plants have orange labels).
We first used phylogenetic information and host-plant use to infer the frequency of shifts between conifer and flowering plant hosts [25], relative to shifts between hosts within each division. An adaptive zones model would be supported by host shifts overall being common, but those between conifers and flowering plants being rare. Second, we tested for an association between the magnitude of a host shift (i.e. within or between plant divisions) and divergence in host preference, a form of premating RI for insects such as Timema that mate on their host plants [37–39]. Our results support the adaptive zones model, and suggest that the net contribution of ecological shifts to RI can depend on the shifts' magnitude. When larger shifts occur less often, their rarity increases waiting times to speciation. Thus, our findings add to emerging evidence that although the evolution of RI is a key component of the speciation process, it may not always be the factor controlling the frequency at which new lineages originate [40–42].
2. Material and methods
(a). Analysis of transition rates between hosts
Our sampling effort covered regions where Timema have been systematically studied over the last two decades [25,43], and searches were done of the known common hosts of each species. Missing host taxa would be problematic for our study only if this sampling was not random (i.e. systematically missing populations on conifers), which is unlikely. Details of the populations studied here are contained in [25] and in electronic supplementary material, table S1.
We first tested whether shifts between conifers and flowering plants occurred multiple times. We used the reduced-representation sequence data from the 57 geographical populations previously studied in [25] to infer phylogenetic relationships among Timema species and populations. This was done using data deposited in the Dryad Digital Repository (https://doi.org/10.5061/dryad.nq67q; linkage-group multiple alignments under the section ‘Phylogenetic inference and molecular dating’) to produce two new multiple alignments: selecting only the sites with at least two different nucleotides (‘strict-ASC’; 5797 variable sites), which allows using ascertainment bias models for inferences, and selecting also the sites with at least one ambiguity (‘relaxed’; 19 556 variable sites). We used IQTREE 1.6.2 [44] to carry out automatic substitution and partitioning model selection and to infer maximum-likelihood trees using topological constraints in order to test five different hypotheses: clustering by host-plant division (‘division’, implies a single shift between conifers and flowering plants), clustering by Timema species (‘species’, allow for multiple shifts within species), clustering by Timema species and host-plant division within Timema species (‘division within species', allows a single shift within each species), using the previous Bayesian inference from [25] (BEAST) and a maximum-likelihood tree inferred with IQTREE for this study (‘free’, no topological constraint whatsoever). Then, we estimated site-wise log-likelihoods and performed Shimodaira–Hasegawa (SH, [45]), weighted Shimodaira–Hasegawa (WSH, [45]) and approximately unbiased (AU, [46]) tests using IQTREE and consel 1.20 [47].
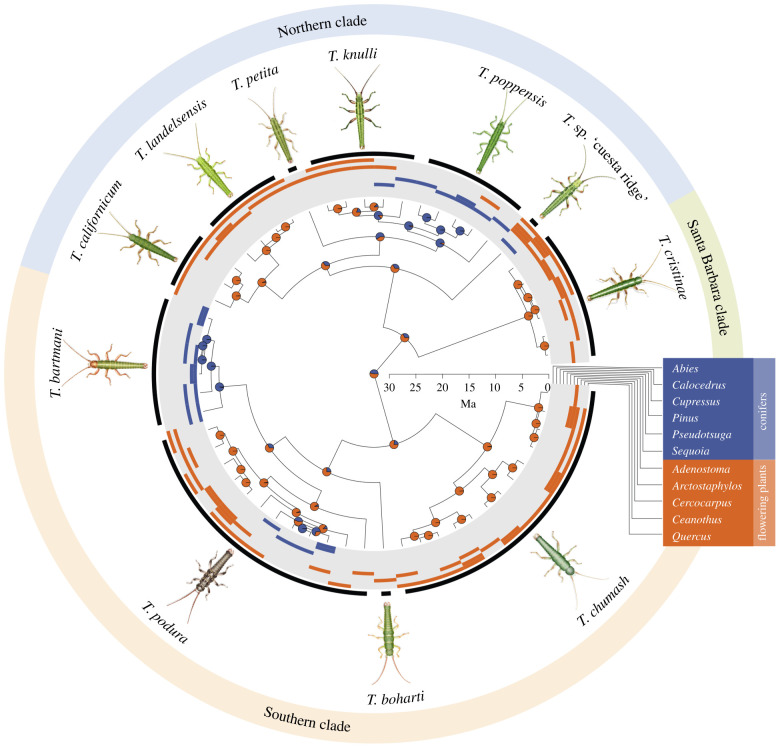
Subsequently, we reconstructed ancestral states using the function rayDISC from the R package corHMM 1.24 [48] in R v. 3.4.4 [49]. This function allows estimates of transition rates and ancestral states for multistate traits given a tree, allowing for polymorphism on the tips (i.e. assigning equal likelihoods to several hosts for a given population in our case), and recognizing both gains and losses of host-plant genera. First, we estimated ancestral states using the Bayesian maximum credibility tree from [25], coding the hosts of each of the 57 geographical populations as conifer, flowering plant or both, and estimating the root probability with the method described in [50,51]. This allowed us to visualize ancestral state marginal probabilities on the nodes of the tree (figure 3).
Figure 3.
Timema species phylogenetic relationships and host-plant use, including ancestral use inferences using conifers and flowering plants as states. The bars on the periphery depict the host-plant use of each population. The pie charts on the internal nodes represent the proportional likelihoods of conifers (blue) and flowering plants (orange) for reconstructed ancestral states. Ma, million years.
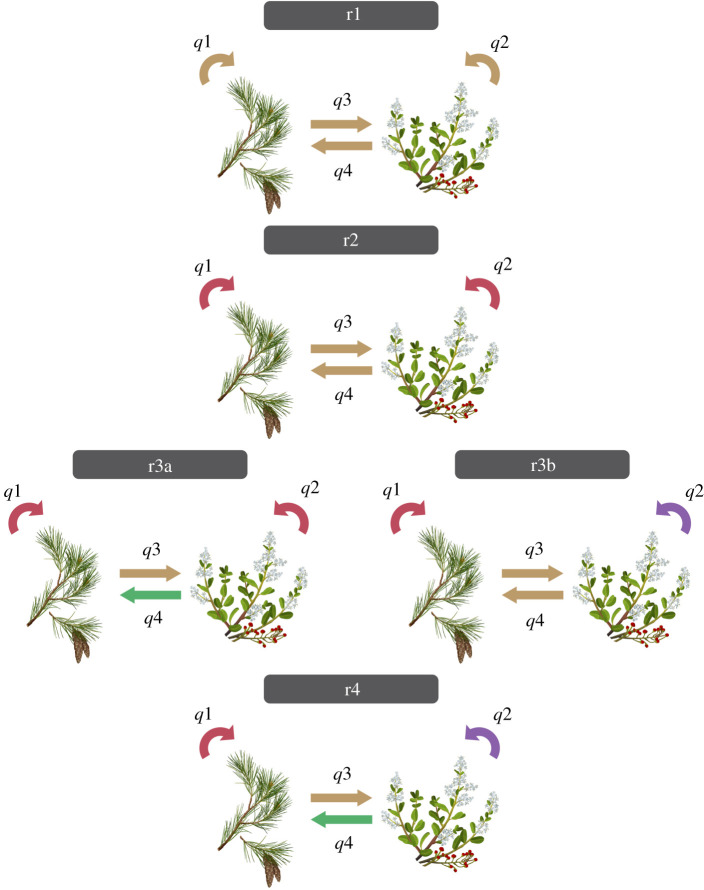
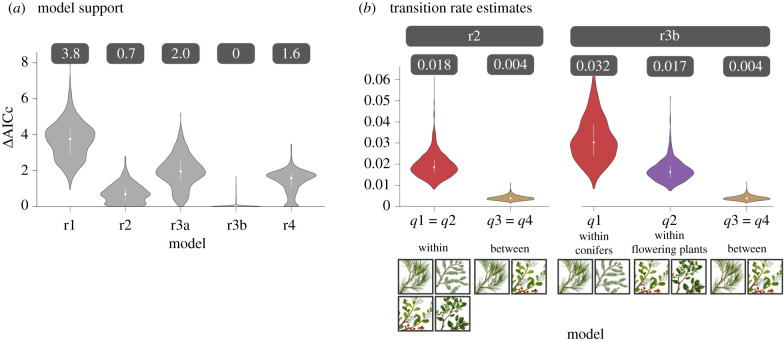
Additionally, however, we were interested in comparing the transition rates between hosts in different genera or families belonging to the same division (‘within’, i.e. conifer to conifer or flowering plant to flowering plant) to those between different divisions (‘between’, i.e. conifer to flowering plant or vice versa). Therefore, we used the host genera as states and fit five different models (figure 4): (i) r1: all transitions forced to have the same rate, (ii) r2: one rate from transitions between conifers and flowering plants and vice versa, and another rate for transitions within conifers or within flowering plants; (iii) r3a: one rate for transitions within either conifers or flowering plants, one rate for transitions from conifer to flowering plant and another rate for flowering plant to conifer; (iv) r3b: one rate for transitions within conifers, one rate for transitions within flowering plants and another rate for transitions between conifers and flowering plants and vice versa; and (v) r4: one rate for transitions from conifer to flowering plant, another rate for flowering plant to conifer, another rate for transitions within conifers and another rate for within flowering plants.
Figure 4.
The models tested in transition-rate analyses. Graphical representation of the transition-rate models built considering four rates: from conifer to conifer (q1), from flowering plant to flowering plant (q2), from conifer to flowering plant (q3) and from flowering plant to conifer (q4). The one-rate model (r1) assumes a single rate for all shifts (q1 = q2 = q3 = q4). The two-rate model (r2) assumes a single rate for within-division shifts and another one for between-division shifts (q1 = q2 ≠ q3 = q4). The three-rate model ‘a’ (r3a) assumes a single rate within divisions and different rates for shifts from conifers to flowering plants and vice versa (q1 = q2 ≠ q3 ≠ q4). The three-rate model ‘b’ (r3b) assumes a single rate for between-division shifts and different rates within conifers and within flowering plants (q1 ≠ q2 ≠ q3 = q4). The four-rate model (r4) assumes different rates for all kinds of shifts (q1 ≠ q2 ≠ q3 ≠ q4).
To assess the robustness of our results, we used five priors for the root: same probability for all host genera (flat), root probabilities weighted using estimated transition rates following [52] (yang) or [50,51] (madd), same probability for all conifer host genera (con) and same probability for all flowering plant genera (flo).
The results were similar in most of the cases, but we focus our description of the results on the inferences using the method of [50,51]. We ran the analyses on 1000 trees taken randomly from the posterior distribution of time-calibrated trees (from [25]) to account for phylogenetic uncertainty. Rates and states were estimated jointly (node.states = ‘joint’, state.recon = ‘estimate’), because such an approach is less prone to getting stuck in local optima [53]. However, most other studies commonly carry out marginal reconstructions (i.e. rate inference followed by ancestral states estimation), and we also did that here for comparative purposes (node.states = ‘marginal’, state.recon = ‘subsequently’).
(b). Host preference trials
We carried out host preference experiments with 3492 individuals from 70 populations (35 pre-determined population/species pairs; see electronic supplementary material, table S2 for details of species, populations, sample sizes, hosts tested, etc.). Tested individuals were captured with sweep nets and placed in plastic cups containing cuttings of two plant species: (i) the plant species that individual was collected on (native host) and (ii) a different plant species, usually the plant species used by the alternate paired population (alternative host). In the morning, the plant species that each individual was found to be resting on after overnight incubation was recorded as the preferred plant species. Each individual was used only once and trials where individuals did not choose a host were excluded from analysis. We quantified host preference differences between paired populations using different host plants in nature (electronic supplementary material, table S2).
(c). Host preference differentiation as a function of host-plant use
The goal was to test if population pairs on more phylogenetically distant hosts (i.e. conifers versus flowering plants) exhibited greater divergence in host preferences than those using more similar hosts. The pairs were chosen primarily to represent a range of divergence in host-plant use, including pairs using the same genus, different genera in the same plant division and different plant divisions. In addition, the taxa compared were generally not distantly related to one another, encompassing also the practical component of access to taxon pairs across disparate parts of the widespread species ranges. Accordingly, 24 pairs were analysed (the remainder used the same host, and were thus not relevant here, but were used for tests of phylogenetic conservatism using individual populations described below). These pairs represented both those where one population used a conifer host and one used a flowering plant host (n = 8 pairs, mean number of individuals tested per population = 37) and those using two different flowering plants hosts (n = 16 pairs, mean number of individuals tested per population = 46).
The mean preference for individual populations was calculated as the proportion of trials that one of the offered hosts was chosen, a value that ranges from zero (focal host never chosen) to one (focal host always chosen). We then calculated host preference divergence between pairs as the absolute difference in the mean preference between pairs, a value that also ranges from zero (identical preferences of the two populations) to one (completely divergent preferences between the two populations, i.e. 100% preference divergence). Note that this value of host preference divergence is identical when either of the two offered hosts is used to calculate the mean preference for individual populations. Phylogenetic distance between hosts was grouped into two categories: moderate for the pairs on two different flowering plants and large for the pairs on a conifer versus a flowering plant host (see Introduction for justification of these categories).
Because preference divergence was bounded between zero and one, we employed β regression to model the influence of divergence in host-plant use on divergence in host-plant preference (dependent variable) using the betareg function in the package betareg 3.1-2 [54] in R, specifying ‘logit’ as link function. We performed analyses with the complete dataset of 24 comparisons, and two subsets including either only conspecific population pairs (n = 18 pairs) or only conspecific population pairs from the same geographical site (n = 13 pairs). We obtained congruent results from all three analyses (see Results).
(d). Phylogenetic conservatism of host preference
Phylogenetic relatedness of taxa generates non-independence of data points obtained from multiple populations or species. To assess the need to account for this effect in the analysis of host preference in Timema, we tested for the presence of phylogenetic signal in the strength of the preference for the native host. We analysed the host preference data described above (see also electronic supplementary material, table S2) in combination with the population-level Bayesian time-calibrated maximum credibility tree from [25] pruned to represent the 48 populations for which host preference data were available (corresponding to 28 geographical localities on the tree). Host preference of each of these individual populations was estimated as the proportion of trials the native host was picked over the alternative host. In cases where a population with host preference data was not represented in our phylogeny (n = 25), we chose the geographically closest population of the same species as its representative in the phylogeny (mean distance to the nearest population was not overly large, being 17.7 km).
As several populations with host preference data mapped to the same tip in the phylogeny, we used two approaches to assign trait values to those tips: (i) we used the mean of host preference for populations mapping to the same tip and (ii) for each tip represented by multiple populations, we sampled a trait value randomly from those populations 1000 times to generate a sample distribution of host preference for those populations. Results were congruent between the two approaches (see Results). We used the function phylosig in the package phytools 0.6–99 [55] in R to calculate Bloomberg's K statistic [56] and its statistical significance. Higher values of K indicate successively stronger phylogenetic signal, with K = 0 in the absence of phylogenetic signal, K = 1 under a Brownian motion model of trait evolution and K > 1 when trait evolution is more constrained. In general, we found a lack of phylogenetic signal for the strength of host preference (details below).
3. Results
(a). Frequency and magnitude of host shifts
We found that large ecological shifts between conifer and flowering plant hosts have occurred multiple times among our study populations, including shifts within species (electronic supplementary material, table S3, AU test, p < 0.001 for both clustering by host divisions, and clustering by host divisions within Timema species). Indeed, host shifts in general appear common in Timema, likely facilitated by standing genetic variation in the ability to use novel hosts [30].
Phylogenetic analyses of transition rates between hosts support a key prediction of the adaptive zones model, i.e. that large host shifts between conifer and flowering plant hosts are relatively rare. Specifically, we compared the fit of five transition-rate models by maximum likelihood (figure 4). Our main interest was on the support for a ‘one-rate model’ that enforced a single transition rate irrespective of whether shifts were within or between plant divisions, relative to models that allowed rates to vary between different types and magnitudes of host shifts. Our analyses revealed that the one-rate model was consistently the least supported one (figure 5; electronic supplementary material, table S4, difference in Akaike information criterion values corrected for sample size (ΔAICc)). The best-fit model allowed for transitions within each division and between them (three-rate model b). However, the most notable increase in support was observed when moving from the one-rate model to a two-rate model that allowed the rates for transitions between divisions to differ from those within divisions (figure 5). Notably, these results were robust to using multiple root probabilities and inference methods (electronic supplementary material, figure S1 and table S4). Thus, we found that transition rates within divisions were approximately five times higher than those between divisions (figure 5; range approx. 3–10× higher using different inference methods and root probabilities, electronic supplementary material, figure S2–S3 and table S5).
Figure 5.
Support for each transition-rate model (see also figure 3). (a) Violin plots depict the distribution of ΔAICc values (difference in sample-size-corrected AIC between the focal model and the best model) after fitting the models by maximum likelihood to 1000 trees randomly taken from the Bayesian posterior distribution of trees from [25]. The median ΔAICc for each model is shown above each violin plot. (b) Distribution of transition-rate estimates for the most supported models r2 and r3b. Colours of violin plots follow the same scheme as rates in figure 3. The median of each rate is shown above each violin plot. The white diamonds and bars represent the median and the interquartile range, respectively.
(b). The evolution of premating isolation due to host preference
Next, we were interested in the evolutionary consequences of extreme host shifts. For 24 population pairs (2252 trials) that use different hosts, we found that host preference was approximately two to three times more differentiated between Timema taxa feeding on different divisions than between those feeding on different families within divisions (figure 6). This result was robust to whether we considered all 24 taxon-pairs (z = 3.50, p = 0.0005, β regression; electronic supplementary material, table S6), the 18 within-species comparisons (z = 2.72, p = 0.0065) or the 13 within-species within-locality comparisons (z = 2.49, p = 0.0128).
Figure 6.
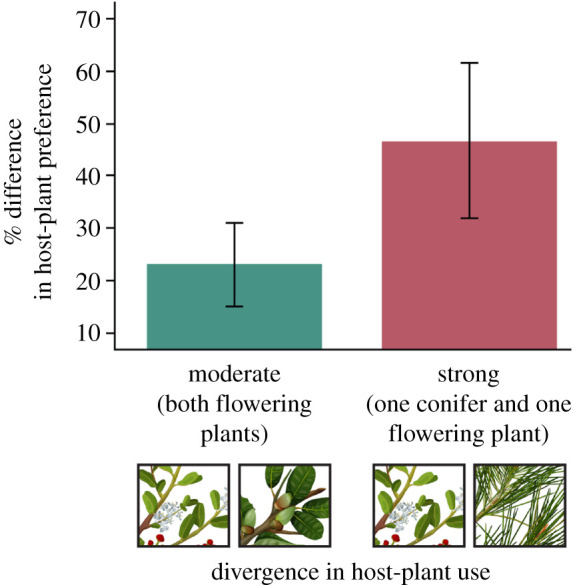
Behavioural host preference experiments. Shown is the mean divergence in host preference (±2 s.e.) between population pairs differing in the degree to which their hosts differ.
(c). Phylogenetic conservatism of host preference
We found little to no evidence for phylogenetic conservatism of host preference, justifying the population-level analyses above. Specifically, Blomberg's K was found to be low and non-significant in the core analysis using the mean as the trait value for tips represented by more than one population (K = 0.101, p = 0.747). This result was robust to the approach of considering the mean, because randomizing the trait value to be equal to that from one of the populations when multiple populations represented one tip always resulted in a K smaller than 0.2. Furthermore, only 7 out of 1000 permutations yielded a p-value lower than 0.05, all being non-significant after correction for multiple comparisons.
4. Discussion
We use ecological and behavioural data in a phylogenetic comparative framework to test general predictions about adaptive zones and the speciation process. Although Timema stick insects use a wide breadth of host plants, we found that host use is dominated by moderate shifts between families within flowering plants or conifers, with only a few extreme shifts between these plant divisions. When extreme shifts do occur, however, they likely generate greater premating isolation (via host preference) than do moderate shifts (figure 1). These results are consistent with the adaptive zones model and suggest that the net contribution of ecological shifts to diversification can reflect a balance between their magnitude and frequency.
As in many correlational or comparative studies, which abound in evolutionary biology, causation is difficult to definitively infer. Thus, it is possible that host preference itself affects the frequency of extreme shifts. However, we consider it more likely that preference evolution is a consequence (rather than cause) of extreme shifts because: (i) host preferences are generally quite modest in absolute terms such that they are unlikely to strongly constrain host shifts (figure 6; electronic supplementary material, table S2) and (ii) they appear evolutionarily labile, with no evidence for phylogenetic conservatism. Below we discuss the causes of observed patterns of host shift, the completion of speciation and limits to diversification (i.e. the net result of the speciation and extinction processes over time). Future work could usefully consider whether this pattern applies to other forms of RI.
(a). Causes of observed patterns of host shift
We have shown that several large ecological shifts between conifers and flowering plants have occurred during the diversification of Timema, although their frequency is much lower than shifts within the divisions. There are at least two core factors that could limit the frequency of shifts between evolutionary distant host plants. First, there could be inherent adaptive constraints, as highly different host plants are likely to constitute distant adaptive peaks. For example, specialization can involve trade-offs resulting in metabolic constraints, in turn making shifts to new hosts more difficult [31,57]. Performance experiments indicate that this is not strongly the case in Timema in terms of the physiological ability to digest new hosts [30], but the existence of trade-offs associated with crypsis and predation is likely [58,59].
Second, the geographical distribution of the plants can put constraints on the colonization of new hosts. Opportunities to shift between conifers and flowering plants may have been ample for Timema, as both kinds of host plants are found commonly intermixed throughout California currently, and were so during most of the Timema diversification history [60,61]. Nonetheless, this geographical overlap has not been formally quantified for the populations studied here. Further work is thus required to quantify the contribution of inherent biological constraints versus the geographical arrangement of plants on the host shifts, but either way shifts between divisions are rare. Future insights on the role of syntopy of host plants would need to consider their past distributions over long time periods, and at a fine geographical scale. One methodological consideration is that most Timema species not included in our analysis feed on conifers (i.e. those outside of California) [29,62–64]. If most of these constitute a sister group to Californian species, our results would hold valid, but could be limited to the Californian lineage.
Interestingly, when each plant division is considered, transition rates between conifers were higher than rates between flowering plants. Gymnosperms are known to have lower morphological and chemical diversity than angiosperms, as well as lower morphological and genomic evolutionary rates [31,65,66]. This could translate into different conifers representing relatively closer adaptive peaks when compared with flowering plants, thus making shifts between them easier. In addition, mixed conifer forests are common in California, but tend to be restricted to particular altitudinal bands and separated in geographical space [66–68], which may have favoured repeated parallel shifts between conifers. Lastly, we cannot discard a potential effect derived from our choice of taxonomic level (i.e. genus). For example, for most of the conifer genera that Timema use, the use is restricted to a single species, whereas for flowering plant genera, there are usually several species per genus used [30,62]. This could result in reduced transition rates within flowering plants. In other words, our conclusions hold well for transition rates between genera, but further work on transitions between species is warranted.
(b). Evolution of host preferences, premating isolation and completing speciation
Studies of T. cristinae have shown that host preferences are likely partially heritable, with ‘hybrids’ between host-plant ecotypes exhibiting preferences intermediate between the parental forms [69–71]. However, further work is required to determine the relative contribution of genetic versus induced environmental factors to this form of RI during the diversification of Timema, and to RI in general across taxa. Induced effects on RI have been reported for imprinting of song in birds [72], cultural differences among killer whale ecotypes [73], and host or mate preference in other insects [74,75]. On the other hand, if environmental effects can be reversed, this could decrease RI. Further work on the role of genetic and environmental effects in speciation is warranted.
In contrast with previous work on patterns of host use in nature [29], we did not find evidence for phylogenetic conservatism of behavioural host preference. This is likely because phylogenetic relationships in past work were based on a single marker (mitochondrial DNA) and not as accurate as those used here, and because host use in nature does not necessarily correspond to behavioural host preference (i.e. less preferred hosts may be used in nature due to availability, necessity or convenience) [70]. Moreover, our results are in agreement with recent experiments showing that most populations retain plasticity in host use [30].
Finally, we note that even the most extreme host preferences documented here were not perfectly divergent between any of the tested taxon pairs (i.e. we never observed a 100% difference between a pair). Thus, RI due to host preference does not appear to reach completion. In part, this could reflect that our experimental design in the laboratory underestimates host preferences in nature, but even so it seems unlikely that host preferences alone can complete speciation in Timema. Moreover, the frequency of shifts between very different hosts is very low such that they alone are unlikely to explain late stages of speciation and the diversification of Timema. Thus, the completion of speciation likely requires other factors, such as periods of geographical isolation and restricted gene flow [25], and the evolution of additional forms of RI. Indeed, there is evidence for RI due to chemical-mediated mate choice [25,71,76–78], selection against immigrants onto new hosts and hybrids [58,59,76,78], and postmating, prezygotic isolation [71,79]. Further work is required to test how moderate versus extreme host shifts affect these forms of RI, if they do at all.
(c). Limits to the rate of speciation
The evolution of RI is generally thought to be a key component of the speciation process [9,80–84]. However, several recent studies suggest that the evolution of RI is not the step limiting speciation rates, particularly over long timescales. For example, the rate of the evolution of RI in birds and flies, estimated experimentally, is uncoupled from speciation rates estimated using phylogenies [42]. Likewise, the diversification of Himalayan songbirds appears limited by the rate of niche filling, not the acquisition of RI [40]. In insects, host shifts usually result in an increase in RI and can initiate speciation processes, but their relative contribution to insect diversification is unclear [3,38,57,85,86].
Our results inform this issue by showing how a key factor other than RI, i.e. the rate at which new niches are colonized, can be important for understanding diversification. In particular, ecological shifts large enough to generate substantial RI may be rare. Thus, the total contribution of an ecological shift of particular magnitude to the diversification of a clade might be the net result of the amount of RI it confers and its frequency. These two factors might be opposing and are potentially interlinked, and consequently, the empirical role of ecological shifts in speciation requires further work. Further studies that examine a range of closely related taxa that vary both in RI and the magnitude of ecological shifts they underwent are warranted.
5. Conclusion
Our results provide evidence that the ecological magnitude of a host shift can affect levels of RI. Shifts themselves appear common in Timema and this is likely facilitated by standing genetic variation in the ability to use novel hosts [29,30], which is likely maintained at least in part by gene flow [71,78,87,88] and balancing selection [43,89,90]. Finally, the results inform limits to divergence, as they show that shifts between ecologically distant host plants are rare, and therefore unlikely to explain diversification on their own. Thus, the rate of species formation could largely be the result of the waiting time for shifts between distant adaptive peaks coupled with events that create geographical isolation.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
The support and resources from the Center for High Performance Computing at the University of Utah are gratefully acknowledged, as well as access to the High Performance Computing Facilities, particularly to the Iceberg HPC cluster, from the Corporate Information and Computing Services at the University of Sheffield. Rosa Marín Ribas drew all the figures.
Data accessibility
The data reported in this paper, including custom code written for analyses, have been archived in the Dryad Digital Repository (https://doi.org/10.5061/dryad.41ns1rnb6) [91]. Code used for the analyses have been deposited in bitbucket repository: https://bitbucket.org/visoca/eco_shifts_timema.
Authors' contributions
M.M. and V.S.-C. carried out phylogenetic hypothesis testing, transition-rate inferences and phylogenetic conservatism analysis. P.N. performed the feeding preference trials. M.M. and Z.G. carried out the host preference differentiation analysis. M.M., V.S.-C. and P.N. participated in the design of the study and drafted the manuscript. All authors critically revised the manuscript, gave final approval for publication and agree to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
The work was funded by a grant from the European Research Council (grant no. NatHisGen R/129639) to P.N. and a Natural Sciences and Engineering Research Council grant to B. J. Crespi (grant no. NSERC 06505). V.S.-C. was supported by a Leverhulme Trust Early Career Fellowship (grant no. ECF-2016-367). M.M. was supported by an Advanced Postdoc.Mobility fellowship from the Swiss National Science Foundation (grant no. P300P3_147888).
References
- 1.Simpson GG. 1944. Tempo and mode in evolution. New York, NY: Columbia University Press. [Google Scholar]
- 2.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press. [Google Scholar]
- 3.Winkler IS, Mitter C. 2008. The phylogenetic dimension of insect–plant interactions: a review of recent evidence. In The evolutionary biology of herbivorous insects: specialization, speciation, and radiation (ed. Tilmon K.), pp. 240–263. Berkeley, CA: University of California Press. [Google Scholar]
- 4.Mitter C, Farrell B, Wiegmann B. 1988. The phylogenetic study of adaptive zones—has phytophagy promoted insect diversification? Am. Nat. 132, 107–128. ( 10.1086/284840) [DOI] [Google Scholar]
- 5.Hendry AP. 2017. Eco-evolutionary dynamics. Princeton, NJ: Princeton University Press. [Google Scholar]
- 6.Harmon LJ, Schulte JA, Larson A, Losos JB. 2003. Tempo and mode of evolutionary radiation in iguanian lizards. Science 301, 961–964. ( 10.1126/science.1084786) [DOI] [PubMed] [Google Scholar]
- 7.Goldschmidt R. 1940. The material basis of evolution. New Haven, CT: Yale University Press. [Google Scholar]
- 8.Wu C. 2001. The genic view of the process of speciation. J. Evol. Biol. 14, 851–865. ( 10.1046/j.1420-9101.2001.00335.x) [DOI] [Google Scholar]
- 9.Coyne JA, Orr HA. 2004. Speciation, 1st edn Sunderland, MA: Sinauer Associates. [Google Scholar]
- 10.Nosil P, Harmon LJ, Seehausen O. 2009. Ecological explanations for (incomplete) speciation. Trends Ecol. Evol. 24, 145–156. ( 10.1016/j.tree.2008.10.011) [DOI] [PubMed] [Google Scholar]
- 11.Nosil P. 2012. Ecological speciation. Oxford, UK: Oxford University Press. [Google Scholar]
- 12.Seehausen O, et al. 2014. Genomics and the origin of species. Nat. Rev. Genet. 15, 176–192. ( 10.1038/nrg3644) [DOI] [PubMed] [Google Scholar]
- 13.Mallet J. 2008. Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Phil. Trans. R. Soc. B 363, 2971–2986. ( 10.1098/rstb.2008.0081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallet J, Beltran M, Neukirchen W, Linares M. 2007. Natural hybridization in heliconiine butterflies: the species boundary as a continuum. BMC Evol. Biol. 7, 28 ( 10.1186/1471-2148-7-28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nosil P, Feder JL, Flaxman SM, Gompert Z. 2017. Tipping points in the dynamics of speciation. Nat. Ecol. Evol. 1, 0001 ( 10.1038/s41559-41016-40001) [DOI] [PubMed] [Google Scholar]
- 16.Brawand D, et al. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513, 375–381. ( 10.1038/nature13726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feulner PGD, et al. 2013. Genome-wide patterns of standing genetic variation in a marine population of three-spined sticklebacks. Mol. Ecol. 22, 635–649. ( 10.1111/j.1365-294X.2012.05680.x) [DOI] [PubMed] [Google Scholar]
- 18.Feulner PGD, et al. 2015. Genomics of divergence along a continuum of parapatric population differentiation. PLoS Genet. 11, e1004966 ( 10.1371/journal.pgen.1004966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berner D, Grandchamp A-C, Hendry AP. 2009. Variable progress toward ecological speciation in parapatry: stickleback across eight lake–stream transitions. Evolution 63, 1740–1753. ( 10.1111/j.1558-5646.2009.00665.x) [DOI] [PubMed] [Google Scholar]
- 20.Twomey E, Vestergaard JS, Venegas PJ, Summers K. 2016. Mimetic divergence and the speciation continuum in the mimic poison frog Ranitomeya imitator. Am. Nat. 187, 205–224. ( 10.1086/684439) [DOI] [PubMed] [Google Scholar]
- 21.Burri R, et al. 2015. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 25, 1656–1665. ( 10.1101/gr.196485.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrew RL, Rieseberg LH. 2013. Divergence is focused on few genomic regions early in speciation: incipient speciation of sunflower ecotypes. Evolution 67, 2468–2482. ( 10.1111/evo.12106) [DOI] [PubMed] [Google Scholar]
- 23.Powell THQ, Hood GR, Murphy MO, Heilveil JS, Berlocher SH, Nosil P, Feder JL. 2013. Genetic divergence along the speciation continuum: the transition from host race to species in Rhagoletis (Diptera: Tephritidae). Evolution 67, 2561–2576. ( 10.1111/evo.12209) [DOI] [PubMed] [Google Scholar]
- 24.Martin SH, et al. 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23, 1817–1828. ( 10.1101/gr.159426.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riesch R, et al. 2017. Transitions between phases of genomic differentiation during stick-insect speciation. Nat. Ecol. Evol. 1, 0082 ( 10.1038/s41559-017-0082) [DOI] [PubMed] [Google Scholar]
- 26.Price TD. 2007. Speciation in birds. Woodbury, NY: Roberts and Company. [Google Scholar]
- 27.Funk DJ, Nosil P. 2007. Comparative analyses and ecological speciation in herbivorous insects. In Specialization, speciation, and radiation: the evolutionary biology of herbivorous insects (ed. Tilmon K.), pp. 117–135. Berkeley, CA: California University Press. [Google Scholar]
- 28.Funk DJ, Nosil P, Etges WJ. 2006. Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proc. Natl Acad. Sci. USA 103, 3209–3213. ( 10.1073/pnas.0508653103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandoval C. 2000. Phylogenetic evidence for the evolution of ecological specialization in Timema walking-sticks. J. Evol. Biol. 13, 249–262. ( 10.1046/j.1420-9101.2000.00164.x) [DOI] [Google Scholar]
- 30.Larose C, Parker DJ, Schwander T. 2018. Fundamental and realized feeding niche breadths of sexual and asexual stick insects. Proc. R. Soc. B 285, 20181805 ( 10.1098/rspb.2018.1805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forister ML, et al. 2015. The global distribution of diet breadth in insect herbivores. Proc. Natl Acad. Sci. USA 112, 442–447. ( 10.1073/pnas.1423042112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiz-Palacios O, Schneider H, Heinrichs J, Savolainen V. 2011. Diversification of land plants: insights from a family-level phylogenetic analysis. BMC Evol. Biol. 11, 341 ( 10.1186/1471-2148-11-341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottlieb OR, Kubitzki K. 1984. Chemosystematics of the Gnetatae and the chemical evolution of seed plants. Planta Med. 50, 380–385. ( 10.1055/s-2007-969743) [DOI] [PubMed] [Google Scholar]
- 34.Kubitzki K, Gottlieb OR. 1984. Phytochemical aspects of angiosperm origin and evolution. Acta Bot. Neerland. 33, 457–468. ( 10.1111/j.1438-8677.1984.tb01837.x) [DOI] [Google Scholar]
- 35.Kubitzki K, Gottlieb OR. 1984. Micromolecular patterns and the evolution and major classification of angiosperms. Taxon 33, 375–391. ( 10.2307/1220975) [DOI] [Google Scholar]
- 36.Leitch AR, Leitch IJ. 2012. Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytol. 194, 629–646. ( 10.1111/j.1469-8137.2012.04105.x) [DOI] [PubMed] [Google Scholar]
- 37.Feder JL, Opp SB, Wlazlo B, Reynolds K, Go W, Spisak S. 1994. Host fidelity is an effective premating barrier between sympatric races of the apple maggotfly. Proc. Natl Acad. Sci. USA 91, 7990–7994. ( 10.1073/pnas.91.17.7990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Funk DJ. 1998. Isolating a role for natural selection in speciation: host adaptation and sexual isolation in Neochlamisus bebbianae leaf beetles. Evolution 52, 1744–1759. ( 10.1111/j.1558-5646.1998.tb02254.x) [DOI] [PubMed] [Google Scholar]
- 39.Hirai Y, Kobayashi H, Koizumi T, Katakura H. 2006. Field-cage experiments on host fidelity in a pair of sympatric phytophagous ladybird beetles. Entomol. Exp. Appl. 118, 129–135. ( 10.1111/j.1570-7458.2006.00365.x) [DOI] [Google Scholar]
- 40.Price TD, et al. 2014. Niche filling slows the diversification of Himalayan songbirds. Nature 509, 222–225. ( 10.1038/nature13272) [DOI] [PubMed] [Google Scholar]
- 41.Rabosky DL. 2016. Reproductive isolation and the causes of speciation rate variation in nature. Biol. J. Linnean Soc. 118, 13–25. ( 10.1111/bij.12703) [DOI] [Google Scholar]
- 42.Rabosky DL, Matute DR. 2013. Macroevolutionary speciation rates are decoupled from the evolution of intrinsic reproductive isolation in Drosophila and birds. Proc. Natl Acad. Sci. USA 110, 15 354–15 359. ( 10.1073/pnas.1305529110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nosil P, Villoutreix R, de Carvalho CF, Farkas TE, Soria-Carrasco V, Feder JL, Crespi BJ, Gompert Z. 2018. Natural selection and the predictability of evolution in Timema stick insects. Science 359, 765–770. ( 10.1126/science.aap9125) [DOI] [PubMed] [Google Scholar]
- 44.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. ( 10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16, 1114–1116. ( 10.1093/oxfordjournals.molbev.a026201) [DOI] [Google Scholar]
- 46.Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508. ( 10.1080/10635150290069913) [DOI] [PubMed] [Google Scholar]
- 47.Shimodaira H, Hasegawa M. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246–1247. ( 10.1093/bioinformatics/17.12.1246) [DOI] [PubMed] [Google Scholar]
- 48.Beaulieu JM, O'Meara BC, Donoghue MJ. 2013. Identifying hidden rate changes in the evolution of a binary morphological character: the evolution of plant habit in campanulid angiosperms. Syst. Biol. 62, 725–737. ( 10.1093/sysbio/syt034) [DOI] [PubMed] [Google Scholar]
- 49.R Core Team. 2018. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See https://www.R-project.org/. [Google Scholar]
- 50.FitzJohn RG, Maddison WP, Otto SP. 2009. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58, 595–611. ( 10.1093/sysbio/syp067) [DOI] [PubMed] [Google Scholar]
- 51.Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol. 56, 701–710. ( 10.1080/10635150701607033) [DOI] [PubMed] [Google Scholar]
- 52.Yang Z. 2006. Bayesian methods. In Computational molecular evolution (eds Harvey PH, May RM), pp. 145–184. Oxford, UK: Oxford University Press. [Google Scholar]
- 53.Joy JB, Liang RH, McCloskey RM, Nguyen T, Poon AFY. 2016. Ancestral reconstruction. PLoS Comput. Biol. 12, e1004763 ( 10.1371/journal.pcbi.1004763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cribari-Neto F, Zeileis A. 2010. Beta regression in R. J. Stat. Softw. 34, 1–24. ( 10.18637/jss.v034.i02) [DOI] [Google Scholar]
- 55.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 56.Blomberg SP, Garland T, Ives AR. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745. ( 10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 57.Fordyce JA. 2010. Host shifts and evolutionary radiations of butterflies. Proc. R. Soc. B 277, 3735–3743. ( 10.1098/rspb.2010.0211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nosil P. 2004. Reproductive isolation caused by visual predation on migrants between divergent environments. Proc. R. Soc. B 271, 1521–1528. ( 10.1098/rspb.2004.2751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nosil P, Crespi BJ. 2006. Experimental evidence that predation promotes divergence in adaptive radiation. Proc. Natl Acad. Sci. USA 103, 9090–9095. ( 10.1073/pnas.0601575103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forister ML, Dyer LA, Singer MS, Stireman Iii JO, Lill JT. 2011. Revisiting the evolution of ecological specialization, with emphasis on insect–plant interactions. Ecology 93, 981–991. ( 10.1890/11-0650.1) [DOI] [PubMed] [Google Scholar]
- 61.Millar CI. 2012. Geologic, climatic, and vegetation history of California. In The Jepson manual: vascular plants of California (eds Baldwin BG, Goldman DH, Keil DJ, Patterson R, Rosatti TJ, Wilken DH), pp. 49–67, 2nd edn Berkeley, CA: University of California Press. [Google Scholar]
- 62.Law JH, Crespi BJ. 2002. The evolution of geographic parthenogenesis in Timema walking-sticks. Mol. Ecol. 11, 1471–1489. ( 10.1046/j.1365-294X.2002.01547.x) [DOI] [PubMed] [Google Scholar]
- 63.Law JH, Crespi BJ. 2002. Recent and ancient asexuality in Timema walkingsticks. Evolution 56, 1711–1717. ( 10.1111/j.0014-3820.2002.tb01484.x) [DOI] [PubMed] [Google Scholar]
- 64.Sandoval C, Carmean DA, Crespi BJ. 1998. Molecular phylogenetics of sexual and parthenogenetic Timema walking-sticks. Proc. R. Soc. Lond. B 265, 589–595. ( 10.1098/rspb.1998.0335) [DOI] [Google Scholar]
- 65.Sarkar P, Bosneaga E, Auer M. 2009. Plant cell walls throughout evolution: towards a molecular understanding of their design principles. J. Exp. Bot. 60, 3615–3635. ( 10.1093/jxb/erp245) [DOI] [PubMed] [Google Scholar]
- 66.Griffin JR, Critchfield WB. 1972. The distribution of forest trees in California. Res. Paper PSW-RP-82. Berkeley, CA: Pacific Southwest Forest and Range Experiment Station, Forest Service, US Department of Agriculture; vol. 60, p. 082.
- 67.Heusser LE, Lyle M, Mix A. 2000. Vegetation and climate of the northwest coast of North America during the last 500 ky. High-resolution pollen evidence from the northern California Margin. In Proceedings of the Ocean Drilling Program, Scientific Results, Vol. 167 (eds. Lyle M, Koizumi I, Richter C), pp. 217–226. College Station, TX: Ocean Drilling Program. [Google Scholar]
- 68.Minnich R. 2007. Southern California conifer forests. In Terrestrial vegetation of California, 3rd edn (eds Barbour M, Keeler-Wolf T, Schoenherr AA), pp. 502–538. Berkeley, CA: University of California Press. [Google Scholar]
- 69.Nosil P, Crespi BJ, Sandoval CP, Kirkpatrick M. 2006. Migration and the genetic covariance between habitat preference and performance. Am. Nat. 167, E66–E78. ( 10.1086/499383) [DOI] [PubMed] [Google Scholar]
- 70.Nosil P, Sandoval CP, Crespi BJ. 2006. The evolution of host preference in allopatric vs. parapatric populations of Timema cristinae walking-sticks. J. Evol. Biol. 19, 929–942. ( 10.1111/j.1420-9101.2005.01035.x) [DOI] [PubMed] [Google Scholar]
- 71.Nosil P. 2007. Divergent host plant adaptation and reproductive isolation between ecotypes of Timema cristinae walking sticks. Am. Nat. 169, 151–162. ( 10.1086/510634) [DOI] [PubMed] [Google Scholar]
- 72.Grant BR, Grant PR. 2008. Fission and fusion of Darwin's finches populations. Phil. Trans. R. Soc. B 363, 2821–2829. ( 10.1098/rstb.2008.0051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Riesch R, Barrett-Lennard LG, Ellis GM, Ford JKB, Deecke VB. 2012. Cultural traditions and the evolution of reproductive isolation: ecological speciation in killer whales? Biol. J. Linnean Soc. 106, 1–17. ( 10.1111/j.1095-8312.2012.01872.x) [DOI] [Google Scholar]
- 74.Wood TK, Guttman SI. 1982. Ecological and behavioral basis for reproductive isolation in the sympatric Enchenopa binotata complex (Homoptera, Membracidae). Evolution 36, 233–242. ( 10.1111/j.1558-5646.1982.tb05036.x) [DOI] [PubMed] [Google Scholar]
- 75.Wood TK, Keese MC. 1990. Host-plant induced assortative mating in Enchenopa treehoppers. Evolution 44, 619–628. ( 10.1111/j.1558-5646.1990.tb05942.x) [DOI] [PubMed] [Google Scholar]
- 76.Nosil P, Crespi BJ, Gries R, Gries G. 2007. Natural selection and divergence in mate preference during speciation. Genetica 129, 309–327. ( 10.1007/s10709-006-0013-6) [DOI] [PubMed] [Google Scholar]
- 77.Nosil P, Crespi BJ, Sandoval CP. 2002. Host-plant adaptation drives the parallel evolution of reproductive isolation. Nature 417, 440–443. ( 10.1038/417440a) [DOI] [PubMed] [Google Scholar]
- 78.Nosil P, Crespi BJ, Sandoval CP. 2003. Reproductive isolation driven by the combined effects of ecological adaptation and reinforcement. Proc. R. Soc. Lond. B 270, 1911–1918. ( 10.1098/rspb.2003.2457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nosil P, Crespi BJ. 2006. Ecological divergence promotes the evolution of cryptic reproductive isolation. Proc. R. Soc. B 273, 991–997. ( 10.1098/rspb.2005.3359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coyne JA, Orr HA. 1989. Patterns of speciation in Drosophila. Evolution 43, 362–381. ( 10.1111/j.1558-5646.1989.tb04233.x) [DOI] [PubMed] [Google Scholar]
- 81.Mayr E. 1942. Systematics and the origin of species. New York, NY: Columbia University Press. [Google Scholar]
- 82.Mayr E. 1947. Ecological factors in speciation. Evolution 1, 263–288. ( 10.1111/j.1558-5646.1947.tb02723.x) [DOI] [Google Scholar]
- 83.Mayr E. 1954. Change of genetic environment and evolution. In Evolution as a process (eds Huxley J, Hardy AC, Ford EB). London, UK: Allen & Unwin. [Google Scholar]
- 84.Mayr E. 1963. Animal species and evolution. Harvard, MA: Harvard University Press. [Google Scholar]
- 85.Funk DJ, Filchak KE, Feder JL. 2002. Herbivorous insects: model systems for the comparative study of speciation ecology. Genetica 116, 251–267. ( 10.1023/A:1021236510453) [DOI] [PubMed] [Google Scholar]
- 86.Ehrlich PR, Raven PH. 1964. Buttferlies and plants: a study in coevolution. Evolution 18, 586–608. ( 10.1111/j.1558-5646.1964.tb01674.x) [DOI] [Google Scholar]
- 87.Nosil P. 2009. Adaptive population divergence in cryptic color-pattern following a reduction in gene flow. Evolution 63, 1902–1912. ( 10.1111/j.1558-5646.2009.00671.x) [DOI] [PubMed] [Google Scholar]
- 88.Nosil P, Crespi BJ. 2004. Does gene flow constrain adaptive divergence or vice versa? A test using ecomorphology and sexual isolation in Timema cristinae walking-sticks. Evolution 58, 102–112. ( 10.1111/j.0014-3820.2004.tb01577.x) [DOI] [PubMed] [Google Scholar]
- 89.Lindtke D, Lucek K, Soria-Carrasco V, Villoutreix R, Farkas TE, Riesch R, Dennis SR, Gompert Z, Nosil P. 2017. Long-term balancing selection on chromosomal variants associated with crypsis in a stick insect. Mol. Ecol. 26, 6189–6205. ( 10.1111/mec.14280) [DOI] [PubMed] [Google Scholar]
- 90.Comeault AA, et al. 2015. Selection on a genetic polymorphism counteracts ecological speciation in a stick insect. Curr. Biol. 25, 1–7. ( 10.1016/j.cub.2015.05.058) [DOI] [PubMed] [Google Scholar]
- 91.Muschick M, Soria-Carrasco V, Feder JL, Gompert Z, Nosil P. 2020. Data from: Adaptive zones shape the magnitude of premating reproductive isolation in Timema stick insects Dryad Digital Repository. ( 10.5061/dryad.41ns1rnb6) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Muschick M, Soria-Carrasco V, Feder JL, Gompert Z, Nosil P. 2020. Data from: Adaptive zones shape the magnitude of premating reproductive isolation in Timema stick insects Dryad Digital Repository. ( 10.5061/dryad.41ns1rnb6) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The data reported in this paper, including custom code written for analyses, have been archived in the Dryad Digital Repository (https://doi.org/10.5061/dryad.41ns1rnb6) [91]. Code used for the analyses have been deposited in bitbucket repository: https://bitbucket.org/visoca/eco_shifts_timema.